Abstract
The purpose of the present study was to investigate the biological role of microRNA-761 (miR-761) in colorectal cancer (CRC) and the underlying mechanisms by which miR-761 regulates CRC cell proliferation and migration. Quantitative polymerase chain reaction was performed to measure miR-761 expression in CRC tumor tissues and cell lines. It was demonstrated that miR-761 expression was dramatically reduced in CRC tumor tissues and cell lines compared with in normal tissues and cell lines. Overexpression of miR-761 significantly decreased CRC cell growth and migration. Using bioinformatics analysis and luciferase reporter assays, Rab3D was identified as a novel target of miR-761. In addition, it was demonstrated that Rab3D expression was negatively correlated with miR-761. Furthermore, overexpression of Rab3D could reverse the inhibitory effects of miR-761 on cell proliferation and migration. Collectively, the present study demonstrated that miR-761 overexpression could inhibit the proliferation and migration of CRC cell lines, partly at least, via directly targeting Rab3D.
Keywords: miR-761, Rab3D, colorectal cancer, proliferation, migration
Introduction
It is hypothesized that the rise of colorectal cancer (CRC) incidence and mortality rate in China is due to the prevalence of the westernized lifestyle, obesity and physical inactivity (1,2). However, the mechanisms underlying CRC tumorigenesis have not been fully understood until now (3). Importantly, the lack of efficient biomarkers for CRC diagnosis and treatment has resulted in tumors reaching an advanced tumor stage prior to diagnosis and consequently a worse overall survival (4). Therefore, the identification of novel biomarkers will help us to improve the survival quality of CRC patients.
microRNAs (miR/miRNAs) are a family of non-coding RNAs that can regulate target genes expression through 3′-untranslated region (UTR), 5′-UTR or open reading frame binding (5–7). Extensive studies have demonstrated that miRNA expression is dysregulated in cancer, functioning as either tumor suppressors or oncogenes (8,9). miR-761 was demonstrated to be closely associated with the initiation and progression of several cancer types (10–12). For instance, Zhou et al (10) demonstrated that miR-761 overexpression could impact the function of mitochondrial through targeting Mitofusin-2 and resulted in impaired tumor growth and metastasis. The oncogenic role of miR-761 was also validated in non-small cell lung cancer (NSCLC) by Yan et al (11). They demonstrated miR-761 targets inhibitor of growth family, member 4 and tissue inhibitor of metalloproteinase 2 to promote NSCLC progression and metastasis (11). On the contrary, Shi et al (12) reported that miR-761 expression was significantly downregulated in ovarian cancer tissues compared with in their paired noncancerous tissues. Functional assays revealed that miR-761 suppressed ovarian cancer proliferation and invasion by targeting MSI1 (12). In particular, Cao et al (13) demonstrated miR-761 regulates chemo-sensitive CRC cells by targeting forkhead box protein M1. However, more effort is required to fully elucidate the function of miR-761 in CRC.
In this present study, Rab3D was identified as a direct target of miR-761 as it contains a binding sequence in its 3′-UTR. The biological role of miR-761 in regulating CRC cell proliferation and migration was also investigated. It was also demonstrated that miR-761 expression was significantly downregulated in CRC and could regulate cell proliferation and migration through targeting Rab3D. Identification this connection will provide novel therapeutic approaches for CRC.
Materials and methods
Clinical specimens
A total of 113 pairs of CRC tissues and adjacent normal tissues were obtained from CRC patients who underwent treatment between May 2010 and December 2012 at Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute (Shenyang, China). All collected tissues were snap-frozen and stored at −80°C until RNA or protein extraction. None of these patients have ever received any anticancer treatments prior to surgery. Furthermore, the protocol was approved by the Research Ethics Committee of Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute and written informed consent was obtained from all patients. The clinicopathological features of these enrolled patients were summarized in Table I.
Table I.
Correlations of miR-761 and clinicopathological features of CRC patients.
| Clinicopathological features | No. | Low miR-761 (n=58) | High miR-761 (n=55) | P-value |
|---|---|---|---|---|
| Age (years) | ns | |||
| >60 | 53 | 30 | 23 | |
| <60 | 60 | 28 | 32 | |
| Sex | ns | |||
| Male | 51 | 27 | 24 | |
| Female | 62 | 31 | 31 | |
| Tumor size (cm) | 0.023 | |||
| >5 | 64 | 34 | 30 | |
| <5 | 49 | 24 | 25 | |
| Tumor stage | 0.016 | |||
| I–II | 48 | 22 | 26 | |
| III | 65 | 36 | 29 |
miR-761, microRNA-761; CRC, colorectal cancer; ns, not significant.
Cell culture and transfection
Human normal colon epithelial cell line (FHC) and CRC cell lines (HT29, SW480 and SW620) were purchased from the American Type Culture Collection (Manassas, VA, USA). CRC cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was used to culture the FHC cells. These cells were maintained in a 37°C humidified atmosphere containing 5% CO2.
Transfection was conducted using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, cells were cultured until about 70% confluence. miR-761 mimic (miR-mimic) for miR-761 upregulation, negative control (NC) miRNA and pCMV3-Rab3D plasmid for Rab3D overexpression was mixed with Lipofectamine® 2000 and incubated with cell suspensions. Following 48-h transfection, cells were collected and used for subsequent experimentation. miRNAs were purchased from Shanghai GenePharma Co., Ltd., (Shanghai, China). pCMV3-Rab3D plasmid was purchased from Sino Biological Inc., (Beijing, China).
Cell proliferation assay
Cell proliferation was measured by Cell Counting kit-8 (CCK-8; Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. Briefly, cells were seeded at a density of 5×103 cells/well and incubated for 0, 24, 48 and 72 h. Following this the CCK-8 reagent was added to each well and further incubated for 4 h. Absorbance was detected at 450 nm using an ELISA plate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Wound healing assay
Cells were plated in 6-well plates and allowed to grow until ~90% confluence. A pipette tip (200 µl) was used to create wound in cell surface following serum starvation for 24 h. Images was captured 0 and 24 h after the wound was created using a CX31 light microscope (Olympus Corporation, Tokyo, Japan).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from tissues and cells using TRIzol® reagent (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). The expression level of miR-761 was assessed by qPCR using SYBR® Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.) on an ABI 7500 real-time PCR System (Applied Biosystems, Foster City, CA, USA). The following primer pairs were used for qPCR: miR-761 forward, 5′-ACAGCAGGCACAGAC-3′ and reverse, 5′-GAGCAGGCTGGAGAA-3′; U6 small nuclear (sn)RNA forward, 5′-CTGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The following thermocyclinf conditions were used for the qPCR: Initial denaturation at 95°C for 10 min; 40 cycles of 95°C for 30 sec and 58°C for 30 sec. miR-761 expression levels were quantified using the 2−ΔΔCq method and normalized to the internal reference gene U6 (14).
Western blot analysis
Radioimmunoprecipitation assay protein lysis buffer (Beyotime Institute of Biotechnology) was used to extract total protein from cells and tissues. Total protein concentration was analyzed by bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology) and 50 µg protein/lane was separated via SDS-PAGE on a 10% gel. The separated proteins were transferred onto polyvinlyidene difluoride membranes (Beyotime Institute of Biotechnology) and blocked for 1 h at room temperature with 5% fat-free milk blocking. The membranes were incubated with primary antibodies (rabbit anti-Rab3D; 1:1,000; cat. no. ab128997; Abcam, Cambridge, MA, USA and rabbit anti-GAPDH; 1:1,000; cat. no. 5174, Cell Signaling Technology, Inc., Danvers, MA, USA) overnight at 4°C. Following washing, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:10,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h at room temperature. Protein bands were developed using the BeyoECL Plus kit (Beyotime Institute of Biotechnology).
Luciferase activity reporter assay
TargetScan (www.targetscan.org/vert_72) was used to predict target genes of miR-761. Rab3D was identified as a potential target of miR-761 with a putative miR-761 binding site in the 3′-UTR. Wild-type (wt) and mutant (mut) Rab3D 3′-UTR was cloned into pMIR-Report (Promega Corporation, Madison, WI, USA). For luciferase activity analysis, cells were seeded into 24-well plates at a density of 5×103 cells/well and co-transfected with wt or mut Rab3D construct along with miR-mimic or NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were collected at 48 h following transfection for luciferase activities measurement using a Dual Luciferase Assay system (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation from at least three independent experiments. Difference in groups was analyzed using Student's t-test or one-way analysis of variance and Tukey test at GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Kaplan-Meier method and log-rank test was used to calculate overall survival. Correlation of miR-761 and Rab3D was analyzed using Spearman's correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-761 is downregulated in CRC tissues and cell lines
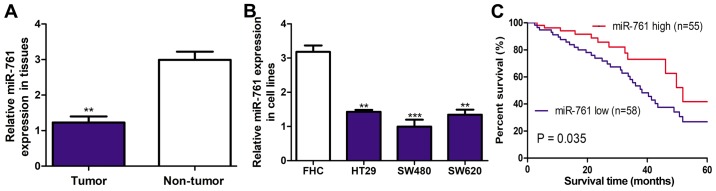
It was demonstrated that miR-761 expression level was significantly downregulated in CRC tissues compared with the adjacent normal tissues using RT-qPCR (P<0.01 Fig. 1A). Furthermore, miR-761 expression level in CRC cell lines was examined and demonstrated that miR-761 expression was significantly downregulated in CRC cell lines compared with the FHC cell line (P<0.01; Fig. 1B). Furthermore, miR-761 expression was the lowest in the SW480 cell line out of the three CRC cell lines investigated (Fig. 1B). Therefore, SW480 cell line was selected for further functional assays.
Figure 1.
miR-761 expression is downregulated in CRC tissues and cell lines. RT-qPCR was used to analyze miR-761 expression in (A) CRC tissues and nontumor tissues. P<0.01 vs. the nontumor group. (B) miR-761 expression in CRC cell lines and (HT29, SW480 and SW620) and FHC. (C) Low expression of miR-761 predicts poor 5-year overall survival of CRC patients. **P<0.01 and ***P<0.001 vs. the FHC group. miR-761, microRNA-761; CRC, colorectal cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Downregulation of miR-761 is correlated with poor 5-year overall survival
The association of miR-761 expression and 5-year overall survival was further investigated to evaluate the clinical significance of miR-761 expression in CRC progression. The median expression of miR-761 level was used to subdivide into miR-761 low (n=58) or high (n=55) expression groups. It was demonstrated that miR-761 low expression was correlated with worse 5-year overall survival (P=0.035; Fig. 1C). The analysis of association between miR-761 and clinicopathological features demonstrated that low miR-761 expression was correlated with tumor size (P=0.023) and tumor stage (P=0.016; Table I). However, no significant association between miR-761 expression and age or sex was observed (Table I).
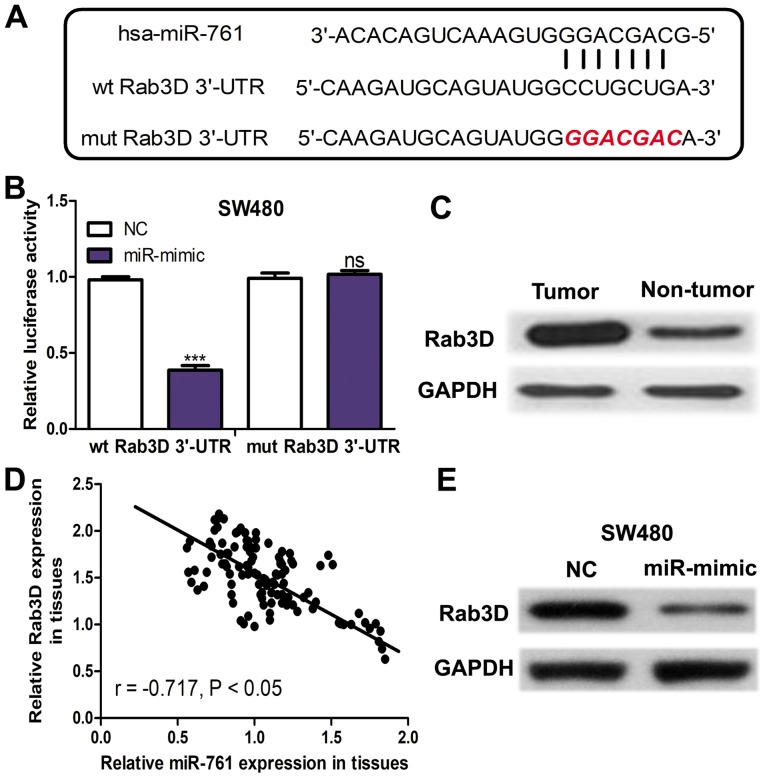
Rab3D is a direct target of miR-761
TargetScan was used to predict potential targets of miR-761. The binding site between miR-761 and Rab3D 3′-UTR was presented in Fig. 2A. The Luciferase activity reporter assay revealed that miR-mimic significantly decreased the luciferase activity of wt Rab3D 3′-UTR (P<0.001) but not mut Rab3D 3′-UTR (Fig. 2B). The protein expression of Rab3D was measured in the collected tissues and it was demonstrated that Rab3D expression was upregulated in CRC tissues compared with in the adjacent normal tissues (Fig. 2C). Furthermore, an inverse correlation between miR-761 and Rab3D expression was identified in CRC tissues (Fig. 2D). Furthermore, miR-761 expression in miRNAs transfected SW480 cell line was detected. It was demonstrated that Rab3D expression was downregulated in the SW480 cell line transfected with miR-mimic compared with the NC (Fig. 2E).
Figure 2.
Rab3D is a direct target of miR-761 in CRC. (A) A schematic description of the base-pairing interaction between miR-761 and 3′-UTR of Rab3D. (B) miR-761 overexpression significantly decreased luciferase activity of wt Rab3D 3′-UTR but not mut Rab3D 3′-UTR. (C) Western blotting to analyze Rab3D expression in CRC tissues and nontumor tissues. (D) Correlation analysis was performed for miR-761 and Rab3D expression in CRC tissues. (E) Western blotting to analyze Rab3D expression in the SW480 cell line with the miR-mimic or NC transfection. ***P<0.001 vs. the wt NC. ns vs. the wt NC group. miR-761, microRNA-761; ns, not significant; CRC, colorectal cancer; UTR, untranslated region; wt, wild-type; mut, mutant; NC, negative control.
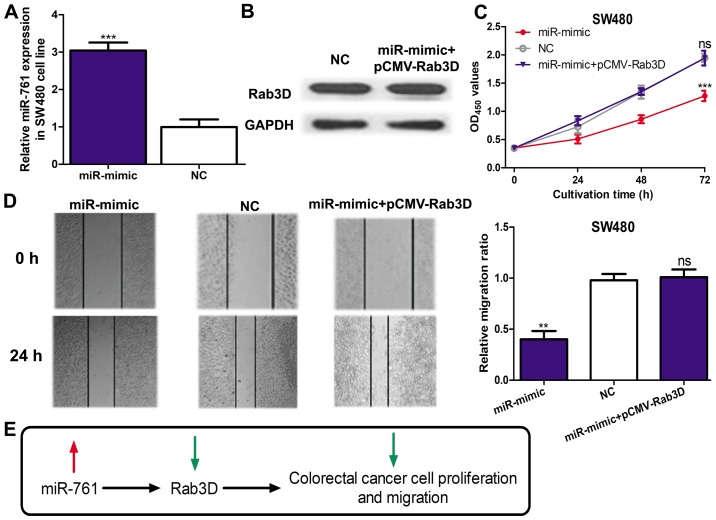
miR-761 regulates cell proliferation and migration through targeting Rab3D
To investigate whether Rab3D was a mediator for the biological role of miR-761, an miR-mimic and pCMV3-Rab3D was co-transfected into the SW480 cell line. miR-761 expression was significantly elevated by the miR-mimic (P<0.001; Fig. 3A). In addition, Rab3D expression in the miR-mimic and pCMV-Rab3D co-transfected cells was similar to that in the NC transfected cell, which suggested pCMV-Rab3D could efficiently abolish the effect of the miR-mimic on Rab3D expression (Fig. 3B). A cell proliferation assay revealed that miR-mimic could significantly downregulate SW480 cell proliferation (P<0.001) but this effect can be partially reversed by pCMV-Rab3D (Fig. 3C). Similarly, the cell migration assay also demonstrated that the inhibition effect of miR-mimic on cell migration could be reversed by pCMV-Rab3D (Fig. 3D). These results confirmed overexpression of miR-761 inhibited the proliferation and migratory abilities of CRC through regulating the expression of Rab3D (Fig. 3E).
Figure 3.
miR-761 regulates CRC cell proliferation and migration through targeting Rab3D. (A) Reverse transcription-quantitative polymerase chain reaction was used to analyze miR-761 expression in the SW480 cell line with the miR-mimic or NC transfection. (B) Western blotting was used to analyze Rab3D expression in the SW480 cell line with miR-mimic and pCMV-Rab3D co-transfection. (C) Cell Counting kit-8 assay was used to analyze cell proliferation in the SW480 cell line with the miR-mimic and pCMV-Rab3D transfection. (D) Wound-healing assay to analyze cell migration following miR-mimic and pCMV-Rab3D transfection. (E) A schematic diagram demonstrating the miR-761/Rab3D axis in CRC progression. **P<0.01 and ***P<0.001 vs. the NC group. ns vs. the NC group. ns, not significant; miR-761, microRNA-761; CRC, colorectal cancer.
Discussion
CRC development has been reported to be accompanied by numerous genetic and epigenetic alterations (4). However, the molecular mechanisms underlying CRC pathogenesis are still not fully understood and therefore the therapeutic methods for treating CRC remain limited (3,4). Therefore, the identification of novel biomarkers to facilitate the development of novel therapeutic methods for CRC is in urgent demand.
miR-761 was aberrantly expressed in several human types of cancer including hepatocellular carcinoma, NSCLC and ovarian cancer (10–12). miR-761 was also capable of regulating cell behaviors through regulating numerous target genes (10–12). For example, it was reported that miR-761 regulated the sensitivity of CRC cell to chemotherapy but little is known regarding the function of miR-761 in mediating cell behaviors (13). In addition, the current study demonstrated that miR-761 expression was downregulated in CRC tissues compared with in matched normal tissues. Furthermore, low miR-761 expression was a predictor for advancer tumor stage, large tumor size and even poorer 5-year overall survival of CRC patients. Collectively, miR-761 expression may serve as a prognostic predictor for CRC.
miRNAs are important regulators for tumorigenesis and it was proved that miRNAs have the potential to be used as novel therapeutic methods for cancer (10–13,15). Rab3D, one member of the Rab GTPase family, serves an oncogenic role in several cancer types including osteosarcoma, esophageal squamous cell carcinoma and CRC (16–18). Evidence demonstrated that Rab3D is a key player in regulating tumor metastasis, which suggested that silencing Rab3D expression may be a way to restrain cancer metastasis (19). However, the molecule that regulates Rad3D expression in the progression of cancer is poorly understood. In the present study, it was demonstrated Rab3D was a direct target of miR-761 by bioinformatics analysis and luciferase activity reporter assay. Importantly, the present study demonstrated the inverse correlation between miR-761 and Rab3D expression in CRC tissues. In vitro functional assays revealed overexpression of miR-761 inhibited CRC cell proliferation and migration. In addition, it was demonstrated that overexpression of Rab3D attenuated the suppressive effects of miR-mimic on CRC cell proliferation and migration.
In conclusion, the present study demonstrated that miR-761 which was downregulated in CRC inhibited CRC cell proliferation and migration partially through regulating Rab3D. The present study also demonstrate that low miR-761 was correlated with worse 5-year overall survival of CRC patients. Furthermore, the present study provides a novel insight into the mechanism regarding CRC progression and a promising future for miR-761/Rab3D axis-oriented treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YR, GS, PJ and QM participated in the study design, carried out experiments, collected and analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute.
Patient consent for publication
Written informed consent was obtained from all patients prior to enrollment in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Goss PE, Strasser-Weippl K, Lee-Bychkovsky BL, Fan L, Li J, Chavarri-Guerra Y, Liedke PE, Pramesh CS, Badovinac-Crnjevic T, Sheikine Y, et al. Challenges to effective cancer control in China, India, and Russia. Lancet Oncol. 2014;15:489–538. doi: 10.1016/S1470-2045(14)70029-4. [DOI] [PubMed] [Google Scholar]
- 2.Varghese C, Shin HR. Strengthening cancer control in China. Lancet Oncol. 2014;15:484–485. doi: 10.1016/S1470-2045(14)70056-7. [DOI] [PubMed] [Google Scholar]
- 3.Chakradhar S. Colorectal cancer: 5 big questions. Nature. 2015;521:S16. doi: 10.1038/521S16a. [DOI] [PubMed] [Google Scholar]
- 4.Brody H. Colorectal cancer. Nature. 2015;521:S1. doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 5.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA processing and human cancer. J Clin Med. 2015;4:1651–1667. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: Playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Zhang L, Zheng B, Yan Y, Zhang Y, Xie H, Zhou L, Zheng S, Wang W. MicroRNA-761 is upregulated in hepatocellular carcinoma and regulates tumorigenesis by targeting Mitofusin-2. Cancer Sci. 2016;107:424–432. doi: 10.1111/cas.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan A, Yang C, Chen Z, Li C, Cai L. MiR-761 promotes progression and metastasis of non-small cell lung cancer by targeting ING4 and TIMP2. Cell Physiol Biochem. 2015;37:55–66. doi: 10.1159/000430333. [DOI] [PubMed] [Google Scholar]
- 12.Shi C, Zhang Z. miR-761 inhibits tumor progression by targeting MSI1 in ovarian carcinoma. Tumour Biol. 2016;37:5437–5443. doi: 10.1007/s13277-015-4377-z. [DOI] [PubMed] [Google Scholar]
- 13.Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget. 2017;9:321–331. doi: 10.18632/oncotarget.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Jiashi W, Chuang Q, Zhenjun Z, Guangbin W, Bin L, Ming H. MicroRNA-506-3p inhibits osteosarcoma cell proliferation and metastasis by suppressing RAB3D expression. Aging (Albany NY) 2018;10:1294–1305. doi: 10.18632/aging.101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Kong RR, Sun LZ. Silencing of Rab3D suppresses the proliferation and invasion of esophageal squamous cell carcinoma cells. Biomed Pharmacother. 2017;91:402–407. doi: 10.1016/j.biopha.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Luo Y, Ye GY, Qin SL, Mu YF, Zhang L, Qi Y, Qiu YE, Yu MH, Zhong M. High expression of Rab3D predicts poor prognosis and associates with tumor progression in colorectal cancer. Int J Biochem Cell Biol. 2016;75:53–62. doi: 10.1016/j.biocel.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Liu W, Lu X, Fu Y, Li L, Luo Y. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget. 2015;6:11125–11138. doi: 10.18632/oncotarget.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used/or analyzed during the current study are available from the corresponding author on reasonable request.