Abstract
The increased speed and sensitivity in mass spectrometry-based proteomics has encouraged its use in biomedical research in recent years. Large-scale detection of proteins in cells, tissues, and whole organisms yields highly complex quantitative data, the analysis of which poses significant challenges. Standardized proteomic workflows are necessary to ensure automated, sharable, and reproducible proteomics analysis. Likewise, standardized data processing workflows are also essential for the overall reproducibility of results. To this purpose, we developed PaDuA, a Python package optimized for the processing and analysis of (phospho)proteomics data. PaDuA provides a collection of tools that can be used to build scripted workflows within Jupyter Notebooks to facilitate bioinformatics analysis by both end-users and developers.
Keywords: proteomics, high-throughput, data analysis, python library
Introduction
Data analysis in (phospho)proteomics is constantly evolving. State of the art mass spectrometers are able to identify and quantify thousands of proteins in a single shot-gun experiment, generating large volumes of data. The era of next-generation proteomics has further driven the use of mass spectrometry (MS) in biomedical research by allowing biological samples to be processed in high-throughput fashion.1 The need to cope with complex experimental designs and big data has driven the search for more efficient approaches for proteomics data analysis.
Bioinformatics has already dealt with the challenges of large-scale data processing in other “omics” fields. An illustration of high-throughput analysis in genomics and transcriptomics is given by Galaxy.2,3 This established web-based platform allows data mining and workflow construction from standalone scripts. Moreover, Galaxy offers an open and collaborative environment, which facilitates genomics research through improved accessibility, reproducibility, and transparency. Quantitative (phospho)proteomics can also benefit from such platforms, and their advancement is reliant on the availability of scriptable analysis tools. For instance, Röst et al. developed the OpenMS software, which offers both standard workflows and individual tools that together with a Python scripting interface allow high-throughput MS data analysis.4 Reproducibility of analyses is dependent on stored workflow files containing complete records of the analysis history, and allows different users to apply them on their own data.5
Lately, the combination of programming language alongside documentation language is gaining interest. This concept, first introduced by Donald Knuth as Literate Programming in the 1980s, promotes the use of descriptive documented pipelines to make analyses more robust, more portable, more easily maintained, and eventually pieces of literature.6 The open source Jupyter Notebooks system has been developed in this context with the aim to share and reproduce interactive data analysis.7 Notably, Jupyter supports over 40 programming languages popular in data science (e.g., Python, R, or Julia), and can leverage big data tools for high-throughput analysis. By combining explanatory text, raw code, charts, and figures, Jupyter Notebooks can be used by scientists as complete and detailed program documentation alongside publication.8
To perform the analysis of quantified (phospho)proteomic data in Jupyter, we have developed PaDuA, a Python package first optimized for MaxQuant output data.9,10 Of the available proteomics quantification software, MaxQuant is the most commonly used freely available software package for analyzing large-scale mass-spectrometric data sets.10 Modeled on established (phospho)proteomics analysis methods, PaDuA provides tools for data processing, filtering, and statistical analysis both within the Jupyter notebook environment and in other scriptable systems. Results are read and written in tabular format so that further analysis with other platforms like Perseus11 or R12 is possible. Since the analysis procedure is split up in small blocks of code, it is possible to repeat and optimize the analysis as a whole but also partially. The final analysis can be easily shared as a notebook file, guaranteeing reproducibility of results over time. It also allows researchers to reuse and adapt the workflows for their own analysis, supporting standardization of methods.
We have already applied PaDuA for investigating molecular responses of a large-scale (phospho)proteomics experiment upon drug treatments.13 In this study, we demonstrate the versatility of PaDuA on two published phospho- and proteomics data sets and the reproducibility of these analyses using Jupyter notebooks.
Experimental Procedures
PaDuA Development
PaDuA source code is freely available for download from https://github.com/mfitzp/padua and available under the BSD 2-clause (Simplified) license. The software is released as a standard Python package, and it is compatible with both Python 2.7 and 3.4+ and made available via the Python Package Index (PyPi). It features a complete set of standard proteomics processing, analysis, and visualization tools accessible via the fully documented (http://padua.readthedocs.io/en/latest/) application programming interface (API). PaDuA makes extensive use of other open source libraries including the Python scientific and numerical computing libraries SciPy and NumPy for data analysis,14,15 pandas DataFrame objects for internal data representations,16 and scikit-learn for machine learning algorithms.17 Publication quality figures are generated via Matplotlib with export in vector and high resolution formats.18 PaDuA is designed to perform analysis by selecting columns from output tables generated by MaxQuant. This software package is available in different versions, which may slightly differ in the columns’ header, affecting the performance of PaDuA. The use of a template containing standard labeled columns matching the ones listed in the quantified MaxQuant tables could overcome this limitation.
PaDuA Workflow Strategy
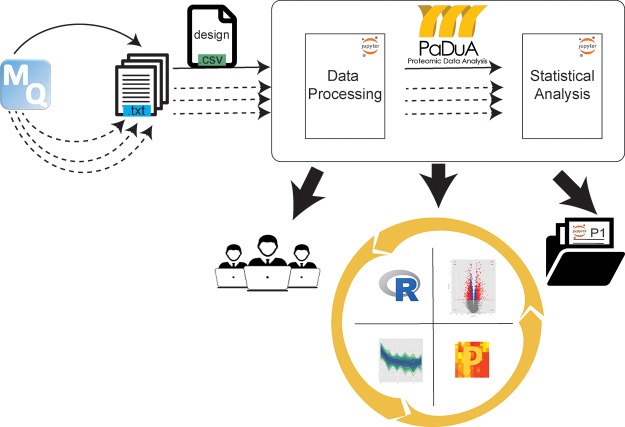
The PaDuA analysis workflow is illustrated in Figure 1. Search output files generated by MaxQuant are imported into a running Jupyter Notebook environment together with the experimental design and then processed through two consecutive steps: Data Processing and Statistical Analysis, each represented by a separate Jupyter notebook. The final output provides a complete list of publication-quality figures and tables that can be exported in a number of formats. Analyses can be quickly updated in case of reprocessed MaxQuant inputs simply by rerunning the workflow. Existing notebooks can be shared among other users and stored as recorded documentation for past projects.
Figure 1.
PaDuA works within the Jupyter Notebook environment and uses MaxQuant output search files and the experimental design table as input. Data Processing and Statistical Analysis notebooks are used for filtering and analyzing data, respectively. Results can be exported to other platforms like R or Perseus, shared among different users or stored with back-up projects. The full analysis can be reprocessed infinite times (dot lines).
Input and Output
PaDuA supports input from all file types offered by the Pandas library, including CSV, Excel, HDF, SQL, JSON, and Python pickle format. Standardized tab-delimited formats are used as input for data processing, and as output for R,12 Phosphopath,19 and Perseus.11 A table labeled as design in CSV format is required for mapping individual samples to experimental conditions. This table contains at least two columns: “Label” as for sample labels derived from MaxQuant output, and “Group” as for categorical column corresponding to classification of samples according to the treatment. Depending on the experimental workflow, more columns could be listed in design: “Timepoint” as numeric column corresponding to the time point, “Replicate” as for numeric column corresponding to the number of biological replicate, and “Technical” as for numeric column corresponding to the number of technical replicates. These group types are not restricted, and other groups can be set if required by an experiment. Moreover, in the included workflows, the pickle format is used as input for Statistical Analysis to simplify reloading of processed data.
Data Processing
Initial steps for (phospho)proteomics analysis are focused on refining data sets to the final format needed for statistical analysis. This is achieved through standard processing and filtering steps that can be consistently and rapidly applied with PaDuA. Either intensity (or LFQ) or ratio columns can be selected for quantification analysis. In addition, PaDuA supports basic data normalization strategies and log2 transformation, which are commonly applied before statistical analysis, while more complicated normalization strategies are possible using Python libraries specialized for this purpose. Filter tools can be used to simplify the overall data set, and each analysis step generates DataFrame objects, which can be further inspected within the notebook environment or exported in various output formats. Finally, PaDuA supports two data imputation strategies to automatically fill missing values with estimated quantities based on statistical models including (i) random sampling from a normal distribution and (ii) least-squares modeling of present values based on structural equation modeling (SEM), as already described by Webb-Robertson et al.20 The data processing workflow concludes with export of the final DataFrame, both as CSV and Python pickle format.
Statistical Analysis
PaDuA data analysis is structured around two included submodules: Analysis and visualize. The former performs statistical analysis returning the numerical results of the operation, while the latter generates plots for the same analysis. Supported statistical analysis tools include quality control tools, which evaluate the quality of each sample (i.e., sample-wise Pearson correlation and enrichment analysis), and several multivariate methods that are well suited to isolate important variation in large data sets such as principal component analysis (PCA), partial least-squares regression (PLS-R), partial least-squares discriminant analysis (PLS-DA), and analysis of variance (ANOVA). Plot visualizations include mainly volcano plots and clustering analysis such as hierarchical clustering, Venn diagrams, and KEGG pathways. All standard data plotting functions from the Pandas library may be also used.
Results and Discussion
To benchmark PaDuA as a versatile and reproducible data analysis tool, two different data sets publicly available in Proteomics Identifications Database (PRIDE) were selected. The first (PXD000293) was generated using a label-free quantification approach on a large-scale Ti4+-IMAC phosphopeptide enrichment.21 In this study, de Graaf et al. demonstrated the qualitative and quantitative reproducibility of such approach in monitoring the temporal phosphorylation signaling of Jurkat T-cells upon stimulation of the G protein coupled receptors with their ligand Prostaglandin E2 (PGE2). The binding between G protein coupled receptors and PGE2, indeed, leads to the activation of intracellular signaling transduction cascades including cAMP/PKA as well as the PI3K-dependent ERK1/2 pathways. For this experiment, Jurkat cells were cultured in three biological replicates and harvested after 0, 5, 10, 20, 30, and 60 min of PGE2 stimulation. Phosphopeptides were enriched using three independent Ti4+-IMAC enrichment columns for every biological replicate, and each column was analyzed twice by nanoliquid chromotography–tandem mass spectrometry (nLC–MSMS). For the second data set (PXD000497), Smit et al. used a dimethyl labeling strategy to quantify (phospho)proteome changes in melanoma cells after drug treatment.22 The subsequent integration with next generation sequencing data obtained by melanoma cell transduced with shRNA library allowed the authors to identify ROCK1 as novel therapeutic target that can be used in the treatment of melanoma patients. For the proteomics experiment, melanoma cells were cultured in three biological replicates and treated without drug (control) and with PLX4720 (BRAF inhibitor). Both control and treated samples derived from 1 and 3 days were collected and labeled as “Light” (L), “Medium” (M), and “Heavy” (H), respectively. Jupyter notebooks showing the workflow analyses for both data sets are further provided as.ipynb format together with the design tables in the Supporting Information.
Demonstration data: phospho-data
Data Processing
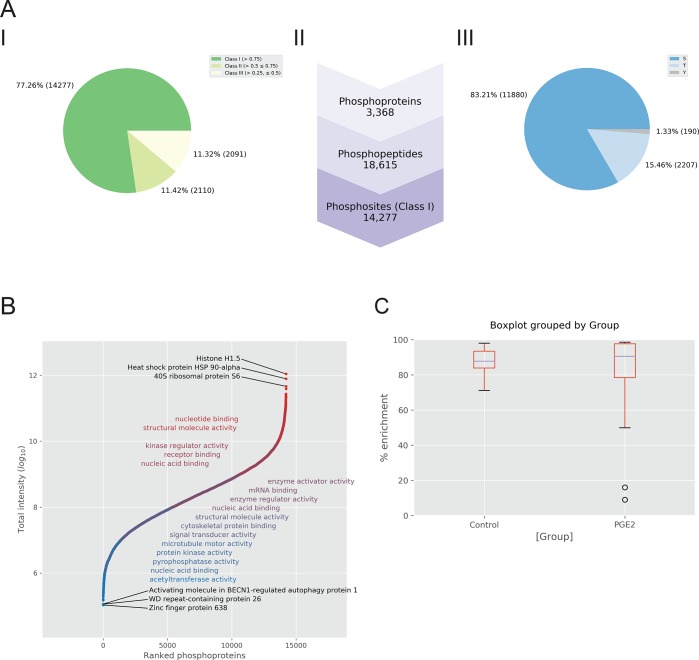
Phospho(STY)Sites, modificationSpecificPeptides, and Evidence are the .txt files selected from the phosphoproteomics data set PXD000293. These are the output tables generated by MaxQuant containing the list of quantified phosphosites, modified peptides, and identified peptides, respectively. Both Phospho(STY)Sites and its design table (Supporting Information) are initially imported as input files. A filtering step is immediately performed using MaxQuant metadata annotations to remove peptides flagged as “contaminants” and “reverse”. Next, identified phosphopeptides are further filtered to ensure confident site localization of the modification with a probability typically at 0.75. PaDuA also calculates relative percentage of phosphorylations in different localization probability groups, displaying these as pie charts. In the current phosphopeptide data set, 77% of the phosphosites are Class I (>0.75), while Class II (>0.5 ≤ 0.75) and III (>0.25 ≤ 0.5) each contain around 11% (Figure 2A, panel I). A useful overview of the quality of the experimental data is provided by a summary list of the total number of phosphoproteins, phosphopeptides, and phosphosites (Class I) as shown in Figure 2A (panel II). Relative abundances of modified amino acids are also rapidly calculated in PaDuA, and in this data set, over 83% of phosphorylated amino acid sites are serine, 15% are threonine, while just 1.33% are tyrosine (Figure 2A, panel III). A global overview of biological function of the identified phosphoproteins, in combination with their intensity distribution, can be observed in PaDuA using the rank-intensity plot, containing Gene Ontology (GO) annotations queried from the PANTHER database23,24 (Figure 2B). PaDuA emulates the expand side table process of Perseus:11 All the columns containing 1, 2, and 3 modifications for the same phosphopeptide are folded into rows, obtaining a unique column containing up to three modifications for each peptide. This step is necessary to facilitate the subsequent normalization step, which is based on the subtraction of the median of the column for each sample. Moreover, this simplifies the following quantification steps where each column corresponds to a sample condition. After normalizing intensity columns, a final multi-index table (DataFrame) can be obtained by matching the design table with selected columns from the input search. This DataFrame contains sample annotations arranged horizontally, and quantified values arranged vertically (Figure S-1). The use of this multi-index matrix allows easy filtering of the number of quantified values based on either time points, or number of biological or technical replicates. For these phosphoproteomics data, PaDuA calculates 10 732 phosphorylation events in at least two out of three biological replicates.
Figure 2.
(A) Data Processing notebook illustrates summaries of the phosphoproteomics identification data as standard graphs. Panel I shows the percentage of phosphosites belonging to different localization probability groups; panel II displays the list of identified phosphoproteins, phosphopeptides and phosphosites (Class I); panel III represents the percentage of modified phosphosites on serine, threonine and tyrosine (Class I). (B) Rank intensity plot shows phosphoprotein intensity values versus their corresponding ranks. Annotation of phosphophoproteins can be visualized by overlaying on the S curve the results of GO enrichment analysis. (C) Box plots of percentage of phosphopeptide enrichment for both unstimulated (control) and stimulated samples with PGE2.
Statistical Analysis
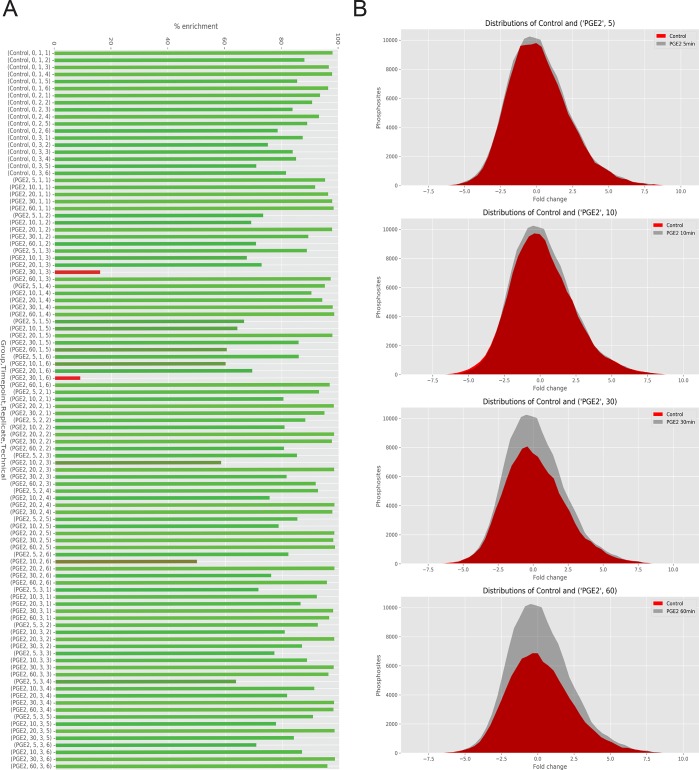
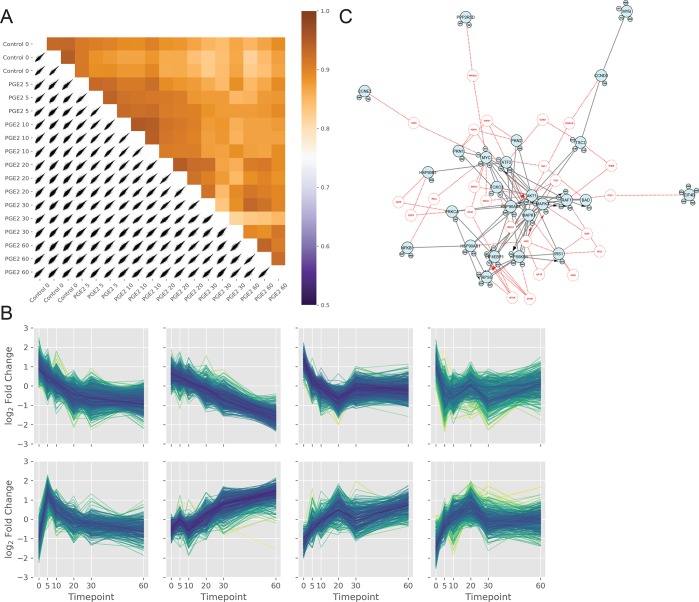
The .pickle file resulting from the data processing is then used for the next analysis step. The percentage of phosphopeptide enrichment in the data set can be calculated dividing the phosphopeptide relative abundances through the nonmodified peptide relative abundances from the MaxQuant modificationSpecificPeptides or Evidence files, annotated with the same experimental design of design table. Bar-plots and box-plots are used to visualize the phosphopeptide enrichment trend and to detect potential outliers. Enrichment scores can be calculated per group or per single sample, and percentage values correspond to the number of quantified phosphorylated peptides with respect to the total number of peptides. Figure 2C shows the average phosphopeptide enrichment being higher than 90% for both control and samples stimulated with PGE2 with two outliers for PGE2 stimulated samples. These outliers can be visualized in a bar-plot as shown in Figure 3A, displaying in red the technical replicates 1 and 6 of biological replicate 1 at 30 min after stimulation with PGE2. This feature in PaDuA allows the user to quickly recognize the two failed enrichments, which can be removed from the multi-index DataFrame to ensure quality of the data. Another informative function is given by “comparedist”, which calculates and compares the number of phosphorylation events happening in different samples or conditions. In the data used here, the number of phosphorylation events was found to be reduced over time after PGE2 stimulation compared to the control (Figure 3B). To gain further insight into the data set, PaDuA allows the construction of multiscatter plots based on Pearson correlation analysis. The heat-map visualization of these plots allows a rapid check of data integrity (Figure 4A). For studying temporal regulation patterns, PaDuA provides a hierarchical clustering function, illustrated in Figure 4B, where eight clusters are used to display the temporal dynamics of the significantly regulated phosphorylated sites. Further GO enrichment analysis of any of the clusters can be performed selecting ‘function’, ’process’, ‘cellular_location’, ‘protein_class’, or ‘pathway’ from the PANTHER database. Finally, PaDuA can export filtered lists of significant phosphosites to PhosphoPath formats19 for subsequent temporal signaling network and enrichment analyses in Cytoscape.25 As already shown by de Graaf et al.,21 PI3K-AKT signaling is one of the most significantly enriched pathways in this phosphorylation data set (p-value = 5.49 × 10–78), and its network is illustrated in Figure 4C.
Figure 3.
(A) Bar plot of phosphopeptide enrichment analysis for each single sample. Red bars display a phosphopeptide enrichment percentage below 20%. (B) Distribution of phosphosite events plotted as a Gaussian curve area at each time-point. Stimulated samples (red) show reduction of phosphorylation respect to the control (gray) over time.
Figure 4.
(A) Correlation plot of the independent phosphoproteomics experiments shows Pearson coefficient correlation values as a heat-map. (B) Hierarchical clustering of samples across the time course experiment. Samples are z-scored along the 0-axis (y) by default. (C) PI3K/AKT network visualized in PhosphoPath using the PaDuA output containing the significant regulated phosphosites and their quantitative ratios.
Demonstration Data: Proteomics Data
Data Processing
For the proteomics workflow, ProteinGroups is the .txt file containing the quantified protein groups from MaxQuant, and therefore the one selected from the proteomic-data set PXD000497 for further analysis. Both ProteinGroups and its design table (Supporting Information) are imported as input files, followed by common filtering steps as removing reverse database identifications and contaminants. Moreover, to ensure all proteins are quantified according to 1% FDR, peptides only identified because containing post- translational modifications are removed. In this way, PaDuA allows the selection of ratio intensity columns to further process isotopically labeled proteomics data. After building the annotated multi-index table DataFrame, a final filtering step can be performed to select protein groups quantified in at least two out of three biological replicates. For this proteomics data set, PaDuA calculates 4785 protein groups over the three sampled time-points.
Statistical Analysis
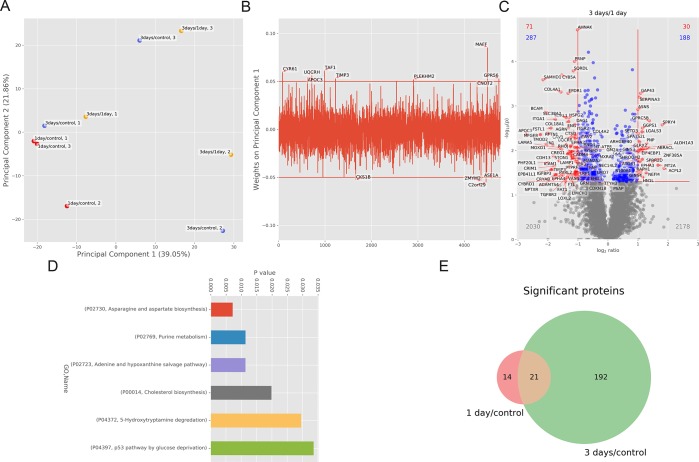
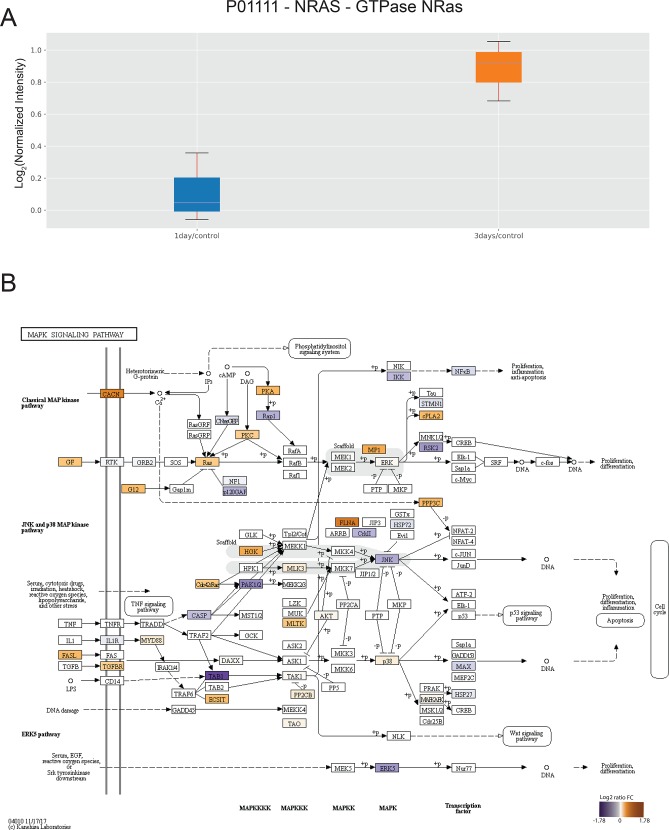
The resulting .pickle file is then used as input for the data analysis notebook (Supporting Information). Principal component analysis (PCA) can be used as quality control tool to capture differences between groups while identifying possible outliers. Moreover, PCA allows to select interesting proteins from the input data on the basis of the relationship between experimental groups and features. PaDuA supports PCA with sample annotations, emphasizing the visualization of clusters and variation. Figure 5A shows a separation of samples between 1 and 3 days drug treatment versus control (1 day/control and 3 days/control) along principal component 1 (PC1), revealing a poor clustering of biological replicates at 3 days, which is further reflected in the inability to cluster biological replicates of 3 days/1 day. In addition, as a result from the PCA analysis, PaDuA generates the score and weight plots, which can be used to interpret the main biological response causing the difference between clusters. An example of weight plot related to PC1 is visualized in Figure 5B. Selecting an arbitrary cutoff on the weight axis allows researchers to identify proteins that contribute most (weights > 0.05) or less (weights < 0.05) to the separation along the PC1 axis. Among the proteins with weights >0.05, we can observe the transcription factors TAF1 and MAFF, which possess DNA-binding activity, and CYR61 and GPR56, which play active roles in cell adhesion. One-sample or two-sample independent t tests can be used to calculate proteins significantly regulated after drug treatment. These analyses are visualized as volcano plots, which may be annotated with regulated proteins or gene names, together with information on total number of up, down, and significantly regulated values. As an example, we show a two-sample t-test analysis of 3 days versus 1 day treatment, revealing 30 and 71 proteins significantly up- and downregulated, respectively, with a p-value < 0.05 and a fold change cutoff of 2 (red dots in Figure 5C). Enrichment analysis of significant up-regulated proteins–calculated in PaDuA with PANTHER database and using ‘Homo sapiens’ as default background reveals metabolic pathways significantly upregulated (p-value <0.05), as shown in Figure 5D. To classify common regulated proteins under different conditions, PaDuA can display Venn diagrams, form which the identified subsets of proteins can be easily exported as CSV file for further analysis. Figure 5E displays 227 significantly regulated proteins of which 21 are in common between 1 and 3 days drug treatment versus control. Quantitative expression of these proteins can be further visualized through basic plotting tools such as box-plots. Figure 6A illustrates ratio expression of the protein NRAS at both 1 and 3 days versus control. As reported by Smit et al.,22 NRAS is up-regulated after 3 days drug treatment compared to control and 1 day treatment. Finally, PaDuA is able to map protein quantitation values onto signaling pathways with a built-in script that generates a gradient-colored KEGG pathway26 (Figure 6B). Thanks to this feature, it is possible to rapidly evaluate the regulation of the cellular response after 3 days of drug treatment by mapping it onto the MAPK pathway, which easily visualizes the upregulated proteins which may play a role in melanoma BRAF inhibitor resistance such as RAS and Cdc42.22
Figure 5.
(A) PCA analysis of quantitative proteome data with sample annotations: Colors distinguish early treated (red) from late-treated samples (blue). In yellow, the third experimental group is indicated, which consists of the ratio between 3 days and 1 day treatment. For each sample, the biological replicate number is reported. (B) Weight of principal component 1 identifies key proteins, which affect the separation between the early and late-treated samples. (C) Volcano plot as visualization of one-sample t-test of protein expression levels at 3 days versus control. Statistically significant values with p-value < 0.05 and fold change ≥ 2 are labeled in red. Values with p-value < 0.05 and fold change ≤ 2 are labeled in blue. All the values with p-value > 0.05 are labeled in gray. (D) Bar plot of GO enrichment analysis of significant up-regulated pathways at 3 days treatment. (E) Venn diagram of significantly regulated proteins at 1 day and 3 days of treatment versus control.
Figure 6.
(A) Box plot of NRAS protein expression at both 1 day and 3 days of treatment versus control. (B) KEGG pathway shows protein regulation after 3 days of drug treatment in MAPK signaling.
Conclusions
We have presented PaDuA, a new Python library for large-scale (phospho)proteomics data analysis. We primarily developed PaDuA with the idea to propose a new concept of standardized data analysis and data sharing. There is a constantly growing need in the proteomics community for such workflow especially in project-based environment. Nowadays, MaxQuant represents one of the most well-known and freely available quantification platform currently used in proteomics. Therefore, our proof of concept for PaDuA is based on MaxQuant output, with the intent that both users and programmers can contribute to further development of PaDuA in an interactive manner.
We have shown the versatility of the tool by applying standard workflows strategies to two example data sets. Built in Python, PaDuA benefits from the existing ecosystem of data analysis tools including Jupyter Notebooks. Users with only basic Python programming knowledge can work with standardized notebooks, while more proficient programmers can integrate and customize the analysis within other tools and environments. PaDuA is a valuable platform for rapid and automatable analysis of both isotopically labeled and label-free MS data.
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (NWO) through the Gravity Program CGC.nl, Proteins At Work embedded in The Netherlands Proteomics Center, as part of the National Roadmap Largescale Research Facilities of The Netherlands (Project No. 184.032.201) and through a VIDI grant for M.A. (723.012.102).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00576.
Author Contributions
‡ These authors contributed equally.
The authors declare no competing financial interest.
Notes
PaDuA is made available at https://github.com/mfitzp/padua. A manual with a complete description of its installation and use is available at https://padua.readthedocs.io/en/latest/. Proteomics and phosphoproteomics data were downloaded from the Pride database ProteomeXchange ID: PXD000497 and PXD000293, respectively. Jupyter notebooks and table input file for the phosphoproteomics workflow and Jupyter notebooks and table input file for the proteomics workflow are available via github at https://github.com/mfitzp/padua.
Supplementary Material
References
- Altelaar A. F. M.; Munoz J.; Heck A. J. R. Next-Generation Proteomics: Towards an Integrative View of Proteome Dynamics. Nat. Rev. Genet. 2013, 14 (1), 35–48. 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- Giardine B.; Riemer C.; Hardison R. C.; Burhans R.; Elnitski L.; Shah P.; Zhang Y.; Blankenberg D.; Albert I.; Taylor J.; et al. Galaxy: A Platform for Interactive Large-Scale Genome Analysis. Genome Res. 2005, 15 (10), 1451–1455. 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J.; Nekrutenko A.; Taylor J.; Galaxy: A Comprehensive Approach for Supporting Accessible, Reproducible, and Transparent Computational Research in the Life Sciences. Genome Biol. 2010, 11 (8), R86. 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst H. L.; Sachsenberg T.; Aiche S.; Bielow C.; Weisser H.; Aicheler F.; Andreotti S.; Ehrlich H.-C.; Gutenbrunner P.; Kenar E.; et al. OpenMS: A Flexible Open-Source Software Platform for Mass Spectrometry Data Analysis. Nat. Methods 2016, 13 (9), 741–748. 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- Pfeuffer J.; Sachsenberg T.; Alka O.; Walzer M.; Fillbrunn A.; Nilse L.; Schilling O.; Reinert K.; Kohlbacher O. OpenMS – A Platform for Reproducible Analysis of Mass Spectrometry Data. J. Biotechnol. 2017, 261, 142–148. 10.1016/j.jbiotec.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Knuth D. E. Literate Programming. Comput. J. 1984, 27 (2), 97–111. 10.1093/comjnl/27.2.97. [DOI] [Google Scholar]
- Kluyver T.; Ragan-kelley B.; Pérez F.; Granger B.; Bussonnier M.; Frederic J.; Kelley K.; Hamrick J.; Grout J.; Corlay S.; et al. Jupyter Notebooks - a Publishing Format for Reproducible Computational Workflows. Position. Power Acad. Publ. Play. Agents Agendas 2016, 87–90. 10.3233/978-1-61499-649-1-87. [DOI] [Google Scholar]
- Shen H. Interactive Notebooks: Sharing the Code. Nature 2014, 515 (7525), 151–152. 10.1038/515151a. [DOI] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26 (12), 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Cox J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11 (12), 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Sinitcyn P.; Carlson A.; Hein M. Y.; Geiger T.; Mann M.; Cox J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13 (9), 731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- R Development Core Team.. R: A language and environment for statistical computing; The R Foundation, 2018. http://www.r-project.org/.
- Ressa A.; Bosdriesz E.; de Ligt J.; Mainardi S.; Maddalo G.; Prahallad A.; Jager M.; de la Fonteijne L.; Fitzpatrick M.; Groten S. A System-Wide Approach to Monitor Responses to Synergistic BRAF and EGFR Inhibition in Colorectal Cancer Cells. Mol. Cell. Proteomics 2018, 17, 1892. 10.1074/mcp.RA117.000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T. E. Python for Scientific Computing. Comput. Sci. Eng. 2007, 9 (3), 10–20. 10.1109/MCSE.2007.58. [DOI] [Google Scholar]
- van der Walt S.; Colbert S. C.; Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 2011, 13 (2), 22–30. 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- McKinney W.Data Structures for Statistical Computing in Python. Proc. 9th Python Sci. Conf., 2016.
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V.; et al. Scikit-Learn: Machine Learning in Python Gaël Varoquaux. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hunter J. D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9 (3), 90–95. 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- Raaijmakers L. M.; Giansanti P.; Possik P. A.; Mueller J.; Peeper D. S.; Heck A. J. R.; Altelaar A. F. M. PhosphoPath: Visualization of Phosphosite-Centric Dynamics in Temporal Molecular Networks. J. Proteome Res. 2015, 14 (10), 4332–4341. 10.1021/acs.jproteome.5b00529. [DOI] [PubMed] [Google Scholar]
- Webb-Robertson B.-J. M.; Wiberg H. K.; Matzke M. M.; Brown J. N.; Wang J.; McDermott J. E.; Smith R. D.; Rodland K. D.; Metz T. O.; Pounds J. G.; et al. Review, Evaluation, and Discussion of the Challenges of Missing Value Imputation for Mass Spectrometry-Based Label-Free Global Proteomics. J. Proteome Res. 2015, 14 (5), 1993–2001. 10.1021/pr501138h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf E. L.; Giansanti P.; Altelaar A. F. M.; Heck A. J. R. Single-Step Enrichment by Ti4+-IMAC and Label-Free Quantitation Enables in-Depth Monitoring of Phosphorylation Dynamics with High Reproducibility and Temporal Resolution. Mol. Cell. Proteomics 2014, 13 (9), 2426–2434. 10.1074/mcp.O113.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit M. A.; Maddalo G.; Greig K.; Raaijmakers L. M.; Possik P. A.; Van Breukelen B.; Cappadona S.; Heck A. J.; Altelaar A. M.; Peeper D. S. ROCK1 Is a Potential Combinatorial Drug Target for BRAF Mutant Melanoma. Mol. Syst. Biol. 2014, 10, 772. 10.15252/msb.20145450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H.; Lazareva-Ulitsky B.; Loo R.; Kejariwal A.; Vandergriff J.; Rabkin S.; Guo N.; Muruganujan A.; Doremieux O.; Campbell M. J.; et al. The PANTHER Database of Protein Families, Subfamilies, Functions and Pathways. Nucleic Acids Res. 2005, 33, D284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H.; Muruganujan A.; Casagrande J. T.; Thomas P. D. Large-Scale Gene Function Analysis with the PANTHER Classification System. Nat. Protoc. 2013, 8 (8), 1551–1566. 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13 (11), 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H.; Goto S.; Sato K.; Fujibuchi W.; Bono H.; Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27 (1), 29–34. 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.