Abstract
Hypothalamic GnRH together with gonadal steroids and activins/inhibin regulate its receptor gene (Gnrhr) expression in vivo, which leads to crucial changes in GnRHR numbers on the plasma membrane. This is accompanied by alterations in the gonadotroph sensitivity and responsiveness during physiologically relevant situations. Here we investigated basal and GnRH-regulated Gnrhr expression in rodent pituitary gonadotrophs in vitro. In pituitary cells from adult animals cultured in the absence of GnRH and steroid hormones, the Gnrhr expression was progressively reduced but not completely abolished. The basal Gnrhr expression was also operative in LβT2 immortalized gonadotrophs never exposed to GnRH. In both cell types, basal transcription was sufficient for the expression of functional GnRHRs. Continuous application of GnRH transiently elevated the Gnrhr expression in cultured pituitary cells followed by a sustained fall without affecting basal transcription. Both basal and regulated Gnrhr transcriptions were dependent on the protein kinase C signaling pathway. The GnRH-regulated Gnrhr expression was not operative in embryonal pituitary and LβT2 cells and was established neonatally, the sex-specific response patterns were formed at the juvenile-peripubertal stage and there was a strong correlation between basal and regulated gene expression during development. Thus, the age-dependent basal and regulated Gnrhr transcription could account for the initial blockade and subsequent activation of the reproductive system during development.
Keywords: Gonadotrophs, LβT2 cells, GnRH, Gnrhr, protein kinase C, ERK1/2
1. Introduction
The expression of GnRH receptors (GnRHRs) in pituitary gonadotrophs and their activation by hypothalamic GnRH are critical to the neuroendocrine regulation of reproduction in all vertebrates [1]. The GnRHRs are seven-transmembrane domain receptors [2] that signal through heterotrimeric Gq and/or G11 proteins. This activates phospholipase C-β1, which cleaves phosphatidylinositol-4,5-bisphosphate to generate inositol-1,4,5-trisphosphate and diacylglycerol [3]. In gonadotrophs, the binding of inositol-1,4,5-trisphosphate to inositol-1,4,5-trisphosphate receptor channels causes oscillatory calcium release from endoplasmic reticulum coupled with modulation of electrical activity and calcium influx through voltage-gated calcium channels [4]. Diacylglycerol and other lipophilic molecules that stay in the plasma membrane activate a family of protein kinase C (PKC) enzymes alone or together with calcium [5]. Signaling molecules downstream of PKC and calcium include mitogen-activated protein kinases (MAPK) [6], phospholipase D [7], and phospholipase A2 [8]. In LβT2 immortalized gonadotrophs, but not in cultured rat pituitary cells, GnRH also stimulates cAMP production through Gs signaling pathway [9].
The number of GnRHRs on the plasma membrane of gonadotrophs determines their responsiveness and varies during development, estrous cycle, pregnancy, lactation, and after gonadectomy. These modulations take place—at least in part—at the transcriptional level [10]. Transcription of the GnRHR gene (Gnrhr) promoters expressed in immortalized cells exhibit basal and regulated activities [1]. The gene structures accounting for regulated Gnrhr expression have been studied in numerous species including mouse [11], rat [12], human [13, 14], ovine [15], and porcine [16]. GnRH, estradiol, and progesterone are generally accepted as major regulators of Gnrhr expression in pituitary gonadotrophs [17–20]. The role of activin signaling in Gnrhr promoter activity is also established [21–23], as well as that glucocorticoids [24, 25] and pituitary adenylate cyclase activating polypeptide [26] contribute to the regulation of Gnrhr expression. In contrast, basal Gnrhr transcription, the signaling pathways accounting for it, and the developmental aspects of basal and regulated Gnrhr expression have not been systematically investigated. This probably reflects the common belief that the pituitary can respond to GnRH at any age and that the postnatal pattern of GnRH secretion is the main factor determining blockade and activation of the reproductive system [27].
To address these questions, we used cultured anterior pituitary cells from neonatal to adult female and male rats and developed a practical system to separately study basal and GnRH-stimulated Gnrhr expression during maturation. We also used cultured mouse pituitary cells to evaluate the species difference in Gnrhr expression. The mouse immortalized male LβT2 gonadotrophs [24] were also used to study basal and regulated Gnrhr expression.
2. Materials and methods
2.1. Chemicals
Fura 2-AM, medium 199, DMEM, as well as horse and fetal calf sera were purchased from Life Technologies (Grand Island, NY). GnRH, cetrorelix, BayK 8644, nifedipine, and 2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one (PD98059) were from Tocris Bioscience (Bristol, UK). The 3-(1-(3-(dimethylamino)propyl)-5-methoxy-1H-indol-3-yl)-4-(1H-indol-3-yl)-1H-pyrrole-2,5-dione (Gö6983), 1,4-diamino-2,3-dicyano-1,4-bis[2-a minophenylthio]butadiene (U0126), 4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4- pyridyl)imidazole (SB203580), 2H-dibenzo[cd,g]indazol-6-one SP600125, 2-[N-(4’-methoxybenzenesulfonyl)]amino-N-(4’-chlorophenyl)-2-propenyl-N-methylbenzyla mine phosphate (KN92), and N-[2-[[[3-(4-chlorophenyl)-2-propen yl]methylamino]methyl]phenyl]-N-(2-hydroxyethyl)-4-methoxybenzenesulphonamide (KN93) were from Calbiochem (La Jolla, CA). The (3Z)-3-[[[3-[(dimethylamino)methyl] phenyl]amino]phenylmethylene]-2,3-dihydro-N,N-dime thyl-2-oxo-1H-indole-6-carboxamide (BIX02189) was from Selleckchem (Houston, TX). Forskolin, phorbol 12-myristate 13-acetate PMA, staurosporine, a rabbit polyclonal anti-ACTB, and a protease inhibitor cocktail were from Sigma Aldrich (St. Louis, MO). The polyvinylidene difluoride (PVDF) blotting membranes and a rabbit polyclonal IgG Anti-PKCα, β, γ were from EMD Millipore Corporation (Temecula, CA). The donkey anti-rabbit IgG-HRP was from Santa Cruz Biotechnology (Santa Cruz, CA). Protein molecular weight marker was obtained from Bionexus (Oakland, CA). SuperSignal West Femto Chemiluminescent Substrate Kit was purchased from Thermo Fisher Scientific (Waltham, MA).
2.2. Cell cultures
Experiments were performed with cultured anterior pituitary cells from normal 6–7 week old female mice of different strains as well as infant to adult Sprague-Dawley male and female rats. All animals were from Taconic Farm (Germantown, NY). Animals were housed under constant temperature and humidity with the light on between 6 AM and 8 PM. Euthanasia was performed via asphyxiation with CO2, and the anterior pituitary glands were removed after decapitation. The experiments were approved by the National Institute of Child Health and Human Development Animal Care and Use Committee (14–041). The anterior pituitary cells were mechanically dispersed after treatment with trypsin and EDTA as described [28]. The cells were plated on poly-D-lysine coated 24-well plates—1.5 million per well. These were cultured in medium 199 containing Earle’s salts, sodium bicarbonate, and 10% heat-inactivated horse serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). If not otherwise stated, experiments were performed in cells bathed in 0.1% BSA-containing medium 199 Hank’s solution. The LβT2 immortalized pituitary cells were cultured in DMEM supplemented with 10% heat inactivated fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). The cell types were plated on polystyrene tissue culture treated 24-well plates and cultured until 70% confluence. If not otherwise stated, the cells were kept in 0.1% BSA-containing medium with 199 Hank’s solution overnight. The experiments were performed in the same type of medium, which was replaced at the beginning of treatment.
2.3. Quantitative RT-PCR (qRT-PCR) analysis
The total RNA was extracted from the pituitary tissue of cultured cells using RNeasy Plus Mini Kit purchased from Qiagen (Valencia, Ca). The amount of RNA was estimated using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE) and reverse transcribed with Transcriptor First Stand cDNA Synthesis Kit obtained from Roche Applied Science (Indianapolis, IN). An analysis of the relative gene expression was performed using quantitative real-time PCR and the comparative Ct method [29, 30]. For this, the LightCycler TaqMan Master mix and Lightcycler 2.0 Real-time PCR (Roche Applied Science, Indianapolis, IN) system were used. To compare the relative expression levels of the transcripts, the levels were calibrated against Gapdh and shown as percentage values with Gapdh expressed as 100%. Applied Biosystems predesigned Taq-Man Gene Expression Assays were used for Gapdh Mm99999915_g1 and Gnrhr Mm00439143_m1. The target gene expression levels were determined by comparative 2^(− δC(T)) quantification method using GAPDH as the reference gene. Linear regression analysis with mean amplification C(T) values showed no effects of age, gender or GnRH treatment on the expression of GAPDH mRNA in the anterior pituitary tissue and cells. This justified the use of GAPDH as a reference gene for analysis of mRNA expression.
2.4. Western blot analysis
A NuPAGE Electrophoresis System from Life Technologies (Grand Island, NY) was used for Western blot analysis. The cultured LβT2 cells were lysed using RIPA buffer (ready-to-use solution containing 150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) supplemented with a protease inhibitor cocktail. The samples were separated on NuPAGE Novex with 10% Bis-Tris gel and transferred onto a PVDF membrane. The membrane was blocked for 2 h at room temperature and was then incubated overnight at 4 °C with one of the primary antibodies: anti-PKCα, β, γ (1:1000) or anti-ACTB (actin beta) (1:10000). All studies were performed in 3% BSA in TBST (0.5 M Tris Base, 9% NaCl, 1.5% Tween 20, pH 8.4). After incubation with peroxidase-conjugated secondary antibody diluted to 1:10000, the blots were incubated with the SuperSignal West Femto Chemiluminescent Substrate, and the bands were visualized on a FluorChem E Digital Imaging System (ProteinSimple, San Jose, CA).
2.5. Intracellular calcium ion measurements
Measurements of the intracellular calcium ion concentrations ([Ca2+]i) in single cells were performed as previously described [31]. Briefly, the dispersed rat or mice anterior pituitary cells or LβT2 gonadotrophs were plated on poly-L-lysine-coated coverslips and cultured for 20 h. Next, the cells were washed and bathed in Krebs-Ringer-like medium containing 2.5 μM Fura 2 AM (Thermo Fisher Scientific, Waltham, MA) for 1 h at room temperature. Afterwards, the coverslips were washed in Krebs-Ringer-like medium and were mounted on the stage of an inverted Observer-D1 microscope (Carl Zeiss, Oberkochen, Germany) attached to an ORCA-ER camera (Hamamatsu Photonics, Hamamatsu City, Japan) and a Lambda DG-4 wavelength switcher (Sutter, Novato, CA). The hardware control and image analysis were performed using Metafluor software (Molecular Devices, Downingtown, PA). Experiments were performed with a 40x oil immersion objective during exposure to alternating 340 and 380 nm excitation beams. The intensity of light emission at 520 nm was followed simultaneously in several single cells. The changes in [Ca2+]i are presented as the ratio of fluorescence intensities (F340/F380).
2.6. Intracellular cAMP measurements
Cyclic nucleotide production was monitored using LβT2 gonadotrophs. Briefly, cells (1 million per well) were plated in 24-well plates and incubated overnight at 37 °C in 5% CO2-air and saturated humidity. The next day, the medium was removed, and the cells were washed and stimulated at 37 °C under 5% CO2-air and saturated humidity for 30 min in the presence (experimental groups) and absence of 1 μM forskolin (controls). In both groups, the medium was supplemented with 1 mM isobutylmethylxantine. Cyclic nucleotides were measured in cell extracts using our stock of specific antisera that were characterized previously [32]. The 125cAMP tracer was purchased from Perkin-Elmer Life Sciences (Boston, MA).
2.7. Data analysis
All numerical values in the text are reported as the mean ± SEM from one of at least three similar experiments. KaleidaGraph Program (Synergy Software, Reading, Pennsylvania) was used for calculation of significant differences between means determined by a Student’s t-test or an ANOVA accompanied with the post hoc Student-Newman-Keuls test as well as for regression/correlation analyses and calculation of the half time of decay in gene expression. P-values of less than 0.05 were considered significant.
3. Results
3.1. Gnrhr expression is down- and up-regulated in cultured rat pituitary cells
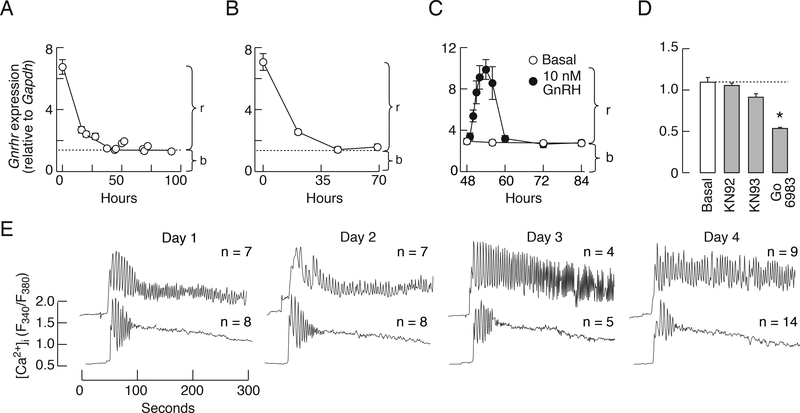
After cell dispersion, the Gnrhr expression progressively decreased as a function of time in both female and male pituitary cell cultures. It reached steady-state at about 20% of transcription observed in freshly dispersed cells over four days of incubation. Figure 1A illustrates the time course of decay in Gnrhr expression in pituitary cells from 7-week old female rats cultured in medium 199 containing 10% horse serum for 96 h without any change of the medium during the study period. Similar decay profiles in Gnrhr expression were also observed in cells cultured in medium 199 containing fetal calf serum (data not shown) as well as in pituitary cells bathed in 0.1% BSA-containing medium 199 that was replaced every 12 h (Fig. 1B). Thus, the kinetics of decay in Gnrhr expression in vitro and the existence of substantial residual gene transcription are independent of the sera origin and concentration; the estimated half time of the decay is 17.5 ± 2.4 h (n = 4).
Figure 1.
Basal and regulated rat pituitary Gnrhr transcriptions are functional in vitro. A and B, Downregulation of Gnrhr expression in pituitary cells from females cultured in medium 199 containing 10% horse serum (A) or 0.1% BSA (B). C, Time course of GnRH-stimulated Gnrhr expression in 2-day old cultures of female pituitary cells. GnRH was continuously present during 36 h incubation. b, basal transcription, r, regulated transcription. D, Inhibition of basal Gnrhr expression by 10 μM Gö6983—a PKC-specific inhibitor. Notice the lack of effect of 1 μM isoquinolonesulfonamide KN93—a calmodulin kinase II specific inhibitor—and KN92—an inactive analog—on basal Gnrhr expression. Data shown in this and following figures are mean ± SEM values from one of at least three similar experiments each performed in 4–6 replicates if not otherwise stated. The ANOVA analysis revealed significantly higher levels of Gnrhr expression in GnRH-stimulated cells for 1, 2 3, 4, 6, and 8 h (C). Asterisks indicate significant differences (P < 0.05) between untreated and treated groups determined by t-test (D). E, GnRH-induced calcium signaling in single gonadotrophs; representative traces. Cells were cultured for 1, 2, 3, or 4 days without GnRH, loaded with Fura 2 AM, and stimulated with 1 nM GnRH. The time and [Ca2+]i scales are identical for all records and basal [Ca2+]i was around 0.5 units. Numbers above traces indicate number of cells exhibiting (top panel) or not (bottom panel) sustained calcium oscillations. In all experiments, the pituitaries were derived from 7-week old female rats.
In subsequent experiments, the dispersed pituitary cells from 7-week old female rats were cultured for two days to down-regulate Gnrhr expression, washed, and stimulated with 10 nM GnRH for up to 36 h. This time course study revealed a progressive increase in Gnrhr expression during continuous GnRH application. It reached a peak response at around 6 h of incubation followed by decay to a steady-state level over the next 6 h; the steady-state level of transcription was identical to that observed in untreated cells. It was not affected by the continuous presence of GnRH during the following 24 h of incubation (Fig. 1C). These results imply that the decay in gene expression in vitro (Fig. 1A and B) reflects at least in part the loss of regulated transcription mediated by GnRH, that Gnrhr expression occurs in the absence of any stimuli, and is not affected by continuous GnRH application (we termed this basal gene expression).
Basal Gnrhr expression was significantly reduced in female cells bathed in medium containing 1 μM Gö6983, a specific inhibitor of PKC [33] (Fig. 1D), but not affected by isoquinolonesulfonamide KN-93 or KN-92 (Fig. 1D). KN-93 is a selective calmodulin-kinase II inhibitor and KN-92, an inactive analog [34]; both of these block voltage-gated potassium channels in a calmodulin-kinase II-independent manner [35]. Basal Gnrhr transcription was also not affected by cetrorelix (data not shown), which acts as GnRHR antagonist and inverse agonist [1]. Thus, it is reasonable to conclude that the intrinsic activity of PKC at least partially accounts for basal Gnrhr expression.
We used single cell [Ca2+]i measurements to examine how the decay in Gnrhr transcription after cell dispersion influences the expression of functional GnRHRs in rat gonadotrophs. Cultured female rat pituitary cells were loaded with Fura 2 and stimulated with 1 nM GnRH. At that concentration, the GnRH induced sustained (top panels) or transient (bottom panels) calcium oscillations. Both patterns of signaling were observed in gonadotrophs cultured for 1, 2, 3, or 4 days (Fig. 1E). This suggests that basal Gnrhr expression is sufficient to maintain expression of functional GnRHR during the prolonged period without GnRH and endogenous steroids.
3.2. Basal but not regulated Gnrhr transcription is operative in immortalized gonadotrophs
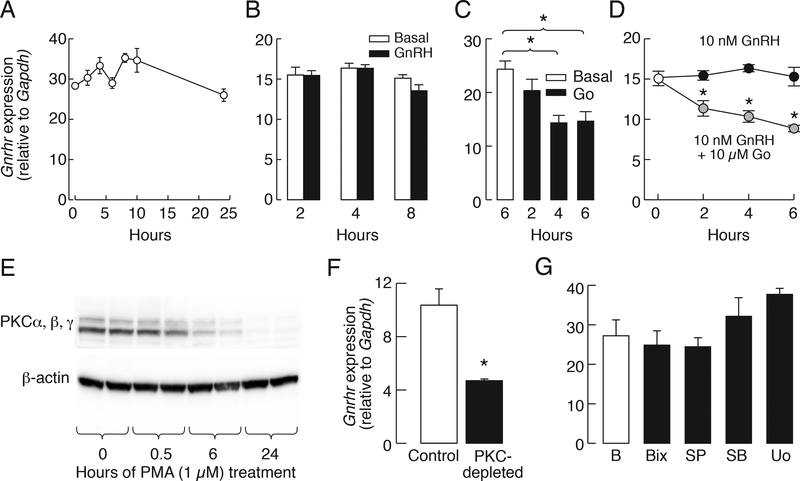
Gonadotrophs represent only around 10% of the cells in primary cultures of female pituitary cells [36]. This limits the capacity for detailed studies of basal Gnrhr expression. To overcome this limitation, we used LβT2 pituitary gonadotrophs. In naïve cells, i.e. never stimulated with GnRH and cultured in fetal calf-containing medium, the Gnrhr transcription was operative. Replacement of fetal calf serum from incubation medium with 0.1% BSA-containing medium did not affect Gnrhr expression significantly during the first 24 h (Fig. 2A). In further experiments, cells were kept in 0.1% BSA-containing medium 199 overnight and washed in the morning; all treatments were done in the same type of medium.
Figure 2.
Basal but not regulated Gnrhr expression is operative in immortalized mouse LβT2 cells. A, Removing fetal calf serum does not affect on Gnrhr expression in LβT2 cells during the first 24 h of incubation. B, The inability of GnRH (10 nM) to stimulate Gnrhr expression during 2–8 h continuous stimulation. C and D, Time-course effects of 10 μM Gö6983 on Gnrhr expression in the presence and absence of 10 nM GnRH. E, PMA (1 μM)-induced progressive depletion of immunoreactive PKC isozymes during 24 h of incubation. F, Reduction of basal Gnrhr expression in PKC-depleted cells. G, The lack of effect of MAPK inhibitors, Bix02189 (Bix)—a MEK5/ERK5 inhibitor—SP600126 (SP)—a JNK inhibitor—SB203580 (SB)—a p38-MAPK inhibitor—and U0126 (Uo)—an ERK1/2 inhibitor— on basal Gnrhr expression during 6 h of treatment. In the experiments shown in B – D and G, cells were kept in 0.1% BSA medium 199 overnight before experiments were performed. Asterisks indicate significant differences between pairs, P < 0.05, determined by ANOVA (C and D) and t-test (F).
In contrast to cultured rat pituitary cells, the GnRH could not alter Gnrhr expression in LβT2 cells during 2, 4, and 8 h incubation (Fig. 2B), despite the expression of functional GnRHR as documented by 1 nM GnRH-induced increase in [Ca2+]i (data not shown). Consistent with the literature [37], our experimental conditions show that coupling of the GnRHR signaling pathway to transcription in LβT2 cells was preserved for several other genes. This includes a time-dependent stimulation of early response genes c-Fos and c-Jun expression (data not shown).
Similar to cultured pituitary cells, the application of 10 μM Gö6983 induced a time-dependent inhibition of basal Gnrhr expression in LβT2 cells (Fig. 2C). In the presence of GnRH, Gö6983 also inhibited Gnrhr expression in a time-dependent manner (Fig. 2D). To further examine the dependence of basal Gnrhr expression of PKC signaling pathway, the LβT2 cells were exposed to 1 μM PMA for 24 h to deplete endogenous PKC. Figure 2E shows time-course of depletion of PKC using an antibody specific for α, β, and γ isosymes. Basal Gnrhr expression was significantly reduced in PKC-depleted cells (Fig. 2F). In contrast, inhibition of MEK1/2 with U0126, p38 MAPK with SB203580 [38], big MAPK with BIX02189 [39], and JNK with SP600125 [40], did not affect basal gene expression (Fig. 2G). These results indicate that basal Gnrhr expression is influenced at least partially by PKC signaling pathway in a MAPK-independent manner.
3.3. Calcium and phorbol ester PMA stimulate Gnrhr expression in rat but not in LβT2 gonadotrophs
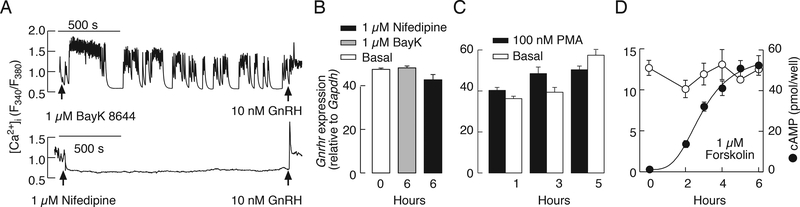
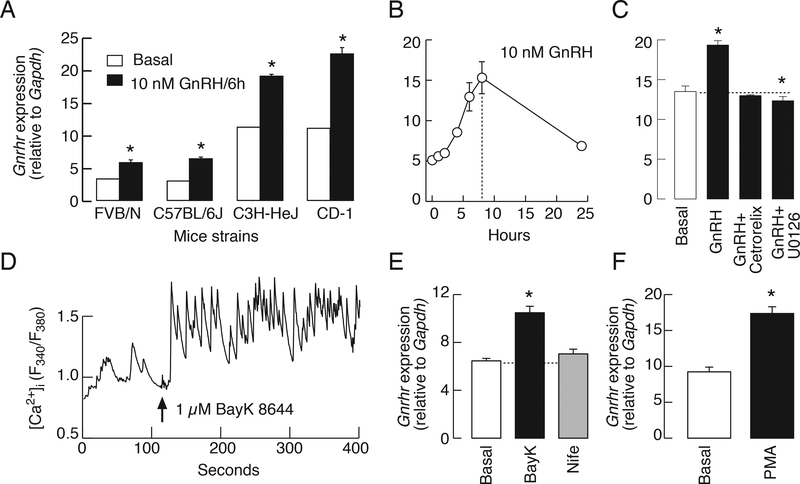
In female rat pituitary cells derived from seven week-old animals and cultured for two days, high potassium-induced elevation in [Ca2+]i (Fig. 3A) also stimulated Gnrhr expression (Fig. 3B). In contrast, 1 μM forskolin did not increase Gnrhr expression (basal: 3.7 ± 0.3, forskolin: 2.6 ± 0.1); moreover, treatment with 10 μM H-89—an inhibitor of cAMP dependent protein kinase—did not alter basal- (basal: 3.7 ± 0.2, H89: 4.1 ± 0.3) or GnRH-induced Gnrhr expression (GnRH: 11.9 ± 1.1, GnRH + H89: 11.8 ± 0.8). The Gnrhr mRNA levels were also elevated in a time- (Fig. 3C) and concentration (Fig. 3D)-dependent manner via treatment with PMA, which is a PKC-specific activator. The time-course of 100 nM PMA-induced Gnrhr expression was highly comparable to that observed during GnRH application. These results are consistent with the hypothesis that GnRH-induced Gnrhr expression in cultured pituitary gonadotrophs is mediated by calcium and PKC signaling pathways.
Figure 3.
Activation of calcium influx and PKC triggers regulated Gnrhr expression in cultured rat pituitary cells. A, High potassium-induced [Ca2+]i in a gonadotroph identified by subsequent application of GnRH (representative trace). B, Concentration-dependent effects of KCl on Gnrhr expression. Asterisks indicate significant differences between pairs, P < 0.05, determined by ANOVA. C, A time-course study of 100 nM PMA-induced Gnrhr expression. D, Concentration dependence of PMA on Gnrhr expression over 6 h of stimulation.
Similar to rat gonadotrophs, high potassium-induced depolarization in LβT2 cells facilitated calcium influx (data not shown), but the rise in [Ca2+]i was not accompanied with an increase in Gnrhr mRNA levels (controls: 12.6 ± 1.3; KCl: 12.2 ± 1.7). The L-type calcium channel agonist BayK 8644 also facilitated calcium influx whereas nifedipine—a blocker of these channels—inhibited spontaneous calcium signaling in LβT2 cells (Fig. 4A). However, BayK 8644 could not stimulate and nifedipine did not down-regulate Gnrhr expression in these cells (Fig. 4B). In further contrast to rat pituitary cells, PMA could not stimulate Gnrhr expression in LβT2 cells (Fig. 4C). As in primary culture of rat pituitary cells, elevation in cAMP production by 1 μM forskolin in LβT2 cells had no effect on Gnrhr expression (Fig. 4D). These results further confirmed that regulated Gnrhr expression in LβT2 gonadotrophs could not be triggered by activating calcium, PKC, and/or cAMP signaling pathways.
Figure 4.
The ineffectiveness of calcium, PKC, and cAMP on the regulation of Gnrhr mRNA levels in mouse LβT2 gonadotrophs. A and B, Effects of BayK 8644 and nifedipine on calcium signaling (A) and Gnrhr expression (B). C, The lack of effects of PMA on Gnrhr expressions during 1, 3, and 5 h of incubation. D, Forskolin-induced cAMP production, and the lack of effect on basal Gnrhr expression for 6 h incubation.
3.4. Regulated rat Gnrhr expression depends on MAPK signaling pathway
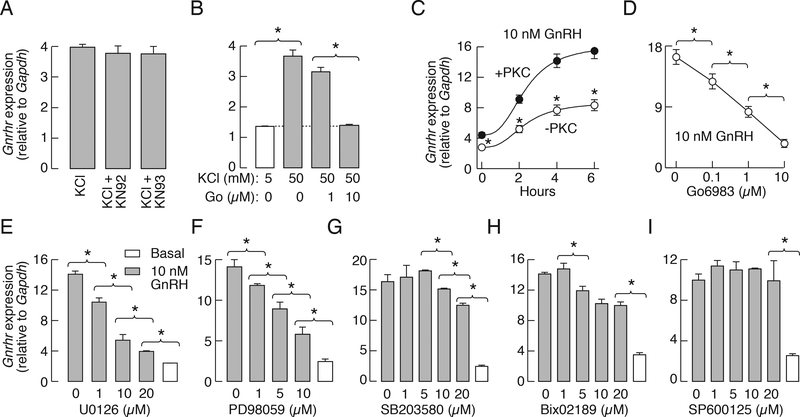
Several observations indicated that the PKC-ERK1/2 signaling pathway plays an important role in calcium and GnRH-induced Gnrhr expression in the primary culture of pituitary cells that were derived from seven week-old female rats and cultured for 48 h prior to experiments: i. Application of Gö6983, but not KN92 or KN93, inhibited high potassium-stimulated Gnrhr expression in a concentration-dependent manner (Fig. 5A and B). ii. In PKC-depleted cells induced by prolonged (24 h) treatment with 1 μM PMA, GnRH-stimulated Gnrhr expression was significantly attenuated. Fig. 5C illustrates the time-course of GnRH-induced Gnrhr expression in controls (+PKC) and PKC-depleted pituitary cells (−PKC). iii. Application of Gö6983 also inhibited GnRH-stimulated Gnrhr expression in a concentration-dependent manner (Fig. 5D). iv. Inhibition of MEK1/2 with U0126 and PD98059 attenuated GnRH-induced Gnrhr expression in a concentration-dependent manner (Fig. 5E and F). Inhibition of p38 MAPK with SB203580 and of big MAPK with BIX02189 was less effective (5G and H, respectively), while inhibition of JNK with SP600125 did not affect GnRH-stimulated gene expression (Fig. 5I). None of these treatments affected the expression of secreted phosphoprotein 1 gene, indicating that effects of U0126 and PD98059 on Gnrhr expression do not reflect the toxicity of these drugs in concentrations used.
Figure 5.
Regulated rat Gnrhr expression in cultured pituitary cells occurs through PKC-MAPK signaling pathways. A, The lack of effect of 1 μM KN-93 and 1 μM KN-92 on 50 mM K+-induced Gnrhr expression. B, Inhibition of high potassium-induced Gnrhr expression by Gö6983. C, GnRH-induced Gnrhr expression in PKC-containing (+PKC) and -depleted (−PKC) cells. D, Concentration-dependent effect of Gö6983, a PKC-specific inhibitor, on GnRH-stimulated Gnrhr expression. E and F, Concentration-dependent effects of MEK1/2 inhibitors U0126 (E) and PD98059 (F) on GnRH-stimulated Gnrhr expression. G-I, Partial inhibition of GnRH-stimulated Gnrhr transcription by SB203580, a p38-MAPK inhibitor (G), and Bix02189, a MEK5/ERK5 inhibitor (H), and the lack of effect of SP600126, a JNK inhibitor (I). In all experiments, pituitaries were derived from seven week-old female rats. In panels A, B, and D-I, the cells were incubated with inhibitors for 30 min followed by 6 h incubation with or without 25 mM KCl (A and B) and 10 nM GnRH (C-I). In all panels, asterisks indicate significant differences between pairs, determined by ANOVA.
3.5. Regulated Gnrhr expression is operative in cultured mouse pituitary cells
There are substantial differences among species, including rat and mouse, in the Gnrhr promoter and in tissue-specific factors that control gene expression [10, 41], which could implicate differential regulation of transcription and/or could account for the lack of regulated Gnrhr expression in mouse LβT2 cells. Using four different strains of mice, however, we show here that continuous application of 10 nM GnRH for 6 h stimulated Gnrhr expression in dispersed female pituitary cells cultured for two days (Fig. 6A). The time-course study was done with C57BL/6J mice and further showed the transient nature of GnRH-induced up-regulation of Gnrhr expression (Fig. 6B). GnRH was ineffective in the presence of cetrorelix and U0126, indicating the dependence of agonist action on GnRHRs signaling through MEK1/2 pathway; neither treatment affected basal Gnrhr transcription (Fig. 6C).
Figure 6.
GnRH also stimulates mouse pituitary Gnrhr expression through calcium and PKC-ERK1/2 signaling pathways. A, Basal and GnRH-stimulated Gnrhr expression in cultured pituitary cells from different mice strains. Asterisks indicate significant differences between pairs. B-F, Cultured pituitary cells from C57BL/6J mice. B, A time course study of GnRH-stimulated Gnrhr expression. C, The lack of effect of 10 nM GnRH on Gnrhr expression in the presence of 1 μM cetrorelix acetate, a GnRHR antagonist, and inhibition of GnRH-stimulated Gnrhr expression by U0126. D and E, Effect of BayK 8644, an L-type voltage-gated calcium channel agonist, on calcium influx (D) and Gnrhr expression (E). Notice the lack of effects of nifedipine—an L-type calcium channel antagonist—on Gnrhr expression. In calcium measurements, the identification of gonadotrophs was done by application of GnRH at the end of recording (not shown). The effects of PMA (100 nM) on Gnrhr expression (F). In all experiments Gnrhr expression was evaluated after 6 h. Experiments were performed in mouse pituitary cells 24 h after dispersion. In A, C, E, and F, asterisks indicate significant differences (P < .05) when compared to basal gene expression, determined by t-test.
BayK 8644 also stimulated calcium influx in mouse gonadotrophs (Fig. 6D). This was accompanied with an increase in Gnrhr expression, whereas nifedipine did not affect basal gene expression (Fig. 6E). In further agreement with work using rat pituitary cells, the application of 100 nM PMA for 6 h also stimulated Gnrhr expression (Fig. 6F). Thus, GnRH-induced Gnrhr expression in mice gonadotrophs in vitro also depends on calcium and PKC signaling pathways. This shows that other reasons account for the lack of regulated Gnrhr expression in LβT2 cells.
3.6. Basal and regulated rat Gnrhr expressions are age- and sex-specific
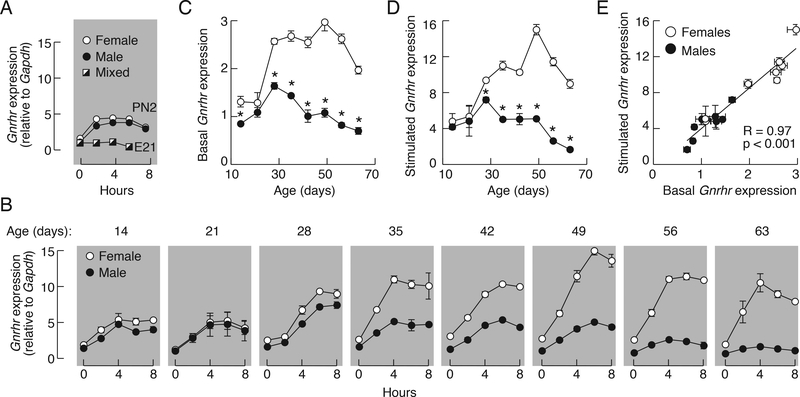
In further work, we analyzed the dependence of basal and GnRH-induced Gnrhr expression in vitro on the age and sex of animals using embryonal, neonatal, infant, juvenile, peripubertal, and postpubertal females and male rats as donors for anterior pituitaries. After dispersion, pituitary cells were cultured for two days to down-regulate Gnrhr expression. In all experiments, basal Gnrhr expression was estimated at time 0, cells were stimulated with 10 nM GnRH for up to 8 h, and samples were collected every 2 h. Figure 7 summarize these time-course studies.
Figure 7.
Basal and GnRH-regulated rat pituitary Gnrhr transcriptions are age and sex dependent. A and B, Time-course studies of 10 nM GnRH-induced Gnrhr expression in embryonal pituitaries (E21), neonatal (PN2) and infant to adult pituitary cells derived from 7 to 20 animals per age group were done 48 h after dispersion. The age of animals in panel B is indicated on the top of gray panels. Adult female groups were composed of animals in different stages of estrous cycle. C-E, The relationship between basal and regulated Gnrhr expression in males (closed circles), females (open circles). Data are derived from Fig. 7B; the 0 time points were used for basal (C) and the peak responses were used as values for GnRH-stimulated gene expression (D). Asterisks indicate significant differences between pairs, determined by ANOVA. E, Correlation of basal and regulated rat pituitary Gnrhr expression. R, Coefficient of correlation.
Basal Gnrhr expression (time = 0) was detectable in embryonal (ED21) and two day-old (PN2) pituitaries (Fig. 7A) as well as in all preparations from postnatal animals (Fig. 7B). In pituitaries from postnatal animals, basal Gnrhr expression varied and was significantly lower in cells from male pituitaries (Fig. 7C). The GnRH-dependent Gnrhr transcription was not observed in the embryonal pituitary but was already established in two day-old animals—the amplitude of response was relatively low (~2.5 fold increase from basal expression) and comparable in female and male cells (Fig. 7A). The GnRH also stimulated Gnrhr gene transcription in all postnatal age groups (Fig. 7B). Similar to cells from neonatal animals, the responsiveness of gonadotrophs from 14 and 21 day-old animals to GnRH was relatively low and similar in female and male cultures. Cells from older animals showed increased responsiveness to GnRH and in all age groups the peak amplitude of response was higher in cells from female rats (Fig. 7D). Correlation analysis revealed a linear relationship between basal and stimulated Gnrhr mRNA levels with a highly significant R = 0.97 (Fig. 7E). These results suggest that the ratio between basal and regulated Gnrhr expression in vitro is comparable in both sexes, but that transcriptional activity is elevated in cells from female rats.
4. Discussion
It is well documented that pituitary GnRHR expression is regulated by GnRH. Castration causes an increase in GnRH secretion coupled with an increase in GnRHR binding sites, while blocking GnRH input causes a drop in the receptor number as estimated by radioreceptor assays [42]. In vivo experiments also revealed that pulsatile GnRH delivery up-regulates its receptor, whereas continuous GnRH causes receptor down-regulation [1]. This at least partially reflects the changes in Gnrhr expression because in pituitary cells from adult rats upregulation of GnRHR mRNA occurs through transcriptional activation rather than modulation of mRNA stability [43].
Here we show that Gnrhr expression exponentially decreases after the dispersion of pituitary cells with a half time comparable to that estimated in experiments with actinomycin D in female rat pituitary [43]. The decay in Gnrhr expression was incomplete and reached a steady-state level within 50 h of incubation, which was about 20% of that observed immediately after the dispersion. In contrast, the in vitro decay of Tshb expression is progressive with time and reaches non-detectable levels within 70 h of incubation [28]. Consistent with the hypothesis that the decline in Gnrhr expression in pituitary cells reflects the loss of GnRH, its continuous application caused recovery of Gnrhr expression with a peak in response after 6 to 8 h stimulation. Others also observed stimulatory effect of GnRH on Gnrhr expression in rat pituitary cells during 6 h continuous application [44]. This recovery was transient, followed by decay in expression during continuous GnRH application, which did not go below the steady-state level established during the 2-day incubation, i.e. the transcription was not shut-off. These observations are consistent with the presence of a basal and GnRH-regulated Gnrhr promoter activity expressed in immortalized gonadotrophs [41, 45].
It is well known that GnRH stimulates Gnrhr expression through PKC [18, 46, 47] and that MAPK represents downstream elements of this signaling pathway [48]. In agreement with this, our pharmacologically based experiments point to the critical role of these enzymes in GnRH-induced Gnrhr expression in rat pituitary cells. The role of calcium influx in Gnrhr expression was shown in experiments with GGH3–1 cells expressing mouse Gnrhr promoter [46]. Here we show that voltage-gated calcium influx in cultured rodent pituitary cells also increases rat Gnrhr expression. The findings that pituitary cells express calcium-regulated adenylyl cyclases [49], that GnRHR also signals through Gs pathway in LβT2 cells [9], and that cAMP through protein kinase A stimulates Gnrhr expression in transfected αT3–1 pituitary cells [45], could indicate that this signaling pathway accounts for stimulation of Gnrhr expression. However, neither cultured rat pituitary cells nor LβT2 cells responded with stimulation of Gnrhr expression when stimulated with forskolin at a concentration that induced significant increases in cAMP production, which could indicate difference in responses between αT3–1 and LβT2 cells. Furthermore, Gö6983 inhibited voltage-gated calcium influx-induced Gnrhr expression in a dosedependent manner, indicating that both GnRH and calcium influx utilize the same pathway to facilitate transcription. Further studies are needed to clarify whether the decay of GnRH-induced Gnrhr expression in the presence of prolonged GnRH application reflects desensitization of signaling pathways accounting for upregulation of transcription or the transient nature of activation.
The current data also indicate that basal Gnrhr expression at least in part depends on PKC signaling pathways, indicating that the activity of these enzymes in dispersed pituitary cells and LβT2 cells is elevated in the absence of GnRH. This could reflect a GnRHR independent or dependent process—the latter driven by the constitutive/intrinsic activity of these receptors. The intrinsic receptor activity is well established for several GPCRs, including histaminic, b-adrenergic, GABA, 5-HT, and dopaminergic receptors [50]. It has been well established that hundreds of GnRHR mutants show no constitutive activity and some of these mutants are retained in endoplasmic reticulum and unable to traffic to plasma membrane. Furthermore, some pharmacoperones were able to rescue their trafficking problem, leading to re-establishment of basal receptor activity [51]. It has also been suggested that cetrorelix may have dual actions—both as a pure antagonist but also as an inverse agonist [1]. In our experiment, cetrorelix did not inhibit basal Gnrhr expression. However, further work is needed to dissociate between these two hypotheses.
To the best of our knowledge, the role of basal Gnrhr expression in pituitary cells has not been studied. We previously observed the expression of functional receptors in cultured pituitary cells for a prolonged period when the intracellular pool of LH was practically depleted [52]. The present data are consistent with this conclusion. It is of physiological and evolutionary importance that gonadotrophs keep expressing some functional GnRHR and eliminate dedifferentiation until endogenous or exogenous GnRH could up-regulate Gnrhr to stimulate gonadotropin subunit genes and resuscitate reproduction. For example, high water temperature induces termination of spawning in female red seabrem through downregulation of brain Gnrh1 expression, pituitary Gnrhr and Lhb expression and serum estradiol levels [53]. In Kallmann syndrome, pulsatile GnRH administration restores gonadal function [54], suggesting that this mechanism is also operative in humans. Knobil’s group showed that LH and FSH secretion during sustained GnRH application gradually recovered with pulsatile GnRH injection after suppression [55]. These findings are consistent with a hypothesis that basal Gnrhr expression in the prolonged absence or continuous presence of GnRH protected gonadotrophs from the loss of functional GnRHRs. This hypothesis should be addressed by examining the GnRHRs protein levels in these experimental conditions, which does not only reflect de novo synthesis but also the rate of receptor trafficking and the rate of degradation.
Although little is known about GnRH release during maturation, it was assumed to be minimal before the later stages of puberty; the pituitary gland responds to GnRH application at any developmental stage [56]. However, it was recently reported that the frequency of GnRH release in the late embryonal stage was high and reached a maximum in newborn male mice and remained elevated during the first seven days of life [57]. This suggests that the lack of pituitary secretory response blocks downstream activation of the reproductive functions by elevated GnRH. Our results are consistent with this later hypothesis; GnRH was ineffective in triggering Gnrhr expression in embryonal rat pituitaries and low effective in neonatal pituitary cells. This is in general agreement with findings that GnRHR presence in embryonal rats is very low [58], but functional in terms of GnRH-induced LH and FSH release [59]. In vivo, pituitary PACAP and follistatin levels were high in the fetal (E19) pituitary and decline after parturition [60]. This could suggest that elevated follistatin attenuates upregulation of Gnrhr by activin and that this process begins to reverse at the time of birth.
Like embryonal pituitaries, the LβT2 mouse gonadotrophs showed basal but not regulated Gnrhr expression. Others also observed no increase in transcriptional activity in GnRH-stimulated αT3–1 cells [61], and very low response of βT2 cells to GnRH application, but high response to glucocorticoids [24]. In αT3–1 cells, homologous upregulation of GnRHR reflects modulation of the capacity of cellular RNA to direct the biosynthesis of GnRH receptors [61, 62]. In general, there is an agreement in the field that αT3–1 cells, which express Gnrhr and Cga only, are progenitor gonadotrophs, i.e. to represent embryonal cell type, whereas LβT2 cells represent differentiated male gonadotroph cell model because express Fshb and Lhb as well. However, LβT2 cells express several progenitor markers, including SOX9, E-cadherin, S100b, SF-1, and Pit-1 [63]. The same group also generated immortal gonadotrophs that express LHβ and SOX2 and concluded that they represent progenitor like cells [63]. We have shown that both αT3–1 and LβT2 cells do not generate oscillatory calcium and electrical signals when stimulated with GnRH in contrast to postnatal gonadotrophs [64, 65]. Also, LβT2 cells do not express Dmp1, another postnatal gonadotroph-specific marker [66]. Together with the finding that GnRH could not trigger Gnrhr expression, it is reasonable to suggest that LβT2 cells are not fully differentiated gonadotrophs and may better represent embryonal gonadotrophs.
The time course and the amplitude of GnRH-induced Gnrhr expression were comparable in pituitary cells from both sexes during neonatal and infantile periods. The sex-specific pattern of the response was established during the juvenile-peripubertal period. Variations in GnRHR mRNA levels were detected during the estrus cycle and after ovariectomy and orchidectomy in various species [67–69]. Our experiments with cells from adult females were done using animals from different stages. Here we also show that basal Gnrhr expression varied in pituitary cells from developing animals, and there was a strong correlation between basal and GnRH-stimulated Gnrhr expression. The relationship between basal and regulated Gnrhr expression was comparable in pituitary cells from both sexes, but the higher basal gene expression in females was accompanied by amplified regulated expression. This suggests that transcriptional activity is elevated in cells from female rats of juvenile to adult age.
In conclusion, these data indicate that basal Gnrhr transcription is an intrinsic property of gonadotrophs established during embryogenesis and maintained throughout development and adult periods. It is controlled by the PKC signaling pathways in a MAPK-independent manner. Basal Gnrhr transcription secures the presence of a sufficient number of receptors to preserve functionality in gonadotrophs independent of the status of GnRH secretion. It also determines the sex- and age-specific up-regulation by GnRH and steroids. GnRH-regulated Gnrhr transcription develops postnatally in a PKC-dependent manner and involves MAPK as a downstream signaling pathway. Both basal and regulated Gnrhr transcriptions are age-dependent, and their coordinate actions determine the expression level of functional receptors and responsiveness of gonadotrophs to GnRH during development. The sex-specific basal and regulated Gnrhr expression is preserved in vitro. Further work is needed to clarify the basis for these differences, including dependence of basal and regulated in vitro Gnrhr expression on estrous stage.
Acknowledgment
This work was supported by the Intramural Research Program of the National Institute of Child Health and Human Development (ZIA DH000195-22).
Abbreviations:
- [Ca2+]i
intracellular calcium concentration
- GnRHR
gonadotropin-releasing hormone receptor
- Gnrhr
GnRHR gene
- ERK
extracellular-signal-regulated kinases
- JNK
c-Jun N-terminal kinases
- MAPK
mitogen-activated protein kinases
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate.
Footnotes
Competing Interest: The authors declare no competing financial interests.
References
- 1.McArdle GA, Roberson MS. Gonadotropes and Gonadotropin-releasing hormone signaling Knobil and Neill’s Physiology or Reproduction, Fourth Edition. Elsevier Inc; 2015:335–397. [Google Scholar]
- 2.Millar RP, Lu ZL, Pawson AJ, Flanagan CA, Morgan K, Maudsley SR. Gonadotropin-releasing hormone receptors. Endocr Rev 2004; 25:235–275. [DOI] [PubMed] [Google Scholar]
- 3.Naor Z, Huhtaniemi I. Interactions of the GnRH receptor with heterotrimeric G proteins. Front Neuroendocrinol 2013; 34:88–94. [DOI] [PubMed] [Google Scholar]
- 4.Stojilkovic SS, Tabak J, Bertram R. Ion channels and signaling in the pituitary gland. Endocr Rev 2010; 31:845–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naor Z, Harris D, Shacham S. Mechanism of GnRH receptor signaling: combinatorial cross-talk of Ca2+ and protein kinase C. Front Neuroendocrinol 1998; 19:1–19. [DOI] [PubMed] [Google Scholar]
- 6.Bliss SP, Navratil AM, Xie J, Roberson MS. GnRH signaling, the gonadotrope and endocrine control of fertility. Front Neuroendocrinol 2010; 31:322–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stojilkovic SS, Catt KJ. Novel aspects of GnRH-induced intracellular signaling and secretion in pituitary gonadotrophs. J Neuroendocrinol 1995; 7:739–757. [DOI] [PubMed] [Google Scholar]
- 8.Naor Z Signaling by G-protein-coupled receptor (GPCR): studies on the GnRH receptor. Front Neuroendocrinol 2009; 30:10–29. [DOI] [PubMed] [Google Scholar]
- 9.Tsutsumi R, Mistry D, Webster NJ. Signaling responses to pulsatile gonadotropin-releasing hormone in LbetaT2 gonadotrope cells. J Biol Chem 2010; 285:20262–20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schang AL, Querat B, Simon V, Garrel G, Bleux C, Counis R, Cohen-Tannoudji J, Laverriere JN. Mechanisms underlying the tissue-specific and regulated activity of the Gnrhr promoter in mammals. Front Endocrinol (Lausanne) 2012; 3:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albarracin CT, Kaiser UB, Chin WW. Isolation and characterization of the 5’-flanking region of the mouse gonadotropin-releasing hormone receptor gene. Endocrinology 1994; 135:2300–2306. [DOI] [PubMed] [Google Scholar]
- 12.Reinhart J, Xiao S, Arora KK, Catt KJ. Structural organization and characterization of the promoter region of the rat gonadotropin-releasing hormone receptor gene. Mol Cell Endocrinol 1997; 130:1–12. [DOI] [PubMed] [Google Scholar]
- 13.Fan NC, Peng C, Krisinger J, Leung PC. The human gonadotropin-releasing hormone receptor gene: complete structure including multiple promoters, transcription initiation sites, and polyadenylation signals. Mol Cell Endocrinol 1995; 107:R1–8. [DOI] [PubMed] [Google Scholar]
- 14.Kakar SS. Molecular structure of the human gonadotropin-releasing hormone receptor gene. Eur J Endocrinol 1997; 137:183–192. [DOI] [PubMed] [Google Scholar]
- 15.Campion CE, Turzillo AM, Clay CM. The gene encoding the ovine gonadotropin-releasing hormone (GnRH) receptor: cloning and initial characterization. Gene 1996; 170:277–280. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z, Gibson JP, Archibald AL, Haley CS. The porcine gonadotropin-releasing hormone receptor gene (GNRHR): genomic organization, polymorphisms, and association with the number of corpora lutea. Genome 2001; 44:7–12. [DOI] [PubMed] [Google Scholar]
- 17.Yasin M, Dalkin AC, Haisenleder DJ, Kerrigan JR, Marshall JC. Gonadotropin-releasing hormone (GnRH) pulse pattern regulates GnRH receptor gene expression: augmentation by estradiol. Endocrinology 1995; 136:1559–1564. [DOI] [PubMed] [Google Scholar]
- 18.White BR, Duval DL, Mulvaney JM, Roberson MS, Clay CM. Homologous regulation of the gonadotropin-releasing hormone receptor gene is partially mediated by protein kinase C activation of an activator protein-1 element. Mol Endocrinol 1999; 13:566–577. [DOI] [PubMed] [Google Scholar]
- 19.Norwitz ER, Cardona GR, Jeong KH, Chin WW. Identification and characterization of the gonadotropin-releasing hormone response elements in the mouse gonadotropin-releasing hormone receptor gene. J Biol Chem 1999; 274:867–880. [DOI] [PubMed] [Google Scholar]
- 20.Davis TL, Whitesell JD, Cantlon JD, Clay CM, Nett TM. Does a nonclassical signaling mechanism underlie an increase of estradiol-mediated gonadotropin-releasing hormone receptor binding in ovine pituitary cells? Biol Reprod 2011; 85:770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama T, Conn PM. Modulation of activin A action and specificity in the rat gonadotrope by protein kinase C. Endocrinology 1993; 133:496–504. [DOI] [PubMed] [Google Scholar]
- 22.Duval DL, Ellsworth BS, Clay CM. Is gonadotrope expression of the gonadotropin releasing hormone receptor gene mediated by autocrine/paracrine stimulation of an activin response element? Endocrinology 1999; 140:1949–1952. [DOI] [PubMed] [Google Scholar]
- 23.Norwitz ER, Xu S, Xu J, Spiryda LB, Park JS, Jeong KH, McGee EA, Kaiser UB. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both gonadotropin-releasing hormone (GnRH)- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J Biol Chem 2002; 277:37469–37478. [DOI] [PubMed] [Google Scholar]
- 24.Turgeon JL, Kimura Y, Waring DW, Mellon PL. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol 1996; 10:439–450. [DOI] [PubMed] [Google Scholar]
- 25.Maya-Nunez G, Conn PM. Transcriptional regulation of the GnRH receptor gene by glucocorticoids. Mol Cell Endocrinol 2003; 200:89–98. [DOI] [PubMed] [Google Scholar]
- 26.Pincas H, Laverriere JN, Counis R. Pituitary adenylate cyclase-activating polypeptide and cyclic adenosine 3’,5’-monophosphate stimulate the promoter activity of the rat gonadotropin-releasing hormone receptor gene via a bipartite response element in gonadotrope-derived cells. J Biol Chem 2001; 276:23562–23571. [DOI] [PubMed] [Google Scholar]
- 27.Moenter SM. Leap of Faith: Does Serum Luteinizing Hormone Always Accurately Reflect Central Reproductive Neuroendocrine Activity? Neuroendocrinology 2015; 102:256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bargi-Souza P, Kucka M, Bjelobaba I, Tomic M, Janjic MM, Nunes MT, Stojilkovic SS. Loss of basal and TRH-stimulated Tshb expression in dispersed pituitary cells. Endocrinology 2015; 156:242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He ML, Zemkova H, Koshimizu TA, Tomic M, Stojilkovic SS. Intracellular calcium measurements as a method in studies on activity of purinergic P2X receptor channels. Am J Physiol Cell Physiol 2003; 285:C467–479. [DOI] [PubMed] [Google Scholar]
- 32.Kostic TS, Andric SA, Stojilkovic SS. Spontaneous and receptor-controlled soluble guanylyl cyclase activity in anterior pituitary cells. Mol Endocrinol 2001; 15:1010–1022. [DOI] [PubMed] [Google Scholar]
- 33.Gschwendt M, Dieterich S, Rennecke J, Kittstein W, Mueller HJ, Johannes FJ. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett 1996; 392:77–80. [DOI] [PubMed] [Google Scholar]
- 34.Means AR. Regulatory cascades involving calmodulin-dependent protein kinases. Mol Endocrinol 2000; 14:4–13. [DOI] [PubMed] [Google Scholar]
- 35.Popovic MA, Stojilkovic SS, Gonzalez-Iglesias AE. Effects of isoquinolonesulfonamides on action potential secretion coupling in pituitary cells. Horm Mol Biol Clin Investig 2010; 1:35–42. [DOI] [PubMed] [Google Scholar]
- 36.Van Goor F, Zivadinovic D, Martinez-Fuentes AJ, Stojilkovic SS. Dependence of pituitary hormone secretion on the pattern of spontaneous voltage-gated calcium influx. Cell type-specific action potential secretion coupling. J Biol Chem 2001; 276:33840–33846. [DOI] [PubMed] [Google Scholar]
- 37.Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem 2001; 276:47195–47201. [DOI] [PubMed] [Google Scholar]
- 38.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 2000; 351:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatake RJ, O’Neill MM, Kennedy CA, Wayne AL, Jakes S, Wu D, Kugler SZ Jr., Kashem MA, Kaplita P, Snow RJ. Identification of pharmacological inhibitors of the MEK5/ERK5 pathway. Biochem Biophys Res Commun 2008; 377:120–125. [DOI] [PubMed] [Google Scholar]
- 40.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J 2003; 371:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hapgood JP, Sadie H, van Biljon W, Ronacher K. Regulation of expression of mammalian gonadotrophin-releasing hormone receptor genes. J Neuroendocrinol 2005; 17:619–638. [DOI] [PubMed] [Google Scholar]
- 42.Clayton RN, Catt KJ. Gonadotropin-releasing hormone receptors: characterization, physiological regulation, and relationship to reproductive function. Endocr Rev 1981; 2:186–209. [DOI] [PubMed] [Google Scholar]
- 43.Cheon M, Park D, Park Y, Kam K, Park SD, Ryu K. Homologous upregulation of gonadotropin-releasing hormone receptor mRNA occurs through transcriptional activation rather than modulation of mRNA stability. Endocrine 2000; 13:47–53. [DOI] [PubMed] [Google Scholar]
- 44.Cheon M, Park D, Kim K, Park SD, Ryu K. Homologous upregulation of GnRH receptor mRNA by continuous GnRH in cultured rat pituitary cells. Endocrine 1999; 11:49–55. [DOI] [PubMed] [Google Scholar]
- 45.Kakar SS, Malik MT, Winters SJ. Gonadotropin-releasing hormone receptor: cloning, expression and transcriptional regulation. Prog Brain Res 2002; 141:129–147. [DOI] [PubMed] [Google Scholar]
- 46.Lin X, Conn PM. Transcriptional activation of gonadotropin-releasing hormone (GnRH) receptor gene by GnRH: involvement of multiple signal transduction pathways. Endocrinology 1999; 140:358–364. [DOI] [PubMed] [Google Scholar]
- 47.Cheng KW, Ngan ES, Kang SK, Chow BK, Leung PC. Transcriptional down-regulation of human gonadotropin-releasing hormone (GnRH) receptor gene by GnRH: role of protein kinase C and activating protein 1. Endocrinology 2000; 141:3611–3622. [DOI] [PubMed] [Google Scholar]
- 48.Haisenleder DJ, Cox ME, Parsons SJ, Marshall JC. Gonadotropin-releasing hormone pulses are required to maintain activation of mitogen-activated protein kinase: role in stimulation of gonadotrope gene expression. Endocrinology 1998; 139:3104–3111. [DOI] [PubMed] [Google Scholar]
- 49.Kucka M, Bjelobaba I, Tomic M, Stojilkovic SS. The Role of Cyclic Nucleotides in Pituitary Lactotroph Functions. Front Endocrinol (Lausanne) 2013; 4:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kenakin T Principles: receptor theory in pharmacology. Trends Pharmacol Sci 2004; 25:186–192. [DOI] [PubMed] [Google Scholar]
- 51.Janovick JA, Conn PM. Use of pharmacoperones to reveal GPCR structural changes associated with constitutive activation and trafficking. Methods Enzymol 2010; 485:277–292. [DOI] [PubMed] [Google Scholar]
- 52.Tomic M, Cesnajaj M, Catt KJ, Stojilkovic SS. Developmental and physiological aspects of Ca2+ signaling in agonist-stimulated pituitary gonadotrophs. Endocrinology 1994; 135:1762–1771. [DOI] [PubMed] [Google Scholar]
- 53.Okuzawa K, Gen K. High water temperature impairs ovarian activity and gene expression in the brain-pituitary-gonadal axis in female red seabream during the spawning season. Gen Comp Endocrinol 2013; 194:24–30. [DOI] [PubMed] [Google Scholar]
- 54.Chryssikopoulos A, Gregoriou O, Vitoratos N, Rizos D, Papadias K. The predictive value of double Gn-RH provocation test in unprimed Gn-RH-primed and steroid-primed female patients with Kallmann’s syndrome. Int J Fertil Womens Med 1998; 43:291–299. [PubMed] [Google Scholar]
- 55.Belchetz PE, Plant TM, Nakai Y, Keogh EJ, Knobil E. Hypophysial responses to continuous and intermittent delivery of hypopthalamic gonadotropin-releasing hormone. Science 1978; 202:631–633. [DOI] [PubMed] [Google Scholar]
- 56.Ojeda SR, Skinner MK. Puberty in the rats In: Knobil and Neill’s Physiology of Reproduction 2006; Elsevier:2061–2126. [Google Scholar]
- 57.Glanowska KM, Burger LL, Moenter SM. Development of gonadotropin-releasing hormone secretion and pituitary response. J Neurosci 2014; 34:15060–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aubert ML, Begeot M, Winiger BP, Morel G, Sizonenko PC, Dubois PM. Ontogeny of hypothalamic luteinizing hormone-releasing hormone (GnRH) and pituitary GnRH receptors in fetal and neonatal rats. Endocrinology 1985; 116:1565–1576. [DOI] [PubMed] [Google Scholar]
- 59.Schafer SJ, McShan WH. Gonadotropic hormone release from fetal and adult rat pituitary glands after in vitro exposure to synthetic LH-FSH-RH. Neuroendocrinology 1974; 16:332–341. [DOI] [PubMed] [Google Scholar]
- 60.Moore JP Jr, Villafuerte BC, Unick CA, Winters SJ. Developmental changes in pituitary adenylate cyclase activating polypeptide expression during the perinatal period: possible role in fetal gonadotroph regulation. Endocrinology 2009; 150:4802–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsutsumi M, Laws SC, Sealfon SC. Homologous up-regulation of the gonadotropin-releasing hormone receptor in alpha T3–1 cells is associated with unchanged receptor messenger RNA (mRNA) levels and altered mRNA activity. Mol Endocrinol 1993; 7:1625–1633. [DOI] [PubMed] [Google Scholar]
- 62.Tsutsumi M, Laws SC, Rodic V, Sealfon SC. Translational regulation of the gonadotropin-releasing hormone receptor in alpha T3–1 cells. Endocrinology 1995; 136:1128–1136. [DOI] [PubMed] [Google Scholar]
- 63.Kim GL, Wang X, Chalmers JA, Thompson DR, Dhillon SS, Koletar MM, Belsham DD. Generation of immortal cell lines from the adult pituitary: role of cAMP on differentiation of SOX2-expressing progenitor cells to mature gonadotropes. PLoS One 2011; 6:e27799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merelli F, Stojilkovic SS, Iida T, Krsmanovic LZ, Zheng L, Mellon PL, Catt KJ. Gonadotropin-releasing hormone-induced calcium signaling in clonal pituitary gonadotrophs. Endocrinology 1992; 131:925–932. [DOI] [PubMed] [Google Scholar]
- 65.Naidich M, Shterntal B, Furman R, Pawson AJ, Jabbour HN, Morgan K, Millar RP, Jia J, Tomic M, Stojilkovic S, Stern N, Naor Z. Elucidation of mechanisms of the reciprocal cross talk between gonadotropin-releasing hormone and prostaglandin receptors. Endocrinology 2010; 151:2700–2712. [DOI] [PubMed] [Google Scholar]
- 66.Kucka M, Bjelobaba I, Clokie SJ, Klein DC, Stojilkovic SS. Female-specific induction of rat pituitary dentin matrix protein-1 by GnRH. Mol Endocrinol 2013; 27:1840–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaiser UB, Jakubowiak A, Steinberger A, Chin WW. Regulation of rat pituitary gonadotropin-releasing hormone receptor mRNA levels in vivo and in vitro. Endocrinology 1993; 133:931–934. [DOI] [PubMed] [Google Scholar]
- 68.Kakar SS, Grantham K, Musgrove LC, Devor D, Sellers JC, Neill JD. Rat gonadotropin-releasing hormone (GnRH) receptor: tissue expression and hormonal regulation of its mRNA. Mol Cell Endocrinol 1994; 101:151–157. [DOI] [PubMed] [Google Scholar]
- 69.Winters SJ, Kawakami S, Sahu A, Plant TM. Pituitary follistatin and activin gene expression, and the testicular regulation of FSH in the adult Rhesus monkey (Macaca mulatta). Endocrinology 2001; 142:2874–2878. [DOI] [PubMed] [Google Scholar]