Abstract
Zebrafish transgenic lines provide valuable insights into gene functions, cell lineages and cell behaviors during development. Spatiotemporal control over transgene expression is a critical need in many experimental approaches, with applications in loss- and gain-of-function expression, ectopic expression and lineage tracing experiments. The Cre/loxP recombination system is a powerful tool to provide this control and the demand for validated Cre and loxP zebrafish transgenics is high. One of the major challenges to widespread application of Cre/loxP technology in zebrafish is comparatively small numbers of established tissue-specific Cre or CreERT2 lines. We used Tol2-mediated transgenesis to generate Tg(CrymCherry;−1.9mylz2:CreERT2) which provides an inducible CreERT2 source driven by muscle-specific mylz2 promoter. The transgenic specifically labels the trunk and tail skeletal muscles. We assessed the temporal responsiveness of the transgenic by screening with a validated loxP reporter transgenic ubi:Switch. Further, we evaluated the recombination efficiency in the transgenic with varying concentrations of 4-OHT, for different induction time periods and at different stages of embryogenesis and observed that higher recombination efficiency is achieved when embryos are induced with 10μM 4-OHT from 10-somites or 24 hpf till 48 or 72 hpf. The transgenic is an addition to currently available zebrafish transgenesis toolbox and a significant tool to advance muscle biology studies in zebrafish.
Keywords: Cre/loxP, transgenic, zebrafish, muscles, tissue-specific, inducible
INTRODUCTION
The zebrafish is widely used as a model system to study development and disease due to several advantages (Lieschke and Currie 2007). Zebrafish have a short generation time and produce a large number of transparent embryos that develop externally (Roberts et al. 2014). Further, they can be easily subjected to large-scale mutagenesis and chemical screens (Amsterdam et al. 1999; Driever et al. 1996). Zebrafish transgenic lines have provided valuable insights into gene functions, cell lineages and cell behaviors directing normal development and in pathologic states. Many experimental approaches such as loss- and gain-of-function expression, ectopic expression and lineage tracing experiments require spatiotemporal control over transgene expression. In the mouse model, spatiotemporal control of transgene expression can be obtained through site-specific recombination systems such as Cre/loxP and Flp/FRT (van der Weyden et al. 2002). In zebrafish, Cre/loxP system has become increasingly effective to study tissue specific overexpression, ectopic expression and lineage tracing experiments where Cre recombinase catalyzes the site-specific recombination of two loxP sites (Metzger et al. 1995; Mongera et al. 2013; Mosimann et al. 2011; Yoshikawa et al. 2008). Cre recombinase fused to the mutated human ligand-binding domain of the estrogen receptor (ER) has been shown to possess excellent ligand sensitivity and inducible recombination efficiency (Feil et al. 1996; Metzger et al. 1995). The most commonly used version of CreER is the CreERT2 which is inducible with exposure to tamoxifen (TAM) or its active metabolite, 4-hydroxytamoxifen (4-OHT) (Feil et al. 1996; Feil et al. 1997), and has been effectively applied for tightly regulated spatiotemporal control during lineage tracing in zebrafish embryogenesis (Hans et al. 2009).
The advent of Tol2-mediated transgenesis has triggered an increase in Cre/loxP applications in zebrafish (Hans et al. 2009; Kawakami 2007). Multiple zebrafish lines carrying single-insertion loxP cassettes generated through Tol2-mediated transgenesis are now available (Mosimann et al. 2011; Yoshikawa et al. 2008). The field of zebrafish genetics has also seen a growing number of lines with tissue- specific Cre/CreERT2 expression, including zp3:Cre, pax2a:CreERT2, myl7:CreERT2, gata4:CreERT2, lmo2:Cre, sox10:Cre, nkx2.5:CreERT2, sox10:CreERT2, krt1c19e:CreERT2 and kdrl:CreERT2 to name a few. (Guner-Ataman et al. 2013; Hans et al. 2009; Jopling et al. 2010; Kikuchi et al. 2010; Lee et al. 2014; Liu et al. 2008; Rodrigues et al. 2012b; Zhao et al. 2014; Zhou et al. 2011). Muscle-specific CreERT2 lines such as cmlc2:CreERT2, car:CreERT2 and msgn1:CreERT2 have also been reported (Kikuchi et al. 2010; Nguyen et al. 2014; Rodrigues et al. 2012a). While cmlc2:CreERT2 is restricted to cardiomyocytes only, car:CreERT2 expresses in craniofacial, heart and trunk skeletal muscles (Kikuchi et al. 2010; Rodrigues et al. 2012a). msgn1:CreERT2 is shown to express in the dorsal aorta cells in the somites along with the trunk skeletal muscles (Nguyen et al. 2014). A recent genome-wide gene trap screen yielded four CreERT2 lines that express in the somites at early developmental time points (Jungke et al. 2015). While these CreERT2 lines have the potential to be useful in zebrafish muscle studies in future, further work needs to be performed for validation, by crossing with loxP reporter lines and resulting in successful Cre-mediated recombination.
Fast skeletal muscle myosin light chain 2 (mylz2) is a muscle-specific protein that is activated during zebrafish somitogenesis and is specifically expressed in fast skeletal muscles (Storer et al. 2013; Xu et al. 1999; Xu et al. 2000). It can be detected in somites as early as 16 hours post fertilization (hpf) by whole mount in situ hybridization (WISH) (Xu et al. 2000). We report the generation, validation and characterization of a new CreERT2 transgenic line, Tg(CrymCherry;−1.9mylz2:CreERT2) driven by mylz2. The transgenic specifically labels the trunk and tail skeletal muscles upon induction with 4-OHT. The presence of a transgenesis marker that indicates the presence of the insertion makes the maintenance of the line easier. We further analyze the recombination efficiency in the transgenic with changing concentration, induction stage and exposure time of 4-OHT to elucidate the effects of these conditions. The transgenic Tg(CrymCherry;−1.9mylz2:CreERT2) is an addition to the available muscle-specific CreERT2 lines. The line will be of use in experimental designs where expression is needed specifically in trunk and tail skeletal muscles and excludes the heart and the hematopoietic cells where the currently available muscle-specific CreERT2 transgenics, cmlc2:CreERT2, car:CreERT2 and msgn1:CreERT2 are expressed.
MATERIALS AND METHODS
Plasmid construction
pENTR5’_mylz2 (Ablain et al. 2015) and pENTRD_CreERT2 (Mosimann et al. 2011) plasmids were kind gifts from Leonard I. Zon, Boston, MA, while pDestTol2A2-CrymCherry (Berger and Currie 2013) was kindly shared by Geoffrey and Caroline Burns, Massachusetts General Hospital, Boston, MA. The construct (CrymCherry−1.9mylz2:CreERT2) is a multisite gateway assembly of pENTR5’_mylz2, pENTRD_CreERT2 and p3E_SV40polyA (Tol2 kit# 302, (Kwan et al. 2007)) with pDestTol2A2-CrymCherry. The assembly was accomplished via MultiSite Gateway in vitro cloning system (Invitrogen, Carlsbad, CA) according to standard protocols. The plasmid (CrymCherry;−1.9mylz2:CreERT2) was confirmed by Sanger DNA sequencing with multiple primers. Sanger DNA sequencing was performed by the CCIB Core Facility at Massachusetts General Hospital (Cambridge, MA). All constructs and the corresponding digital plasmid maps are available upon request.
Tol2 mRNA synthesis
To generate Tol2 mRNA, pCS3FA-transposase (Tol2 kit# 396, (Kwan et al. 2007)) was linearized with NotI (New England Biolabs, Ipswich, MA, USA). The linearized plasmid was purified by QIAquick PCR purification kit (Qiagen, Valencia, CA, USA), 1μg of the purified, linearized plasmid was used as a template to transcribe capped Tol2 mRNA using the SP6 mMESSAGE mMACHINE RNA Synthesis Kit (Ambion, Foster City, CA, USA) according to the manufacturer’s instructions. Tol2 mRNA was purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and stored at −80°C for future use.
Zebrafish husbandry
All embryos and fish were raised and cared (Westerfield et al. 1999), using established protocols in accordance with the Subcommittee on Research Animal Care, Massachusetts General Hospital. Tg(ubi:loxP-EGFP-loxP-mCherry) (ubi:Switch) (Mosimann et al. 2011) was kindly shared by Jenna Galloway, Massachusetts General Hospital, Boston, MA. For 4-OHT induction experiments, embryos were collected and kept in 0.003% phenylthiourea (PTU) in 1×E3.
Microinjection
2 nl of 25 ng/μl of (CrymCherry;−1.9mylz2:CreERT2) plasmid with 25 ng/μl Tol2 mRNA was injected into one-cell stage eggs derived from wildtype (Tü) crosses. Positive injected embryos were selected at 3 days post injection (dpi) on the basis of the presence of transgenesis marker CrymCherry and grown to adulthood.
Transgenic Screening
Multiple positive F0s were out-crossed with the wildtype. F1 embryos were selected at 3 days post fertilization (dpf) on the basis of presence of the transgenesis marker CrymCherry and raised to adulthood. Each F1 was screened by crossing with ubi:Switch and inducing the resulting embryos at 10 somites (14 hpf) with 10μM 4-OHT. Screened F1s were out-crossed with the wildtype to generate independent lines.
4-OHT treatment for CreERT2 induction
For 4-OHT (Sigma, St. Louis, MO; H7904) treatments, a 10mM stock solution was made in ethanol and stored in dark at −20°C. An induction medium with 4-OHT at 0.5, 5, 10 or 20μM final concentration in 0.003% PTU in 1×E3 (as per the requirement of the experimental design) was freshly prepared immediately before the start of experiment. About 20 ubi:Switch -positive embryos were obtained from a cross between males of the F2 generation of CrymCherry;−1.9mylz2:CreERT2 line B with ubi:Switch females at each of the desired developmental stage: 10 somites, 24 hpf or 48 hpf. These were placed in fresh 0.003% PTU in 1×E3 in separate wells of a 6-well plate. The E3 medium was aspirated out and 10ml of the induction medium was immediately applied to each well to elicit recombination. The embryos were kept in the induction medium at 28°C. At the desired time point, the embryos were washed three times with 0.003% PTU in 1×E3 to remove 4-OHT. If the embryos were needed to be grown further until 72 hpf for imaging, they were grown in 0.003% PTU in 1×E3.
Control ethanol treatments
For ethanol treatments in control experiments, 10μM 4-OHT was volumetrically replaced by 100% ethanol in 0.003% PTU in 1×E3. About 20 ubi:Switch -positive embryos at 10 somites (12-14 hpf) were placed in fresh 0.003% PTU in 1×E3 per well of a 6-well plate. The E3 medium was aspirated out and 10 ml of the ethanol-containing medium was immediately applied to each well. The embryos were kept in the ethanol-containing medium at 28°C. At 72 hpf, the embryos were washed three times with 0.003% PTU in 1×E3 and imaged.
Imaging and data processing
All images were obtained at 72 hpf when the live embryos were washed with 1×E3 and mounted in 3% methylcellulose. Whole body images were obtained at 10× on Nikon 80i compound microscope (Nikon Instruments). High magnification images of somites 11-13 were obtained at 20× using confocal microscope (Nikon A1R Si Confocal Eclipse Ti series). All confocal and software settings were kept unchanged during image acquisition. Images were processed with an NIS-Elements advanced research image acquisition and analysis system (Nikon Instruments), using the maximum intensity projection feature applied to z-stacks for greater depth of clarity. Image files were exported as TIFF files. Figures were composed with Adobe Photoshop CS6 (Adobe, USA).
Measurement of EGFP and mCherry fluorescence
EGFP and mCherry fluorescence in somites 11-13 were measured using Image J (vl.49, NIH). Briefly, the AREA, INTEGRATED DENSITY and MEAN GRAY VALUE were selected in the “set measurements” from the Analyze menu. Each half somite was selected to measure the fluorescence in both EGFP and mCherry channels. The process was repeated for all the three somites (from 11-13) resulting in six measurements from six half somites. For measurement of the background, three regions with no fluorescence were selected and measured as detailed above. The measurements were performed on somites 11-13 in five different fish in each sample set.
Calculation of percentage recombination
The measurements obtained in Image J were imported into a Microsoft Excel worksheet. The corrected total fluorescence (CTF) for both EGFP and mCherry in each half somite of somites 11-13 in one fish was calculated as:
CTF= Integrated Density − (Area of selected region × Mean fluorescence of background readings) (McCloy et al. 2014)
The average of the six CTFs was taken, each for EGFP and mCherry. The percentage recombination in the fish was calculated as:
Further, the average and standard deviation (S.D.) of percentage recombination efficiencies of the five fish in each sample set were computed. All percentage recombination efficiency data are presented as (av.% recombination efficiency ± S.D.).
RESULTS AND DISCUSSION
We used Tol2-mediated transgenesis to generate a muscle-specific, inducible transgenic Tg(CrymCherry;−1.9mylz2:CreERT2). To assess the spatiotemporal responsiveness of the generated transgenic, CrymCherry;−1.9mylz2:CreERT2 F1 males were crossed to females of a Cre-dependent reporter, ubi:Switch (Mosimann et al. 2011) (Fig.1 A). Males of the CreERT2 transgenic were used in the cross to avoid maternal contribution of CreERT2 (Mosimann and Zon 2011). Recombination was elicited in the resulting embryos by administration of 10μM 4-OHT at 10 somites. The embryos were kept in 0.003% PTU in 1×E3 to prevent formation of melanocytes that obstruct imaging of the muscles at 72 hpf. The generated CreERT2 line carries CrymCherry as a transgenesis marker to indicate the presence of the insertion that manifests as mCherry expression in the lens at 3 dpf. The embryos positive for both transgenes were selected based on the ubiquitously expressed EGFP (Fig.1, B, C) and mCherry expression in the lens at 3 dpf (Fig.1, B’, C’, white arrowhead). Cre-mediated mCherry expression was observed in the trunk somites of the 4-OHT-treated embryos starting at 48 hpf (data not shown). As the transgenesis marker expresses at 72 hpf validating the presence of the CreERT2 transgene, imaging was performed at 72 hpf. Strong mCherry expression was observed in the trunk and tail skeletal muscles in 4-OHT-treated CrymCherry;−1.9mylz2:CreERT2; ubi:Switch embryos, after the Cre-mediated switch from EGFP to mCherry (Fig. 1B). Concurrently, apart from the mCherry expression in the lens from the transgenesis marker, no mCherry expression was observed in the ethanol-treated control CrymCherry;−1.9mylz2:CreERT2; ubi:Switch embryos (Fig.1 C).
FIGURE 1.
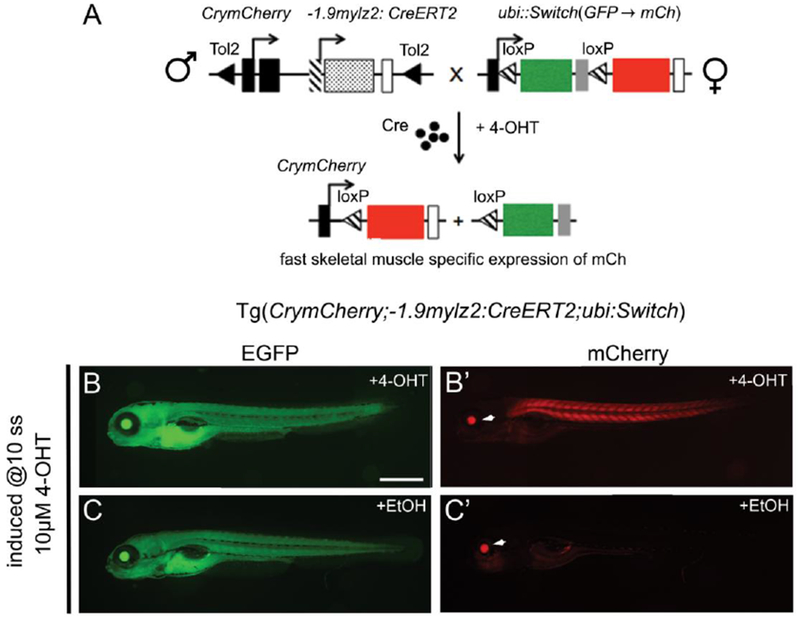
A. Schematic of the CrymCh;−1.9mylz2:CreERT2 and ubi:loxP:EGFP:loxP:mCherry (ubi:Switch) reporter cross shown in panels (B, B’, C, C’). Black arrowheads indicate Tol2 sites, black arrows indicate transcription start positions, black rectangles are CrymCh, shaded rectangle is −1.9mylz2 promoter, dotted rectangle is the CreERT2, shaded arrowheads are loxP sites and open rectangles denote 3’ polyA. (B, B’). Images depict live 72 hpf CrymCh;−1.9mylz2:CreERT2; ubi:Switch embryos treated with 10μM 4-OHT at 10-somites. EGFP expression indicates presence of ubi:Switch transgene (B). White arrowhead indicates CrymCh expression in the eye, which segregates with the −1.9mylz2:CreERT2 transgene and confirms presence of the transgene (B’). Strong and specific mCherry expression is observed in trunk and tail skeletal muscles, confirming successful Cre-mediated switch in tissues expressing mylz2. (C, C’). Images depict live 72 hpf CrymCh;−1.9mylz2:CreERT2; ubi:Switch embryos treated with ethanol at 10-somites. EGFP expression indicates presence of ubi:Switch transgene (C). White arrowhead indicates CrymCh expression in the eye and confirms presence of the transgene (C’). No mCherry expression is observed in skeletal muscles. Scale bar (B, B’ C, C’): 200μm.
The Cre-mediated mCherry expression observed in 4-OHT-treated embryos mimics the previously reported expression pattern of mylz2 in the trunk and tail skeletal muscles (Storer et al. 2013), indicating that CreERT2 is tightly regulated in a skeletal muscle-specific manner in the generated transgenic. However, the transgenic was unable to label the craniofacial skeletal muscles where mylz2 is expressed at 72 hpf (Storer et al. 2013). This is probably due to position effects arising due to the environment of the locus where the CreERT2 transgene integrated during transgenesis (Roberts et al. 2014; Wilson et al. 1990). Out of the eight F1 Crymcherry;−1.9mylz2:CreERT2 males screened by crossing to ubi:Switch females, four produced >50% transgene-positive and recombinant embryos. All four were out-crossed with the wildtype to generate four independent zebrafish lines (lines A, B, C and D). The F2 males from Crymcherry;−1.9mylz2:CreERT2 line B produced ~50% transgene-positive and recombinant embryos with the administration of 10μM 4-OHT at 10 somites (Felker et al. 2016). Hence, it is likely that the line contains a single or two or more closely linked transgene insertion. Line B was selected as the line to be propagated to future generations.
For a given CreERT2 transgenic, the efficiency of recombination is linked to the concentration of 4-OHT used for induction and the timing of the induction. These factors are to be optimal to activate sufficient CreERT2 to recombine a given loxP switch. Towards investigating these factors in the generated transgenic, CrymCherry;−1.9mylz2:CreERT2; ubi:Switch embryos were treated with 0.5, 5, 10 and 20μM 4-OHT at 10 somites and the Cre-mediated recombination efficiency was determined in each case. The embryos induced with 20μM 4-OHT were malformed by 24 hpf (data not shown), indicating that 20μM 4-OHT is toxic to the embryos. The recombination efficiencies at 5 and 5μM 4-OHT were calculated to be 40.63 ± 4.13 % and 48.02 ± 7.47 % respectively (Fig. 2 A, B, Table 1), and increased to 78.70 ± 8.73 % with 10μM 4-OHT (Fig. 2 C, Table 1). The computed recombination efficiencies indicate that 5μM and 10μM are respectively the optimal and the high/saturating concentration of 4-OHT needed for successful Cre-mediated recombination in the transgenic.
FIGURE 2.
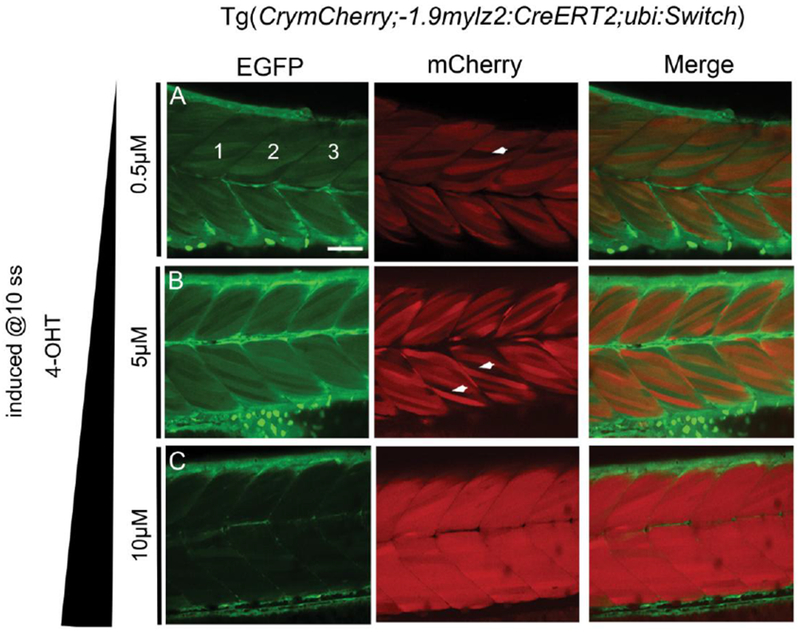
Confocal images of somites 11-13 (marked 1, 2, 3 in EGFP channel, A) in live 72 hpf CrymCh;−1.9mylz2:CreERT2; ubi:Switch embryos treated with 0.5μM (A), 5μM (B) and 10μM 4-OHT (C) at 10-somites. White arrowheads in the mCherry images (A, B) highlight non-recombined muscle fibers. With increase in the induction concentration of 4-OHT, the recombination efficiency in the trunk skeletal muscles are increased. Abbreviation used: ss- somites. Scale bar: 20μm.
TABLE 1.
Percentage recombination efficiency in CrymCh;−1.9mylz2:CreERT2; ubi:Switch embryos treated with 0.5μM, 5μM and 10μM 4-OHT at 10-somites. EGFP and mCherry fluorescence in somites 11-13 (marked 1, 2, 3) was quantified in five embryos for each concentration of 4-OHT.
| induction at 10-somites and somites 11-13 imaged at 72 hpf | |||
|---|---|---|---|
| 4-OHT conc. (μM) | Fish No. | % recombination efficiency | Average (% recombination efficiency) ± SD |
| 0.5 | 1 | 44.15 | 40.63 ± 4.10 |
| 2 | 39 | ||
| 3 | 45.11 | ||
| 4 | 39.91 | ||
| 5 | 35 | ||
| 5 | 1 | 53.90 | 48.02 ± 7.47 |
| 2 | 49.80 | ||
| 3 | 39.34 | ||
| 4 | 41.12 | ||
| 5 | 55.91 | ||
| 10 | 1 | 79.85 | 78.70 ± 8.73 |
| 2 | 91.04 | ||
| 3 | 74 | ||
| 4 | 81.04 | ||
| 5 | 67.59 | ||
As discussed earlier, mylz2 can be detected in somites at around 16 hpf (Xu et al. 2000). Hence, CrymCherry;−1.9mylz2:creERT2; ubi:Switch embryos were induced with saturating concentration of 10μM 4-OHT, at a time point slightly earlier that the expected initiation of CreERT2 expression (10 somites, 14 hpf), at a time point where the CreERT2 expression has just initiated (24 hpf) and at a later time point in embryogenesis (48 hpf), for varying time periods. After induction at 10-somites, removal of 4-OHT at 24, 48 or 72 hpf elicited a recombination efficiency of 45.55 ± 4.85 %, 70.25 ± 5.82 % and 78.70 ± 8.73 % respectively (Fig. 2, C, Fig. 3, A, B, Table 2). On induction at 24 hpf and removal of 4-OHT at 48 hpf or 72 hpf, the recombination efficiency was 71.37 ± 4.73 % and 69.31 ± 7.17 % respectively (Fig. 3, C, D, Table 2). However, if induction with 4-OHT was performed at 48 hpf till 72 hpf, the recombination efficiency in the transgenic was only 21.43 ± 5.14 % (Fig. 3, E, Table 2). Of note, significant variation was observed between embryos treated with 4-OHT under same conditions, as is evident from the S.D. values in Tables 1 and 2. The results suggest that higher recombination efficiency is achieved when embryos are induced with 10μM 4-OHT from 10-somites or 24 hpf till 48 or 72 hpf. When induced around a time point when the Cre expression is expected to begin (16 hpf), the 4-OHT activates the CreERT2 and mediates the EGFP to mCherry switch more robustly leading to higher recombination efficiency. However, if the induction is performed at a later stage (48 hpf), the Cre is activated less robustly leading to lower combination efficiency.
FIGURE 3.
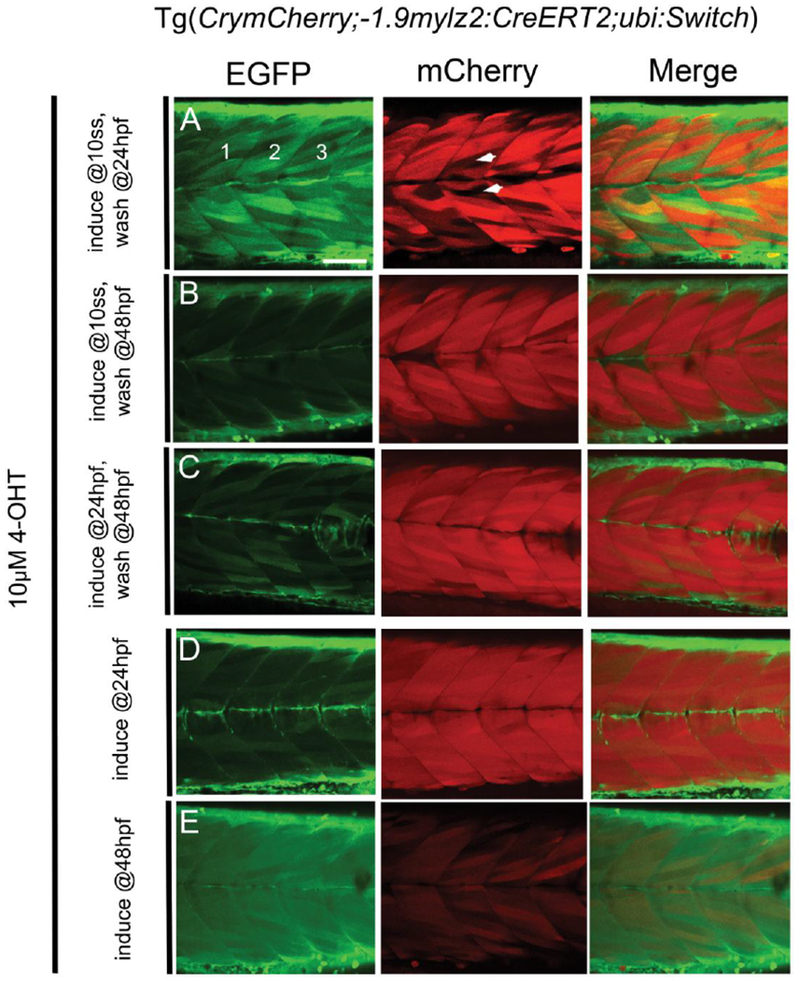
Confocal images of somites 11-13 (marked 1, 2, 3 in EGFP channel, A) in live 72 hpf CrymCh;−1.9mylz2:CreERT2; ubi:Switch embryos induced with 10μM 4-OHT at 10-somites (A, B), 24 hpf (C, D) or 48 hpf (E). 4-OHT was removed at 24 hpf (A) or 48 hpf (B) in embryos induced at 10-somites and removed at 48 hpf (C) or kept until imaging at 72 hpf (D) in embryos induced at 24 hpf. Embryos induced at 48 hpf (E) were kept in 4-OHT until imaging at 72 hpf. White arrowheads in the mCherry image (A) highlight non-recombined muscle fibers. Abbreviation used: ss- somites. Scale bar: 20μm.
TABLE 2.
Percentage recombination efficiency in CrymCh;−1.9mylz2:CreERT2; ubi:Switch embryos treated with 10μM 4-OHT at 10-somites, 24 hpf and 72 hpf for different time periods. EGFP and mCherry fluorescence in somites 11-13 (marked 1, 2, 3) was quantified in five embryos in each sample set. The data set obtained when 4-OHT was introduced at 10-somites and removed at 72 hpf (depicted in bold) is replicated from Table 1.
| induction with 10μM 4-OHT and somites 11-13 imaged at 72 hpf | ||||
|---|---|---|---|---|
| Embryonic stage at induction | Embryonic stage at 4-OHT removal | Fish No. | % recombination efficiency | Average (% recombination efficiency) ± SD |
| 10-somites | 24 hpf | 1 | 45.97 | 45.55 ± 4.85 |
| 2 | 40.09 | |||
| 3 | 52.30 | |||
| 4 | 41.78 | |||
| 5 | 47.62 | |||
| 10-somites | 48 hpf | 1 | 70.13 | 70.25 ± 5.82 |
| 2 | 63.09 | |||
| 3 | 74.62 | |||
| 4 | 77.23 | |||
| 5 | 66.19 | |||
| 10-somites | 72 hpf | 1 | 79.85 | 78.70 ± 8.73 |
| 2 | 91.04 | |||
| 3 | 74 | |||
| 4 | 81.04 | |||
| 5 | 67.59 | |||
| 24 hpf | 48 hpf | 1 | 72.17 | 71.37 ± 4.73 |
| 2 | 69.52 | |||
| 3 | 70.02 | |||
| 4 | 78.93 | |||
| 5 | 66.22 | |||
| 24 hpf | 72 hpf | 1 | 61.34 | 69.31 ± 7.17 |
| 2 | 74.20 | |||
| 3 | 68.24 | |||
| 4 | 78.74 | |||
| 5 | 64.01 | |||
| 48 hpf | 72 hpf | 1 | 23.41 | 21.34 ± 5.14 |
| 2 | 20.11 | |||
| 3 | 15.25 | |||
| 4 | 28.91 | |||
| 5 | 19 | |||
We have monitored the CreERT2 transgene expression through multiple generations (currently in F4) and have not observed transgene silencing. In conclusion, we report the generation of an inducible transgenic Tg(CrymCherry;−1.9mylz2:CreERT2) that expresses specifically in the trunk and tail skeletal muscles. We investigate the factors for optimal recombination in the transgenic and observe that the optimal concentration range and time period of 4-OHT induction is 5- 10μM at 10-somites or 24 hpf till 72 hpf. The Tg(CrymCherry;−1.9mylz2:CreERT2) can be utilized to overexpress or ectopically express genes of interest specifically in trunk and tail skeletal muscles. Further, recently targeted insertion of loxP sites in the zebrafish genome has been made successfully, via homologous recombination-mediated gene editing (Hoshijima et al. 2016). The reported CreERT2 line can be potentially used to generate muscle specific loss-of-function mutants in future. The line is an addition to the currently available zebrafish transgenesis toolbox to advance muscle biology studies and is available from Eric C. Liao, Massachusetts General Hospital, Boston, upon request.
ACKNOWLEDGEMENTS
We thank Dr. Jenna Galloway for discussions and providing valuable feedback towards the manuscript. We thank the Center for Computational and Integrative Biology (CCIB) at Massachusetts General Hospital for the use of the CCIB Core Facility (Cambridge, MA), which provided Sanger DNA sequencing service. This work was supported by the National Institute of General Medical Sciences (NIGMS) P01GM061354, and a research fellowship to K.M. from Shriners Hospitals for Children.
ABBREVIATIONS
- ER
Estrogen Receptor
- TAM
Tamoxifen
- 4-OHT
4-hydroxytamoxifen
- mylz2
Fast skeletal muscle myosin light chain 2
- WISH
Whole mount in situ hybridization
- hpf
Hours post fertilization
- PTU
Phenylthiourea
- F0
Founder animals (injected)
- F1
1st generation animals
- dpi
Days post injection
- dpf
Days post fertilization
- EGFP
Enhanced Green Fluorescent Protein
- CTF
Corrected Total Fluorescence
- SD
Standard Deviation
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI (2015) A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish Dev Cell 32:756–764 doi: 10.1016/j.devcel.2015.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsterdam A et al. (1999) A large-scale insertional mutagenesis screen in zebrafish Genes Dev 13:2713–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger J, Currie PD (2013) 503unc, a small and muscle-specific zebrafish promoter Genesis 51:443–447 doi: 10.1002/dvg.22385 [DOI] [PubMed] [Google Scholar]
- 4.Driever W et al. (1996) A genetic screen for mutations affecting embryogenesis in zebrafish Development 123:37–46 [DOI] [PubMed] [Google Scholar]
- 5.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P (1996) Ligand-activated site-specific recombination in mice Proc Natl Acad Sci U S A 93:10887–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil R, Wagner J, Metzger D, Chambon P (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains Biochem Biophys Res Commun 237:752–757 doi: 10.1006/bbrc.1997.7124 [DOI] [PubMed] [Google Scholar]
- 7.Felker A et al. (2016) In Vivo Performance and Properties of Tamoxifen Metabolites for CreERT2 Control PLoS One 11:e0152989 doi: 10.1371/journal.pone.0152989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guner-Ataman B et al. (2013) Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function Development 140:1353–1363 doi: 10.1242/dev.088351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hans S, Kaslin J, Freudenreich D, Brand M (2009) Temporally-controlled site-specific recombination in zebrafish PLoS One 4:e4640 doi: 10.1371/journal.pone.0004640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshijima K, Jurynec MJ, Grunwald DJ (2016) Precise Editing of the Zebrafish Genome Made Simple and Efficient Dev Cell 36:654–667 doi: 10.1016/j.devcel.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC (2010) Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation Nature 464:606–609 doi: 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jungke P, Hammer J, Hans S, Brand M (2015) Isolation of Novel CreERT2-Driver Lines in Zebrafish Using an Unbiased Gene Trap Approach PLoS One 10:e0129072 doi: 10.1371/journal.pone.0129072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami K (2007) Tol2: a versatile gene transfer vector in vertebrates Genome Biol 8 Suppl 1:S7 doi: 10.1186/gb-2007-8-s1-s7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi K et al. (2010) Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes Nature 464:601–605 doi: 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwan KM et al. (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs Dev Dyn 236:3088–3099 doi: 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 16.Lee RT, Asharani PV, Carney TJ (2014) Basal keratinocytes contribute to all strata of the adult zebrafish epidermis PLoS One 9:e84858 doi: 10.1371/journal.pone.0084858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieschke GJ, Currie PD (2007) Animal models of human disease: zebrafish swim into view Nat Rev Genet 8:353–367 doi: 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Li Z, Emelyanov A, Parinov S, Gong Z (2008) Generation of oocyte-specifically expressed cre transgenic zebrafish for female germline excision of loxP-flanked transgene Dev Dyn 237:2955–2962 doi: 10.1002/dvdy.21701 [DOI] [PubMed] [Google Scholar]
- 19.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A (2014) Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events Cell Cycle 13:1400–1412 doi: 10.4161/cc.28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzger D, Clifford J, Chiba H, Chambon P (1995) Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase Proc Natl Acad Sci U S A 92:6991–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mongera A, Singh AP, Levesque MP, Chen YY, Konstantinidis P, Nusslein-Volhard C (2013) Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells Development 140:916–925 doi: 10.1242/dev.091066 [DOI] [PubMed] [Google Scholar]
- 22.Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI (2011) Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish Development 138:169–177 doi: 10.1242/dev.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosimann C, Zon LI (2011) Advanced zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments Methods Cell Biol 104:173–194 doi: 10.1016/B978-0-12-374814-0.00010-0 [DOI] [PubMed] [Google Scholar]
- 24.Nguyen PD et al. (2014) Haematopoietic stem cell induction by somite-derived endothelial cells controlled by meox1 Nature 512:314–318 doi: 10.1038/nature13678 [DOI] [PubMed] [Google Scholar]
- 25.Roberts JA et al. (2014) Targeted transgene integration overcomes variability of position effects in zebrafish Development 141:715–724 doi: 10.1242/dev.100347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues AM, Christen B, Marti M, Izpisua Belmonte JC (2012a) Skeletal muscle regeneration in Xenopus tadpoles and zebrafish larvae BMC Dev Biol 12:9 doi: 10.1186/1471-213X-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues FS, Doughton G, Yang B, Kelsh RN (2012b) A novel transgenic line using the Cre-lox system to allow permanent lineage-labeling of the zebrafish neural crest Genesis 50:750–757 doi: 10.1002/dvg.22033 [DOI] [PubMed] [Google Scholar]
- 28.Storer NY, White RM, Uong A, Price E, Nielsen GP, Langenau DM, Zon LI (2013) Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis Development 140:3040–3050 doi: 10.1242/dev.087858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Weyden L, Adams DJ, Bradley A (2002) Tools for targeted manipulation of the mouse genome Physiol Genomics 11:133–164 doi: 10.1152/physiolgenomics.00074.2002 [DOI] [PubMed] [Google Scholar]
- 30.Westerfield M, Doerry E, Douglas S (1999) Zebrafish in the Net Trends Genet 15:248–249 [DOI] [PubMed] [Google Scholar]
- 31.Wilson C, Bellen HJ, Gehring WJ (1990) Position effects on eukaryotic gene expression Annu Rev Cell Biol 6:679–714 doi: 10.1146/annurev.cb.06.110190.003335 [DOI] [PubMed] [Google Scholar]
- 32.Xu Y et al. (1999) Fast skeletal muscle-specific expression of a zebrafish myosin light chain 2 gene and characterization of its promoter by direct injection into skeletal muscle DNA Cell Biol 18:85–95 doi: 10.1089/104454999315655 [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, He J, Wang X, Lim TM, Gong Z (2000) Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis Dev Dyn 219:201–215 doi: [DOI] [PubMed] [Google Scholar]
- 34.Yoshikawa S, Kawakami K, Zhao XC (2008) G2R Cre reporter transgenic zebrafish Dev Dyn 237:2460–2465 doi: 10.1002/dvdy.21673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L et al. (2014) Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration Proc Natl Acad Sci U S A 111:1403–1408 doi: 10.1073/pnas.1311705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou T et al. (2011) Dominant-negative C/ebpalpha and polycomb group protein Bmi1 extend short-lived hematopoietic stem/progenitor cell life span and induce lethal dyserythropoiesis Blood 118:3842–3852 doi: 10.1182/blood-2010-12-327908 [DOI] [PubMed] [Google Scholar]


