Abstract
Background:
It is cleared that some probiotic strains inhibit biofilm formation of oral bacteria, but its mechanisms are not clearly understood yet. It is proposed that one of the mechanisms can be biosurfactant production, a structurally diverse group of surface-active compounds synthesized by microorganisms. Hence, this study focused on the evaluation of the anti-biofilm and antiadhesive activities of the L. rhamnosus derived-biosurfactant against Streptococcus mutans and its effect on gtfB/C and ftf genes expression level.
Materials and Methods:
In this in vitro study Lactobacillus rhamnosus ATCC7469 overnight culture was used for biosurfactant production. The biosurfactant effect on the surface tension reduction was confirmed by drop collapse method. Chemical bonds in the biosurfactant were identified by Fourier transform infrared (FTIR). Anti-biofilm and antiadhesive activities of the biosurfactant were determined on glass slides and in 96-well culture plates, respectively. The effect of the biosurfactant on gtfB/C and ftf genes expression level was also investigated after biofilm formation, total RNA extraction, and reverse transcription by quantitative real-time reverse transcriptase polymerase chain reaction (PCR) assay (quantitative PCR). The data were assessed by one-way analysis of variance in the Tukey–Kramer postdeviation test for all pairs. P < 0.05 was considered statistically significant.
Results:
The FTIR results of biosurfactant showed that it was protein rich. It also showed anti-biofilm formation activity on the glass slide and antiadhesive activity till 40% on microtiter plate wells. It also showed a significant reduction (P < 0.05) in gtfB/C and ftf genes expression level.
Conclusion:
L. rhamnosus-derived biosurfactant exhibits a significant inhibitory effect on biofilm formation ability of S. mutans due to downregulation of biofilm formation associated genes, gtfB/C and ftf. L. rhamnosus-derived biosurfactant with substantial antiadhesive activity is suitable candidates for use in new generations of microbial antiadhesive agents.
Key Words: Biofilm, biosurfactant, genes, Lactobacillus rhamnosus, Streptococcus mutans
INTRODUCTION
One of the most widespread chronic multifactorial diseases is dental caries which affects children and adults.[1] Teeth colonization by several strains of oral streptococci, especially Streptococcus mutans initiates the dental plaque formation which plays an important role in caries and also periodontal disease development in humans. Dental plaque is a complex bacterial biofilm community which its composition is determined by factors such as cell adherence, coaggregation, growth, and survival in the oral cavity.[2]
S. mutans ability to synthesize extracellular glucans from the glucose moiety of sucrose using types of glucosyltransferases (GTFB, GTFC, and GTFD) and homopolymers of fructose, the fructans from the fructose moiety of sucrose by fructosyltransferase (FTF), are major virulence factors of this pioneer bacterium.[3] The GTFs and also FTF secreted by S. mutans make specific binding sites available for bacterial colonization of the tooth surface or bacterial attachment to each other, adjusting the strongly adherent biofilms formation, the precursor of dental caries.[4,5,6] In vitro studies have shown that gtfB, gtfC, and also ftf genes are important for the sucrose-dependent attachment of S. mutans cells to hard surfaces, but gtfD is not necessary.[7] Therefore, perfect therapeutics for this disease will aim to selectively inhibit the biofilm formation process along with preserving the normal bacterial flora of the mouth.[8] Hence, the gtfB, gtfC, and also ftf genes have become a potential target for protection against dental caries.
The process of removing or preventing the accumulation of bacteria is called antifouling which can be used to control biofilm formation or biofouling. The best-known and most widely-used anti-plaque substance is chlorhexidine, which is not side-effect free. Various antiplaque compounds such as plant alkaloids, biguanides, phenol and essential oils, fluorinated amines and ammonium compounds, and detergents such as sodium lauryl sulfate, some antibiotics, and triclosan, have been screened for their ability to interfere with dental biofilm formation.[9] However, due to their ineffectiveness or undesirable side effects, the searches for alternative efficient and safe antiplaque agents are in continued demands. One optional approach is using inexpensive, effective, stable, novel, and natural products as anti-biofouling agents.[10,11]
Lactobacilli, as probiotic agents, are thought to interfere with pathogens through different mechanisms,[12] such as reducing biofilm acidogenicity following the short-term consumption of probiotics products in children with certain oral biofilm and risk profile.[13] Although it is cleared that some Lactobacillus strains can inhibit biofilm formation of oral bacteria, the mechanisms by which lactobacilli can do this are not clearly understood. However, one of the mechanisms can be biosurfactant production, a structurally diverse group of surface-active compounds synthesized by microorganisms. Biosurfactants have attracted attention in recent years because having several advantages over synthetic surfactants, such as low toxicity, innate efficient ecological acceptability, and biodegradability.[14]
Therefore, the present study aimed to investigate the effect of L. rhamnosus (ATCC7469)-derived biosurfactant on biofilm formation and gtfB, gtfC, and ftf genes expression level in S. mutans biofilm cells using real-time reverse transcriptase polymerase chain reaction (RT-PCR).
MATERIALS AND METHODS
Bacteria and culture conditions
In this in vitro study The no 22 S. mutans strain was previously isolated from dental plaque in our laboratory S. mutans 22, previously isolated from dental plaque (and selected among 40 isolates due to its the highest ability of biofilm formation)[15] and S. mutans ATCC 35668 were used in this study. The strains were cultured on mitis salivarius agar and blood agar medium and incubated at 37°C in a CO2-enriched atmosphere. Lactobacillus rhamnosus ATCC 7469 as a probiotic source was cultured in MRS broth or agar.
Biosurfactant production
An overnight culture of L. rhamnosus (15 ml) was inoculated into MRS broth (600 ml) and incubated for 24 h. The cells were harvested using centrifugation (10,000 × g, 5 min, 10°C), washed twice with demineralized water and resuspended in phosphate buffered saline (PBS) (100 ml). For biosurfactant production, the lactobacilli were incubated at room temperature (2 h) with gentle stirring. Then, the bacterial cells were removed by centrifugation, and the supernatant was filtered through a 0.22-mm filter (Millipore). A portion of the supernatant (10 ml) was used directly in the adhesion assay, and the remainder was dialyzed against demineralized water at 4°C in a Spectrapor membrane tube (molecular weight cutoff 6000–8000 kDa; Spectrum Medical Industries, Inc.) and freeze-dried.[16]
Drop collapse method
For testing whether the biosurfactant could reduce the surface tension between water and hydrophobic surfaces, its ability to collapse a water droplet was tested as follows: A droplet of 25 μl extracted biosurfactant was placed onto parafilm and the droplet spreading and flattening on the parafilm surface was monitored over seconds or minutes. Then, methylene blue was added to the supernatants and also water (used as a control) for photographic purposes. The droplets were allowed to dry and the diameters of the dried droplets were recorded.
Fourier-transform infrared spectroscopy
The amount of 2 mg freeze-dried biosurfactants were ground with 100 mg of KBr and compressed (at 7500 kg for 3 min) for obtaining translucent pellets. Infrared absorption spectra were recorded with a Nicolet Impact 400 instrument with the spectral resolution and wavenumber accuracy of 4 and 0.01 cm-1, respectively. A pellet of KBr was used as the background reference. A spectral region of interest quantification was determined by normalizing the area under the absorption bands relative to the CH absorption band area at approximately 2932 cm-1.[17]
Biofilm formation assay
To observe the effect of biosurfactant on S. mutans biofilm formation on a glass slide, overnight culture of S. mutans (1 ml, 108 CFUml-1) was inoculated into a flask containing sterile BHI broth +1% sucrose (100 ml) and two slides with and without L. rhamnosus-derived biosurfactant. The glass slides were washed with a detergent solution, rinsed in distilled water, air-dried, and autoclaved at 121°C for 15 min before use. The flasks were incubated in an orbital incubator at 35°C–37°C with shaking at 100 rpm for 18–20 h. Then, the glass slides were rinsed off twice with PBS (10 ml) for removing unattached cells. The glass slides were then stained with 2% crystal violet (5 min), washed, air-dried, and photographed under an optical microscope with a digital camera (Nikon, Eclipse, E200, Japan).
Antiadhesion assay in microtiter plate
To determine the antiadhesive activity of biosurfactant against S. mutans strains, the wells of a sterile 96-well flat-bottomed microtiter plate were filled with the crude biosurfactant (200 μl). The plate was then incubated at 4°C for 18 h and washed twice with PBS. Control wells contain PBS buffer. The amount of 200 μl of S. mutans suspension (according to 0.5 Mc Farland) was added to the wells and incubated for 4 h at room temperature. Unattached cells were washed with PBS three times. The adherent cells of S. mutans were fixed with 200 μl of ethanol per well, and after 15 min, the plates were emptied and left to dry. Then, the plates were stained for 5 min with 200 μl of 2% crystal violet used for Gram staining per well. Excess stain was rinsed off under running tap water. After the plates were air-dried, the dye bound to the adherent microorganisms was resolubilized with 200 μl of 33% (v/v) glacial acetic acid per well and the optical density of each well were read at 492 nm. This assay estimates the percentage of microbial adhesion reduction versus the control wells.[18,19]
Biofilm preparation for gene expression analysis
To generate biofilms on a 24-well polystyrene microtiter plates, the amount of 20 μl S. mutans overnight culture were inoculated in each well and cultivated in brain–heart infusion (BHI) broth (2 ml) supplemented with 1% sucrose. The plates were then incubated in an atmosphere enriched with 5% CO2 at 37°C. After incubation (18-h), the wells were washed with PBS for removing unattached cells. The biofilm was incubated again in fresh BHI supplemented with 1% sucrose; after an 18-h incubation period, the spent medium was aspirated again. The cells were washed and the biofilm was incubated again in fresh BHI broth + 1% sucrose in the presence or absence of 2.5 mg ml-1 of freeze-dried biosurfactant. The cells of the biofilms were taken away after 4-h incubation and transferred to tubes containing PBS solution (2 ml) and vortexed.[20]
Total RNA extraction
For RNA extraction, the S. mutans ATCC 35668 and S. mutans 22 biofilm cells prepared in the presence or absence of biosurfactant (in three replicates) on microtiter plates were used. Cells were disrupted using a ribolyser instrument (Hybaid, UK) and the supplied kit according to the manufacturer's instructions. In brief, RNA containing supernatant from the ribolyser tube was transferred to a new RNase-free microtube, centrifuged, treated with chloroform-isoamyl alcohol (300 μl), vortexed, and centrifuged again. Then, precipitation with isopropanol was performed for total RNA recovering. The recovered total RNA was dried under sterile conditions. Extracted RNA quantitative and qualitative evaluations were done using a spectrophotometer (Biophotometer, Eppendorf, Rs 232-C, Germany) and agarose gel electrophoresis.
Reverse transcription
A reverse transcription reaction mixture (20 μL) containing random hexamers (50 ng), total RNA sample (2 μg), and diethylpyrocarbonate-treated water (up to 12 μL) was incubated at 70°C for 5 min for removing any secondary structure and was placed on ice. Subsequently, 5X RT buffer (4 μL), 20 U μL-1 ribonuclease inhibitor (1 μL) and 10 mM dNTPs mix were added to each reaction mixture and incubated at 37°C for 5 min. Then, 1 μL reverse transcriptase (RT) was added and the mixture was incubated at 42°C for 60 min. The mixture was heated at 70°C for 10 min to terminate the reaction. The samples of cDNA were stored at 20°C for further manipulation.
Real-time quantitative reverse transcriptase polymerase chain reaction
The ABI StepOne Plus (Applied Biosystems, CA, USA) instrument and the SYBR Green PCR Master Mix (Qiagen, GmbH-Germany) were used for real-time quantitative RT-PCR. The gtfB, gtfC, and ftf genes relative quantification were performed using 16S rRNA as a reference gene.[20]
All primers (gtf B, gtf C, ftf genes, and housekeeping gene) for real-time-PCR were controlled with NCBI Primer Blast software and obtained from Takapuzist Company (Bioneer, Korea) [Table 1].
Table 1.
Nucleotide sequences of primers
| Primer | Sequence (5/- 3/) | Fragment location | Accession number |
|---|---|---|---|
| ftf - F | AAATATGAAGGCGGCTACAACG | 1358-1379 | M18954 |
| ftf - R | CTTCACCAGTCTTAGCATCCTGAA | 1435-1458 | M18954 |
| gtfB - F | AGCAATGCAGCCAATCTACAAAT | 1150-1172 | M17361 |
| gtfB -R | ACGAACTTTGCCGTTATTGTCA | 1224-1245 | M17361 |
| gtfC - F | CTCAACCAACCGCCACTGTT | 434-453 | M22054 |
| gtfC - R | GGTTTAACGTCAAAATTAGCTGTATTAGC | 496-524 | M22054 |
| 16S - F | CCTACGGGAGGCAGCAGTAG | 243-262 | X58303 |
| 16S - R | CAACAGAGCTTTACGATCCGAAA | 321-343 | X58303 |
The reaction mixture (20 μL) contained Master Mix (10 μL), 200 ng of template cDNA (2 μL) and 200 nmol l-1 of appropriate forward and reverse PCR primers. Three repeats of the same reaction tubes were used for each test sample and three reaction tubes without cDNA template were used as negative controls for each primer pair to check for DNA contamination. The thermal cycling program consisted of an initial denaturation at 95°C for 5 min (holding stage), 40 cycles of denaturation at 95°C for 15 s followed by annealing and extension at 60°C for 1 min (cycling stage). Negative and positive controls were also included. The critical threshold cycle (CT) values were determined, and the relative expression levels were calculated according to the comparative CT (ΔΔCT) method. The cDNA quantities for gtfB, gtfC, and ftf genes were normalized to that of cDNA synthesized from 16S rRNA gene in the same sample. The values were compared to those obtained from the control (nontreated) for determining the changes in genes expression level in each sample.
Statistical analysis
All real-time PCR assays for target genes (gftB, gtfC, and ftf) and two target samples (S. mutans strain 22 and standard strain ATCC35668) were performed in triplicate. The data were assessed by one-way analysis of variance in the Tukey–Kramer postdeviation test for all pairs. P < 0.05 was considered statistically significant.
RESULTS
The droplets of biosurfactant resulted in a collapsed droplet, indicating the biosurfactant effect on the surface tension reduction. No activity was detected for distilled water, as predicted.
The molecular composition of the biosurfactant derived from L. rhamnosus was analyzed using Fourier-transform infrared (FTIR) spectroscopy [Figure 1]. The most important bands were located at 3423 cm-1 (OH or –NH2 stretching vibration in glycoproteins), 2913 cm-1 (CH band: CH2–CH3 stretching), 1634 cm-1 (AmI band: C = O stretching vibration in proteins), 1412/cm (AmII band: NH bending in proteins), 1237 (PI phosphate bond), and 1105/cm (PII band: polysaccharides). Therefore, the FTIR spectra showed that this biosurfactant consists of a mixture of protein and polysaccharides.
Figure 1.
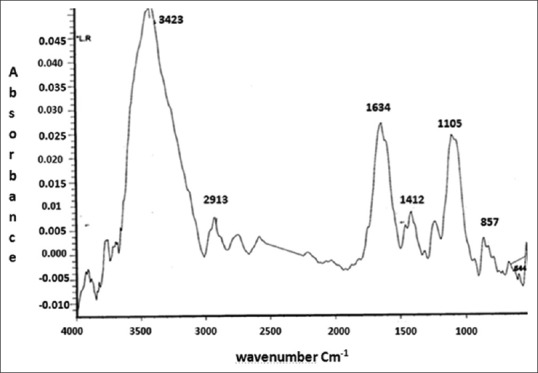
The Fourier-transform infrared spectrum of the freeze-dried biosurfactant released from Lactobacillus rhamnosus ATCC 7469.
Biofilm formation
As shown in Figure 2, 2.5 mg ml-1 of extracted L. rhamnosus biosurfactant showed significant anti-biofouling activity because it reduced the process of attachment and biofilm production [Figure 2a and b].
Figure 2.
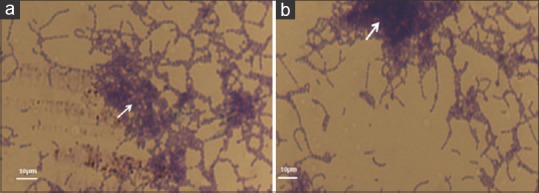
Streptococcus mutans biofilm formation. (a) an experimental group (in the presence of Lactobacillus rhamnosus-derived biosurfactant). (b) a control group (in the absence of biosurfactant). The arrows show the biofilm depth on a glass slide. The magnification of both images is ×400.
Antiadhesive activity
The crude biosurfactant anti-adhesive activity was evaluated against two strains of S. mutans [Figure 3]. The crude biosurfactant showed an antiadhesive effect against both S. mutans strains, but the antiadhesive effect is depending on the strains tested. The crude biosurfactant had the highest antiadhesive activity against S. mutans 22 with inhibition percentages till 40%.
Figure 3.
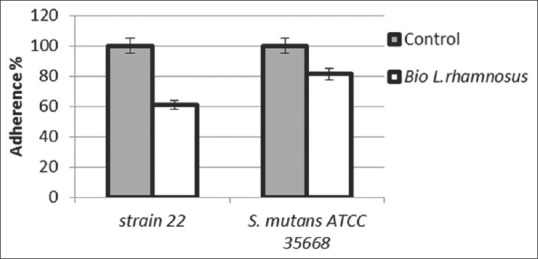
The effect of Lactobacillus rhamnosus derived biosurfactant on the adherence ability of Streptococcus mutans strains. The control treatment was performed in the absence of biosurfactant.
The effect of biosurfactant on gtfB, gtfC, and ftf genes expression
The attached bacterial cells of the formed biofilm on 24-well microtiter plate were collected and used for RNA extraction. The extracted RNAs quality and quantity are presented in Table 2. The effect of L. rhamnosus biosurfactant on gtfB, gtfC, and ftf genes expression in S. mutans ATCC 35668 and S. mutans 22 biofilm cells was quantified by real-time RT-PCR [Figure 4]. An internal reference of 16S rRNA gene was used. In the biofilm condition, biosurfactant significantly reduced the expression level of gtfB, gtfC, and ftf genes (P < 0.05).
Table 2.
The extracted RNAs quality and quantity
| RNA obtained from | µg/ml | OD260/280 ratio |
|---|---|---|
| S. mutans 22 biofilm | 311.2 | 1.8 |
| S. mutans ATCC 35668 biofilm | 165.8 | 1.83 |
| S. mutans 22 + L. rhamnosus biosurfactantbiofilm | 133.3 | 1.9 |
| S. mutans ATCC 35668 + L. rhamnosus biosurfactant biofilm | 280.1 | 1.79 |
S. mutans: Streptococcus mutans; L. rhamnosus: Lactobacillus rhamnosus
Figure 4.
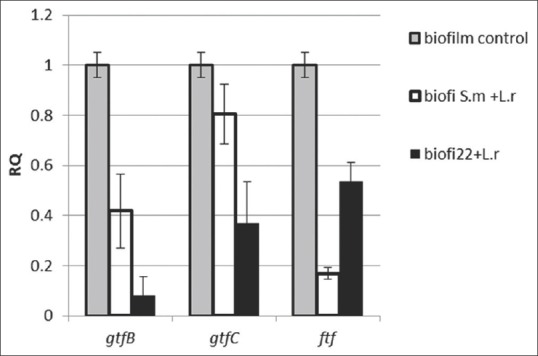
The effect of Lactobacillus rhamnosus-derived biosurfactant on gtfB/C and ftf genes expression level in immobilized biofilm. S. m = Streptococcus mutans ATCC 35668; 22 is Streptococcus mutans strain 22. The mRNA expression levels were calibrated relative to the control group (in the absence of biosurfactant). The results are expressed as the means and standard errors of duplicate experiments using primers specific for gtfB/C, ftf, and 16S rRNA (normalizing gene).
The P values obtained for each of the three genes (P < 0.05), is indicative of the biosurfactant efficiency in downregulation of the adhesive promoting genes of S. mutans strains.
However, the biosurfactant effect on the three genes expression level was not identical, and a more significant decrease in gtfB gene expression was observed. The extent of the decrease in gtfB and gtfC expression levels in S. mutans 22 was more than the standard strain (P < 0.05) due to its higher adherence ability.
DISCUSSION
Dental caries is a multifactorial infectious disease closely related to the presence of cariogenic bacteria such as S. mutans which are embedded in the dental plaque biofilm. As S. mutans plays the major role in dental caries, it should be inhibited. Hence, any strategy which directs toward the reduction of S. mutans count and activity is valuable for maintaining healthy oral cavity. With increasing resistance to synthetic antimicrobials,[21] the researchers have encouraged to find alternative natural products such as probiotic bacteria and their biosurfactants[22,23] due to their negative effect on other microbial species.[19,24] In recent years, the use of biosurfactant to control biofilms has been evaluated extensively.[25,26,27] Hence, in this study, the biosurfactant extracted from L. rhamnosus ATCC 7469 was investigated as a potential control for S. mutans biofilm. The extracted biosurfactant activity was shown by the drop collapse technique.
FTIR analysis of freeze-dried biosurfactant extracted from L. rhamnosus was compared with the reference compounds albumin, salivary glycoprotein, dextran, lipoteichoic acid, and other FTIR spectra of biosurfactants and showed that the biosurfactant from L. rhamnosus had more protein than polysaccharide and phosphate. Moldes et al. observed a 3200–3500 cm-1 peak in the Lactobacillus pentosus biosurfactant spectrum which indicates the presence of OH and NH groups in glycoproteins.[28] Such structures also proposed for the biosurfactants derived from Lactobacillus lactis and Lactobacillus paracasei.[19,29,30] Brzozowski et al. showed that Lactobacillus. fermenti 126 and L. rhamnosus CCM 1825 synthesize biosurfactants which are mixtures of several compounds (protein, polysaccharide, and phosphate).[31] They also stated that the protein component of biosurfactant has an important role in decreasing surface tension, while Rodrigues et al. showed that other compounds than protein in the biosurfactant are responsible for decreasing surface tension.[19]
As seen in Figure 3, the biosurfactant significantly reduced the adhesion of S. mutans strains to the microtiter plate surfaces. Ahmed et al. also demonstrated that Lactobacillus acidophilus, Lactobacillus Salivarius, and L. rhamnosus reduced the biofilm formation by S. mutans.[32] However, the specific mechanisms of such effects have not yet been clearly determined, and they seem to be highly dependent on the biosurfactant type and the properties of the target bacteria. The simplest way to explain biosurfactant antiadhesion activities would be through direct antimicrobial action which has not been observed in all cases,[33] as well as in this case, and it is pH dependent in the case of mutans streptococci.[34] Walencka et al. also stated that antiadhesion mechanism of biosurfactant is more closely related to the changes in surface tension and bacterial cell wall charge which are important in controlling the primary electrostatic repulsion barrier between the bacterial cell surface and its substratum. Surfactants may also affect both cell-to-cell and cell-to-surface interactions. Their results support the idea that lactobacillus-derived biosurfactant has an effect on these interactions.[33] Brzozowski et al. also express that the biosurfactants chemical structure seems to have an important role on their antiadhesive properties. They also stated that biosurfactants with a higher proportion of proteins are more effective on adhesion inhibition.[31]
It is known that the adherence and colonization of S. mutans to the surfaces is facilitated by surface-associated proteins encoded by various genes.[35] Among these genes, gtfs and ftf produce proteins which catalyze the cleavage of sucrose to synthesize extracellular glucan and fructan polysaccharides, respectively.[36]
Water-insoluble glucans, encoded from gtfB/C, are important for the bacterial adherence to the teeth surface and to each other and also provide shelter and food for bacteria. Hence, GTF B and C have been shown to be important virulence factors of mutans streptococci associated with the dental caries pathogenesis.[32] The product of ftf gene, FTF, is supposed to function as extracellular storage polysaccharides also provide a unique microenvironment for bacterial growth, metabolism, and survival. Therefore, inhibition or the reduction of these polysaccharides synthesis by affecting the encoding genes expression level is an attractive route for preventing dental caries and also plaque formation.
Hence, in this study, the effect of L. rhamnosus-derived biosurfactant on gtf B, gtf C, and ftf gene expression level of S. mutans were also investigated. The results showed that the biosurfactant reduced 92% and 58% expression of the gtfB gene in the case of S. mutans 22 and ATCC 35668, respectively. Furthermore, it could downregulate the expression level of gtfC and ftf genes in both strains significantly (P < 0.05). Downregulation of gtfB/C directly leads to less accumulation of insoluble glucans, which in turn reduces the chance of dental caries. Other researchers also showed that the application of biosurfactants derived from Lactobacillus casei and Lactobacillus rutteri caused considerable downregulation of the gtf B, gtf C, and ftf genes expression.[37,38]
The results imply that either the Lactobacillus biosurfactant or a putative signaling molecule in the extract could downregulate the expression level of genes with an important role in the attachment and biofilm formation of S. mutans without having a major effect on the S. mutans viability. Furthermore, downregulating of gtf B and gtf C and ftf may also have an effect on converting the activity. However, the molecular mechanism of such effect and also the clinical application of biosurfactants as safe and selective therapeutic agents for dental caries prevention should be studied in the future.
According to the results of this study and also other researches, it can be concluded that biosurfactants can modify the expression level of virulence genes, the surface properties of bacterial cells, and reduce their adhesive abilities and also interfere with biofilm development and cell to cell communication.
CONCLUSION
The antiadhesive properties of the crude biosurfactant isolated from L. rhamnosus against S. mutans strains along with the downregulation of biofilm formation associated genes, gtfB, gtfC, and ftf was confirmed in this study. The results suggest the possible use of this biosurfactant as an antiadhesive or anti-biofilm agent, with applications against dental plaque formation and also dental caries prevention.
Financial support and sponsorship
Nil.
Conflicts of interest
The authors of this manuscript declare that they have no conflicts of interest, real or perceived, financial or nonfinancial in this article.
Acknowledgments
This study was carried out in Isfahan (Khorasgan) branch, Islamic Azad University and also Medicine School of Isfahan University of Medical Sciences, Isfahan, Iran with no financial support and sponsorship.
REFERENCES
- 1.Yang WY, Kim CK, Ahn CH, Kim H, Shin J, Oh KB, et al. Flavonoid glycosides inhibit sortase A and sortase A-mediated aggregation of Streptococcus mutans, an oral bacterium responsible for human dental caries. J Microbiol Biotechnol. 2016;26:1566–9. doi: 10.4014/jmb.1605.05005. [DOI] [PubMed] [Google Scholar]
- 2.Ferrazzano GF, Amato I, Ingenito A, Zarrelli A, Pinto G, Pollio A, et al. Plant polyphenols and their anti-cariogenic properties: A review. Molecules. 2011;16:1486–507. doi: 10.3390/molecules16021486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa A, Furukawa S, Fujita S, Mitobe J, Kawarai T, Narisawa N, et al. Inhibition of Streptococcus mutans biofilm formation by Streptococcus salivarius fruA. Appl Environ Microbiol. 2011;77:1572–80. doi: 10.1128/AEM.02066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192:3024–32. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murata RM, Branco-de-Almeida LS, Franco EM, Yatsuda R, dos Santos MH, de Alencar SM, et al. Inhibition of Streptococcus mutans biofilm accumulation and development of dental caries in vivo by 7-epiclusianone and fluoride. Biofouling. 2010;26:865–72. doi: 10.1080/08927014.2010.527435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao J, Koo H. Structural organization and dynamics of exopolysaccharide matrix and microcolonies formation by Streptococcus mutans in biofilms. J Appl Microbiol. 2010;108:2103–13. doi: 10.1111/j.1365-2672.2009.04616.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida A, Ansai T, Takehara T, Kuramitsu HK. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl Environ Microbiol. 2005;71:2372–80. doi: 10.1128/AEM.71.5.2372-2380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nijampatnam B, Casals L, Zheng R, Wu H, Velu SE. Hydroxychalcone inhibitors of Streptococcus mutans glucosyl transferases and biofilms as potential anticaries agents. Bioorg Med Chem Lett. 2016;26:3508–13. doi: 10.1016/j.bmcl.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdolhosseini A, Ashourioun A. A study of different antiplaque agentsused in dental practices and their therapeutical advantaged. Int J Biol Pharm Allied Sci. 2016;5:2413–2420. [Google Scholar]
- 10.Sendamangalam V, Choi OK, Kim D, Seo Y. The anti-biofouling effect of polyphenols against Streptococcus mutans. Biofouling. 2011;27:13–9. doi: 10.1080/08927014.2010.535897. [DOI] [PubMed] [Google Scholar]
- 11.Varvaresou A, Iakovou K. Biosurfactants in cosmetics and biopharmaceuticals. Lett Appl Microbiol. 2015;61:214–23. doi: 10.1111/lam.12440. [DOI] [PubMed] [Google Scholar]
- 12.Chew SY, Cheah YK, Seow HF, Sandai D, Than LT. Probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 exhibit strong antifungal effects against vulvovaginal candidiasis-causing Candida glabrata isolates. J Appl Microbiol. 2015;118:1180–90. doi: 10.1111/jam.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YT, Chou CC, Hsu CY. Effects of Lactobacillus casei Shirota intake on caries risk in children. J Dent Sci. 2017;12:179–84. doi: 10.1016/j.jds.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Almeida DG, Soares Da Silva RC, Luna JM, Rufino RD, Santos VA, Banat IM, et al. Biosurfactants: Promising molecules for petroleum biotechnology advances. Front Microbiol. 2016;7:1718. doi: 10.3389/fmicb.2016.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahmourespour A, Kasra-Kermanshahi R, Salehi R, Nabi-Nejad A. The effect of lactobacillus fermentumATCC9338 as a probiotic on the adhesion of oral streptococci in vitro. Iran J Med Microbiol. 2008;2:45–51. [Google Scholar]
- 16.Sharma D, Saharan BS. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol Rep (Amst) 2016;11:27–35. doi: 10.1016/j.btre.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patowary K, Patowary R, Kalita MC, Deka S. Characterization of biosurfactant produced during degradation of hydrocarbons using crude oil as sole source of carbon. Front Microbiol. 2017;8:279. doi: 10.3389/fmicb.2017.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn KB, Baik JE, Park OJ, Yun CH, Han SH. Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans. PLoS One. 2018;13:e0192694. doi: 10.1371/journal.pone.0192694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues LR, Teixeira JA, van der Mei HC, Oliveira R. Isolation and partial characterization of a biosurfactant produced by Streptococcus thermophilus A. Colloids Surf B Biointerfaces. 2006;53:105–12. doi: 10.1016/j.colsurfb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Tam A, Shemesh M, Wormser U, Sintov A, Steinberg D. Effect of different iodine formulations on the expression and activity of Streptococcus mutans glucosyltransferase and fructosyltransferase in biofilm and planktonic environments. J Antimicrob Chemother. 2006;57:865–71. doi: 10.1093/jac/dkl085. [DOI] [PubMed] [Google Scholar]
- 21.Allaker RP, Douglas CW. Novel anti-microbial therapies for dental plaque-related diseases. Int J Antimicrob Agents. 2009;33:8–13. doi: 10.1016/j.ijantimicag.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Strahinic I, Busarcevic M, Pavlica D, Milasin J, Golic N, Topisirovic L, et al. Molecular and biochemical characterizations of human oral lactobacilli as putative probiotic candidates. Oral Microbiol Immunol. 2007;22:111–7. doi: 10.1111/j.1399-302X.2007.00331.x. [DOI] [PubMed] [Google Scholar]
- 23.Kõll P, Mändar R, Marcotte H, Leibur E, Mikelsaar M, Hammarström L, et al. Characterization of oral lactobacilli as potential probiotics for oral health. Oral Microbiol Immunol. 2008;23:139–47. doi: 10.1111/j.1399-302X.2007.00402.x. [DOI] [PubMed] [Google Scholar]
- 24.Uehara S, Monden K, Nomoto K, Seno Y, Kariyama R, Kumon H, et al. A pilot study evaluating the safety and effectiveness of lactobacillus vaginal suppositories in patients with recurrent urinary tract infection. Int J Antimicrob Agents. 2006;28(Suppl 1):S30–4. doi: 10.1016/j.ijantimicag.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Dusane DH, Rajput JK, Kumar AR, Nancharaiah YV, Venugopalan VP, Zinjarde SS, et al. Disruption of fungal and bacterial biofilms by lauroyl glucose. Lett Appl Microbiol. 2008;47:374–9. doi: 10.1111/j.1472-765X.2008.02440.x. [DOI] [PubMed] [Google Scholar]
- 26.Dusane DH, Nancharaiah YV, Zinjarde SS, Venugopalan VP. Rhamnolipid mediated disruption of marine Bacillus pumilus biofilms. Colloids Surf B Biointerfaces. 2010;81:242–8. doi: 10.1016/j.colsurfb.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Rivardo F, Turner RJ, Allegrone G, Ceri H, Martinotti MG. Anti-adhesion activity of two biosurfactants produced by Bacillus spp. Prevents biofilm formation of human bacterial pathogens. Appl Microbiol Biotechnol. 2009;83:541–53. doi: 10.1007/s00253-009-1987-7. [DOI] [PubMed] [Google Scholar]
- 28.Moldes AB, Paradelo R, Vecino X, Cruz JM, Gudiña E, Rodrigues L, et al. Partial characterization of biosurfactant from Lactobacillus pentosus and comparison with sodium dodecyl sulphate for the bioremediation of hydrocarbon contaminated soil. Biomed Res Int 2013. 2013:961842. doi: 10.1155/2013/961842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues LR, Teixeira JA, van der Mei HC, Oliveira R. Physicochemical and functional characterization of a biosurfactant produced by Lactococcus lactis 53. Colloids Surf B Biointerfaces. 2006;49:79–86. doi: 10.1016/j.colsurfb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Gudiña EJ, Rocha V, Teixeira JA, Rodrigues LR. Antimicrobial and antiadhesive properties of a biosurfactant isolated from Lactobacillus paracasei ssp. paracasei A20. Lett Appl Microbiol. 2010;50:419–24. doi: 10.1111/j.1472-765X.2010.02818.x. [DOI] [PubMed] [Google Scholar]
- 31.Brzozowski B, Bednarski W, Gołek P. The adhesive capability of two Lactobacillus strains and physicochemical properties of their synthesized biosurfactants. Food Technol Biotechnol. 2011;49:177–86. [Google Scholar]
- 32.Ahmed A, Dachang W, Lei Z, Jianjun L, Juanjuan Q, Yi X, et al. Effect of lactobacillus species on Streptococcus mutans biofilm formation. Pak J Pharm Sci. 2014;27:1523–8. [PubMed] [Google Scholar]
- 33.Walencka E, Rózalska S, Sadowska B, Rózalska B. The influence of Lactobacillus acidophilus-derived surfactants on staphylococcal adhesion and biofilm formation. Folia Microbiol (Praha) 2008;53:61–6. doi: 10.1007/s12223-008-0009-y. [DOI] [PubMed] [Google Scholar]
- 34.Söderling EM, Marttinen AM, Haukioja AL. Probiotic lactobacilli interfere with Streptococcus mutans biofilm formation in vitro. Curr Microbiol. 2011;62:618–22. doi: 10.1007/s00284-010-9752-9. [DOI] [PubMed] [Google Scholar]
- 35.Decker EM, Klein C, Schwindt D, von Ohle C. Metabolic activity of Streptococcus mutans biofilms and gene expression during exposure to xylitol and sucrose. Int J Oral Sci. 2014;6:195–204. doi: 10.1038/ijos.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasfi R, Abd El-Rahman OA, Zafer MM, Ashour HM. Probiotic lactobacillus sp. Inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J Cell Mol Med. 2018;22:1972–83. doi: 10.1111/jcmm.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savabi O, Kazemi M, Kamali S, Salehi AR, Eslami G, Tahmourespour A, et al. Effects of biosurfactant produced by Lactobacillus casei on gtfB, gtfC, and ftf gene expression level in S. mutans by real-time RT-PCR. Adv Biomed Res. 2014;3:231. doi: 10.4103/2277-9175.145729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salehi R, Savabi O, Kazemi M, Kamali S, Salehi AR, Eslami G, et al. Effects of lactobacillus reuteri-derived biosurfactant on the gene expression profile of essential adhesion genes (gtfB, gtfC and ftf) of Streptococcus mutans. Adv Biomed Res. 2014;3:169. doi: 10.4103/2277-9175.139134. [DOI] [PMC free article] [PubMed] [Google Scholar]


