Abstract
Predicting gene functions from genome sequence alone has been difficult, and the functions of a large fraction of plant genes remain unknown. However, leveraging the vast amount of currently available gene expression data has the potential to facilitate our understanding of plant gene functions, especially in determining complex traits. Gene coexpression networks—created by integrating multiple expression datasets—connect genes with similar patterns of expression across multiple conditions. Dense gene communities in such networks, commonly referred to as modules, often indicate that the member genes are functionally related. As such, these modules serve as tools for generating new testable hypotheses, including the prediction of gene function and importance. Recently, we have seen a paradigm shift from the traditional “global” to more defined, context-specific coexpression networks. Such coexpression networks imply genetic correlations in specific biological contexts such as during development or in response to a stress. In this short review, we highlight a few recent studies that attempt to fill the large gaps in our knowledge about cellular functions of plant genes using context-specific coexpression networks.
Keywords: context specific, coexpression networks, gene function prediction, network analysis, modules, clusters
The most important utility of gene coexpression networks (GCNs) is in expanding the current state of functional annotations of plant genes. The time-honored technique of genetic screens and loss-of-function mutant analyses for gene function characterization has obvious limitations 1 and has mapped the functions of only about 24% of all genes in the model plant Arabidopsis thaliana (The Arabidopsis Information Resource (TAIR) portal: https://bit.ly/2Ak85Zu). The most alarming fact is that less than 1% of all known genes in important crops like maize and rice have experimentally identified functions 2, 3. The less precise orthology-based function assignments are still the default method for newly sequenced plant genomes 4. However, despite having a great degree of similarity in sequence or protein domains, genes can evolve for divergent functions 5, especially those involved in specialized metabolism (SM) 6, leaving younger plant genes less annotated 7. Nevertheless, homology-based function annotations are coupled with other experimentally derived manual annotations 8 and made available as Gene Ontologies (GOs) for several plant species 9. The plant GO and other function annotation catalogs 10, 11 provide excellent leads, or a priori evidence, to enhance the development of GCNs that offer a scalable and dynamic framework for prediction of gene functions in silico.
GCNs are constructed by connecting pairs of genes if they have high (statistically determined) correlation in their expression profiles across a large set of samples ( Figure 1 legend). Coexpressed gene pairs often represent functional coupling, such as through coordinate regulation of specific pathways. Identifying and connecting gene pairs at the genome-wide level are ultimately what reconstruct a network, which is a graphical and mathematical abstraction of functional associations among genes in the cellular states being examined. Much like in community analysis of human social networks 12, extraction of densely connected gene communities becomes the next step in gene network analysis. The general idea is that because the genes in each community have a higher degree of coexpression, they are more likely to be under the same regulatory program that dictates their expression and therefore might also be functionally related 13. These communities are broadly referred to as “clusters” or “modules” of coexpressed genes.
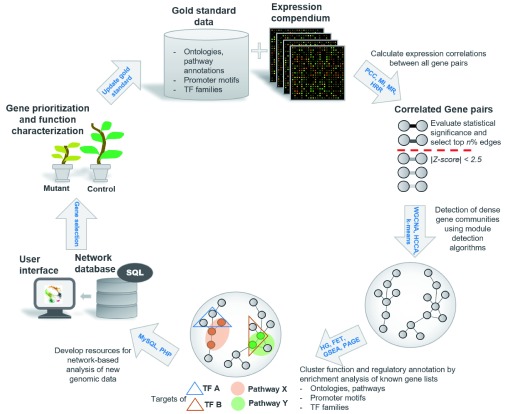
Figure 1. Coexpression network analysis workflow.
A gene coexpression network is constructed by integrating gene expression profiles from a large compendium of datasets. The datasets can be sampled from public repositories like the Gene Expression Omnibus 42 and quite often are chosen in a manner that represents a unifying biological context (for example, response to abiotic stress or specific tissues/organs of the plant). Correlations in expression profiles of all gene pairs across all samples then are calculated by using a similarity measure such as PCC (Pearson’s correlation coefficient), MI (mutual information), MR (mutual rank) 30, or HRR (highest reciprocal rank) 43. Statistically significant gene pairs then are linked to each other, and the resulting network is clustered by using a module detection algorithm such as WGCNA (weighted gene coexpression network analysis) in R, HCCA (heuristic cluster chiseling algorithm) 44, or the k-means clustering, where k determines the number of clusters. These algorithms identify densely connected network neighborhoods, or modules, that may harbor genes of a common function, pathway, or regulon of a transcription factor (TF) complex. These functional and regulatory attributes of gene modules can be statistically tested by using gene sets from “gold standard” function annotation data. Most often, not all genes within a predicted module have a function annotation, but if the module is significantly enriched with genes known for certain biological process, the functions of other unknown genes can be imputed. Quite often, these data are organized as databases and presented as webtools. (MySQL is a Structured Query Language–based database management system, and PHP is a server-side scripting language.) A community-driven approach is taken to use the data and predict the function(s) of uncharacterized genes. Experimentally validated gene functions then are added to the existing gold standard to further refine the computational predictions in future experiments. The whole process tends to accelerate the process of identifying uncharacterized genes for specific biological processes. FET, Fisher’s exact test; GSEA, gene set enrichment analysis; HG, hypergeometric test; PAGE, parametric analysis of geneset enrichment.
Biological insights from the resulting clustering data can be gained by using well-established computational approaches in gene set analysis (GSA). A wide variety of GSA methods are available that can be used to evaluate whether a predicted module is significantly over-represented with genes already annotated for certain functional categories representing biological processes or pathways 14– 17. These predefined functional gene sets for plants can be obtained from annotation catalogs like the GO 18, the MapMan bins 19, and other pathway databases 20. GSA therefore enables propagation of known function information from already annotated genes within each module to genes yet unannotated. However, this technique has several caveats 21, and the strength of gene function prediction in this manner depends largely on the quality and expanse of available annotations for the species under consideration. Additionally, a well-designed statistical framework for detecting familiar DNA motifs, or a de novo motif search, in the promoters of module genes potentially links transcription factors (TFs) as putative regulators of module expression 22, 23.
Coexpressed gene modules: a treasure trove for plant development and stress biologists
The typical steps involved in construction of GCNs are shown in Figure 1. Some studies report pre-computed global plant gene networks in the form of web applications that provide excellent resources and tools for users to query and visualize subnetworks of interest 24– 30. The popular methods, challenges, and caveats associated with construction and use of GCNs are reviewed by others 31– 33.
Whereas the construction of a GCN is fairly simple and straightforward, partitioning of a coexpression network into seemingly diverse functional modules is not a trivial task. Several approaches of module detection from gene expression are proposed in the literature and were comprehensively evaluated recently 34. Of these methods, weighted GCN analysis (WGCNA) 35 has been embraced by plant biologists as the most popular method to identify and analyze gene modules in specific developmental phases of a variety of plant species.
In the context of seed development, Zhan et al. 36 profiled gene expression in different compartments of maize kernels at the filling stage and identified a module specific to the basal endosperm transfer layer (a tissue layer important for nutrient transport during grain filling). This study also found a link to MRP-1 as a likely regulator of this module by analyzing cis-regulatory elements enriched in the promoter of genes in this module 36. Similarly, an analysis of modules in developing seeds of two contrasting cultivars of soybean led to the identification of the role of a cytochrome P450 family gene which increases seed size and weight upon overexpression 37. In wheat, Wang et al. 38 identified a module that showed strong association with spike-related traits. The authors of this study validated this module by characterizing three new member genes that showed altered spike complexity when overexpressed in an elite variety of wheat 38. An attempt to identify modules of seed development has also been made by using two cultivars of chickpea with contrasting seed size and weight 39 and seeds of two different species of cotton 40.
To study the development of woodland strawberry ( Fragaria vesca) fruits, Shahan et al. 41 created three separate coexpression networks: one represented early-stage floral tissues, another spanned the development of pre-fertilized flower to fruits, and the third represented fruits at the ripening stages 41. These networks were helpful in establishing the role of the ghost tissue in iron transport post-fertilization 41. To mine these networks, the authors of this study built an interactive web interface, which is of value to other users in the field ( www.fv.rosaceaefruits.org).
These recent studies have highlighted the efficacy of the WGCNA as a quick and easy method to process new, high-resolution developmental transcriptomes and recover plant gene modules. However, assessing the stability of predicted modules and performance in terms of accuracy is highly desirable for generating new hypotheses 45, 46. For example, Shahan et al. 41 found little overlap between clusters obtained by multiple WGCNA runs each with a different subsampling of genes. Through various numerical experiments, the authors suggest that a consensus clustering scheme is much more robust in terms of predicting true functional relationships between genes 41. Also, the WGCNA method implicitly decides on the number of modules to produce from the dataset and, as evident from the aforementioned studies, typically retrieves very few modules (about 10–20 modules per dataset) in a non-exhaustive manner under default settings. It is still unclear whether this is due to the nature of underlying datasets or whether other relevant modules (or subnetworks within the identified WGCNA modules) still exist in these development networks. We believe that a vast majority of plant transcriptomes are still under-utilized in the retrieval of biologically relevant information. Secondary analyses of these datasets can reflect on new knowledge pertaining to the originally assayed biological processes, especially the role of regulatory genes 47, 48, which is often overlooked in terms of the potential in typical GCN analysis.
In another type of module identification setting, the goal is to cluster a GCN by using a graph clustering algorithm optimized to detect a large number of dense modules. This type of study aims to partition the GCN into many functionally cohesive gene modules, typically in the range of a few hundred modules per network. This is a desirable property of biological network analysis, and several algorithms exist that allow the user to define module size as desired 44, 49, 50. A large number of dense modules would likely separate large metabolic pathways and biological processes into their constituent parts. Intuitively, an important cautionary aspect of such an analysis is controlling the granularity of resulting modules 51. Very large modules will fail to answer the fundamental question of why the genes are clustered that way. It becomes difficult to explain the regulatory cohesiveness of large modules, as these will incorporate genes that lie on distant bifurcated branches of large metabolic pathways that show significant levels of correlation in expression 52. Conversely, a finer granularity will break the network into too many very small modules (fewer than 10 genes), essentially hampering the statistical process of establishing any functional or regulatory context for these modules, rendering them unusable.
A balance between the number of modules, average module size, and other network topological features such as separation (how functionally isolated each module is from other modules) and density (how well genes within each module form a clique) should be carefully monitored by tuning parameters of the clustering algorithm 34, 53. For example, modules in the Rice Environment Coexpression Network (RECoN) 26 were identified by optimizing the parameters of a graph-based clustering algorithm that works under the principle that functionally related genes have denser connections within a network 50. With controlled minimum cluster size and density threshold to partition the network, 1744 dense abiotic stress–associated rice modules were extracted from RECoN. Most of these modules contain genes sharing related GO terms and thus can be used as gene sets (in addition to gene sets from GO and other function databases) in enrichment analysis of new differential expression profiles. The RECoN webtool (available at https://bit.ly/2BOky7x) also provides a window to mine undiscovered novel modules associated with tolerance to abiotic stresses in rice 26.
An example of a graph-based clustering algorithm applied to a developmental context in rice is the RiceAntherNet, which delineated 545 modules related to anther and pollen development 54. Of these, 29 modules that contain differentially expressed genes in nine previously known male sterile mutant lines are regarded as important to anther development 54. This study also compared the rice anther modules to modules in FlowerNet, a GCN for anther and pollen development in Arabidopsis 55, and found a significant amount of conservation of gene coexpression between rice and Arabidopsis during anther development.
GCNs have also been instrumental in the discovery of novel biological phenomena. For example, the conservation of a longevity module between Medicago truncatula and Arabidopsis suggests conserved genetic pathways related to defense mechanisms 56. Phylogenetic conservation of coexpression modules was examined in greater detail recently for several model plants 7. One of the observations of this study was that genes from the same phylostratum tend to be more frequently connected in GCNs 7, suggesting new uses of existing GCNs in extrapolating gene functions of lesser-studied plant species. Moreover, it has been shown that coexpression, rather than physical proximity on chromosomes (biosynthetic gene clusters), is a more reliable signal for predicting genes involved in SM 57. Understanding the mechanisms of SM is of great interest in the study of medicinal plants 58.
The curious case of transcription factors
Another desirable characteristic of coexpression network modeling is incorporating information about regulatory interactions in the process of module identification. This type of study leads to the discovery of modular gene regulatory networks (GRNs). GRNs are different from GCNs in the sense that GCNs treat TF and non-TF nodes (genes) similarly whereas GRN involves sophisticated reverse-engineering algorithms that operate on TFs differently. The objective of a GRN is to capture direct, causal edges between TF and their putative targets and filter spurious indirect correlations that naturally arise in coexpression data.
Biological interpretations of predicted GRNs depend largely on the type of transcriptional dataset used. A meta-analysis by Marbach et al. 59 shows that expression data from TF knockouts or overexpression experiments can be very informative in predicting targets but this type of data is useful mostly for capturing downstream regulatory effects 60, 61. On the other hand, time-series expression data can capture temporal dynamics of regulatory networks more systematically 62, 63. For example, a time-series transcriptome dataset was used to predict GRNs active for lateral root initiation in Arabidopsis, which revealed genetic cascades involved in positive and negative feedback loops as well as target genes of the AUXIN RESPONSE FACTOR 7 64. Steady-state expression data have also been very useful in regulatory network analysis and predicting TF function. For example, using a collection of gene expression datasets from Arabidopsis seeds, subnetworks associated with desiccation tolerance (DT) were predicted, leading to the identification of three novel TFs now confirmed for their roles in mediating seed DT 65.
Recently, de Luis Balaguer et al. 66 developed a dynamic Bayesian network (DBN)-based framework which showed us a broader picture of the spatiotemporal dynamics of stem cell differentiation in the roots of Arabidopsis. Although DBNs are able to unfold cyclic processes and dependencies in time-course expression data, they are inherently limited by the computational complexity which increases with the number of genes 61. The algorithm proposed by de Luis Balaguer et al. 66 seems to circumvent this limitation by applying DBNs to smaller sets of genes within each spatially distinct coexpressed cluster within the roots. This combination of spatial and temporal expression data in GRN inference established a new role of PERIANTHIA (PAN) as a stem cell regulator 66.
There are more than a dozen other published algorithms for inference of GRNs from large-scale expression data. Several concepts in GRN inference, available algorithms, and their limitations and applications in plant studies are well summarized by others as a primer to interested researchers 61, 63, 67, 68. An earlier meta-analysis of some of the popular approaches suggests integrating predictions from different algorithms to boost the accuracy of the consensus GRN 59. This technique was later implemented in Arabidopsis stress datasets to predict an oxidative-stress GRN 69 and was further explored in the development of a secondary cell wall biosynthesis network 70. The authors of the former study used “coregulated” gene pairs instead of “coexpressed” gene pairs into the k-means clustering framework and recovered 572 stress-related modules 69.
Because a large sample size can be obtained for Arabidopsis, it has been a favorite model in many currently reported GRNs. However, rapidly expanding data repositories allow selection of interest-appropriate transcriptional datasets 71, encouraging the application of standard GRN inference techniques to relatively lesser-studied crop genomes. For example, Xiong et al. 47 re-analyzed the maize embryo and endosperm development dataset 72 and predicted modules and regulators involved in the transport of nutrients to the developing seed 47.
Inference of GRNs can be further enhanced in a direct network framework (non-modular GRNs), where the primary goal is to explore regulatory targets of a few TFs of interest in more detail as focused small-scale subnetworks. For example, a combination of transcriptional and chromatin immunoprecipitation data (ChIP-seq) revealed GRN components involved in coordinate regulation of root hair growth in Arabidopsis 73. Furthermore, an integrative analysis of ChIP-seq, mRNA-seq, and miRNA-seq data revealed SEP3 as an upstream regulator of MIR319a and TCP4, which together form a feed-forward loop to regulate petal development 74. In the context of GRNs that manifest during abiotic stresses, Wilkins et al. 75 employed the concept of ERGINs (environmental gene regulatory influence networks) in rice. They integrated chromatin accessibility data (ATAC-seq) with the current state of knowledge about putative regulatory interactions in rice and used these data points as priors to learn a GRN from expression dataset of five tropical Asian rice cultivars grown under abiotic-stress conditions in the field as well as greenhouses. The study identified regulatory interactions between 113 TFs and 4052 target genes of rice 75.
Conclusions
The current state of accumulated plant gene expression data has immense potential for the discovery of components involved in complex traits. The property of modularity in gene networks can be exploited and gene modules treated as fundamental biological units with dynamic expression and regulatory properties. The techniques of module extraction have proven to be very effective in experimental validations while also suggesting a vast scope for improvement in terms of not only statistical methods used but also how gene networks are perceived and evaluated for research. As noticed by Gillis and Pavlidis 21, one has to consider that there are several caveats associated with using prior knowledge from GO (and other such annotation catalogs) for function prediction from GCN modules. Predictions can be very biased toward genes and categories that are extremely well annotated and can be driven solely by other computationally predicted annotations rather than empirical evidence.
Transcriptome datasets integrated in a global manner capture broad, constitutive functional relationships that might not vary much with different tissues or organs, developmental phases, or environmental cues like biotic or abiotic stress 67, 76. On the other hand, specifying an overarching biological theme in selection of datasets offers intuitive concepts that can be objectively tested. For example, just like individual transcriptomes, GCNs created to study one particular biological process (for example, seed development or response to abiotic stress) can be considered static. Comparison of GCNs constructed from conditionally distinct samples, or differential coexpression analysis, will provide valuable information on how plant systems alter their mechanisms in response to different developmental cues and environmental perturbations 77, 78. Moreover, a comparison of sets of modules derived under different contexts should potentially map and distinguish modules that are conserved throughout growth and development from those that are under constant rewiring.
One major question that remains is how to systematically produce a ranked list of genes most relevant to a given trait/process of interest from these complex interconnected gene relationships in networks. Information buried within hundreds of thousands to sometimes millions of predicted functional relationships is not intuitively tractable for researchers interested in selecting a few actionable candidate genes relevant to a biological process of interest. Research toward development of computational tools capable of using gene networks to systematically enrich gene prioritization pipelines 79 would be extremely useful for integration with gene lists from genome-wide association study (GWAS) datasets in a systems genetics approach to probe complex agricultural traits 80, 81.
It is important to recognize that gene expression data by itself could have limited potential in deciphering cellular organization, regulated at various levels. However, we are optimistic about the future, as integrating signals from heterogeneous molecular datasets will enable training of smart algorithms to identify genomic patterns of components already known to be associated with a phenotype of interest. The trained models then can be used as predictive tools to discover new genes associated with the phenotype 82 and study crop genetics as outlined in recent reviews 83, 84.
Acknowledgements
The authors would like to thank Julie Thomas, of the University of Arkansas, for critical reading of the manuscript. The authors thank the reviewers of this manuscript for providing valuable comments that helped improve it.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Marek Mutwil, Max-Planck Institute for Molecular Plant Physiology, Potsdam, Germany
Ross Sozzani, Department of Plant and Microbial Biology, North Carolina State University, North Carolina, USA
Natalie Clark, Department of Plant and Microbial Biology, North Carolina State University, North Carolina, USA
Funding Statement
This work was supported by funding from National Science Foundation award NSF#1716844 on “Systems genetics analysis of photosynthetic carbon metabolism in rice”.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Provart NJ, Alonso J, Assmann SM, et al. : 50 years of Arabidopsis research: highlights and future directions. New Phytol. 2016;209(3):921–44. 10.1111/nph.13687 [DOI] [PubMed] [Google Scholar]
- 2. Wang J, Qi M, Liu J, et al. : CARMO: a comprehensive annotation platform for functional exploration of rice multi-omics data. Plant J. 2015;83(2):359–74. 10.1111/tpj.12894 [DOI] [PubMed] [Google Scholar]
- 3. Wimalanathan K, Friedberg I, Andorf CM, et al. : Maize GO Annotation-Methods, Evaluation, and Review (maize-GAMER). Plant Direct. 2018;2(4):e00052 10.1002/pld3.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lohse M, Nagel A, Herter T, et al. : Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014;37(5):1250–8. 10.1111/pce.12231 [DOI] [PubMed] [Google Scholar]
- 5. Gerlt JA, Babbitt PC: Can sequence determine function? Genome Biol. 2000;1(5):REVIEWS0005. 10.1186/gb-2000-1-5-reviews0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chae L, Kim T, Nilo-Poyanco R, et al. : Genomic signatures of specialized metabolism in plants. Science. 2014;344(6183):510–3. 10.1126/science.1252076 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Ruprecht C, Proost S, Hernandez-Coronado M, et al. : Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 2017;90(3):447–65. 10.1111/tpj.13502 [DOI] [PubMed] [Google Scholar]
- 8. Balakrishnan R, Harris MA, Huntley R, et al. : A guide to best practices for Gene Ontology (GO) manual annotation. Database (Oxford). 2013;2013:bat054. 10.1093/database/bat054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yi X, Du Z, Su Z: PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41(Web Server issue):W98–103. 10.1093/nar/gkt281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mueller LA, Zhang P, Rhee SY: AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132(2):453–60. 10.1104/pp.102.017236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanehisa M, Goto S, Kawashima S, et al. : The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Girvan M, Newman ME: Community structure in social and biological networks. Proc Natl Acad Sci U S A. 2002;99(12):7821–6. 10.1073/pnas.122653799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Usadel B, Obayashi T, Mutwil M, et al. : Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009;32(12):1633–51. 10.1111/j.1365-3040.2009.02040.x [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Castillo-Davis CI, Hartl DL: GeneMerge--post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19(7):891–2. 10.1093/bioinformatics/btg114 [DOI] [PubMed] [Google Scholar]
- 15. Kim SY, Volsky DJ: PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. 10.1186/1471-2105-6-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Subramanian A, Tamayo P, Mootha VK, et al. : Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Irizarry RA, Wang C, Zhou Y, et al. : Gene set enrichment analysis made simple. Stat Methods Med Res. 2009;18(6):565–75. 10.1177/0962280209351908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian T, Liu Y, Yan H, et al. : agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45(W1):W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Usadel B, Poree F, Nagel A, et al. : A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32(9):1211–29. 10.1111/j.1365-3040.2009.01978.x [DOI] [PubMed] [Google Scholar]
- 20. Naithani S, Preece J, D'Eustachio P, et al. : Plant Reactome: a resource for plant pathways and comparative analysis. Nucleic Acids Res. 2017;45(D1):D1029–D1039. 10.1093/nar/gkw932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillis J, Pavlidis P: "Guilt by association" is the exception rather than the rule in gene networks. PLoS Comput Biol. 2012;8(3):e1002444. 10.1371/journal.pcbi.1002444 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Vandepoele K, Quimbaya M, Casneuf T, et al. : Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150(2):535–46. 10.1104/pp.109.136028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma S, Shah S, Bohnert HJ, et al. : Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet. 2013;9(10):e1003840. 10.1371/journal.pgen.1003840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yim WC, Yu Y, Song K, et al. : PLANEX: the plant co-expression database. BMC Plant Biol. 2013;13:83. 10.1186/1471-2229-13-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schaefer RJ, Briskine R, Springer NM, et al. : Discovering functional modules across diverse maize transcriptomes using COB, the Co-expression Browser. PLoS One. 2014;9(6):e99193. 10.1371/journal.pone.0099193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krishnan A, Gupta C, Ambavaram MMR, et al. : RECoN: Rice Environment Coexpression Network for Systems Level Analysis of Abiotic-Stress Response. Front Plant Sci. 2017;8:1640. 10.3389/fpls.2017.01640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Proost S, Mutwil M: PlaNet: Comparative Co-Expression Network Analyses for Plants. Methods Mol Biol. 2017;1533:213–27. 10.1007/978-1-4939-6658-5_12 [DOI] [PubMed] [Google Scholar]
- 28. You Q, Xu W, Zhang K, et al. : ccNET: Database of co-expression networks with functional modules for diploid and polyploid Gossypium. Nucleic Acids Res. 2017;45(D1):D1090–D1099. 10.1093/nar/gkw910 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Ferrari C, Proost S, Ruprecht C, et al. : PhytoNet: comparative co-expression network analyses across phytoplankton and land plants. Nucleic Acids Res. 2018;46(W1):W76–W83. 10.1093/nar/gky298 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Obayashi T, Aoki Y, Tadaka S, et al. : ATTED-II in 2018: A Plant Coexpression Database Based on Investigation of the Statistical Property of the Mutual Rank Index. Plant Cell Physiol. 2018;59(1):e3. 10.1093/pcp/pcx191 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Rhee SY, Mutwil M: Towards revealing the functions of all genes in plants. Trends Plant Sci. 2014;19(4):212–21. 10.1016/j.tplants.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 32. Li Y, Pearl SA, Jackson SA: Gene Networks in Plant Biology: Approaches in Reconstruction and Analysis. Trends Plant Sci. 2015;20(10):664–75. 10.1016/j.tplants.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 33. Serin EA, Nijveen H, Hilhorst HW, et al. : Learning from Co-expression Networks: Possibilities and Challenges. Front Plant Sci. 2016;7:444. 10.3389/fpls.2016.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Saelens W, Cannoodt R, Saeys Y: A comprehensive evaluation of module detection methods for gene expression data. Nat Commun. 2018;9(1):1090. 10.1038/s41467-018-03424-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Langfelder P, Horvath S: WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Zhan J, Thakare D, Ma C, et al. : RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell. 2015;27(3):513–31. 10.1105/tpc.114.135657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du J, Wang S, He C, et al. : Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J Exp Bot. 2017;68(8):1955–1972. 10.1093/jxb/erw460 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Wang Y, Yu H, Tian C, et al. : Transcriptome Association Identifies Regulators of Wheat Spike Architecture. Plant Physiol. 2017:175(2):746–757. 10.1104/pp.17.00694 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Garg R, Singh VK, Rajkumar MS, et al. : Global transcriptome and coexpression network analyses reveal cultivar-specific molecular signatures associated with seed development and seed size/weight determination in chickpea. Plant J. 2017;91(6):1088–107. 10.1111/tpj.13621 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Hu G, Hovav R, Grover CE, et al. : Evolutionary Conservation and Divergence of Gene Coexpression Networks in Gossypium (Cotton) Seeds. Genome Biol Evol. 2016;8(12):3765–83. 10.1093/gbe/evw280 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Shahan R, Zawora C, Wight H, et al. : Consensus Coexpression Network Analysis Identifies Key Regulators of Flower and Fruit Development in Wild Strawberry. Plant Physiol. 2018;178(1):202–16. 10.1104/pp.18.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Barrett T, Troup DB, Wilhite SE, et al. : NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–5. 10.1093/nar/gkl887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mutwil M, Klie S, Tohge T, et al. : PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23(3):895–910. 10.1105/tpc.111.083667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mutwil M, Usadel B, Schütte M, et al. : Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 2010;152(1):29–43. 10.1104/pp.109.145318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langfelder P, Luo R, Oldham MC, et al. : Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7(1):e1001057. 10.1371/journal.pcbi.1001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shannon CP, Chen V, Takhar M, et al. : SABRE: a method for assessing the stability of gene modules in complex tissues and subject populations. BMC Bioinformatics. 2016;17(1):460. 10.1186/s12859-016-1319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiong W, Wang C, Zhang X, et al. : Highly interwoven communities of a gene regulatory network unveil topologically important genes for maize seed development. Plant J. 2017;92(6):1143–56. 10.1111/tpj.13750 [DOI] [PubMed] [Google Scholar]
- 48. Gupta C, Krishnan A, Schneider A, et al. : SANe: The Seed Active Network for Discovering Transcriptional Regulatory Programs of Seed Development. bioRxiv. 2017. 10.1101/165894 [DOI] [Google Scholar]
- 49. Blondel VD, Guillaume JL, Lambiotte R, et al. : Fast unfolding of communities in large networks. J Stat Mech. 2008;2008:P10008 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- 50. Jiang P, Singh M: SPICi: a fast clustering algorithm for large biological networks. Bioinformatics. 2010;26(8):1105–11. 10.1093/bioinformatics/btq078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Dongen S, Abreu-Goodger C: Using MCL to extract clusters from networks. Methods Mol Biol. 2012;804:281–95. 10.1007/978-1-61779-361-5_15 [DOI] [PubMed] [Google Scholar]
- 52. Uygun S, Peng C, Lehti-Shiu MD, et al. : Utility and Limitations of Using Gene Expression Data to Identify Functional Associations. PLoS Comput Biol. 2016;12(12):e1005244. 10.1371/journal.pcbi.1005244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Horvath S, Dong J: Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4(8):e1000117. 10.1371/journal.pcbi.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin H, Yu J, Pearce SP, et al. : RiceAntherNet: a gene co-expression network for identifying anther and pollen development genes. Plant J. 2017;92(6):1076–91. 10.1111/tpj.13744 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Pearce S, Ferguson A, King J, et al. : FlowerNet: a gene expression correlation network for anther and pollen development. Plant Physiol. 2015;167(4):1717–30. 10.1104/pp.114.253807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Righetti K, Vu JL, Pelletier S, et al. : Inference of Longevity-Related Genes from a Robust Coexpression Network of Seed Maturation Identifies Regulators Linking Seed Storability to Biotic Defense-Related Pathways. Plant Cell. 2015;27(10):2692–708. 10.1105/tpc.15.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wisecaver JH, Borowsky AT, Tzin V, et al. : A Global Coexpression Network Approach for Connecting Genes to Specialized Metabolic Pathways in Plants. Plant Cell. 2017;29(5):944–59. 10.1105/tpc.17.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Chakraborty P: Herbal genomics as tools for dissecting new metabolic pathways of unexplored medicinal plants and drug discovery. Biochim Open. 2018;6:9–16. 10.1016/j.biopen.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Marbach D, Costello JC, Küffner R, et al. : Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9(8):796–804. 10.1038/nmeth.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mejia-Guerra MK, Pomeranz M, Morohashi K, et al. : From plant gene regulatory grids to network dynamics. Biochim Biophys Acta. 2012;1819(5):454–65. 10.1016/j.bbagrm.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 61. Banf M, Rhee SY: Computational inference of gene regulatory networks: Approaches, limitations and opportunities. Biochim Biophys Acta Gene Regul Mech. 2017;1860(1):41–52. 10.1016/j.bbagrm.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 62. Krouk G, Lingeman J, Colon AM, et al. : Gene regulatory networks in plants: learning causality from time and perturbation. Genome Biol. 2013;14(6):123. 10.1186/gb-2013-14-6-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gaudinier A, Brady SM: Mapping Transcriptional Networks in Plants: Data-Driven Discovery of Novel Biological Mechanisms. Annu Rev Plant Biol. 2016;67:575–94. 10.1146/annurev-arplant-043015-112205 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Lavenus J, Goh T, Guyomarc'h S: Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell. 2015;27(5):1368–88. 10.1105/tpc.114.132993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. González-Morales SI, Chávez-Montes RA, Hayano-Kanashiro C, et al. : Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2016;113(35):E5232–41. 10.1073/pnas.1610985113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. de Luis Balaguer MA, Fisher AP, Clark NM, et al. : Predicting gene regulatory networks by combining spatial and temporal gene expression data in Arabidopsis root stem cells. Proc Natl Acad Sci U S A. 2017;114(36):E7632–E7640. 10.1073/pnas.1707566114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Zhu F, Panwar B, Guan Y: Algorithms for modeling global and context-specific functional relationship networks. Brief Bioinform. 2016;17(4):686–95. 10.1093/bib/bbv065 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Haque S, Ahmad JS, Clark NM, et al. : Computational prediction of gene regulatory networks in plant growth and development. Curr Opin Plant Biol. 2018;47:96–105. 10.1016/j.pbi.2018.10.005 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Vermeirssen V, De Clercq I, van Parys T, et al. : Arabidopsis ensemble reverse-engineered gene regulatory network discloses interconnected transcription factors in oxidative stress. Plant Cell. 2014;26(12):4656–79. 10.1105/tpc.114.131417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Taylor-Teeples M, Lin L, de Lucas M, et al. : An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517(7536):571–5. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Papatheodorou I, Fonseca NA, Keays M, et al. : Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46(D1):D246–D251. 10.1093/nar/gkx1158 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Chen J, Zeng B, Zhang M, et al. : Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014;166(1):252–64. 10.1104/pp.114.240689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shibata M, Breuer C, Kawamura A, et al. : GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development. 2018;145(3): pii: dev159707. 10.1242/dev.159707 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Chen D, Yan W, Fu LY, et al. : Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat Commun. 2018;9(1):4534. 10.1038/s41467-018-06772-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Wilkins O, Hafemeister C, Plessis A, et al. : EGRINs (Environmental Gene Regulatory Influence Networks) in Rice That Function in the Response to Water Deficit, High Temperature, and Agricultural Environments. Plant Cell. 2016;28(10):2365–84. 10.1105/tpc.16.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Myers CL, Troyanskaya OG: Context-sensitive data integration and prediction of biological networks. Bioinformatics. 2007;23(17):2322–30. 10.1093/bioinformatics/btm332 [DOI] [PubMed] [Google Scholar]
- 77. Ma C, Xin M, Feldmann KA, et al. : Machine learning-based differential network analysis: a study of stress-responsive transcriptomes in Arabidopsis. Plant Cell. 2014;26(2):520–37. 10.1105/tpc.113.121913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jiang Z, Dong X, Li ZG, et al. : Differential Coexpression Analysis Reveals Extensive Rewiring of Arabidopsis Gene Coexpression in Response to Pseudomonas syringae Infection. Sci Rep. 2016;6:35064. 10.1038/srep35064 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Liseron-Monfils CV, Olson A, Ware D: NECorr, a Tool to Rank Gene Importance in Biological Processes using Molecular Networks and Transcriptome Data. bioRxiv. 2018. 10.1101/326868 [DOI] [Google Scholar]
- 80. Lee T, Kim H, Lee I: Network-assisted crop systems genetics: network inference and integrative analysis. Curr Opin Plant Biol. 2015;24:61–70. 10.1016/j.pbi.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 81. Schaefer R, Michno JM, Jeffers J, et al. : Integrating Coexpression Networks with GWAS to Prioritize Causal Genes in Maize. Plant Cell. 2018;30(12):2922–2942. 10.1105/tpc.18.00299 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Lloyd JP, Seddon AE, Moghe GD, et al. : Characteristics of Plant Essential Genes Allow for within- and between-Species Prediction of Lethal Mutant Phenotypes. Plant Cell. 2015;27(8):2133–47. 10.1105/tpc.15.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ma C, Zhang HH, Wang X: Machine learning for Big Data analytics in plants. Trends Plant Sci. 2014;19(12):798–808. 10.1016/j.tplants.2014.08.004 [DOI] [PubMed] [Google Scholar]
- 84. Hu H, Scheben A, Edwards D: Advances in Integrating Genomics and Bioinformatics in the Plant Breeding Pipeline. Agriculture. 2018;8(6):75 10.3390/agriculture8060075 [DOI] [Google Scholar]; F1000 Recommendation