Abstract
Background
Despite the advances in therapy, the occurrence of drug-resistant human immunodeficiency virus type 1 (HIV-1) is a major obstacle to successful treatment. This study aimed to characterize the genetic diversity and to determine the prevalence of transmitted drug resistance mutations (TDRM) between individuals recently or chronically diagnosed with HIV-1 from Paraná, Brazil.
Methods
A total of 260 HIV-1 positive antiretroviral therapy-naïve patients were recruited to participate on the study, of which 39 were recently diagnosed. HIV-1 genotyping was performed using sequencing reaction followed by phylogenetic analyses to determine the HIV-1 subtype. TDRM were defined using the Calibrated Population Resistance Tool program.
Results
The HIV-1 subtypes frequency found in the studied population were 54.0% of subtype B, 26.7% subtype C, 6.7% subtype F1 and 12.7% recombinant forms. The overall prevalence of TDRM was 6.7%, including 13.3% for recently diagnosed subjects and 5.9% for the chronic group.
Conclusions
The prevalence of resistance mutations found in this study is considered moderate, thus to perform genotyping tests before the initiation of antiretroviral therapy may be important to define the first line therapy and contribute for the improvement of regional prevention strategies for epidemic control.
Electronic supplementary material
The online version of this article (10.1186/s12981-019-0219-1) contains supplementary material, which is available to authorized users.
Keywords: HAART, Genotype, Epidemiology, Mutation, Drug resistance
Background
Brazil has reported the largest number of acquired immune deficiency syndrome (AIDS) in Latin America [1]. The estimated number of people living with human immunodeficiency virus (HIV) in Brazil is 926,742, with sexual exposure being the main transmission path (96.9%), and predominance of the homo/bisexual category (48.7%). In the state of Paraná, in 2017, the detection rate was 17.1 per 100 thousand inhabitants, and, in the last 10 years, the total of accumulated cases was 15,699 [2].
Nevertheless, the rate of AIDS detection and mortality has been decreasing steadily in Brazil in recent years, mainly due to the antiretroviral therapy (ART), which is distributed by public health programs since the 1990s [2]. However, one unavoidable consequence of the widespread use of ART is the selection of mutations associated with drug resistance, which can be transmitted to uninfected individuals. Transmitted drug resistance mutations (TDRM) is associated with impaired outcomes from ART, including first line of highly active antiretroviral therapy (HAART) with two nucleoside reverse transcriptase inhibitors (NRTI) plus one nonnucleoside reverse transcriptase inhibitors (NNTRI) or one protease inhibitor (PI) [3] and other therapeutic interventions, such as pre-exposure prophylaxis (PrEP), post-exposure prophylaxis (PEP) and prevention of vertical transmission [4]. Detection of resistance mutations before the initiation of ART is considered cost-effective in areas where TDRM prevalence is 1% or higher, because it avoids spending public funds on drugs which will certainly result in virological failure [5].
In developed countries, guidelines recommend baseline genotypic testing in all patients newly diagnosed with HIV infection to ensure the effectiveness of first line ART [6, 7]. Indeed, Hofstra et al. demonstrated that in a setting without adequate surveillance, the prevalence of TDRM involving NNRTI resistant HIV can increase to worrisome levels, compromising the efficacy of standard first-line NNRTI-based regimen [8]. Unfortunately, although moderate levels of TDRM have been reported in Brazil [9–14], ranging from 3.8% in Maranhão [10] to 13.9% in São Paulo [12], the genotyping test is not available for most treatment-naïve patients [15].
Surveillance of TDRM among treatment-naïve recently diagnosed patient and of HIV-1 diversity is important, since it may contribute to the improvement of regional prevention strategies for epidemic control. Also, about 2 years after initial infection by HIV-1, the reversion of resistance mutations may occur; in other words, strains with TDRM can become undetectable in blood sample due to the growing of wild virus that has higher fitness than the mutated virus [16]. However, most studies developed in Brazil did not consider whether the patient was recently or chronically infected/diagnosed.
To the best of our knowledge, this was the first study that investigated HIV-1 diversity and evaluated TDRM between recently or chronically diagnosed ART-naïve patients with HIV-1 from Paraná State, Brazil.
Materials and methods
Study population
This is a cross-sectional study with consecutive sampling. A total of 281 individuals older than 18 years, seropositive for HIV-1 and treatment-naïve, who were attended at the Specialized Service STD/AIDS (SAE) of the municipalities of Maringá and Londrina, Paraná State, Brazil, from February 2010 to January 2013 were included in the study. The exclusion criteria were: co-infection with hepatitis B and/or C and history of ARV use. Based on this, the study population was formed by 260 subjects, which were divided into two groups, Recent and Chronic. The Recent group was composed by individuals whose first positive serology was dated less than 1 year from the date of collection for this study. To minimize the possibility of late diagnosis, patients with a CD4+ T cell count smaller than 400 cells/mm3 were excluded from the Recent group.
Sample processing
Peripheral blood samples were collected with EDTA anticoagulant from all patients to determine viral and immunological parameters including counts of CD4+ T cells, viral load quantification and genotyping of HIV-1. Plasma was separated and kept at − 80 °C until use.
CD4+ T cells counts
CD4+ T cells counts were performed using fresh total blood by flow cytometry (BD Trucount™ Tubes) using FACS Calibur device (Becton–Dickinson, NJ, USA). The results were expressed as cells/mm3.
Viral load determination
Viral load was determined using plasma samples by branched DNA methodology (VERSANT® HIV-1 RNA 3.0 Assay) with a System 340 bDNA Analyzer (Bayer Health Care, NY, USA). The results were expressed on a logarithmic scale in base 10.
Genotyping of HIV-1
For HIV-1 genotyping, plasma viral RNA was extracted using the QIAmp® Viral RNA Mini Kit (Qiagen, Courtaboeuf, France) extraction kit, according to manufacturer’s instructions, followed by retrotranscription reaction in order to obtain cDNA. The genes for protease (PR) and reverse transcriptase (RT) were amplified as described before [17] and the purified products of the polymerase chain reaction (PCR) were sequenced with ABI PRISM® Kit BigDye™ Terminator version 3.1 (Applied Biosystems, Foster City, CA), according to manufacturer’s instructions, using the ABI 3500XL sequencer (Applied Biosystems, Foster City, CA).
Quality analysis and editing of the sequences obtained were performed in the SeqMan DNAStar Lasergene v7.0 program [18].
Phylogenetic analysis and subtype determination
Subtypes were initially determined using Rega HIV Subtyping Tool v. 2.0 [19] and then confirmed based on the phylogenetic tree constructed in MEGA 5.0 software [20]. For the tree construction, sequences obtained from the Los Alamos HIV Reference Database were used [21]. Phylogenetic analysis was performed by a neighbor-joining algorithm using the Tamura-Neinucleotide substitution model, with 1000 replicates, the tree obtained is available as Additional file 1. The profiles of recombination were confirmed by bootscanning analysis with the Recombinant Identification Program v. 3.0—RIP 3.0 [22]. An additional file provide bootscanning results of the recombinants found in the study population (Additional file 2). Once HIV-1 has high genetic plasticity, sequences that presented ≥ 85% of similarity in phylogenetic analysis were excluded from the study to avoid genotyping error due to cross-contamination between samples.
Transmitted drug resistance determination
Transmitted drug resistance was defined as the presence of at least one major mutation listed at Drug Surveillance Mutation Resistance (SDRM-2009). For this, the Calibrated Population Resistance Tool (CPR) v. 6.0 beta program was used [23]. The susceptibility of HIV to antiretrovirals was determined using the National Agency for AIDS Research (ANRS) algorithm v.22 [24].
Statistical analysis
The prevalence of transmitted resistance was calculated based on the number of subjects in each group. Categorical variables were compared by two-tailed Chi square test or Fisher’s exact test, when applicable, with a confidence interval (CI) of 95%, using OpenEpi version 3.1 [25]. Quantitative variables were compared using the Mann–Whitney test using GraphPad Prism v. 5 (GraphPad Software Inc., San Diego, CA). All results of p value < 0.05 were considered statistically significant.
Results
A total of 281 volunteers agreed to participate in the study; 91 of them were attended at the Maringá SAE and 190 at the Londrina SAE. Ten subjects were excluded due to co-infection and/or previous use of ARVs, and 11 were excluded due to the lack of data allowing classification of their infections as recent or chronic. Of the remaining 260 participants, 39 (15%) were classified as recently diagnosed and 221 (85%) as chronic carriers. There was no statistical difference between groups for gender, age and mode of transmission (Table 1).
Table 1.
Epidemiological characteristics in treatment naïve individuals with HIV-1 from Paraná State, Brazil, 2010–2013
| Recent group (n = 39) n (%) |
Chronic group (n = 221) n (%) |
p valuea | |
|---|---|---|---|
| Sex | |||
| Female | 11 (28.21) | 92 (41.63) | – |
| Male | 28 (71.80) | 129 (58.37) | 0.1141 |
| Age | |||
| 13–24 | 7 (17.95) | 22 (9.96) | 0.1133 |
| 25–49 | 28 (71.80) | 163 (73.76) | 0.4379 |
| > 50 | 4 (10.26) | 36 (16.29) | – |
| Transmission | |||
| Sexual | 36 (92.31) | 208 (94.12) | 0.7613 |
| IDUs | 1 (2.56) | 8 (3.62) | – |
| Indeterminate | 2 (5.13) | 5 (2.26) | 0.3747 |
HIV-1 human immunodeficiency virus type 1, IDUs Injecting drug users
aTwo-tailed Chi square test with confidence interval of 95%
The viral load levels found among individuals in the Chronic group (median = 4.61 log10; interquartile range = 3.79 to 5.05 log10) were significantly higher (p = 0.0004) than those found in individuals whose infection was considered recent (median = 3.88 log10; interquartile range = 3.54 to 4.37 log10). Regarding CD4+ T cell count, the average found in the Recent group (median = 583 cells/mm3; interquartile range = 502 to 807 cells/mm3) was significantly higher (p < 0.0001) than in the Chronic group (median = 317 cells/mm3; interquartile range = 95 to 503 cells/mm3).
The samples of some subjects (93/260, 35.8%) were not amplified and sequenced. Additionally, to avoid genotyping error due to cross-contamination between samples, patients whose HIV-1 sequence presented ≥ 85% of similarity in phylogenetic analysis were excluded from statistical analyses. Thus, 150 samples were evaluated according to subtype and presence of ARV resistance mutations, 15 (10.0%) from the Recent group and 135 (90.0%) from the Chronic group (Fig. 1).
Fig. 1.
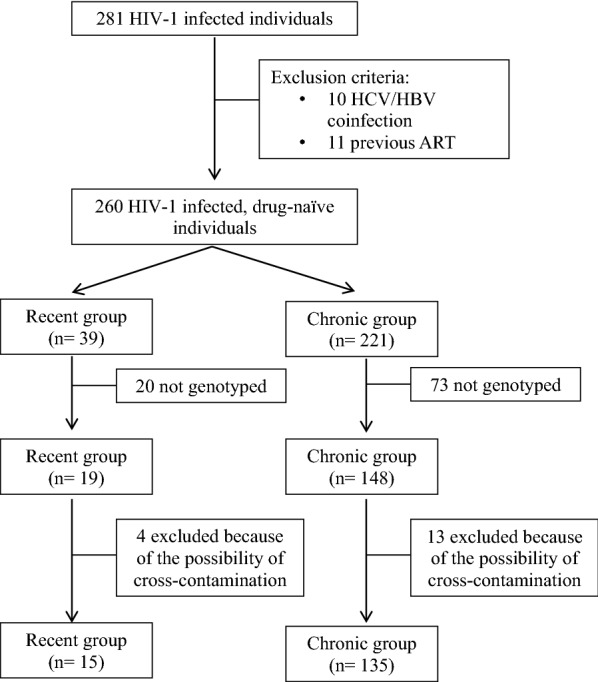
Flowchart of the study population
The phylogenetic analyses showed that most of the samples were subtype B (81/150, 54.0%), followed by subtype C (40/150, 26.7%), F (10/150, 6.7%) and recombinant forms (19/150, 12.7%) (Table 2), including BF (13/19, 68.4%), BC (1/19, 5.3%), BCF (3/19, 15.8%), BDF (1/19, 5.3%) and BCH (1/19, 5.3%).
Table 2.
Frequency of HIV-1 subtypes and mutations associated with drug-resistance in drug-naïve individuals from Paraná State, Brazil, 2010-2013
| Recent group (n = 15) | Chronic group (n = 135) | All (n = 150) | |
|---|---|---|---|
| Subtype a | |||
| B | 10 (66.7; 41.7 to 84.8) | 71 (52.6; 44.2 to 60.8) | 81 (54.0; 46.0 to 61.8) |
| C | 4 (26.7; 10.9 to 52.0) | 36 (26.7; 19.9 to 34.7) | 40 (26.7; 20.2 to 34.3) |
| F | 0 (0.0; 0.0 to 20.4) | 10 (7.4; 4.1 to 13.1) | 10 (6.7; 3.7 to 11.8) |
| CRFs | 1 (6.7; 1.2 to 29.8) | 18 (13.3; 8.6 to 20.1) | 19 (12.7; 8.3 to 18.9) |
| Resistance mutations by class b | |||
| PR region with PI mutation | 0 (0.0; 0.0 to 20.4) | 1 (0.7; 0.1 to 4.1) | 1 (0.7; 0.1 to 3.7) |
| RT region with NRTI mutation | 1 (6.7; 1.2 to 29.8) | 3 (2.2; 0.8 to 6.3) | 4 (2.7; 1.0 to 6.7) |
| RT region with NNRTI mutation | 1 (6.7; 1.2 to 29.8) | 4 (3.0; 1.2 to 7.4) | 5 (3.3; 1.4 to 7.6) |
| Any resistance mutation | 2 (13.3; 3.7 to 37.9) | 8 (5.9; 3.0 to 11.3) | 10 (6.7; 3.7 to 11.8) |
Results are presented as N (%; 95% CI). HIV-1, human immunodeficiency virus type 1; PR, protease; RT, reverse transcriptase; PI, protease inhibitors; NRTI, nucleoside analogue reverse transcriptase inhibitor; NNRTI, non-nucleoside analogue reverse transcriptase inhibitors
aComparisons performed using two-tailed Chi square test with confidence interval of 95%
bComparisons performed using two-tailed Fisher’s exact test with confidence interval of 95%
All comparisons had p values > 0.05
The overall prevalence of TDRM was 6.7% (10/150), including 13.3% (2/15) in recently diagnosed patients and 5.9% (8/135) in the chronic group, with no significant difference between groups (p = 0.2764). Mutations associated with resistance to NRTIs were most frequently observed, followed by mutations associated with resistance to NNRTIs and only one sample presented TDRMs associated with resistance to PIs (Table 2). The mutations found in the study population are listed on Table 3 according to the HIV-1 subtype and time of diagnosis. One newly diagnosed patient showed two TDRMs for NRTIs and one subject of the chronic group had two TDRMs for PIs. No sequence was related to resistance to both NRTIs and NNRTIs or triple-class resistance.
Table 3.
Drugs-resistance mutations according to HIV-1 subtypes and time of diagnosis in drug-naïve individuals from Paraná State, Brazil, 2010–2013
| Sample identification | Diagnosis | HIV-1 subtype | NRTI mutations | NNRTI mutations | PI mutations | Resistance [23, 24] |
|---|---|---|---|---|---|---|
| L19 | Chronic | B | – | K101E | – | EFV, NVP, ETR, RPV |
| L39 | Chronic | C | – | K103 N | – | EFV, NVP |
| L97 | Chronic | B | T215S | – | – | ZDV, d4T |
| L99 | Recent | B | D67 N, T215S | – | – | ZDV, d4T, ABC |
| L150 | Chronic | B | D67G | – | – | – |
| L156 | Chronic | B | K70R | – | – | DDI, ZDV, d4T |
| L168 | Chronic | C | – | G190E | – | EFV, NVP |
| M6 | Chronic | C | – | K103 N | – | EFV, NVP |
| M18 | Recent | B | – | K103 N | – | EFV, NVP |
| M19 | Chronic | B | – | – | I54 V, V82A | IDV, NFV, LPV/r |
HIV-1 human immunodeficiency virus type 1, PR protease, RT reverse transcriptase, PI protease inhibitors, NRTI nucleoside analogue reverse transcriptase inhibitor, NNRTI non-nucleoside analogue reverse transcriptase inhibitors, EFV efavirenz, NVP nevirapine, ETR etravirine, RPV rilpivirine, ZDV zidovudine, d4T stavudine, ABC abacavir, DDI didanosine, IDV indinavir, NFV nelfinavir, LPV/r lopinavir/ritonavir
Discussion
The transmission of resistant HIV-1 has important implications for the success of ART, since it restricts therapeutic options and increases the possibility of therapy failure. For this reason, this phenomenon has attracted the attention of researchers around the world. To our knowledge, this is the first study on the prevalence of transmitted resistance in the state of Paraná which considered the time of HIV-1 infection, discriminating patients recently diagnosed from those who have chronically established infections. The overall prevalence of transmitted resistance found in this study was 6.7%, with a prevalence of 13.3% among individuals with recent infection and 5.9% in chronically infected individuals. Regarding subtypes of HIV-1, subtype B was the most frequently found (54.0%), followed by subtype C (26.7%), and recombinant forms (12.7%). Subtype F had the lowest frequency in the study population (6.7%).
The increasing access to ART has caused a significant rise in rates of TDRMs [26]. The prevalence of TDRMs found in different studies varies according to the region and the population studied [27–31]. Based on the WHO HIVDR classification [32], the present study demonstrated a moderate level of resistance, which is in accordance to a recent survey conducted by Arruda et al. that revealed moderate rate of TDRM in the five Brazilian macroregions [33]. Sax et al. reported that in places where the prevalence of transmitted resistance is 1% or higher, testing for resistance before starting ART is considered cost-effective [5]. The Ministry of Health of Brazil recently published a document recommending the conduction of pretreatment genotyping only for HIV infected people with a current or previous partner taking ART, pregnant women, people co-infected with tuberculosis and children [15]. While in developed countries the prevalence of transmitted resistance is well surveilled, in Brazil the tests are not available for every HIV infected subject and studies on this aspect are few and conflicting.
This study found a TDRM prevalence of 6.7% among treatment-naïve individuals from the northern and northwestern regions of Paraná. This prevalence was similar to that found by Gaspareto et al. (4.2%) in northwestern Paraná in 2012 [17] and to that found by other Brazilian studies [10, 11, 14, 34–37]. Besides local social factors as the access to ART, the differences between the results reported in Brazilian studies for the prevalence of transmitted resistance among treatment-naïve individuals is due, mainly, to heterogeneity in study design, which occurs in the selection of the target population (patients with acute, recent or chronic infections), the methodology used to evaluate resistance mutations, and the definition of resistance mutations [16].
Among the TDRMs (5.9%) detected in this study, the most frequent were NNRTIs (2.9%), then for NRTIs (2.4%) and PIs (0.6%), which is similar to the ratio observed in other studies [35, 38]. However, these results differed from those of Sprinz et al. and Murillo et al., who found a higher frequency of TDRMs for NNRTIs [11] and from other studies that found a higher frequency of TDRMs for NRTIs, then for NNRTIs and for PIs [13, 34]. The K103N TDRM was the most frequently found in this study and has been described in other studies conducted in different regions of Brazil [13, 33–35, 38]. The presence of this mutation is associated with high levels of resistance to NNRTI Nevirapine and Efavirens, which were part of the recommended regimen for initial therapy in HIV-1 infections at the time of sample collection. Other mutations found for NNRTIs, K101E, also described by Pilotto et al. [13], and G190E are also associated with resistance to Nevirapine and Efavirens. All of the mutations that confer resistance to NRTI found in the present study, T215S, D67 N, D67G and K70R, are related to resistance to one of the first anti-HIV drugs approved in Brazil, Zidovudine, and have been observed in other Brazilian studies [13, 35, 38]. The I54 V and V82A mutations found in this study confer resistance to the PIs Indinavir, Nelfinavir and Lopinavir/Ritonavir. The current Brazilian therapeutic guideline recommends the association of two NRTI/NRTIt–lamivudine (3TC) and tenofovir (TDF)—with an integrase inhibitor (INI)—dolutegravir (DTG)—in the basis of the initial treatment scheme as an attempt to avoid the transmitted resistance problem [15].
Studies on the molecular epidemiology and genetic diversity of HIV-1 are essential to better understand the HIV-1 epidemic in Brazil. The frequency of HIV-1 subtypes varies according to the geographical region [39, 40]. The epidemiological profile of the different subtypes of HIV-1 in the country shows a clear predominance of subtype B, corresponding to about 75% of infected individuals [33, 36, 41–43]. Subtypes F and C are present in frequencies that differ from one region to another [36, 42, 43]. Subtypes A and D were also identified in isolated cases in Rio de Janeiro, although their epidemiological importance is not known yet [44, 45]. The frequencies for the different subtypes found in this study are in agreement with those found in other studies in southern Brazil [17, 46].
The present study has some limitations. The sampling of patients in care rather than consecutively of newly diagnosed individuals may have reduced the representativeness of the newly infected patients. Regarding the procedure used to classify the groups, no laboratory technique was used to determine if the infection was recent or chronic. However, viral and immunological parameters related to the course of infection, such as CD4+ T cells and HIV viral load, differed significantly between the groups, with a higher CD4+ T cell count in the Recent group and a higher viral load in the Chronic group, confirming that the criteria used were effective. Another limitation was the large number of unamplifiable samples, resulting in a high failure rate (36%). All the failed samples presented low or undetectable virus load. Additionally, the “in house” PCR methodology may have collaborated to the low sensitivity of the test.
Conclusion
The present study demonstrated a moderate prevalence of TDRMs in the study population, which highlights the need to perform genotyping pretreatment tests to optimize the prescription of ART and prevent treatment failure due to a pre-existing mutation. Prospective studies are still needed to assess the true effectiveness of combination antiretroviral therapy for the treatment of infections with different subtypes of HIV-1 in Brazil. Knowledge on the resistance and cross-resistance of different subtypes of HIV-1 profiles will be critical to the choice of possible therapeutic options, particularly in Brazil, which currently provides antiretroviral therapy to nearly 90% of infected individuals. Additional investigations, including other gene targets, will be necessary to detect possible resistance mutations against new classes of antiretrovirals.
Additional files
Additional file 1. Maximum likelihood phylogenetic tree of HIV pol (PR/RT) sequences obtained from ART-naïve patients from North and Northeast of Paraná, Brazil.
Additional file 2. Bootscanning analysis of recombinant HIV-1 sequences in protease and reverse transcriptase regions from ART-naïve patients from North and Northeast of Paraná, Brazil.
Authors’ contributions
LDB collaborated to sample and data collection, performed molecular biology experiments and statistical analyses, and contributed to the writing of the manuscript. PHC helped to perform DNA extraction. EMVR, JRCD, VAFM and LTU collaborated to sample and data collection. JCCF, EMVR, and DAB contributed for the study conception and design. DAB analyzed and interpreted the data and revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Acknowledgements
We appreciate all the employees of SAE Maringá and Londrina, Paraná State, Brazil, for their help with the collection of samples and epidemiological data. We also appreciate the working group of the Division of Clinical Immunology, Clinical Analysis Laboratory of the State University of Maringá, for processing and analyzing of samples. We are grateful to the Laboratory of AIDS and Molecular Immunology of FIOCRUZ for their technical support and CAPES for the postgraduate scholarship.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study was approved by the Standing Committee on Ethics in Research involving Humans of the State University of Maringá, Paraná State, Brazil (Opinion No. 5642/2012). A free and informed consent form was obtained from all study participants.
Consent for publication
Not applicable.
Availability of data and materials
Sequence data generated and analyzed during the current study are available in the GenBank repository, accession numbers: KM851047–KM851209. Epidemiological data generated and analyzed during the current study are not publicly available due to ethical impediment, but they are available with the corresponding authors on reasonable request.
Funding
This work was supported by the Araucaria Foundation grant funded by the Paraná State Government (05/2011-22721).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- TDRM
transmitted drug resistance mutations
- AIDS
acquired immune deficiency syndrome
- ART
antiretroviral therapy
- HAART
highly active antiretroviral therapy
- NRTI
nucleoside reverse transcriptase inhibitors
- NNTRI
nonnucleoside reverse transcriptase inhibitors
- PI
protease inhibitor
- PrEP
pre-exposure prophylaxis
- PEP
post-exposure prophylaxis
- PR
protease
- RT
reverse transcriptase
- PCR
polymerase chain reaction
Contributor Information
Larissa Danielle Bahls, Phone: +55 44 3011-5388, Email: laribahls@gmail.com.
Pedro Henrique Canezin, Email: pedro_canezin@hotmail.com.
Edna Maria Vissoci Reiche, Email: reiche@sercomtel.com.br.
José Carlos Couto Fernandez, Email: coutofer@ioc.fiocruz.br.
José Ricardo Colleti Dias, Email: ricardocolleti71@gmail.com.
Vera Alice Fernandes Meneguetti, Email: verameneguetti@hotmail.com.
Luis Toshio Ueda, Email: luiz.ueda@sesa.pr.gov.br.
Dennis Armando Bertolini, Email: dabertolini@uem.br.
References
- 1.WHO. UNAIDS Data 2017. Geneva: Joint United NationsProgramme on HIV/AIDS; 2017.
- 2.Brasil. Boletim epidemiológico HIV/Aids 2018. Brasilia: Ministério da Saúde; 2018.
- 3.Wittkop L, Gunthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, de Luca A, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11(5):363–371. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- 4.Pennings PS. HIV drug resistance: problems and perspectives. Infect Dis Rep. 2013;5(Suppl 1):e5. doi: 10.4081/idr.2013.s1.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sax PE, Islam R, Walensky RP, Losina E, Weinstein MC, Goldie SJ, et al. Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin Infect Dis. 2005;41(9):1316–1323. doi: 10.1086/496984. [DOI] [PubMed] [Google Scholar]
- 6.Ryom L, Boesecke C, Gisler V, Manzardo C, Rockstroh JK, Puoti M, et al. Essentials from the 2015 European AIDS Clinical Society (EACS) guidelines for the treatment of adult HIV-positive persons. HIV Med. 2016;17(2):83–88. doi: 10.1111/hiv.12322. [DOI] [PubMed] [Google Scholar]
- 7.Günthard HF, Saag MS, Benson CA, del Rio C, Eron JJ, Gallant JE, et al. Antiretroviral drugs for treatment and prevention of HIV Infection in Adults: 2016 Recommendations of the International Antiviral Society-USA Panel. JAMA. 2016;316(2):191–210. doi: 10.1001/jama.2016.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofstra LM, Sánchez Rivas E, Nijhuis M, Bank LEA, Wilkinson E, Kelly K, et al. High rates of transmission of drug-resistant HIV in Aruba resulting in reduced susceptibility to the WHO Recommended First-line Regimen in Nearly Half of Newly Diagnosed HIV-infected Patients. Clin Infect Dis. 2017;64(8):1092–1097. doi: 10.1093/cid/cix056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moura ME, da Guarda Reis MN, Lima YA, Eulálio KD, Cardoso LP, Stefani MM. HIV-1 transmitted drug resistance and genetic diversity among patients from Piauí State, Northeast Brazil. J Med Virol. 2015;87(5):798–806. doi: 10.1002/jmv.24087. [DOI] [PubMed] [Google Scholar]
- 10.Moura ME, Reis MN, Lima YA, Eulálio KD, Cardoso LP, Stefani MM. Low rate of transmitted drug resistance may indicate low access to antiretroviral treatment in Maranhão State, northeast Brazil. AIDS Res Hum Retroviruses. 2015;31(2):250–254. doi: 10.1089/aid.2014.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sprinz E, Netto EM, Patelli M, Lima JS, Lima MP, Furtado JJ, et al. Primary antiretroviral drug resistance among HIV type 1-infected individuals in Brazil. AIDS Res Hum Retroviruses. 2009;25(9):861–867. doi: 10.1089/aid.2009.0012. [DOI] [PubMed] [Google Scholar]
- 12.Sanabani SS, Pastena É, da Costa AC, Martinez VP, Kleine-Neto W, de Oliveira AC, et al. Characterization of partial and near full-length genomes of HIV-1 strains sampled from recently infected individuals in São Paulo, Brazil. PLoS ONE. 2011;6(10):e25869. doi: 10.1371/journal.pone.0025869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilotto JH, Grinsztejn B, Veloso VG, Velasque LS, Friedman RK, Moreira RI, et al. Moderate prevalence of transmitted drug resistance mutations among antiretroviral-naive HIV-infected pregnant women in Rio de Janeiro. Brazil. AIDS Res Hum Retroviruses. 2013;29(4):681–686. doi: 10.1089/aid.2011.0333. [DOI] [PubMed] [Google Scholar]
- 14.Inocencio LA, Pereira AA, Sucupira MC, Fernandez JC, Jorge CP, Souza DF, et al. Brazilian network for HIV drug Resistance Surveillance: a survey of individuals recently diagnosed with HIV. J Int AIDS Soc. 2009;12:20. doi: 10.1186/1758-2652-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brasil. Protocolo Clínico e Diretrizes Terapêuticas para Manejo da Infecção pelo HIV em Adultos. In: saúde Md, editor. Brasilia, 2017.
- 16.Yerly S, Junier T, Gayet-Ageron A, Amari EB, von Wyl V, Günthard HF, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23(11):1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 17.Gaspareto KV, Mello FM, Dias JR, Meneguetti VA, Storti ME, Ferreira JL, et al. Genetic diversity and primary resistance among HIV-1-positive patients from Maringá, Paraná, Brazil. Rev Inst Med Trop Sao Paulo. 2012;54(4):207–213. doi: 10.1590/S0036-46652012000400005. [DOI] [PubMed] [Google Scholar]
- 18.Swindell SR, Plasterer TN. SEQMAN. Contig assembly. Methods Mol Biol. 1997;70:75–89. [PubMed] [Google Scholar]
- 19.de Oliveira K, Deforche S, Cassol M, Salminem D, Paraskevis C, Seebregts J, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinfomatics. 2005;21(19):3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Los Alamos HIV sequence database. (http://www.hiv.lanl.gov/) Accessed 15 Aug 2013.
- 22.Siepel AC, Halpern AL, Macken C, Korber BT. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11(11):1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 23.Gifford RJ, Liu TF, Rhee SY, Kiuchi M, Hue S, Pillay D, et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics. 2009;25(9):1197-8. (http://cpr.stanford.edu/cpr.cgi). Accessed 15 Aug 2013. [DOI] [PMC free article] [PubMed]
- 24.HIV-1 genotypic drug resistence interpretation’s algorithms. French National Agency for AIDS Research. (http://www.hivfrenchresistance.org/index.html). Accessed 15 Aug 2013.
- 25.Sullivan KM, Dean A, Soe MM. OpenEpi: a web-based epidemiologic and statistical calculator for public health. Public Health Rep. 2009;124(3):471–474. doi: 10.1177/003335490912400320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shet A, Berry L, Mohri H, Mehandru S, Chung C, Kim A, et al. Tracking the prevalence of transmitted antiretroviral drug-resistant HIV-1: a decade of experience. J Acquir Immune Defic Syndr. 2006;41(4):439–446. doi: 10.1097/01.qai.0000219290.49152.6a. [DOI] [PubMed] [Google Scholar]
- 27.Booth CL, Geretti AM. Prevalence and determinants of transmitted antiretroviral drug resistance in HIV-1 infection. J Antimicrob Chemother. 2007;59(6):1047–1056. doi: 10.1093/jac/dkm082. [DOI] [PubMed] [Google Scholar]
- 28.Lama JR, Sanchez J, Suarez L, Caballero P, Laguna A, Sanchez JL, et al. Linking HIV and antiretroviral drug resistance surveillance in Peru: a model for a third-generation HIV sentinel surveillance. J Acquir Immune Defic Syndr. 2006;42(4):501–505. doi: 10.1097/01.qai.0000221677.29693.dd. [DOI] [PubMed] [Google Scholar]
- 29.Smith D, Moini N, Pesano R, Cachay E, Aiem H, Lie Y, et al. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin Infect Dis. 2007;44(3):456–458. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 30.Hofstra LM, Sauvageot N, Albert J, Alexiev I, Garcia F, Struck D, et al. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis. 2016;62(5):655–663. doi: 10.1093/cid/civ963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mungati M, Mhangara M, Gonese E, Mugurungi O, Dzangare J, Ngwende S, et al. Pre-treatment drug resistance among patients initiating antiretroviral therapy (ART) in Zimbabwe: 2008-2010. BMC Res Notes. 2016;9:302. doi: 10.1186/s13104-016-2101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO. The HIV drug resistance report-2012. 2012.
- 33.Arruda MB, Boullosa LT, Cardoso CC, da Costa CM, Alves CR, de Lima ST, et al. Brazilian network for HIV drug resistance surveillance (HIV-BresNet): a survey of treatment-naive individuals. J Int AIDS Soc. 2018;2:3. doi: 10.1002/jia2.25032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira AS, Cardoso LP, Stefani MM. Moderate prevalence of transmitted drug resistance and high HIV-1 genetic diversity in patients from Mato Grosso State, Central Western Brazil. J Med Virol. 2011;83(8):1301–1307. doi: 10.1002/jmv.22128. [DOI] [PubMed] [Google Scholar]
- 35.Carvalho BC, Cardoso LP, Damasceno S, Stefani MM. Moderate prevalence of transmitted drug resistance and interiorization of HIV type 1 subtype C in the inland North State of Tocantins, Brazil. AIDS Res Hum Retroviruses. 2011;27(10):1081–1087. doi: 10.1089/aid.2010.0334. [DOI] [PubMed] [Google Scholar]
- 36.Gräf T, Passaes CP, Ferreira LG, Grisard EC, Morgado MG, Bello G, et al. HIV-1 genetic diversity and drug resistance among treatment naïve patients from Southern Brazil: an association of HIV-1 subtypes with exposure categories. J Clin Virol. 2011;51(3):186–191. doi: 10.1016/j.jcv.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Sucupira MC, Caseiro MM, Alves K, Tescarollo G, Janini LM, Sabino EC, et al. High levels of primary antiretroviral resistance genotypic mutations and B/F recombinants in Santos, Brazil. AIDS Patient Care STDS. 2007;21(2):116–128. doi: 10.1089/apc.2006.0079. [DOI] [PubMed] [Google Scholar]
- 38.de Medeiros RM, Junqueira DM, Matte MC, Barcellos NT, Chies JA, Matos Almeida SE. Co-circulation HIV-1 subtypes B, C, and CRF31_BC in a drug-naïve population from Southernmost Brazil: analysis of primary resistance mutations. J Med Virol. 2011;83(10):1682–1688. doi: 10.1002/jmv.22188. [DOI] [PubMed] [Google Scholar]
- 39.Morgado MG, Guimarães ML, Galvão-Castro B. HIV-1 polymorphism: a challenge for vaccine development—a review. Mem Inst Oswaldo Cruz. 2002;97(2):143–150. doi: 10.1590/S0074-02762002000200001. [DOI] [PubMed] [Google Scholar]
- 40.Soares MA, De Oliveira T, Brindeiro RM, Diaz RS, Sabino EC, Brigido L, et al. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS. 2003;17(1):11–21. doi: 10.1097/00002030-200301030-00004. [DOI] [PubMed] [Google Scholar]
- 41.Almeida FJ, Berezin EN, Rodrigues R, Sáfadi MA, Arnoni MV, Oliveira C, et al. Diversity and prevalence of antiretroviral genotypic resistance mutations among HIV-1-infected children. J Pediatr (Rio J). 2009;85(2):104–109. doi: 10.2223/JPED.1877. [DOI] [PubMed] [Google Scholar]
- 42.Silva MM, Telles FQ, da Cunha CA, Rhame FS. HIV subtype, epidemiological and mutational correlations in patients from Paraná, Brazil. Braz J Infect Dis. 2010;14(5):495–501. doi: 10.1016/S1413-8670(10)70099-4. [DOI] [PubMed] [Google Scholar]
- 43.Stefani MM, Pereira GA, Lins JA, Alcantara KC, Silveira AA, Viegas AA, et al. Molecular screening shows extensive HIV-1 genetic diversity in Central West Brazil. J Clin Virol. 2007;39(3):205–209. doi: 10.1016/j.jcv.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Caride E, Brindeiro R, Hertogs K, Larder B, Dehertogh P, Machado E, et al. Drug-resistant reverse transcriptase genotyping and phenotyping of B and non-B subtypes (F and A) of human immunodeficiency virus type I found in Brazilian patients failing HAART. Virology. 2000;275(1):107–115. doi: 10.1006/viro.2000.0487. [DOI] [PubMed] [Google Scholar]
- 45.Couto-Fernandez JC, Eyer-Silva WA, Guimarães ML, Chequer-Fernandez SL, Grinsztejn B, Delaporte E, et al. Phylogenetic analysis of Brazilian HIV type 1 subtype D strains: tracing the origin of this subtype in Brazil. AIDS Res Hum Retroviruses. 2006;22(2):207–211. doi: 10.1089/aid.2006.22.207. [DOI] [PubMed] [Google Scholar]
- 46.Guimarães ML, Moreira A, Loureiro R, Galvão-Castro B, Morgado MG. Characterization BNfHIa. High frequency of recombinant genomes in HIV type 1 samples from Brazilian southeastern and southern regions. AIDS Res Hum Retroviruses. 2002;18(17):1261–1269. doi: 10.1089/088922202320886307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Maximum likelihood phylogenetic tree of HIV pol (PR/RT) sequences obtained from ART-naïve patients from North and Northeast of Paraná, Brazil.
Additional file 2. Bootscanning analysis of recombinant HIV-1 sequences in protease and reverse transcriptase regions from ART-naïve patients from North and Northeast of Paraná, Brazil.
Data Availability Statement
Sequence data generated and analyzed during the current study are available in the GenBank repository, accession numbers: KM851047–KM851209. Epidemiological data generated and analyzed during the current study are not publicly available due to ethical impediment, but they are available with the corresponding authors on reasonable request.


