Abstract
Proteins and microRNAs (miRNAs) within the axon locally regulate axonal development. However, protein profiles of distal axons of cortical neurons have not been fully investigated. In particular, networks of genes encoding axonal proteins and their related miRNAs in sub compartments of neurons such as axons remain unknown. Using embryonic cortical neurons cultured in a microfluidic device and proteomic approaches, we found that distal axons contain 883 proteins. Bioinformatics analysis revealed that 94 out of these 883 proteins are related to regulating axonal growth. Of the 94 genes encoding these proteins, there were 56 candidate genes that can be putatively targeted by axon-enriched 62 miRNAs with 8mer sites that exactly match these target genes. Among them, we validated 11 proteins and 11 miRNAs, by means of Western blot and RT-PCR, respectively. Treatment of distal axons with chondroitin sulfate proteoglycans (CSPGs) that inhibit axonal growth elevated miR-133b, −203a, −29a and −92a, which were associated with reduced protein level of AKT, MTOR, PI3K, DPYSL2, MAP1B and PPP2CA. In contrast, reduction of miR-128, −15b, −195, −26b, −34b, −376b, −381 by CSPGs was accompanied by increased EZR, KIF5A, DCX, GSK3B, and ROCK2 proteins. In silico pathway analysis revealed an interconnected network of these miRNAs and protein coding genes that is highly related to regulating axonal growth. Our data provide new insights into networks of miRNAs and their related proteins in distal axons in mediating axonal growth.
Keywords: Axonal Growth, Axonal proteins and MiRNAs, Bioinformatics
Introduction
In addition to dendrites, the axon contains mRNAs encoding proteins that locally regulate axonal development [1,2]. In contrast to mRNAs, protein profiles in the axon compartment have not been intensively investigated. Using a proteomic approach, previous studies have shown that neuronal growth cones in the developing rat forebrain contain up to 2,000 proteins involved in axonal pathfinding, cytoskeletal remodeling, vesicular traffic and carbohydrate metabolism [3–5]. MicroRNAs are single-stranded noncoding RNA molecules of 20 to 25 nucleotides. Mature miRNAs bind to Argonaute (Ago) proteins in the RNA-induced silencing complex (RISC), leading to post-transcriptional gene silencing through mRNA destabilization and/or translational repression [6–9]. miRNAs are enriched in distal axons compared to the parent cell bodies [6]. Emerging data indicate that the miRNA machinery protein Ago2 is present in distal axons and that axonal miRNAs locally regulate multiple axonal functions [7–9]. For example, elevation of miR-19a in axons of cultured embryonic cortical neurons promotes axonal growth by inactivation of PTEN signals, a validated target of miR-19a [8]. However, functional networks of miRNAs and their target genes in the axon are complex because each miRNA putatively targets hundreds of mRNAs, and each gene can be regulated by multiple miRNAs [10]. Identification of such functional networks in the axon will provide new insights into understanding molecular mechanisms of miRNAs and genes encoding proteins to locally regulate axonal functions.
Using embryonic cortical neurons cultured in a microfluidic device and proteomic approach, we identified 883 proteins localized to axons of cortical neurons and demonstrated that genes encoding to some of the identified proteins along with axonal miRNAs formed an intertwined network that is highly related to axonal growth.
Materials and Methods
Primary embryonic cortical neurons cultured in a microfluidic culture device
All experimental procedures were carried out in accordance with the NIH Guide for the Care and Use of Laboratory. Animals were approved by the Institutional Animal Care and Use Committee of Henry Ford Hospital. Cortical neurons were harvested from embryonic day-18 Wistar rats (Charles River Laboratories, Spencerville, OH, USA) and cultured according to published studies [11]. Briefly, embryos were removed, and the cerebral cortex was dissected, stripped of meninges, and dissociated into a combination of Ca2+ and Mg2+ free Hanks balanced salt solution containing 0.125% trypsin (Thermo Fisher Scientific, Waltham, MA, USA) digestion, then mechanically triturated. The triturated cells were passed through a 40mm cell strainer (Fisher Scientific, Hampton, NH, USA) to reach a cell density at 3×107cells/ml. A microfluidic culture device used in the present study contains 450 mm long microgrooves that connect cell body and axonal compartments and permit only distal axons to grow into the axonal compartment (Standard Neuron Device, Cat# SND450, Xona Microfluidics, Temecula, CA, USA). Briefly, sterilized device was affixed to poly-D-lysine-coated (Sigma-Aldrich, St. Louis, MO, USA) dishes (35 mm, Corning, Corning, NY, USA). The cortical neurons were plated at a density of 6×105 cells/device in cell body compartment with growth medium composed by neurobasal medium (Invitrogen, Carlsbad, CA, USA), 2% B-27 supplement (Invitrogen), 1% GlutaMax (Invitrogen), and 1% antibiotic-antimycotic (Invitrogen). On day in vitro (DIV) 3, one-half of the medium was replaced with culture medium containing 5-fluorodeoxyuridine. Thereafter, the growth medium was replaced every other day.
To examine the effect of chondroitin sulfate proteoglycans (CSPGs) on axonal growth, CSPGs (2 mg/ml, Millipore, Cat# CC117, Burlington, MA, USA) were applied to the axonal compartment.
Proteomics Analysis
Total proteins in the cell body and axonal compartments were extracted according to published protocols [11,12]. Briefly, 10 μl of lysis buffer (RIPA with 1% protease inhibitor cocktails plus 1% phosphatase inhibitor cocktail, Sigma-Aldrich) was applied to the axonal compartment. Medium in the somal compartment was kept intact during axonal lysis to prevent lysis buffer in the axonal compartment flowing back into the cell body compartment, which minimizes cell body compartment contamination. Then, proteins in the cell body compartment were extracted. During the entire period, the device was placed horizontally on ice. Cell body and axonal lysis from 6 individual devices were pooled for proteomics analysis. Protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Protein profiles from cell body and axonal samples were measured using a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Briefly, protein extracts from individual samples were loaded per lane on SDS-PAGE with a 4–12% gradient. After that, each lane was cut into 10 equal-sized pieces. Proteins in each gel piece were digested and peptides within the gel were extracted. Then, peptides were separated by reverse phase chromatography and analyzed with a Q-Exactive mass spectrometer. Abundant species were fragmented in higher energy collision induced dissociation mode. Data analysis was performed using Proteome Discoverer 1.4 (Thermo Fisher Scientific) which incorporated the Mascot (Matrix Science, Boston, MA, USA) algorithm. The latest protein UniProtKB database was used for identification of rat protein sequences. Scaffold software (Proteome Software) was used to validate and compare MS/MS based peptide and protein identifications. Protein identifications were accepted if they could be established at greater than 99.0% probability with minimum two unique peptides detected. Proteins are presented by their official encoding gene names (Entrez Gene; NCBI). Spectral counts were calculated by normalizing the number of assigned spectra for each protein to the total number of assigned spectra for each sample.
Quantitative Real-Time Reverse Transcriptase (qRT-PCR)
To analyze miRNAs in the axon, axons growing in the axonal compartment of microfluidic culture devices were lysed in Qiazol reagent (Invitrogen). Then, total RNA was isolated using the miRNeasy Mini kit (Qiagen, Valencia, CA, USA). Briefly, medium in the axonal compartment was removed and washed with pre-warmed PBS, while medium in the cell body compartment was kept to minimize the contamination from the cell body. Then 20 μl of Qiazol reagent was applied to the axonal compartment for no more than 1 min, and was totally collected. Axonal miRNAs were extracted with a miRNeasy Mini kit according to the manufactory’s protocol. miRNAs were reversely transcribed with the miRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) and amplified with the TaqMan miRNA (Applied Biosystems). Ct value of U6 was used as the internal control. For message RNA analysis, RNA was reversely transcribed to cDNA with Superscript First-strand Synthesis System (Invitrogen). A SYBR Green PCR master kit (Invitrogen) was used with the appropriate concentrations (lOnM) of forward and reverse primers in a total volume of 20μ1. PCR reactions contained 1μl cDNA. Quantitative RT-PCR was performed using three-stage program parameters provided by the manufacturer as follows; 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 sec at 95°C and 1 min at 60°C. The following primers for qRT-PCR were designed using Primer Premier 5 software. Gamma actin: Forward 5’-ATC GCC GCA CTC GT CAT-3’; Reverse 5’-GCC GTG TTC GAT AGG GTA-3’. GAPDH: Forward 5’-AAC TCC CAC TCT TCC ACC TTC-3’; Reverse 5’-GGT CCA GGG TTT CTT ACT CCTT-3’. GAPDH was used as an internal control. Quantitative RT-PCR was performed at least three times for individual experiments.
Western Blotting
To measure the protein levels in the distal axons, proteins were extracted from distal axons cultured in the axonal compartment as descried above. Axonal lysis from 6 individual devices was pooled for one Western blot. The following primary antibodies were used: rabbit polyclonal anti-Doublecortin (DCX, 1:1000, Abcam, Cambridge, MA, USA), rabbit polyclonal anti-GSK3B (1:1000, Cell Signaling Technology, Danvers, MA, USA), mouse monoclonal anti-mTOR (1:1000, Santa Cruz Biotechnology, Dallas, TX, USA), mouse monoclonal anti-Protein Phosphatase 2 Catalytic Subunit Alpha (PPP2CA, 1:1000, Santa Cruz Biotechnology), mouse monoclonal anti-AKT (1:1000, Abcam), mouse monoclonal anti-KIF5A (1:1000, Santa Cruz Biotechnology), mouse monoclonal anti-Ezrin (EZR, 1:1000, Santa Cruz Biotechnology), mouse monoclonal anti- Dihydropyrimidinase-related protein 2 (DPYSL2, 1:1000, Santa Cruz Biotechnology), rabbit monoclonal anti-microtubule associated protein 1B (MAP1B, 1:1000, Santa Cruz Biotechnology), mouse monoclonal anti-ROCK2 (1:1000, Santa Cruz Biotechnology), rabbit polyclonal anti-PI3 kinase p85 (1:1000, Millipore) and rabbit polyclonal anti-microtubule associated protein 2 (MAP2, 1:1000, Cell Signaling Technology). For detection, horseradish peroxidase-conjugated secondary antibodies were used (1:2000) and followed by enhanced chemiluminescence development (Pierce Biotechnology). Protein levels of β-actin (mouse monoclonal, 1:2000, Abeam) were used as the internal controls for axons. Western blots were performed at least three times for individual experiments.
Bioinformatics analysis
DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov) and Ingenuity Pathway Analysis (IPA) software (QIAGEN; Hilden, Germany) were used for gene ontology (GO) and signaling pathway analysis. IPA includes a manually annotated database of protein interactions and metabolic reactions obtained from the scientific literature. Gene names of all the identified proteins were imported into IPA and processed using the core analysis tool. Using the IPA knowledge base, networks of experimental proteins were built. Pathway enrichment was assessed using a Fisher’s exact test, and pathway maps were then prioritized according to their statistical significance (p < 0.001). Networks were graphically visualized as hubs (proteins) and edges (the relationship between proteins).
Statistical Analysis
All non-pathway based statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS, version 11.0; SPSS, Chicago, IL). Student’s t-test was used when comparing two groups. Values presented in this study are expressed as mean +/− standard deviation (SD). A p-value<0.05 was considered to be significant.
Results
Characterize axonal proteins acquired from distal axonal samples
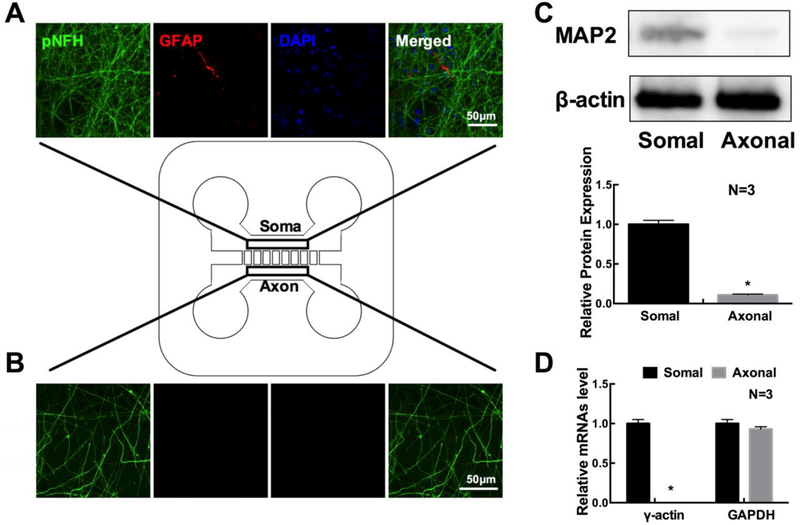
To analyze axonal proteins, primary embryonic cortical neurons from the rat were cultured in microfluidic devices that permit distal axons to grow into the axonal compartment from their parent cell bodies in the soma compartment [8,13]. Double immunofluorescent staining showed that the majority of cells in the cell body compartment were NFH+ neurons and that only few of cells were GFAP+. However, in the axonal compartment, only NFH+ axons were detected (Fig. 1 A, B). Western blot analysis showed that MAP2 proteins were primarily detected in cell body samples, while qRT-PCR analysis showed that mRNA of γ-actin was not detected in axonal samples (Fig. 1 C, D). These data indicate that distal axons in the axonal compartment are not contaminated by neuronal cell bodies and astrocytes, which is consistent with published studies [8,11].
Figure 1.
Confocal microscopic images show the presence of a few GFAP signals (red), abundant of pNFH positive dendrites and axons (green), and DAPI nuclei (blue) in the cell body compartment (A), but only the presence of pNFH positive axons (green) in the distal axon compartment (B). Rectangular boxes in schematic figure of the microfluidic device show areas where confocal images were taken. Panels C and D show quantitative Western blot data of MAP2 and qRT-PCR data of γ-actin, respectively, in distal axons. β-actin and GAPDH were used as internal controls for Western blot and qRT-PCR analysis, respectively. *p<0.05 n=3/group.
Using a mass spectrometer, we then analyzed protein profiles of samples extracted from the cell body or the axonal compartment. The latest protein UniProtKB database was used for identification of rat protein sequences. Scaffold was used to validate and compare MS/MS based peptide and protein identifications. Protein identifications were accepted if they could be established at greater than 99.0% probability with a minimum of two unique peptides detected. A total of 883 and 2,729 proteins were confidently detected from axonal and soma samples, respectively (Suppl 1). The 883 proteins from the axonal samples included several axonal marker proteins such as CFL1, TUBB4B, NFASC, ROBO2, KLC1, TAU and 14–3-3 [14–16]. In axonal samples, astrocyte marker proteins and nuclear proteins in axonal samples were not detected. In contrast, 15% (409/2,729) of total soma proteins were nuclear proteins, the second enriched proteins in the soma cell samples, including NACC1, MBB1A, and NFIA. These data suggest that the axonal samples are unlikely contaminated by samples from the cell body compartment during the sample collection processes.
Overall, 91.8% (811/883) proteins in axonal samples overlapped with proteins detected in the cell body samples, which is expected because the cell body compartment contained many axons that did not pass the microgrooves into the axonal compartment. Thus, protein profiles from the cell body samples represent a combination of proteins from neuron cell bodies and axons. Subsequently, we focus our analysis on protein profiles from the axonal samples.
Axonal protein profiles analysis
Compared to ~2,000 proteins in the growth cone of the developing rat forebrain identified by Estrada-Bernal et al [3], we found that 277 out of 883 proteins in our axonal samples were not included in the growth cone (Suppl 2). Using a microarray approach, Taylor et al have identified the presence of 287 mRNA transcripts in distal axons of the cortical neurons cultured within the microfluidic device [13]. We compared 883 axonal proteins identified in the present study against the 287 axonal mRNAs and found that 43 proteins matched their mRNAs in the microarray data (Suppl 3), suggesting that these proteins could potentially be synthesized in the distal axons.
Using DAVID bioinformatics resources 6.8, we performed GO Term analysis of proteins from axonal samples. The proteins were classified into their annotated biological process, molecular function and cellular component. Significantly enriched biological processes included transport, metabolites and energy (Table 1). Transportation proteins such as vesicle-mediated transportation play an important role in maintaining axonal growth by delivering cellular components to axons from their cell bodies [17–19]. Enriched molecular functions were mainly nucleotide and ribonucleotide binding proteins such as guanyl nucleotide binding and guanyl ribonucleotide binding (Table 1). These proteins regulate RNA trafficking in axons [20–22]. Enriched cellular components were cytosolic and mitochondrial proteins (Table 1). Our GO results are comparable to GO functional analysis in the rat axonal growth cone performed by others [3].
Table 1.
Top 10 GO terms for each of the three annotation categories in axon-enriched proteins
| Category | Term | Count | P-value |
|---|---|---|---|
| Biological Process | vesicle-mediated transport | 97 | 2.6×10−31 |
| protein localization | 106 | 2.2×10−22 | |
| generation of precursor metabolites and energy | 60 | 2.5×10−22 | |
| protein transport | 94 | 1.1×10−21 | |
| establishment of protein localization | 94 | 2.0×10−21 | |
| intracellular transport | 85 | 7.0×10−20 | |
| membrane organization | 63 | 7.4×10−20 | |
| cellular carbohydrate catabolic process | 31 | 6.7×10−19 | |
| alcohol catabolic process | 30 | 7.9×10−18 | |
| glucose catabolic process | 27 | 9.5×10−18 | |
| Molecular Function | cytoskeletal protein binding | 79 | 8.3×10−24 |
| nucleotide binding | 195 | 1.1×10−17 | |
| purine ribonucleotide binding | 164 | 1.9×10−16 | |
| ribonucleotide binding | 164 | 2.0×10−16 | |
| purine nucleotide binding | 169 | 2.5×10−16 | |
| guanyl nucleotide binding | 58 | 7.5×10−15 | |
| guanyl ribonucleotide binding | 58 | 7.5×10−15 | |
| GTP binding | 56 | 1.6×10−14 | |
| identical protein binding | 81 | 5.5×10−14 | |
| actin binding | 46 | 1.5×10−13 | |
| Cellular Component | Cytosol | 220 | 6.0×10−58 |
| Cytoskeleton | 153 | 9.5×10−29 | |
| soluble fraction | 77 | 1.1×10−26 | |
| Mitochondrion | 175 | 3.6×10−25 | |
| pigment granule | 36 | 5.8×10−23 | |
| Melanosome | 36 | 5.8×10−23 | |
| cytoskeletal part | 117 | 6.4×10−23 | |
| neuron projection | 88 | 1.8×10−22 | |
| cell projection | 119 | 2.9×10−22 | |
| Vesicle | 106 | 1.2×10−20 |
Rank was based on the p-value.
Compared to the axonal samples, the cell body protein samples showed some different profiles in the three GO terms (Table 2), which further suggest that proteins detected in the axonal samples are specifically localized to axons.
Table 2.
Top 10 GO terms for each of the three annotation categories in soma-enriched proteins.
| Category | Term | Count | P-value |
|---|---|---|---|
| Biological Process | establishment of protein localization | 242 | 2.4×10−50 |
| protein transport | 240 | 5.5×10−50 | |
| protein localization | 267 | 3.2×10−48 | |
| vesicle-mediated transport | 200 | 6.0×10−45 | |
| intracellular transport | 209 | 4.8×10−41 | |
| translational elongation | 74 | 1.7×10−37 | |
| membrane organization | 127 | 2.0×10−26 | |
| generation of precursor metabolites and energy | 109 | 1.1×10−24 | |
| synaptic transmission | 96 | 3.6×10−20 | |
| regulation of neurotransmitter levels | 51 | 2.3×10−18 | |
| Molecular Function | nucleotide binding | 543 | 8.0×10−45 |
| purine nucleotide binding | 451 | 1.9×10−35 | |
| purine ribonucleotide binding | 430 | 1.1×10−33 | |
| ribonucleotide binding | 430 | 1.3×10−33 | |
| RNA binding | 165 | 1.3×10−26 | |
| cytoskeletal protein binding | 145 | 1.5×10−23 | |
| purine nucleoside binding | 359 | 4.0×10−23 | |
| nucleoside binding | 361 | 4.6×10−23 | |
| adenyl nucleotide binding | 349 | 1.3×10−21 | |
| guanyl ribonucleotide binding | 122 | 2.8×10−20 | |
| Cellular Component | cytosol | 469 | 4.7×10−80 |
| mitochondrion | 429 | 1.4×10−43 | |
| cell fraction | 352 | 2.3×10−41 | |
| mitochondrial part | 222 | 3.4×10−38 | |
| organelle membrane | 330 | 7.1×10−35 | |
| non-membrane-bounded organelle | 532 | 3.6×10−32 | |
| intracellular non-membrane-bounded organelle | 532 | 3.6×10−32 | |
| internal side of plasma membrane | 98 | 3.1×10−31 | |
| organelle envelope | 216 | 1.6×10−30 | |
| cytosolic part | 76 | 3.4×10−30 |
Rank was based on the p-value.
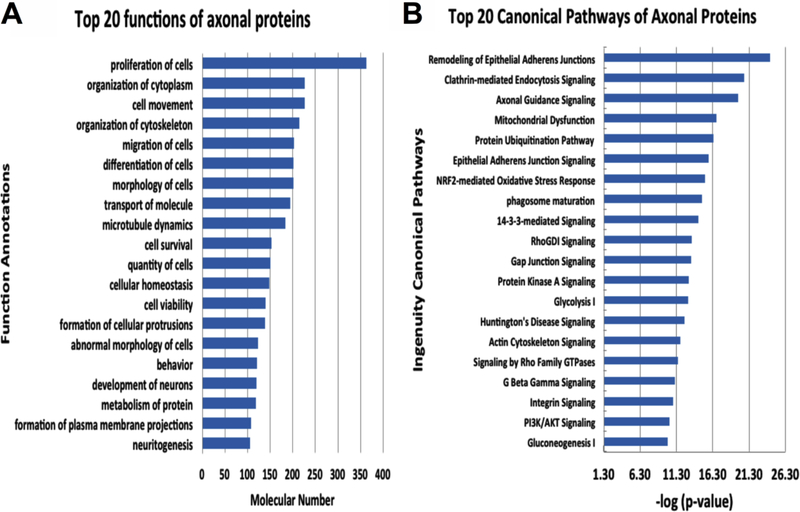
To examine signaling networks that are involved by axonal proteins, we performed pathway map analysis by means of IPA. A total 500 functional and diseases terms were identified by IPA (p <0.000001). We identified the top 20 functional and disease terms and signaling pathways (Fig.2). The remodeling of epithelial adherent junctions was the most significant -log(p-value) =24.20. Moreover, proteins involved in axonal guidance signaling were also highly significant -log(p- value) =19.80. The 14–3-3 mediated signaling pathway and PI3K/AKT signaling pathway proteins were similarly and highly ranked and are known to be important for axonal growth [23–25]. Interestingly, further analyzing 72 proteins that were detected only in axonal samples, we found that 41 out of the 72 (56.9%) proteins are directly associated to central nervous system (CNS) development. A few of these proteins have been shown to regulate axonal growth, and these proteins are selenium binding protein 1 (SBP1) that is located in axons and promotes axonal extending [26], fibromodulin (FMOD) that plays an important role in axonal extension and myelination [27] and LDL receptor related protein 4 (LRP4) that enhances synapse number [28].
Figure 2.
Top 20 functions of axonal enriched proteins (A) and top 20 canonical pathways involved in axonal proteins (B).
A network of axonal proteins and miRNAs in mediating axonal growth
Using IPA, we next identified 94 axonal proteins that were significantly associated with axonal growth and formation (p < 0.0001) (Table 3). These 94 proteins were within 10 functional categories, including growth of axons, guidance of axons, branching of axons, shape change of axons and others (Table 3). Approximately 80% of the 94 proteins (71/94 proteins) were previously detected in the axonal growth cone of rat cortical neurons [3,29–35].
Table 3.
Axon-enriched proteins related to axonal growth
| Diseases or Functions Annotation | p-value | Molecules |
|---|---|---|
| axonogenesis | 1.99×10−26 | ACTB, APC, CASP3, GDI1, LLGL1, LRP4, MAP2K1, MBP, PTPRZ1, RAB10, RAB3A, RAB8A, RAP1B, STMN1, STXBP5, UCHL1, RUFY3, GSK3B, SNAP91 |
| growth of axons | 1.78×10−19 | BRSK1, DCX, NES, ROCK2 |
| guidance of axons | 2.71×10−15 | ALCAM, CDH2, TUBB3, CELSR3, CNTN1, CNTN2, DPYSL5, EPHA5, GLI3, KIF5C, MAPK1, MAPK3, MYH10, NRXN1, NRXN3, TENM2 |
| outgrowth of axons | 2.04×10−14 | PLXNA3, PTPRS, SPRR1A, SYN1, VAMP2, VIM, GPM6A, ROBO1, KIF3C |
| branching of axons | 1.12×10−10 | A2M, ACTR3, APP, CDC42, EZR, GAP43, KALRN, KIF2A, L1CAM, MTOR, NRP1, PALM, PDIA3 |
| elongation of axons | 4.70×10−8 | CDK5, PAFAH1B1, PPP2CA, STMN2 |
| length of axons | 2.97×10−7 | CLASP2, KRAS, DPYSL3, RAC1, RHOA |
| shape change of axons | 1.15×10−6 | RTN4, SOD1, DCC, DPYSL2 |
| morphology of axons | 6.03×10−6 | CD47, CKB, DCLK1, GPM6B, KIDINS220, MAP1B, MAPT, NCAM1, NFASC, NRCAM, PPP3CA, SH3GL1, SH3GL2, VCAN |
| quantity of axons | 6.58×10−6 | APOE, BAX, CANX, NEFM, FYN, KIF5A |
Rank was based on the p-value.
To verify the presence of proteins in the axon, we selected 9 candidates representing differently predicted functional categories from the 94 proteins: GSK3B (axonogenesis), DCX and ROCK2 (growth of axons), EZR and MTOR (branching of axons), DPYSL2 (shape change of axons), KIF5A (quantity of axons), MAP1B (morphology of axons), and PPP2CA (elongation of axons). Western blot analysis demonstrated that these 9 candidate proteins were present in the axonal samples (Suppl 4). In addition, we demonstrated the presence of GSK3B signaling pathway related proteins PI3K and AKT (Suppl 4), suggesting that this signaling pathway may locally mediate axonal function [36,37].
MiRNAs regulate posttranscriptional gene expression and are enriched in the axon compartment [6,38]. Axon enriched miRNAs play an important role in mediating axonal function and growth [39]. Using a miRNA array, we previously reported axonal enriched miRNA profiles within the cultured cortical neurons [11]. To identity miRNAs in the axon that have the potential to locally target the axonal growth genes coding to the 94 proteins, we performed an in-silico miRNA target identification analysis using a seed match criterion that miRNAs with 8mer sites exactly match putative target genes, based on the fact that 8mer sites efficaciously affect miRNA-target interactions [40]. Of the 94 genes, we found that there were 56 candidate genes that can be potentially targeted by the 62 axonal enriched miRNAs detected in our previous miRNA array data [11], including the genes coding 11 aforementioned proteins validated by Western blot. We thus focused on genes that code these 11 proteins and their related miRNAs. Using qRT-PCR, we found 11 miRNAs in the axon, which could putatively target genes coding the 11 proteins (Table.4).
Table 4.
Genes encoding axon-enriched proteins that can be perfectly matched by axon-enriched miRNAs
| Genes | miRNAs |
|---|---|
| DCX | miR-128, miR-34b |
| AKT | miR-34b |
| DPYSL2 | miR-29a |
| MAP1B | miR-92a |
| PPP2CA | miR-133b |
| ROCK2 | miR-381 |
| GSK3B | miR-128 |
| MTOR | miR-26b |
| PI3K | miR-203a, miR-92a |
| EZR | miR-376b |
| KIF5A | miR-195, miR-15b, miR-29a |
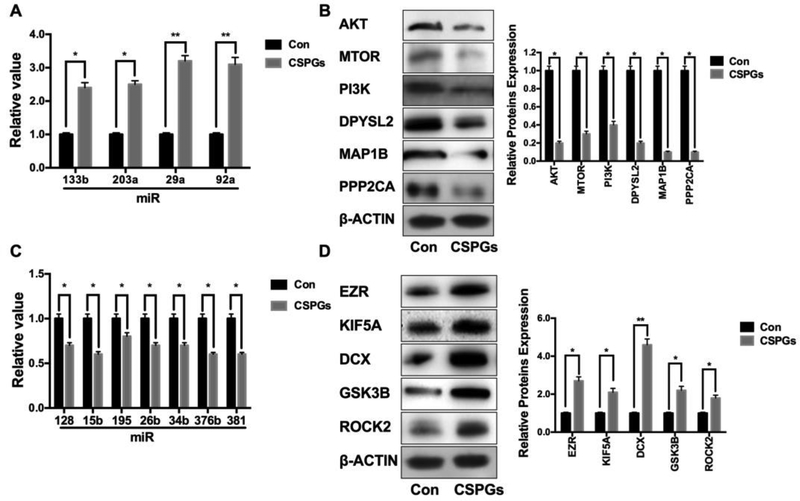
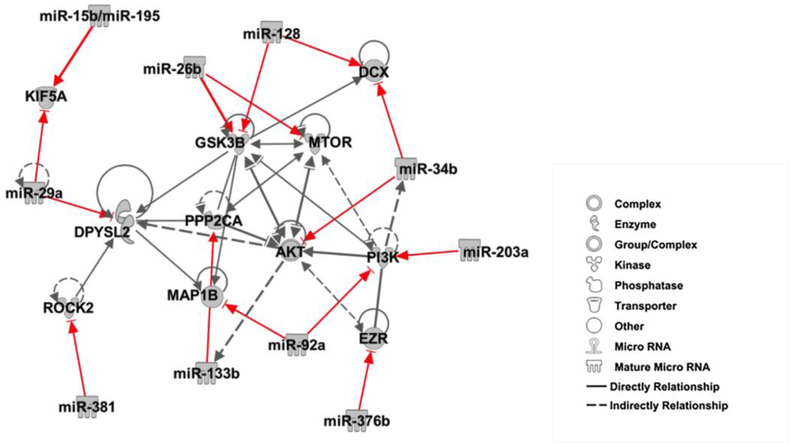
We and others have previously demonstrated that CSPGs inhibit axonal growth [11,41]. We thus examined whether axonal application of CSPGs alters levels of these 11 miRNAs and 11 proteins in distal axons cultured in the axonal compartment of the microfluidic device. Quantitative RT-PCR and Western blot analysis of axonal samples extracted from the axonal compartment revealed that CSPGs elevated miR-133b, −203a, −29a and −92a, which were associated with reduced protein expression of AKT, MTOR, PI3K, DPYSL2, MAP1B, and PPP2CA. In contrast, miR-128, −15b, −195 −26b, −34b, −376b, and −381 decreased by CSPGs were accompanied with elevation of protein expression of EZR, KIF5A, DCX, GSK3B, and ROCK2 (Fig.3 and Table.5). To identify a network of these miRNAs and proteins, we performed an IPA connectivity analysis of these 11 miRNAs and 11 proteins. We found that the 11 miRNAs and genes encoding for the 11 proteins were highly interconnected to axonal growth- related networks (Fig.4), suggesting that the axonal miRNAs altered by CSPGs have the potential to locally regulate their target genes and to act as mediators of the inhibitory effect of CSPGs on axonal growth.
Figure 3.
CSPGs alter levels of miRNAs and proteins in distal axons. Quantitative RT-PCR and Western blot analysis show increased miRNAs (A) and reduced proteins (B), respectively; and decreased miRNAs (C) and augmented proteins (D), respectively, in distal axons treated with CSPGs compared to control group without the CSPGs treatment. *p<0.05, and **p<0.01, n=3/group.
Table 5.
CSPGs-altered axon-enriched miRNAs and their related proteins
| Term | ID |
|---|---|
| Upregulated miRNAs | miR-133b, miR-203a, miR-29a, miR-92a |
| Reduced Proteins | AKT, MTOR, PI3Kp85, DPYSL2, MAP1B, PPP2CA |
| Downregulated miRNAs | miR-128, miR-15b, miR-195, miR-26b, miR-34b, miR-376b, miR-381 |
| Increased Proteins | EZR, KIF5A, DCX, GSK3B, ROCK2 |
Figure 4.
A network of miRNAs and their related proteins is highly associated with axonal growth, which was generated using IPA pathway building tools. Individual proteins are represented as nodes, and the different shapes of the nodes represent the functional class of the proteins, as indicated in the figure. The edges define the relationships of the nodes; the arrows indicate the direction of the interaction. Red lines indicate that miRNAs directly target genes encoding to the proteins which have been experimentally validated.
Discussion
Using a proteomic approach, the present study demonstrated distal axons of cultured embryonic cortical neurons contain 883 proteins including cytosolic, mitochondrial, nucleotide and ribonucleotide binding proteins. In addition, we detected an interconnected network of miRNAs and their related proteins within the distal axon that are directly involved in axonal growth. These data provide a molecular basis for further investigating the roles of proteins and miRNAs within the distal axon in mediating axonal function.
Although the role of individual axonal mRNAs, miRNAs and proteins in regulating axonal function has been demonstrated, proteomics studies of distal axons are relatively challenging because only a limited amount of pure axonal materials can be acquired [4,13]. Using the microfluidic device, we demonstrated that proteins extracted from distal axon samples within the axonal compartment are unlikely contaminated by proteins from neuronal cell bodies and astrocytes, further supporting that the microfluidic device permits us to isolate axons without contamination from non-axonal materials [11,13]. We found that distal axons of cultured embryonic cortical neurons contain 883 proteins that are highly involved in multiple signaling pathways including cellular axonal guidance signaling and mitochondrial function. Compared with proteomics data of the growth cone from the developing rat forebrain [3], the majority of proteins in the distal axon of cortical neurons detected in the present study were not previously reported. Future studies of these proteins will provide a better understanding of their physiological relevance in mediating distal axon signaling and function.
We found that 43 proteins in the distal axon matched their mRNAs in the axons of cortical neurons detected by Taylor et al [13]. However, whether these axonal proteins are locally translated by their mRNAs remains to be investigated, although the presence of intra-axonal protein synthesis machinery has been demonstrated [42].
Axonal enriched miRNAs play an important role in axonal development and function [9]. A particular miRNA may modulate a large number of genes, but genes targeted by a specific miRNA are dependent on the cellular context [43,40]. In addition, 8mer sites efficaciously affect miRNA-target interactions [44,40]. Using genes encoding identified proteins, in particular for proteins highly related to axonal growth, our bioinformatics analysis revealed an axon-specific network of 11 miRNAs and their related 11 genes encoding proteins in regulating distal axon growth, which provides new insights into interconnected networks of miRNAs and their related proteins in distal axons. The miRNAs and proteins within this network have been demonstrated to regulate axonal extension and branches [8,45–47]. For example, miR-9 has been shown to target MAP1B, which regulates axon development [45]. The present study showed that in addition to miR-9, miR-92a may also target MAP1B. Generally, our data show an inverse relation between the individual miRNAs and their target proteins that we examined in distal axons; however, there are exceptions, such as KIF5A that is targeted by miR-15b, and miR-29a and miR-195. CSPGs upregulated miR-29a, but downregulated miR-15b and miR-195, while CSPGs elevated KIF5A protein levels. These data suggest that interactions between miRNAs and their target genes in distal axons are complicated. Furthermore, although our data showed that CSPGs increased multiple axonal inhibitory proteins and substantially reduced axonal promoting proteins, CSPGs augmented DCX protein which is known to promote axonal growth of new neurons. Thus, the net effect of CSPGs-altered network of miRNAs and proteins could lead to inhibition of axonal growth. The present study suggests that proteomics in combination with miRNA array or RNA-seq analysis may identify axon-specific networks of miRNAs and their target genes governing axonal functions. Additional studies to fully validate the miRNAs and their target genes in the network and to investigate biological function of these networks are warranted, which could potentially provide new therapeutic targets for axonal dysfunction related diseases.
Supplementary Material
Significance.
Axonal proteins locally regulate axonal development. However, the protein profile in the distal axon has not been investigated. Using proteomic and quantitative RT-PCR approaches in combination with bioinformatics analysis, the present study identified a network of miRNAs and their target proteins in distal axons, which mediate axonal growth. This finding provides molecular basis for further investigating the roles of proteins and miRNAs within the distal axon in mediating axonal function.
Acknowledgment:
This work was supported by National Institutes of Health (RO1 NS088656 and ROI NS75156), and American Heart Association (16SDG29860003)
Footnotes
Additional Information:
The authors declare no competing financial interests.
References:
- 1.Jung H, Yoon BC, Holt CE (2012) Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nature reviews Neuroscience 13 (5):308–324. doi: 10.1038/nrn3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomes C, Merianda TT, Lee SJ, Yoo S, Twiss JL (2014) Molecular Determinants of the Axonal mRNA Transcriptome. Developmental neurobiology 74 (3):218–232. doi: 10.1002/dneu.22123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estrada-Bernal ASS, Sosa LJ, Simon GC, Hansen KC, et al. (2012) Functional Complexity of the Axonal Growth Cone: A Proteomic Analysis. PLoS ONE 7 (2):e31858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Igarashi M (2014) Proteomic identification of the molecular basis of mammalian CNS growth cones. Neuroscience Research 88:1–15. doi: 10.1016/j.neures.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 5.van Niekerk EA, Tuszynski MH, Lu P, Dulin JN (2016) Molecular and Cellular Mechanisms of Axonal Regeneration After Spinal Cord Injury. Molecular & Cellular Proteomics : MCP 15 (2):394–408. doi: 10.1074/mcp.R115.053751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB (2010) Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16 (8):1516–1529. doi: 10.1261/rna.1833310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB (2008) MICRORNA-338 REGULATES LOCAL CYTOCHROME C OXIDASE IV mRNA LEVELS AND OXIDATIVE PHOSPHORYLATION IN THE AXONS OF SYMPATHETIC NEURONS. The Journal of neuroscience : the official journal of the Society for Neuroscience 28 (47):12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Ueno Y, Liu XS, Buller B, Wang X, Chopp M, Zhang ZG (2013) The microRNA- 17–92 cluster enhances axonal outgrowth in embryonic cortical neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 33 (16):6885–6894. doi: 10.1523/JNEUROSCI.5180-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan BB, Kar AN, Gioio AE, Aschrafi A (2013) MicroRNAs in the axon and presynaptic nerve terminal. Frontiers in Cellular Neuroscience 7:126. doi: 10.3389/fncel.2013.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Li J, Cairns MJ (2014) Identifying miRNAs, targets and functions. Briefings in Bioinformatics 15 (1):1–19. doi: 10.1093/bib/bbs075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Chopp M, Liu XS, Kassis H, Wang X, Li C, An G, Zhang ZG (2015) MicroRNAs in the axon locally mediate the effects of chondroitin sulfate proteoglycans and cGMP on axonal growth. Developmental Neurobiology:n/a-n/a. doi: 10.1002/dneu.22292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee D-C, Hassan SS, Romero R, Tarca AL, Bhatti G, Gervasi MT, Caruso JA, Stemmer PM, Kim CJ, Hansen LK, Becher N, Uldbjerg N (2011) Protein Profiling Underscores Immunological Functions of Uterine Cervical Mucus Plug in Human Pregnancy. Journal of proteomics 74 (6):817–828. doi: 10.1016/jjprot.2011.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL (2005) A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nature methods 2 (8):599–605. doi: 10.1038/nmeth777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teunissen CE, Dijkstra C, Polman C (2005) Biological markers in CSF and blood for axonal degeneration in multiple sclerosis. The Lancet Neurology 4 (1):32–41. doi: 10.1016/S1474-4422(04)00964-0 [DOI] [PubMed] [Google Scholar]
- 15.Guillermina López-Bendito NF, Ma Le, Fouquet Coralie, Di Meglio Thomas, Chedotal Alain, Tessier-Lavigne Marc, Marin Oscar(2007) Robo 1 and Robo 2 Cooperate to Control the Guidance of Major Axonal Tracts in the Mammalian Forebrain. The Journal of neuroscience : the official journal of the Society for Neuroscience 27 (13):3395–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Yélamos AS A, Sanchez-Valle R, Casado V, Ramón JM, Graus F, Arbizu T (2001) 14–3-3 protein in the CSF as prognostic marker in early multiple sclerosis. Neurology 57 (4): 722–724 [DOI] [PubMed] [Google Scholar]
- 17.Skene JHP, Willard M (1981) Axonally transported proteins associated with axon growth in rabbit central and peripheral nervous systems. The Journal of Cell Biology 89 (1):96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vos AJG Kurt J., Ackerley Steven, and Miller Christopher C.J. (2008) Role of Axonal Transport in Neurodegenerative Diseases. Annual Review of Neuroscience 31 (1): 151–173. doi: 10.1146/annurev.neuro.31.061307.090711 [DOI] [PubMed] [Google Scholar]
- 19.George Elvira SW, Blandford Vanessa, Tong Xin-Kang,, Alexandre Serrano XF, del Rayo Sa nchez-Carbente Maria,, Florence Servant AWB, Boismenu Daniel, Lacaille Jean-Claude, McPherson Peter S., DesGroseillers Luc, and Sossin Wayne S.(2006) Characterization of an RNA Granule from Developing Brain. Molecular & Cellular Proteomics 5:635–651 [DOI] [PubMed] [Google Scholar]
- 20.Hömberg H, Holt C (2013) RNA-binding proteins and translational regulation in axons and growth cones. Frontiers in Neuroscience 7:81. doi: 10.3389/fnins.2013.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi VB, Baskaran P, Shaw CE, Guthrie S (2014) Tar DNA-binding protein-43 (TDP-43) regulates axon growth in vitro and in vivo(). Neurobiology of Disease 65 (100):25–34. doi: 10.1016/j.nbd.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sotelo-Silveira JR, Calliari A, Kun A, Koenig E, Sotelo JR (2006) RNA Trafficking in Axons. Traffic 7 (5):508–515. doi: 10.1111/j.1600-0854.2006.00405.X [DOI] [PubMed] [Google Scholar]
- 23.Read D, Gorman A (2009) Involvement of Akt in neurite outgrowth. Cell Mol Life Sei 66 (18):2975–2984. doi: 10.1007/s00018-009-0057-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tohda C, Kuboyama T, Komatsu K (2005) Search for Natural Products Related to Regeneration of the Neuronal Network. Neurosignals 14 (l-2):34–45 [DOI] [PubMed] [Google Scholar]
- 25.Sandeep Vasant More SK, Kim In-Su, Kumar Hemant, Kim Byung-Wook, Choi Dong-Kug (2012) The Role of Bioactive Compounds on the Promotion of Neurite Outgrowth. Molecules 17 (6):6728–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyaguchi K (2004) Localization of selenium-binding protein at the tips of rapidly extending protrusions. Histochemistry and Cell Biology 121 (5):371–376. doi: 10.1007/s00418-004-0623-y [DOI] [PubMed] [Google Scholar]
- 27.Lu W-c, Zhou Y-x, Qiao P, Zheng J, Wu Q, Shen Q (2018) The protocadherin alpha cluster is required for axon extension and myelination in the developing central nervous system. Neural Regeneration Research 13 (3):427–433. doi: 10.4103/1673-5374.228724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosca TJ, Luginbuhl DJ, Wang IE, Luo L (2017) Presynaptic LRP4 promotes synapse number and function of excitatory CNS neurons . eLife 6:e27347. doi: 10.7554/eLife.27347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terada K, Kojima Y, Watanabe T, Izumo N, Chiba K, Karube Y (2014) Inhibition of Nerve Growth Factor-Induced Neurite Outgrowth from PC 12 Cells by Dexamethasone: Signaling Pathways through the Glucocorticoid Receptor and Phosphorylated Akt and ERK1/2. PLoS ONE 9 (3):e93223. doi: 10.1371/journal.pone.0093223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ketschek A, Jones S, Spillane M, Korobova F, Svitkina T, Gallo G (2015) Nerve Growth Factor Promotes Reorganization of the Axonal Microtubule Array at Sites of Axon Collateral Branching. Developmental neurobiology 75 (12): 1441–1461. doi: 10.1002/dneu.22294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higuero AM, Sânchez-Ruiloba L, Doglio LE, Portillo F, Abad-Rodrí guez J, Doth CG, Iglesias T (2010) Kidins220/ARMS Modulates the Activity of Microtubule-regulating Proteins and Controls Neuronal Polarity and Development. The Journal of Biological Chemistry 285 (2): 1343–1357. doi: 10.1074/jbc.M109.024703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liz MA, Mar FM, Santos TE, Pimentel HI, Marques AM, Morgado MM, Vieira S, Sousa VF, Pemble H, Wittmann T, Sutherland C, Woodgett JR, Sousa MM (2014) Neuronal deletion of GSK3β increases microtubule speed in the growth cone and enhances axon regeneration via CRMP-2 and independently of MAP 1B and CLASP2. BMC Biology 12:47–47. doi : 10.1186/1741-7007-12-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams RR, Venkatesh I, Pearse DD, Udvadia AJ, Bunge MB (2015) MASHl/Asclla Leads to GAP43 Expression and Axon Regeneration in the Adult CNS. PLoS ONE 10 (3):e0118918. doi: 10.1371/journal.pone.0118918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sosa LJ, Bergman J, Estrada-Bemal A, Glorioso TJ, Kittelson JM, Pfenninger KH (2013) Amyloid Precursor Protein Is an Autonomous Growth Cone Adhesion Molecule Engaged in Contact Guidance. PLoS ONE 8 (5):e64521. doi: 10.1371/journal.pone.0064521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thelen K, Jaehrling S, Spatz JP, Pollerberg GE (2012) Depending on Its Nano-Spacing, ALCAM Promotes Cell Attachment and Axon Growth. PLoS ONE 7 (12):e40493. doi: 10.1371/journal.pone.0040493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hur E-M, Zhou F-Q (2010) GSK3 signaling in neural development. Nature reviews Neuroscience 11 (8):539–551. doi: 10.1038/nm2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hur E-M, Saijilafu, Lee BD, Kim S-J, Xu W-L, Zhou F-Q (2011) GSK3 controls axon growth via CLASP-mediated regulation of growth cone microtubules. Genes & Development 25 (18): 1968–1981. doi: 10.1101/gad.17015911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldie BJ, Cairns MJ (2012) Post-Transcriptional Trafficking and Regulation of Neuronal Gene Expression. Molecular Neurobiology 45 (1):99–108. doi: 10.1007/sl2035-011-8222-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki Y, Gross C, Xing L, Goshima Y, Basseil GJ (2014) Identification of Axon-Enriched MicroRNAs Localized to Growth Cones of Cortical Neurons. Developmental neurobiology 74 (3):397–406. doi: 10.1002/dneu.22113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nam J-W, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP (2014) Global analyses of the effect of different cellular contexts on microRNA targeting. Molecular cell 53 (6): 1031–1043. doi: 10.1016/j.molcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silver J, Miller JH (2004) Regeneration beyond the glial scar. Nat Rev Neurosci 5 (2): 146–156 [DOI] [PubMed] [Google Scholar]
- 42.Twiss JL, Kalinski AL, Sachdeva R, Houle JD (2016) Intra-axonal protein synthesis - a new target for neural repair? Neural Regeneration Research 11 (9): 1365–1367. doi: 10.4103/1673-5374.191193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irena Ivanovska MAC (2008) Combinatorial microRNAs: Working together to make a difference. Cell Cycle 7 (20):3137–3142 [DOI] [PubMed] [Google Scholar]
- 44.Tarang S, Weston MD (2014) Macros in microRNA target identification: A comparative analysis of in silico, in vitro, and in vivo approaches to microRNA target identification. RNA Biology 11 (4):324–333. doi: 10.4161/rna.28649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N (2012) microRNA-9 regulates axon extension and branching by targeting Map lb in mouse cortical neurons. Nat Neurosci 15 (5):697–699. doi:http://www.nature.com/neuro/iournal/vl5/n5/abs/nn.3082,html#supplementary-information [DOI] [PubMed] [Google Scholar]
- 46.Lai Y-W, Chu S-Y, Wei J-Y, Cheng C-Y, Li J-C, Chen P-L, Chen C-H, Yu H-H (2016) Drosophila microRNA-34 Impairs Axon Pruning of Mushroom Body γ Neurons by Downregulating the Expression of Ecdysone Receptor . Scientific Reports 6:39141. doi: 10.1038/srep39141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun X, Zhou Z, Fink DJ, Mata M (2013) HspB1 silences translation of PDZ-RhoGEF by enhancing miR-20a and miR-128 expression to promote neurite extension. Molecular and cellular neurosciences 57:111–119. doi : 10.1016/j.men.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.