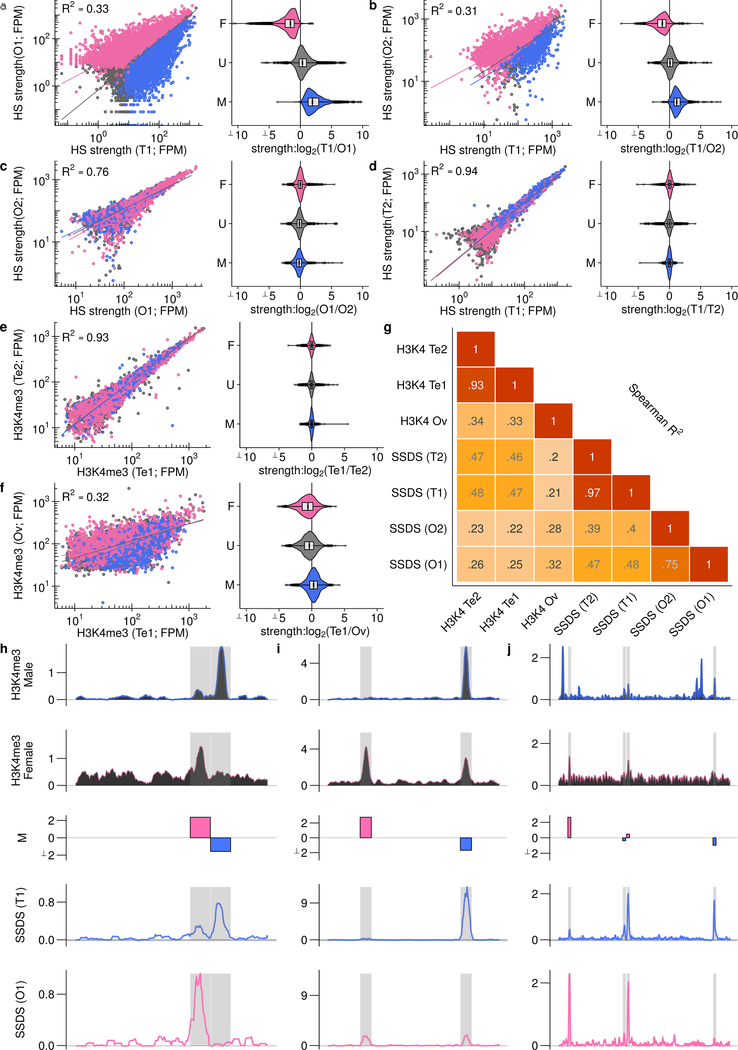
Extended Data Fig 5: Sex biased hotspots are consistent across replicates and are defined before DSB formation.
Hotspot strength was calculated at all autosomal hotspots from the merged O1/T1 DSB maps. The strength of hotspots was re-calculated in two testis (T1, T2) and two ovary (O1, O2) maps. Female-biased (pink), unbiased (grey) and male-biased hotspots (blue) were determined by comparing the T1 and O1 maps. These hotspots are colored the same in all panels. a, Sex-biased hotspots are distributed as expected when comparing the O1 and T1 DSB maps. These data are also plotted in Fig. 2d,e. b, Sex-biased hotspots exhibit the same sex-biases in the O2 sample. c,d, Sex-biased hotspots exhibit no biased usage between samples derived from mice of the same sex. e-g, Sex biases that precede DSB formation were studied by performing H3K4me3 ChIP-Seq in fluorescence assisted cell sorting (FACS)-purified fetal oocytes at 15.5 dpc (see Methods). The H3K4me3 signal at hotspots was quantified and compared to existing maps of H3K4me3 in juvenile mouse testis68. e, The H3K4me3 signal at hotspots is tightly correlated in replicate samples from mouse testis (H3KP1, H3KP2; (Extended Data Fig 1i). f, Akin to what we observe when examining the SSDS signal at DSB hotspots, there is extensive variation in the H3K4me3 signal at hotspots between male and female meiosis. This implies that sex biases are established before DSB formation. Sex biases determined using SSDS remain broadly conserved when we compare H3K4me3 in females to males. g, H3K4me3 at hotspots is better correlated with SSDS from the respective sex. h-j, Sex biases in H3K4me3 ChIP-Seq parallel differences in SSDS signal. H3K4me3 ChIP-Seq coverage is shown in upper panels; Testis (H3KP1; blue) and Ovary (H3OV; pink). The middle panel shows the log2 fold difference (M) between the SSDS signal in testis (T1) and ovary (O1). SSDS coverage for these samples is shown in the lower panels. Grey boxes represent DSB hotspot positions. To allow for quantitative cross comparison, the coverage in each sample is normalized by median signal strength at DSB hotspots in that sample. The genomic coordinates of each window are given underneath.