Abstract
Being at the western fringe of Europe, Iberia had a peculiar prehistory and a complex pattern of Neolithization. A few studies, all based on modern populations, reported the presence of DNA of likely African origin in this region, generally concluding it was the result of recent gene flow, probably during the Islamic period. Here, we provide evidence of much older gene flow from Africa to Iberia by sequencing whole genomes from four human remains from northern Portugal and southern Spain dated around 4000 years BP (from the Middle Neolithic to the Bronze Age). We found one of them to carry an unequivocal sub-Saharan mitogenome of most probably West or West-Central African origin, to our knowledge never reported before in prehistoric remains outside Africa. Our analyses of ancient nuclear genomes show small but significant levels of sub-Saharan African affinity in several ancient Iberian samples, which indicates that what we detected was not an occasional individual phenomenon, but an admixture event recognizable at the population level. We interpret this result as evidence of an early migration process from Africa into the Iberian Peninsula through a western route, possibly across the Strait of Gibraltar.
Keywords: palaeogenome, Africa, Iberia, mitochondrial DNA, gene flow, admixture
1. Introduction
Modern European populations show a southwest-northeast gradient of African diversity with its maximum in Spain [1,2]. It is unclear if this gradient is the consequence of ancient prehistoric contacts or, instead, if it is owing to African migrations into Europe during historical times. Based on genome-wide data from modern populations, African admixture has been estimated to around the time of the Muslim expansion into Iberia [2,3]. However, analyses of mitochondrial DNA (mtDNA) and Y chromosomes in modern individuals suggest a much older admixture event, possibly dated around 10 000–8000 years before present (yBP), [4–6]. One of the strongest pieces of evidence is the existence of mitochondrial haplotypes belonging to the sub-Saharan L macro-haplogroup that form European-specific subclades, suggesting they have evolved locally in Europe [5–7].
Ancient DNA (aDNA) is a powerful resource to reconstruct events in demographic history [8–10]. Studies of prehistoric human remains from Morocco [11,12] and Spain [13] reported genomic evidence of gene flow from Iberia into Late Neolithic Moroccans around 5000 yBP, but none of these works detected admixture in the opposite direction, i.e. from Africa into Iberia. However, all these studies considered a limited number of captured single nucleotide polymorphisms (SNPs), which may not be powerful enough to detect limited levels of gene flow that happened long ago. Also, these studies used ancient Maghrebians as the potential source of African admixture, and hence may not be a good proxy if the gene flow had a sub-Saharan origin.
Here we used a combination of shotgun and whole-genome capture (WGC) strategies to generate whole-genome data from four prehistoric human remains (coverage from 0.4–4.8×) and 13 mitogenomes from the Iberian Peninsula, dated well before any historical African presence in Iberia. We found one sample from Andalusia (southern Spain) to carry the sub-Saharan mitochondrial haplogroup L2a1, to our knowledge never observed before in ancient human remains outside Africa. In addition, the analyses of the ancient nuclear genomes revealed an increased similarity between Middle Neolithic/Chalcolithic (MN/ChL) samples from Spain and ancient sub-Saharan remains. To our knowledge, this study reports for the first time, both at nuclear and mitochondrial level, direct evidence of prehistoric northbound gene flow from Africa into Europe, probably following a trans-Mediterranean western route.
2. Material and methods
(a). Archaeological samples
We sampled 17 ancient individuals from the Iberian Peninsula, originating from: (i) the Mediterranean area in the south of Spain (four individuals from Cueva del Ángel, Lucena, Córdoba; 37°24′22″ N, 4°24′59″ W) and (ii) the Atlantic watershed in the north of Portugal (three individuals from Lorga de Dine; 41°52′30″ N, 6°55′40″ W) and Spain (10 individuals from Galicia). Most samples have been carbon dated to around 3000–4500 yBP, covering the Middle Neolithic, the Chalcolithic and Bronze Age (BA) period in Iberia (table 1; electronic supplementary material, table S1 and data S1).
Table 1.
Details of samples sequenced at nuclear genome level.
| sample ID | site | Cal yBP | material | sequencing Strategy | %end. DNA | Gen. cov | Mt. cov. | Mt. hg. | Biol sex | Y hg | (%) mtDNA cont. (C+MD/C-MD)b | (%) X.cont |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COV20126 | Covacha del Ángel, Lucena (Spain) | 3637 ± 60 | tooth | WGC | 20.3(8.3)a | 0.39× | 84.7× | L2a1 | XY | G2a2b | 3.1/1.4 | 2–3 |
| LU339 | Sima del Ángel, Lucena (Spain) | 4889 ± 68 | petrous bone | shotgun | 31.5 | 4.78× | 102.2× | H3 | — | — | 0.2/0.3 | |
| LD1174 | Lorga de Dine (Portugal) | 4467 ± 61 | petrous bone | shotgun | 45.5 | 3.76× | 145.6× | U5b2b5 | XX | — | 2.3/0.1 | |
| LD270 | Lorga de Dine (Portugal) | 4386 ± 128 | petrous bone | shotgun | 33.7 | 4.20× | 131.1× | U8a | — | — | 2.5/0.3 |
aFor COV20126, the percentage of endogenous DNA is given for the library before capture (8.3% within brackets) and after capture (20.3%). More details about the next-generation sequencing data are given in the electronic supplementary material, table S2.
b(C + MD): percentage contamination including sites with potentially damaged bases. (C − MD): percentage of contamination excluding sites with potentially damaged bases (C to T and G to A transitions).
(b). Laboratory processing
All laboratory steps before polymerase chain reaction (PCR) amplification were carried out in dedicated aDNA facilities at the Universities of Potsdam (Germany) and York (UK). Samples were preferentially taken from teeth and petrous bone, when available, and all remains were decontaminated by physical removal of the surfaces and ultraviolet treatment before extraction. DNA was extracted following the protocols from [14,15]. One Illumina library was built from each sample, either as single or double stranded (following [16] and [17], respectively) depending on their latitude of origin and storage conditions after excavation. Libraries preserving less than 20% of endogenous DNA were subjected to capture enrichment (electronic supplementary material, table S2).
(c). DNA hybridization capture
We followed two different strategies for the capture experiments: (i) we used capture on array as described in [18] to recover complete mitochondrial genomes from eight samples; and (ii) we developed an in-solution WGC protocol to recover whole genomes and complete mitogenomes from five ancient samples, including COV20126. This protocol is based on homemade probes built from commercial male human DNA (PROMEGA). Similar to [19], the commercial DNA is sonicated to an average fragment size of 100–200 bp and then ligated to biotinylated adapters. The sequence of the adapters is different to that of the Illumina oligos, which prevents amplification or sequencing of the baits after capture. Following WGC, we obtained an enrichment in nuclear sequences ranging from 2 up to 12-fold (electronic supplementary material, table S2). Further details in the electronic supplementary material, document S1.
(d). Sequencing and data processing
Libraries LD270 and LD1174 were shotgun sequenced each on one lane of an Illumina HiSeqX platform, using 150 cycles in paired-end (PE) mode. A library from LU339 and the capture products of COV20126 were sequenced each on a whole flow cell of the Illumina platform NextSeq500 using 76 cycles in single-end (SE) mode. The other four libraries subjected to WGC were pooled and sequenced on the NextSeq500 with 76 cycles and PE mode. Finally, the seven libraries captured on array were pooled and sequenced on a single lane of a HiSeq2500 platform using 76 cycles in SE mode. Details in the electronic supplementary material, document S1 and table S2.
Next-generation sequencing reads were trimmed and mapped to the reference hg19 as described in the electronic supplementary material, document S1.
MapDamage [20] was used to assess the patterns of molecular damage in the mapped reads (electronic supplementary material, figure S1). The level of contamination was estimated based on the presence of secondary variants at haploid sites, i.e. in mtDNA and, in males, on chromosome X (electronic supplementary material, document S1).
Finally, we collected raw read data from published ancient samples and mapped them to the reference following the same pipeline (electronic supplementary material, data S2).
(e). Uniparental markers
For mtDNA analysis, we mapped the reads to the revised Cambridge Reference Sequence (rCRS, NC_012920; [21]). Variants were called using Samtools Mpileup [22] at positions covered by at least three reads and having a mapping and base quality more than or equal to 30. All called variants were confirmed by visual inspection using Tablet [23]. Also, in COV20126, we repeated the analysis after using pmdtools, which restricts the haplogroup assignment to reads showing the typical aDNA pattern of damage [24]. We used Haplogrep (http://haplogrep.uibk.ac.at/) to assign the mitochondrial haplogroups (table 1 and electronic supplementary material, tables S1 and S3).
The biological sex of our ancient samples was determined by comparing the genomic coverage of the X chromosome and of the autosomes [25]. The Y chromosome haplogroup of COV20126 was determined following [26] (electronic supplementary material, figure S2a and S2b).
(f). Phylogenetic analysis
We assembled two datasets: (i) an ancient dataset including complete mitogenome sequences from 194 prehistoric samples (17 new mitogenomes from this study), covering most of Europe, the Near East and Africa, and the sequence of a Neanderthal individual (Feld2, [27]) as outgroup; and (ii) a modern dataset including published mitogenomes from 388 individuals with European and African ancestry belonging to the L macro-haplogroup, our ancient sample COV20126, and an African individual of the L0 haplogroup as outgroup (electronic supplementary material, data S1). Details about the reconstruction of the calibrated phylogenies with BEAST 2.4.8 [28] can be found in the electronic supplementary material, document S1.
(g). Datasets for population genetic analysis
We called the autosomal SNPs included in the Human Origins chip [29] in the ancient samples sequenced at nuclear level. We used GATK-3.0-0 pileup for the base calling. At positions covered by more than one read, one allele was randomly chosen with a probability equal to the base frequency at that position. Then, the chosen alleles were duplicated to form homozygous genotypes. We used PLINK [30] to merge these calls to reference ancient and modern datasets from [31]. Also, we realigned bam files and mapped raw read data from recently published ancient remains [10,11,13,32,33]. We called the variants of the Human Origins chip in these alignments and merged the overlapping positions with our ancient samples and those in [31].
After merging, only ancient samples with more than 15 000 called SNPs were kept for downstream analysis. The final dataset included 269 ancient individuals and 1267 modern individuals from Eurasian, North and South African populations (electronic supplementary material, data S3).
(h). Population genetic analysis
For all population genetic analyses, transitions were removed from the datasets (110 532 SNPs left), as well as SNPs in linkage disequilibrium with r2 > 0.2 (79 130 SNPs left). We first performed a principal component analysis (PCA, EIGENSOFT), projecting ancient individuals onto the PC1–PC2 space defined by modern individuals from Eurasia and North Africa, using Procrustes analysis [34]. Model-based clustering of the ancient individuals, together with Eurasian, North and South African populations, was conducted with Admixture [35]. The genotype data were pruned for linkage disequilibrium using PLINK [30] with parameters—indep-pairwise 200 25 0.5 [36], resulting in 85 831 SNPs retained. We tested different numbers of clusters from K = 2 to 20. The results of 10 iterations per K were combined using CLUMPP [37] and plotted with Distruct [38] (see the electronic supplementary material, figures S4a and S4b).
We used outgroup f3 statistics to measure the amount of shared drift between the Eurasian samples in our Human Origins dataset since divergence from an African outgroup. We limited the analysis to samples sharing at least 10 000 SNP. The test was run using qp3Pop from the Admixtools package [29].
D-statistics were used to test for an excess similarity between the ancient Iberian samples and an African source with regard to other ancient samples [39]. To increase the power of the test, we used the whole-genome data of our four Iberian samples and a set of complete ancient genomes mapped to the hg19 reference. All individuals selected for this test have an average genome coverage ≥ 1×, except COV20126, with a genome coverage ≈ 0.4× (electronic supplementary material, data S2 and document S1). We computed D-statistics using the ABBA-BABA tool in ANGSD (version 0.920/0.921) [40]. The last two bases at both ends of the reads were trimmed, and mapping and base quality were set to 30, in order to minimize the possible effects of miscalling and aDNA molecular damage.
3. Results
(a). Laboratory procedures, sequencing and authenticity
A combination of shotgun sequencing and hybridization capture approaches allowed us to recover nuclear genome data for four ancient samples. Two individuals come from northern Portugal, LD270 (4.2× average genome coverage) and LD1174 (3.8×), both 14C dated to around 4400 calibrated years before present (cal yBP, Chalcolithic), and two from Cordoba in southern Spain, LU339 (4.8×) and COV20126 (0.4×), dated to 4889 ± 68 and 3637 ± 60 cal yBP, respectively (figure 1a and table 1). We also sequenced the complete mitochondrial genomes of these individuals and 13 additional ancient human remains from North and South Iberia, also dated to the Chalcolithic (electronic supplementary material, table S1). We could confirm the authenticity of the sequences, based on their deamination rates at the 5′ and 3′ ends of the reads and average read lengths between 50 and 80 bp (electronic supplementary material, figure S1). Based on mtDNA, we estimated contamination levels between 0.2 and 3.1% (0.1–1.4% when only transversions were considered); based on the X chromosome we estimated a ratio of 2–3% of contamination in COV20126, the only individual analysed at genome level that was identified as a male (table 1 and electronic supplementary material, table S1).
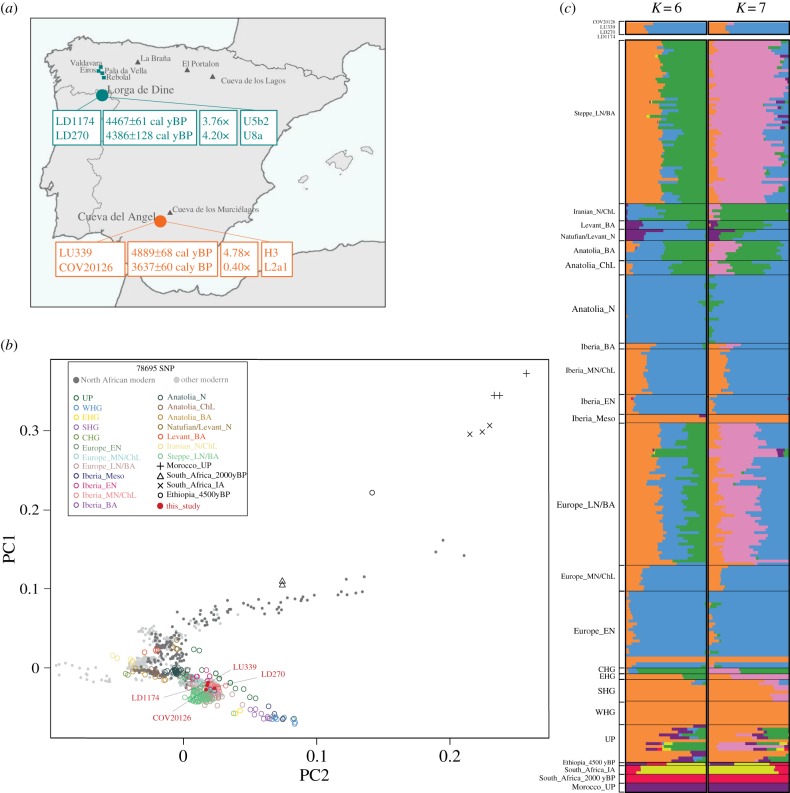
Figure 1.
Geographical and genetic information of the ancient Iberian samples. (a) Archaeological sites included in this study. Sites from which we sequenced complete nuclear genomes are indicated by circles; 14C age (calibrated years before present), average genome coverage and mtDNA haplogroup are reported. Blue squares indicate sampling sites of individuals sequenced only at mitogenome level. Ancient Iberian genomes from published studies included in our analyses are indicated by grey triangles. (b) Principal component analysis (PCA). The nuclear genomes of LD1174, LD270, LU339 and COV20126 were projected onto the first two principal components together with other modern and ancient African, Middle East and Eurasian population samples. (c) The WHG (orange) and Anatolian Neolithic (blue) are the major genome components in all our four Iberian samples, although at K = 7, a component (pink) associated with the Russian Steppes, is already visible in COV20126. UP: Upper Paleolithic; WHG, West hunter–gatherer; SHG, Scandinavian hunter–gatherer; EHG, East hunter–gatherer; CHG, Caucasian hunter–gatherer; EN, Early Neolithic; MN, Middle Neolithic; ChL, Chalcolithic; LN, Late Neolithic; N, Neolithic; BA, Bronze Age.
(b). Mitochondrial and Y chromosome analyses
Most individuals of our study belong to mitochondrial haplogroups previously described in Europe, such as U, H, K, J and V (table 1 and electronic supplementary material, tables S1 and S3) with one striking exception. COV20126, the 3600 yBP individual from Córdoba was assigned to L2a1 l, a typical sub-Saharan haplogroup, to our knowledge never described before in ancient individuals outside Africa. The restriction of the analysis to reads showing typical aDNA molecular damage [24], confirmed the assignation of COV20126 to haplogroup L2a1 l. We followed PhyloTree (http://www.phylotree.org/) to place COV20126 in a parsimonious phylogeny of the L2 haplogroup (electronic supplementary material, figure S5a) and confirmed its assignment to L2a1. Within this clade, the polymorphisms 16189C and 16192 T together with 534 T place COV20126 further down on the tree in subclade L2a1 l, although for a complete assignment to this subclade COV20126 is missing an A at position 143 and a C at position 195. On the same branch of the tree and differing from L2a1 l by only four substitutions, there is L2a1 k, a haplogroup that perhaps has evolved locally in Europe since 13 000 yBP [5,6]. We did not find exact matches to this ancient sequence in a database of modern haplogroup L mtDNAs (n > 2600 haplotypes). However, in present-day populations haplogroup L2a1 l is most frequent in West/West-Central Africa [41], but also in the Caribbean and USA, as a consequence of the transatlantic slave trade, [42]. Most remarkable, the sub-clade, L2a1l2a, comprising five different haplotypes, occurs only in modern DNA samples from Poland [43]. In addition, there are no members belonging to L2a1 l in present-day samples from North Africa.
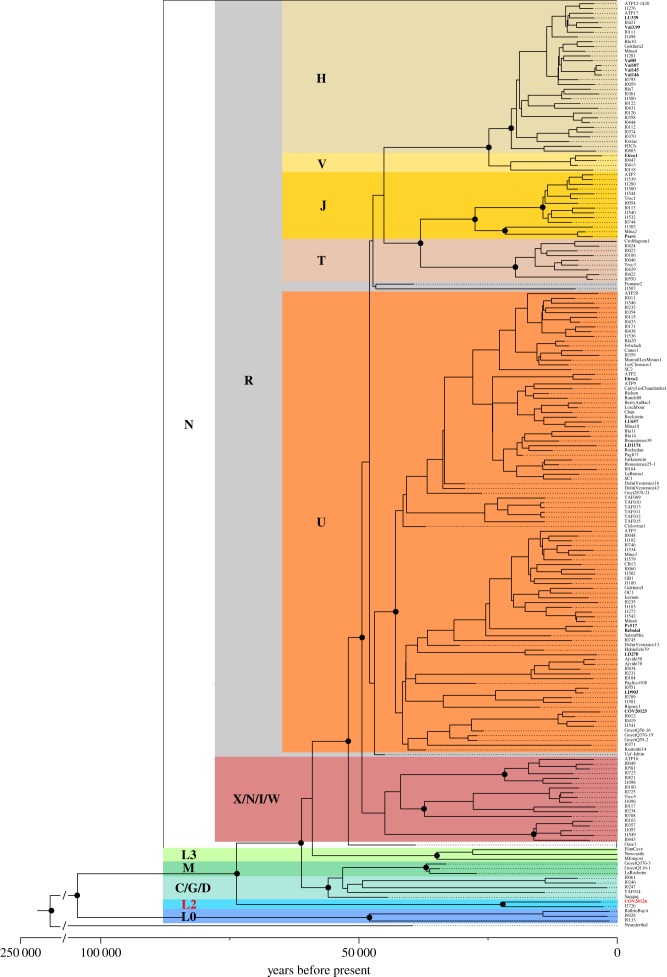
In the phylogeny of figure 2, built on only ancient mtDNAs, COV20126 clusters together with an ancient individual from Tanzania belonging to haplogroup L2a1 (I3 726, dated around 3100 yBP, [32]), basal to all the Eurasian clades (figure 2). Based only on the 14C dates of the individuals in our ancient genomes phylogeny, we estimated the divergence time between COV20126's clade and the Eurasian lineages at ca 73 000 yBP (95% highest posterior density (HPD): 63 400–83 800 yBP).
Figure 2.
Calibrated phylogeny of 194 ancient mitogenomes from Europe, Middle East and Africa. Major mitochondrial haplogroups are identified by different colours. The 17 samples from this study are labelled in bold. They all belong to mitochondrial haplogroups already described in ancient remains from Europe, with the exception of COV20126, which clusters together with an ancient individual from Tanzania (I3726, [32]) belonging to the sub-Saharan haplogroup L2.
The calibrated Bayesian analysis of modern mitochondrial genomes separates the L2 lineages into its major subclades (L2a, L2b, L2c and L2d) with high posterior probability (electronic supplementary material, figure S5b). The L2a branch is further divided into well-supported branches, and COV20126 is assigned to the L2a1 branch with high statistical confidence. The node linking COV20126 with the rest of the L2a1 clade is dated to ca 22 000 yBP (95% HPD: 17 300–28 200 yBP) using a substitution rate as in [44].
COV20126's Y chromosome belongs to the G2a haplogroup, described as typical of Early Neolithic farmers in Europe ([13,31,45,46]; electronic supplementary material, figure S2). Therefore, based on uniparental markers (mtDNA and Y chromosome), COV20126 seems to have ancestors from both Mediterranean shores; the question is to what extent the two ancestral populations contributed to the recombining part of his nuclear genome.
(c). Nuclear genome analysis
To investigate the position of our samples in the context of worldwide genetic diversity, we plotted on a PCA graph the genomes of the four ancient Iberian samples of this study, a subset of ancient samples from the Human Origins dataset [31,46], and recently published ancient genomes from Africa [11,32,33] and Spain ([10,13]; see the electronic supplementary material, data S3). All ancient individuals were projected onto the first two principal components defined by modern genomes (figure 1b). Our Iberian samples cluster together with other Chalcolithic Iberians. COV20126 is just slightly shifted on the plot towards samples previously described as carrying a Caucasian component in their genomes (ancient individuals from the Russian Steppes and Late Neolithic and BA samples from central Europe), and shows no obvious increased affinity with Africa.
Next, we investigated common ancestry among ancient genomes using Admixture and the SNP panel in the Human Origins dataset (figure 1c). Based on cross-validation error, the best-supported value of K is K = 6 (electronic supplementary material, figure S3). At K = 6, this analysis confirmed the similarity between the ancient Iberian samples and other Middle Neolithic and Chalcolithic individuals from Spain (figure 1c), showing the two well-known major genome components related with Western hunter–gatherers (WHG) and Anatolian farmers. At K = 7, a minor fraction of a genome component associated with the Caucasian hunter–gatherers (CHG) and Russian Steppes is evident in COV20126, as well as in two Spanish samples from the same BA period [13,47] (figure 1c; electronic supplementary material, data S3). The presence of this component only in COV20126, i.e. the most recent individual we analysed in the present study, is in agreement with previous work reporting a late arrival of the Pontic Steppe ancestry into the Iberian gene pool [9,46,48,49].
Ancient African samples were included in both analyses, but we did not observe any clear similarity between them, characterized by the red, yellow and purple components in figure 1c, and ancient Iberians, unless for a trace presence (around 0.02%) of sub-Saharan African (red and yellow) components in two Early Neolithic samples from Spain (mur and ATP19; [13]). In short, clustering analyses do not yield any obvious indication of genomic relationships between Africans and COV20126, which could corroborate the findings of the mtDNA analysis.
(d). D-statistic analysis of whole-genome data
The low coverage of some ancient individuals, COV20126 among them, strongly reduced the number of available markers in the previous analysis, and hence our power to detect subtle signals of remote admixture events. Therefore, we decided to use the whole-genome sequences we had generated to formally test for admixture with a sub-Saharan source.
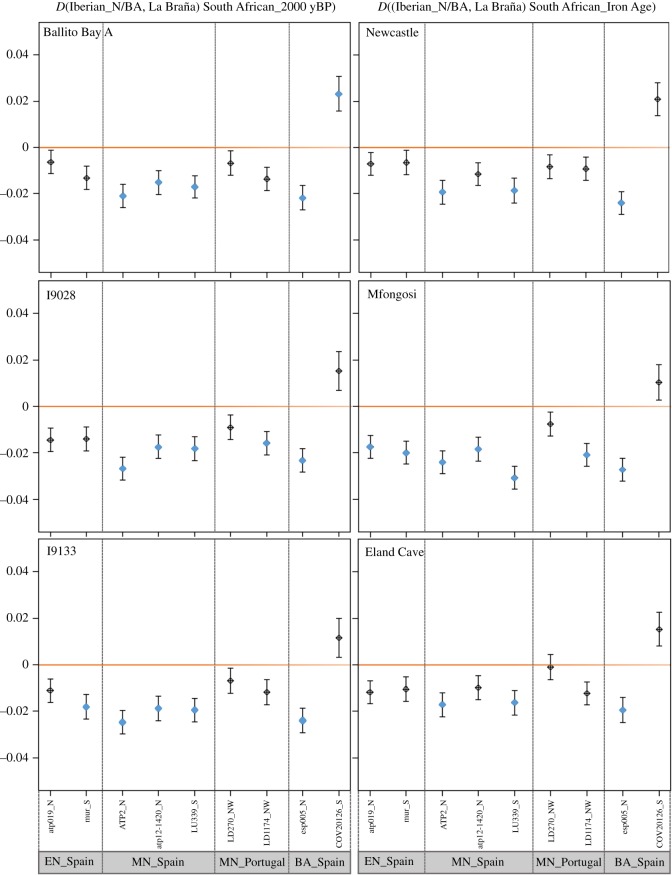
We ran D-statistics [39] in ANGSD [40] using the chimpanzee genome as outgroup. The null hypothesis was that, in the absence of gene flow from Africa, all ancient Iberian samples should form a single cluster, to the Africans' exclusion. Alternatively, African admixture in Iberia after the Mesolithic period would result in negative values of D. Thus, we formulated D-statistics of the form D((Iberian_N/BA, La Braña) African, chimpanzee) (figure 3), where Iberian_N/BA was represented in turn by each of the Early Neolithic, Middle Neolithic and BA samples (electronic supplementary material, data S2) and La Braña is a WHG from Northern Spain, known to have contributed genetically to post-Mesolithic populations in Western Europe [10,13,47,50].
Figure 3.
D-statistics of the form ((Iberian_N/BA, La Braña) African, chimpanzee). The D-statistic values (and standard errors) are given for each of the tests in which Iberian_N/BA was substituted by the Early Neolithic (EN), Middle Neolithic (MN) and Bronze Age (BA) individuals specified at the bottom of the graph. La Braña is a representative WHG from Spain, and the South African source was fixed in each set of comparisons to be one of the ancient sub-Saharan samples (dated around 2000 yBP on the left panels and around the Iron Age (approx. 500 yBP) on the right ones) in our whole-genome dataset (electronic supplementary material, data S2). D-values with ∣Z∣ ≥ 3 are highlighted in blue. We observe a trend to negative values of D, which is indicating gene flow from the African source into the Iberian post-Mesolithic samples. The only exception is COV20126, which is less similar to the African source than La Braña is. (Online version in colour.)
Rather surprisingly, the only positive set of D-values obtained from these analyses was observed for COV20126. Most remarkable is that, in contrast with the null hypothesis, we found most of the tests involving the other ancient Iberian genomes to yield negative values of D (figure 3 and Test1A in the electronic supplementary material, table S4), suggesting the presence of a subtle but significant African component. There is a general trend to negative D-values, with the Spanish Middle Neolithic and Chalcolithic samples showing greater (and statistically significant) similarities with the African genomes, than the Spanish Early Neolithic and Portuguese Middle Neolithic individuals (whose D-values are insignificant). A similar trend towards negative values of D was not observed when we repeated the test with a configuration of the form D((WHG, La Braña) African, chimpanzee) (Test1B in the electronic supplementary material, table S4), neither with a time series of ancient samples from east Europe (WHG and post-Mesolithic samples from Hungary and Romania, Test2 in the electronic supplementary material, table S4). In the latter case, D-values fluctuated around 0, with no visible difference between earlier and later individuals.
In order to test whether the lack of African ancestry in COV20126 could be explained by its lower genome coverage (0.4×) compared with other samples (greater than or equal to 1×), we artificially diminished the quality of the other BA individual (esp005) in our D-statistics test. We subsampled its genome and increased the presence of contaminant sequences to similar values of those in COV20126 (0.4× of genome coverage and ≈ 2,5% of contamination). When we repeated the D-test, despite such modifications, the low-quality version of esp005's genome was still giving signals of African admixture (electronic supplementary material, figure S6). Thus, it seems the low genome coverage cannot by itself explain the apparent absence of African admixture in COV20126's nuclear sequences.
(e). Outgroup f3 statistics
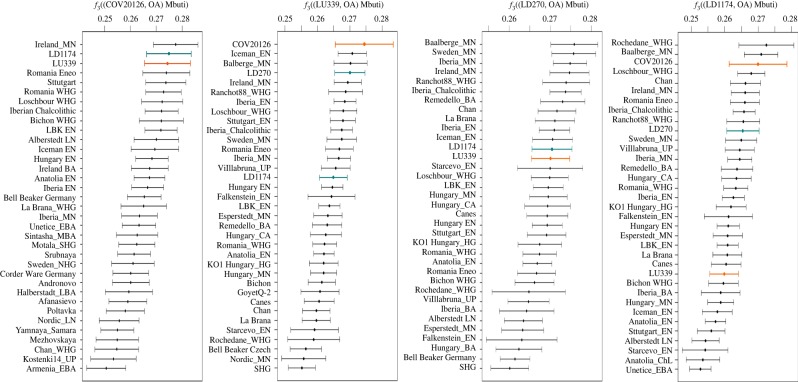
We measured by outgroup f3 statistics the amount of shared genetic drift between pairs of Eurasian samples after separation from an African outgroup (Mbuti) (figure 4; electronic supplementary material, data S4). This analysis confirmed that all our four samples have the highest levels of shared genetic history with other Middle and Early Neolithic samples from Central Europe and Spain, and with Basques and Sardinians among modern populations (electronic supplementary material, figure S7). However, we detected differences concerning the WHG component. While the Spanish Mesolithic Chan and La Braña are within the WHG that share the most genetic drift with the Portuguese LD1174 and LD270 (f3 = 0.271), they are less related with the samples from southern Spain (f3 values around 0.25 for LU339 and COV20126), which share a higher genetic drift with hunter–gatherers from France and Luxembourg than with the Spanish ones (figure 4). This could indicate pre-existing genetic structure within the Iberian hunter-gatherer populations, even though the small differences among the outgroup f3 values call for caution in their interpretation.
Figure 4.
Outgroup f3 statistics. Outgroup f3 statistics as (ancient1, ancient2; Mbuti), where ancient1 is one individual from our study (COV20126, LU339, LD270 and LD1174), and ancient2 is in turn each of the other ancient (OA) samples in the Human Origins dataset. Middle Neolithic samples from Portugal share more genetic drift with Chan (Mesolithic samples from northwest Spain), than the Andalusian COV20126 and LU339 do. Samples from this study are highlighted in orange (southern Iberia) and blue (northern Iberia). (Online version in colour.)
4. Discussion
In this study, we found indisputable evidence of the presence of a mitochondrial sequence of sub-Saharan African origin in a 3600 years-old sample, COV20126, from southern Spain. Considering the absence of any closely related mitogenomes in prehistoric Europe, it is difficult to explain this finding by a process other than cross-Mediterranean gene flow before the BA.
Although COV20216's nuclear genome showed no obvious traces of African admixture, several other samples from Iberia did; in particular, relatively late samples (from the Middle Neolithic and Chalcolithic) collected along the Mediterranean area and on the Spanish plateau. The increased, significant similarity to sub-Saharan African samples shown by these individuals is not matched, as far as we could test, elsewhere in Europe (figure 3 and electronic supplementary material, table S4).
Also, we detected a higher African affinity in the Middle Neolithic Spanish samples than in the Portuguese ones, or in individuals from earlier periods. This fits well with archaeological data, reporting similarities in Neolithic tools and pottery decoration (Almagra and the impressed Oran) between the Andalusian and North African shores [51,52].
Taken together, these findings can be explained by at least one episode of gene flow from Africa to southern Iberia, which apparently did not reach (or had a smaller impact on) the northwest Atlantic fringe. The exact date of this episode is difficult to define with confidence. In principle, if it happened in Middle Neolithic times (i.e. a little more than 3600 years ago, which is COV20126 age), its consequences should be evident in the clustering analysis of samples from that period and geographical area, which was not the case. An alternative possibility is that gene flow may have occurred even earlier in Southern Iberia from a population with Sub-Saharan African features, which left some genetic contribution in the genomes of the people, the local hunter–gatherers, they admixed with. Because hunter–gatherer genomes from Southern Spain are not available yet, the consequences of such gene flow become apparent to us only in samples from Middle Neolithic times, in parallel with the reemergence of the local hunter–gatherer component of ancient European genomes [9,46,47]. This hypothesis implies the existence of some north–south genetic structure in pre-Neolithic Iberia, with hunter–gatherers from the south showing a stronger resemblance with sub-Saharan Africans, and it would account for all findings of the present study, as well as for those of previous studies of modern DNA [1,7]. However, to safely discriminate between the two hypotheses, we would need Mesolithic samples from southern Spain, which are at present unavailable.
Whatever the date of gene flow from Africa might be, the presence of African affinities in several individuals, including a clearly African mitogenome, shows that one or more contacts occurred between prehistoric Iberian populations and a population whose features we can describe as sub-Saharan. Whether that happened in a single episode, or two (accounting for the African affinities in, respectively, Spanish MN/ChL samples, and COV20126), we cannot tell at present. In both cases, the episode(s) we detected left a small, but not negligible, mark at the population level, contributing to a fraction, if minor, of the Iberian gene pool.
With regards to the lack of this signal in the nuclear genome of COV20126, it may be related with his younger age compared to the other samples in the D-statistic test and, possibly, with his geographical origin. COV20126's age (3600 yBP) implies a higher number of generations separating him from the time of the admixture event, which could have further diluted the African component within his genome. Additionally, his geographical proximity to the Mediterranean coast (a major point of arrival of prehistoric migrations [53]), may have increased the presence of DNA variants from different sources in COV20126's ancestors, in comparison with other BA people from the more isolated northern inland regions (esp005). Also, we cannot exclude that technical reasons related with the capture enrichment could have introduced some bias in COV20126's genomic data. The commercial DNA we used to build the probes comes from anonymous donors and the manufacturer does not provide information about their geographical origin. If most of this DNA is of European ancestry, it may have favoured the capture of European-like, rather than African-like, sequences in COV20126's genome.
Finally, our study highlights the informative value of mtDNA as a marker of demographic events, which may be difficult to recognize at the genomic level. Indeed, in the long run, recombination is expected to blur the signals of past admixture events, which may instead be preserved in non-recombining DNA fragments (even though the latter are subjected to a stronger impact of genetic drift). Also, these results show that genetic population structure is often complex, and hence broad generalizations about vast territories or long periods of time are unwarranted, if not adequately supported by detailed geographical and archaeological data. Particularly, in the Iberian Peninsula, the Spanish plateau and the Cantabrian Mountains seem to have played a major role, reducing the possibility of ancient contacts between the Atlantic and Mediterranean watersheds.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Prof. Tom Gilbert and Dr Michael Westbury for their advice on laboratory procedures and statistical analysis. We acknowledge support from Science for Life Laboratory (SciLifeLab) and the Swedish National Genomics Infrastructure (NGI) for assistance in massively parallel sequencing.
Data accessibility
The complete mitogenome sequences are available from GenBank (MK321329-MK321345). The bam files with the genome alignments are available in ENA (PRJEB29189).
Authors' contributions
G.G.F. and G.B. conceived the study. G.G.F., G.B., M.H. and A.M. planned the experiments and the analysis strategy. G.G.F., K.H., J.L.A.P., D.D.M. and H.S. carried out molecular laboratory work. G.G.F., F.T. and E.T. carried out the analysis of genetic data. A.S. and R.R.S. provided support for the analysis and interpretation of mitochondrial DNA data. C.B.R., F.J.B., C.B.M., A.M.S.B., H.A.S., A.G.D., A.d.L-H., R.F.V., M.V., S.A., M.L., X.R.A. and C.F.R. provided samples and input about archaeological context. G.G.F., G.B., M.H., A.M. and E.T. wrote the manuscript with input from all co-authors.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by a Marie Sklodowska-Curie Individual Fellowship to G.G.F (NeoGenHeritage, grant no. 655478); by the European Research Council (ERC) Advanced grant 295733-LanGeLin and the consolidator grant 310763-GeneFlow to G.B. and M.H., respectively; by the research project BIOGEOS (CGL2014-57209-P) of the Spanish MINECO to A.G.; and by the research project HAR2010-21786/HIST of the Spanish MINECO and Xunta de Galicia to R.F.V., M.V., A.L.-H. and X.P.R.-A. A.M. was supported by the European Research Council Consolidator grant 647787—LocalAdaptation.
References
- 1.Auton A, et al. 2009. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 19, 795–803. ( 10.1101/gr.088898.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moorjani P, et al. 2011. The history of African gene flow into southern Europeans, Levantines, and Jews. PLoS Genet. 7, e1001373 ( 10.1371/journal.pgen.1001373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botiguè LR, et al. 2013. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl Acad. Sci. USA 110, 11 791–11 796. ( 10.1073/pnas.1306223110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruciani F, et al. 2007. Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 24, 1300–1311. ( 10.1093/molbev/msm049) [DOI] [PubMed] [Google Scholar]
- 5.Malyarchuk BA, Derenko M, Perkova M, Grzybowski T, Vanecek T, Lazur J. 2008. Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs. Eur. J. Hum. Genet. 16, 1091–1096. ( 10.1038/ejhg.2008.70) [DOI] [PubMed] [Google Scholar]
- 6.Cerezo M, et al. 2012. Reconstructing ancient mitochondrial DNA links between Africa and Europe. Genome Res. 22, 821–826. ( 10.1101/gr.134452.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardiñas AF, Martínez JL, Roca A, García-Vazquez E, López B. 2014. Over the sands and far away: interpreting an Iberian mitochondrial lineage with ancient western African origins. Am. J. Hum. Biol. 26, 777–7783. ( 10.1002/ajhb.22601) [DOI] [PubMed] [Google Scholar]
- 8.Lazaridis I, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413. ( 10.1038/nature13673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haak W, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211. ( 10.1038/nature14317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González-Fortes G, et al. 2017. Paleogenomic evidence for multi-generational mixing between Neolithic farmers and Mesolithic hunter-gatherers in the Lower Danube Basin. Curr. Biol. 27, 1801–1810. ( 10.1016/j.cub.2017.05.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Loosdrecht M, et al. 2018. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552. ( 10.1126/science.aar8380) [DOI] [PubMed] [Google Scholar]
- 12.Fregel R, et al. 2018. Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe. Proc. Natl Acad. Sci. USA 115, 6774–6779. ( 10.1073/pnas.1800851115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdiosera C, et al. 2018. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc. Natl Acad. Sci. USA 115, 3428–3433. ( 10.1073/pnas.1717762115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohland N, Siedel H, Hofreiter M. 2010. A rapid column-based ancient DNA extraction method for increased sample throughput. Mol. Ecol. Resour. 10, 677–683. ( 10.1111/j.1755-0998.2009.02824.x) [DOI] [PubMed] [Google Scholar]
- 15.Dabney J, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15 758–15 763. ( 10.1073/pnas.1314445110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer M, Kircher M.. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. t5448 ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 17.Gansauge MT, Meyer M. 2013. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748. ( 10.1038/nprot.2013.038) [DOI] [PubMed] [Google Scholar]
- 18.Fortes GG, Paijmans JLA. 2015. Analysis of whole mitogenomes from ancient samples. In Whole genome amplification (ed. Kroneis T.), pp. 179–195. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 19.Maricic T, Whitten M, Pääbo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE 5, e14004 ( 10.1371/journal.pone.0014004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 ( 10.1038/13779) [DOI] [PubMed] [Google Scholar]
- 23.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 14, 193–202. ( 10.1093/bib/bbs012) [DOI] [PubMed] [Google Scholar]
- 24.Skoglund P, et al. 2014. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344, 747–750. ( 10.1126/science.1253448) [DOI] [PubMed] [Google Scholar]
- 25.Skoglund P, Storå J, Götherström A, Jakobsson M. 2013. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482. ( 10.1016/j.jas.2013.07.004) [DOI] [Google Scholar]
- 26.van Oven M, Van Geystelen A, Kayser M, Decorte R, Larmuseau MH.. 2014. Seeing the wood for the trees: a minimal reference phylogeny for the human Y chromosome. Hum. Mutat. 35, 187–191. ( 10.1002/humu.22468) [DOI] [PubMed] [Google Scholar]
- 27.Briggs AW, et al. 2009. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321. ( 10.1126/science.1174462) [DOI] [PubMed] [Google Scholar]
- 28.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. 2012. Ancient admixture in human history. Genetics 192, 1065–1093. ( 10.1534/genetics.112.145037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazaridis I, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424. ( 10.1038/nature19310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skoglund P, et al. 2017. Reconstructing prehistoric African population structure. Cell 171, 59–71. ( 10.1016/j.cell.2017.08.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlebusch CM, et al. 2017. Southern African ancient genomes estimate modern human divergence to 350 000 to 260 000 years ago . Science 358, 652–655. ( 10.1126/science.aao6266) [DOI] [PubMed] [Google Scholar]
- 34.Wang C, et al. 2010. Comparing spatial maps of human population-genetic variation using Procrustes analysis. Stat. Appl. Genet. Mol. Biol. 9, 13 ( 10.2202/1544-6115.1493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. ( 10.1101/gr.094052.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tassi F, et al. 2017. Genome diversity in the Neolithic Globular Amphorae culture and the spread of Indo-European languages. Proc. R. Soc. B. 284, 20171540 ( 10.1098/rspb.2017.1540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. ( 10.1093/bioinformatics/btm233) [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg NA. 2004. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138. ( 10.1046/j.1471-8286.2003.00566.x) [DOI] [Google Scholar]
- 39.Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328, 710–722. ( 10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinf. 15, 356 ( 10.1186/s12859-014-0356-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz T, Pala M, Gómez-Carballa A, Richards MB, Salas A. 2017. Updating the African human mitochondrial DNA tree: relevance to forensic and population genetics. For. Sci. Int. Genet. 27, 156–159. ( 10.1016/j.fsigen.2016.12.016) [DOI] [PubMed] [Google Scholar]
- 42.Salas A, et al. 2004. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am. J. Hum. Genet. 74, 454–465. ( 10.1086/382194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa MD, et al. 2013. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nat. Commun. 4, 2543 ( 10.1038/ncomms3543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posth C, et al. 2016. Pleistocene mitochondrial genomes suggest a single major dispersal of non-Africans and a Late Glacial population turnover in Europe. Curr. Biol. 26, 827–833. ( 10.1016/j.cub.2016.01.037) [DOI] [PubMed] [Google Scholar]
- 45.Keller A, et al. 2012. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 3, 698 ( 10.1038/ncomms1701) [DOI] [PubMed] [Google Scholar]
- 46.Mathieson I, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503. ( 10.1038/nature16152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Günther T, et al. 2015. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. USA 112, 11 917–11 922. ( 10.1073/pnas.1509851112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allentoft ME, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522, 167–172. ( 10.1038/nature14507) [DOI] [PubMed] [Google Scholar]
- 49.Olalde I, et al. 2015. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK Cultures. Mol. Biol. Evol. 32, 3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olalde I, et al. 2014. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228. ( 10.1038/nature12960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortés SM, et al. 2012. The Mesolithic–Neolithic transition in southern Iberia. Quat. Res. 77, 221–234. ( 10.1016/j.yqres.2011.12.003) [DOI] [Google Scholar]
- 52.Linstädter J, Medved I, Solich M, Weniger GC. 2012. Neolithisation process within the Alboran territory: models and possible African impact. Quat. Int. 274, 219–232. ( 10.1016/j.quaint.2012.01.013) [DOI] [Google Scholar]
- 53.Isern N, et al. 2017. Modeling the role of voyaging in the coastal spread of the Early Neolithic in the West Mediterranean. Proc. Natl Acad. Sci. USA 114, 897–902. ( 10.1073/pnas.1613413114) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete mitogenome sequences are available from GenBank (MK321329-MK321345). The bam files with the genome alignments are available in ENA (PRJEB29189).