Abstract
The central aim of conservation biology is to understand and mitigate the effects of human activities on biodiversity. To successfully achieve this objective, researchers must take an interdisciplinary approach that fully considers the effects, both direct and indirect, of anthropogenic disturbances on wildlife physiology and health. A recent surge in research has revealed that host-associated microbiota—the archaeal, bacterial, fungal and viral communities residing on and inside organisms—profoundly influence animal health, and that these microbial communities can be drastically altered by anthropogenic activities. Therefore, conservation practitioners should consider the disruption of host-associated microbial diversity as a serious threat to wildlife populations. Despite the tremendous potential for microbiome research to improve conservation outcomes, few efforts have been made to truly integrate these fields. In this review, we call for the microbial renaissance of conservation biology, where biodiversity of host-associated microbiota is recognized as an essential component of wildlife management practices. Using evidence from the existing literature, we will examine the known effects of anthropogenic activities on the diversity of host-associated microbial communities and integrate approaches for maintaining microbial diversity to successfully achieve conservation objectives.
Keywords: microbiome, conservation, wildlife
1. Introduction
Biodiversity—generally defined as the variety of life, genetic material and functional traits—is essential for long-term ecosystem stability [1]. Unfortunately, anthropogenic activities have resulted in dramatic losses of biodiversity worldwide, thereby threatening the functioning of ecosystems, their ability to support robust ecological communities and their resistance to environmental change [1,2]. The primary objectives of conservation biology are to evaluate anthropogenic impacts on biodiversity and to develop practical approaches to prevent the extinction of species [3,4]. To successfully achieve these objectives, we must take an interdisciplinary research approach that fully considers the effects, both direct and indirect, of anthropogenic disturbances on wildlife physiology and health [5–7].
A recent surge in research has demonstrated that host-associated microbiota—the archaeal, bacterial, fungal and viral communities residing on and inside organisms—profoundly influence host health through their impacts on the immune system, digestion, development and even behaviour (reviewed in [8–11]). Host-associated microbial communities are governed by the same ecological principles shaping macro-ecological systems (reviewed in [12]), most notably the influence of extrinsic environmental factors on community composition. The susceptibility of these communities to environmental factors suggests that anthropogenic habitat disturbances (e.g. deforestation, pollution and urbanization) may adversely affect wildlife fitness and survival through disruption of the physiological and performance-related benefits of microbiota, but this mechanism has not been sufficiently explored.
Despite a widely recognized need for microbiome research to be placed in a more ecological context [13–15], especially as it applies to wildlife conservation [16,17], few efforts have been made to truly integrate these fields, especially in a way that might actually address current management practices. In this review, we call for the microbial renaissance of conservation biology, where biodiversity of host-associated microbiota is recognized as an essential component of wildlife management practices. To justify this perspective, we will use evidence from the existing literature to examine (1) the effects of anthropogenic activities on the diversity of host-associated microbiota, (2) approaches for maintaining this microbial diversity, and (3) how conservation practitioners and microbiome researchers can work together to achieve conservation objectives.
2. Threats to host-associated microbial biodiversity
Conservation biologists have identified land-use change, environmental contamination, climate change and infectious disease as some of the most pressing and pervasive threats to biodiversity on our planet [2]. Building on a vast body of literature that demonstrates the direct effect of these threats on wildlife populations, recent research has revealed that these same factors may indirectly affect host health by altering their associated microbial communities. In box 1, we highlight several representative studies to demonstrate how each threat has been shown to affect these communities across a variety of host taxa. In the context of wildlife management, the impact of captivity on host-associated microbial communities is well described, and thus has been included as a threat to microbial biodiversity. While our literature search was not systematic, we have included an expanded reference list (electronic supplementary material, table S1) to further demonstrate the commonplace nature of these effects. Below, we discuss in more detail (1) the mechanisms that may be responsible for the impacts of each threat on microbial communities, and (2) the potential consequences of these alterations for host health, survival and fitness.
Box 1. Documenting the threats to host-associated microbial communities.a.
| land-use change | |
 |
— habitat fragmentation is associated with reductions in diversity and altered community composition of the mammalian gut [18,19] and amphibian skin microbiomes [20] |
| — urbanization decreases bacterial richness and alters the community composition and functional profile of the bird gut microbiome [21] | |
| — amphibians living in agricultural habitats have distinct gut microbial community composition from those living in natural habitats [22] | |
| contamination | |
 |
— heavy metal exposure decreases diversity and alters the community composition of the fish gut microbiome [23] |
| — exposure to polychlorinated biphenyls alters the community composition of the larval amphibian gut microbiome, which persists in the adult life stage [24] | |
| — pesticide and herbicide use are, respectively, linked to compositional changes in the gut microbiome of insects [25] and the skin microbiome of larval and adult amphibians [26] | |
| climate change | |
 |
— increased temperature results in losses of diversity and alterations to the community composition of reptile [27] and amphibian [28] gut microbiomes |
| — warming ocean temperature alters the assemblage of the microbial communities associated with marine sponges [29] | |
| — ocean acidification reduces diversity and alters the community composition of coral associated microbes [30] | |
| infectious disease | |
 |
— parasite infection can decrease diversity of the bird gut microbiome [31], and alter community composition of the mammalian [32–34] and amphibian [35] gut microbiomes |
| — viral infections alter the community composition of the bird gut microbiome [36] and amphibian skin microbiome [37] | |
| — cutaneous fungal infection alters skin microbial communities of amphibians [38], the degree of which increases with pathogen load [39] | |
| captivity | |
 |
— gut microbial communities of fish [40], reptiles [41], birds [42] and mammals [43–47], and skin microbial communities of amphibians [48,49] and reptiles [50], are distinct between captive individuals and their wild conspecifics |
| — mammalian gut microbial communities [47] and amphibian skin microbial communities [48,51] are less diverse in captivity than in the wild |
aRepresentative studies were selected in an effort to incorporate the widest variety of host taxa and specific threats. An expanded reference list further documenting these threats is available in electronic supplementary material, table S1.
(a). Land-use change
Anthropogenic land-use change may primarily alter the gut microbiome through dietary mechanisms, such as shifting food availability, quality or diet composition in degraded habitats. For example, black howler monkeys (Alouatta pigra) living in fragmented forests consume less diverse diets of lower quality than their conspecifics in continuously forested habitats [19]. Lack of variety in available substrates for microbial digestion may explain the concurrent reductions in gut microbiota diversity observed in these animals [19]. Effects of dietary perturbations on gut microbial communities may be less pronounced in generalist host species that can readily exploit new food sources [52], thus buffering these species from anthropogenic habitat degradation, whereas taxa with more specialized foraging ecology may be more vulnerable [18]. Associations between skin microbial community alterations and land-use change are likely to be due to shifts in the availability of microbes that can colonize hosts, as environmental microbial communities can be affected by anthropogenic land-use changes [53]. These patterns could also be driven by increased stress among hosts living in degraded habitats, which can alter host-associated microbial communities [54].
Loss of gut microbiota, whose functions are particularly important for their hosts, may underlie some of the fitness costs incurred by animals occupying disturbed habitats. Amato et al. [19] demonstrated that gut microbial communities of black howler monkeys living in fragmented forests are depleted of microbes that produce butyrate—a short-chain fatty acid that functions as the primary energy source for mammalian colon cells [55]. Thus, loss of these taxa may disrupt energy homeostasis in the gut, with negative consequences for host health. Furthermore, in comparison with conspecifics occupying continuously forested habitat, the gut microbial communities of threatened red colobus monkeys (Procolobus gordonorum) inhabiting fragmented forests harbour significantly fewer microbial taxa involved in the degradation of tannins—toxic xenobiotics present at high abundance in the diet of these folivorous primates [18]. Loss of the ability to detoxify their primary food sources may reduce host fitness.
(b). Environmental contamination
Environmental contaminants can alter host-associated microbial communities through displacement of native bacterial taxa by those capable of withstanding chronic exposure to toxic compounds. For example, the gut of isopods collected from mercury-contaminated habitats harboured significantly greater numbers of mercury-resistant bacteria when compared to those from uncontaminated sites [56]. Similarly, bacteria obtained from the skin of frogs living in habitats contaminated with acid mine drainage exhibited significantly greater tolerance when challenged with toxic effluent in the laboratory than bacteria isolated from frogs living at multiple uncontaminated reference sites [57]. Additionally, contamination alters the composition of environmental microbial communities [58], therefore impacting the reservoir of microbes available in these habitats for ingestion into the gut or inoculation onto the skin.
Environmental contamination may impact host health directly through the loss of functionally important microbes. Honeybees (Apis mellifera) exposed to pesticides harbour gut microbiomes depleted of sugar metabolism and protease activities [25]. These functions are critical for nectar processing, and their loss may therefore lead to downstream effects on honeybee health. The negative physiological consequences of taxa loss may be compounded by the ability of gut microbiota to transform contaminants into more toxic metabolites within the body [59]. For example, Pinyayev et al. [60] reported the transformation of a heavy metal, arsenate, into toxic oxyarsenicals and thioarsenicals by anaerobic bacteria in the mouse caecum, increasing the bioavailability, and potentially the toxicity, of this compound.
(c). Climate change
The impacts of climate change (increased temperature, acidification, etc.) can directly affect microbial reservoirs available for host colonization by altering environmental microbial communities [58,61]. Additionally, these impacts may also indirectly alter host-associated microbial communities through their effects on host physiology. For example, as corals become stressed by warming and acidifying conditions, they can release antibacterial compounds [62], which may impact the diversity or composition of their associated microbial communities. In amphibians, elevated temperature accelerates skin sloughing, which can ultimately reduce the abundance of cultivable cutaneous microbes by up to 100% [63].
Alterations to host-associated microbial communities induced by climate change may negatively impact host health through the breakdown of symbiotic relationships or the loss of important microbial functions. A 2.5°C increase from ambient temperatures dramatically decreased abundance of an obligate insect gut bacterial symbiont, resulting in reduced host growth and body size [64]. In the salamander gut microbiome, Fontaine et al. [28] reported the simultaneous reduction in abundance of protective bacteria, with increases in abundance of potentially pathogenic taxa at elevated temperatures. Corals stressed by ocean warming and acidification have microbiomes that are distinct from healthy hosts, and experience concurrent physiological declines including reductions in calcification rates and algal carbohydrate concentrations [30]. In such cases, the loss of microbial symbionts may not recover even after conditions stabilize. In sponges, Ramsby et al. [29] demonstrated a loss of important microbial taxa at increased water temperatures, and a failure of individuals to regain these symbionts after temperature returned to baseline levels. Furthermore, bacterial taxa that are retained may still lose important functions. For example, protective microbes on amphibian skin lose their ability to inhibit growth of the pathogenic fungus Batrachochytrium dendrobatidis (Bd) at high temperatures [65]. These negative consequences for host health may ultimately scale up to impact animal fitness, as Bestion et al. [27] reported a correlation between the loss of microbial community diversity under simulated climate change conditions and reductions in animal survival.
(d). Infectious disease
Direct interactions between pathogens and symbionts is one mechanism by which infectious disease can impact host-associated microbial communities. In rodents, the gut microbial communities of individuals infected with a parasitic helminth are remarkably similar to those of the adult worms themselves, indicating the potential for transfer of foreign microbes from parasite to host [34]. Furthermore, infection with different species of helminths yields distinct changes to the gut microbiome, indicating that microbiota can respond to the identity of the specific pathogen [32]. Infection by Bd, a fungal pathogen with cell walls composed primarily of chitin, results in opportunistic increases of chitin-degrading bacteria on amphibian skin [38]. Infectious disease may also influence host-associated microbial communities through indirect mechanisms, such as host behavioural changes. In birds, Knutie [31] reported an increase in nest provisioning by parents of offspring infected with parasitic larvae, thereby affecting nestling gut microbial community composition, possibly through a change in food quantity [66].
Disease-altered host-associated microbial communities may disrupt normal physiological functioning, ultimately impacting host health. Knutie [31] demonstrated that birds infected with parasitic larvae exhibit reduced gut microbial diversity, which is linked to diminished antibody levels in these individuals. Through this feedback loop, initial parasitic infection may impact microbial communities such that these individuals are at an increased risk for future infections. In Coquerel's sifakas (Propithecus coquereli), infection with the protozoan Cryptosporidium reduces overall diversity of the gut microbiome, while enriching taxa known to be associated with enteric dysfunction in humans, including inflammation and shortened gut transit time [33]. These obligate folivores depend on long gut transit times and microbial processes to fully extract dietary nutrients, and therefore the resulting effects on host digestion could be debilitating. Furthermore, after infection clearance, the microbiome was slow to recover to its original state, indicating the potential for long-lasting physiological consequences [33].
(e). Captivity
Captivity can profoundly alter host-associated microbiota through a variety of mechanisms, including transitions from natural food sources to less diverse or compositionally different diets, a reduction in environmental microbial reservoirs, co-habitation with other species and antibiotic administration. Clayton et al. [43] reported that non-human primate species harbour distinct gut microbiomes in the wild, but these communities become similar in captivity due to reductions in dietary fibre content. In red-eyed tree frogs (Agalychnis callidryas), dietary carotenoid availability is associated with community composition and diversity of the skin microbiome, and captive individuals fed a carotenoid-enhanced diet harbour a richer community than those fed a carotenoid-free diet [67]. Captive diets may have a greater impact on the microbial communities of specialist host species as opposed to generalists. In woodrats, a dietary specialist (Neotoma stephensi) lost a greater proportion of native gut microbiota in captivity as compared with a generalist (Neotoma albigula) [44]. Furthermore, co-habitation in captivity can homogenize microbiomes between individuals and species [68]. For example, the skin microbial communities of two fruit bat species (Artibeus jamaicensis and Carollia perspicillata) housed together in captivity were more similar to one another than conspecifics housed separately [69]. Including a natural microbial reservoir in captivity may help to ameliorate some of these effects. For example, housing captive red-backed salamanders (Plethodon cinereus) in soil from their natural environment enabled greater retention of native microbes on the skin as opposed to conspecifics in typical laboratory media [49].
Lack of a robust, native microbial community may underlie poor animal health in captivity, and the low success rate of some reintroduction programmes. For example, cheetahs (Acinonyx jubatus) experience reduced reproductive rates and increased mortality in captivity [45]. The most common cause of mortality is bacterial infection [70], which may be due to the significant increases of pathogenic taxa and the enrichment of disease-associated pathways in the functional profile of gut microbiota in captivity [45]. Similarly, in the endangered western capercaillie (Tetrao urogallus), captive individuals exhibit increased richness of bacterial taxa associated with intestinal dysfunction, which has been suggested as a factor contributing to failed reintroductions [42]. Attempts to eliminate problematic taxa with antibiotics may actually result in further adverse health outcomes as these compounds also reduce the abundance of beneficial microbiota [71]. For example, Chlamydia infections are highly prevalent in captive koalas (Phascolarctos cinereus), which are treated with antibiotics. Dahlhausen et al. [46] report that individuals that underwent antibiotic therapy and subsequently died harboured gut microbial communities reduced in diversity and abundance of tannin-detoxifying bacteria essential to koala nutrition and survival. Finally, captive environments may reduce populations of beneficial host-associated microbes directly, with downstream effects on host health. For example, the skin microbiome of boreal toads (Anaxyrus boreas) reared in captivity lacks the diversity of Bd-inhibiting bacteria harboured by wild conspecifics [51]. Upon experimental inoculation with Bd, 100% of captive individuals became infected, which may have been driven by their lack of protective bacteria, as survival in these individuals increased by 40% after subsequent treatment with a probiotic [51].
(f). Summary
Although different mechanisms may be responsible for their effects, all aforementioned threats are capable of affecting the diversity, community composition and function of host-associated microbial communities. While many studies have correlated these impacts with changes in host physiology and impaired health, identification of causal links between specific microbial perturbations, reduced animal fitness and population level declines will be essential to preserve host-associated microbial biodiversity as a means to achieve desired conservation objectives.
3. Maintaining host-associated microbial biodiversity to achieve conservation objectives
The advent of culture-independent microbial inventories and metagenomics enables the characterization of ‘core’ microbial taxa and genes across animal hosts, and the investigation of these potentially essential functions in an ecological and evolutionary context. It is thought that host-associated microbiota provide the means for rapid ecological adaptation in response to changes in local environmental conditions [72]. Therefore, it may be useful to expand the focus of conservation biology beyond preserving taxonomic and genetic diversity to include host-associated microbial diversity. This may be challenging, as geographically separated animal populations may harbour distinct microbial communities that are adapted to their local environments [73]. Additionally, microbial communities and their associated functions change over the lifetime of hosts [74]. The geographical and temporal specificity of host–microbe interactions should be taken into consideration when conducting animal translocations or reintroductions, as there may be mismatching between an individual's microbiome and its new environment [73] or between native and introduced conspecifics [75].
Host-associated microbial communities could also provide valuable baseline data for the detection of dysbioses (health problems caused by an imbalance in gut microbiota) caused by environmental disturbances, or as a new metric for assessing the efficacy of habitat restorations. The idea of using the microbiome as an indicator or index of host health has received attention in some systems [76]. However, even in the medical field it is currently extremely challenging to recognize or identify a ‘healthy’ baseline microbial community, distinguish between cause and effect in community changes, and determine whether these changes have functional consequences for the host [77]. Therefore, conservation practitioners and microbiome researchers must work together to identify reliable microbial indicators that reflect the specific conservation needs of the host.
As discussed above, host-associated microbial communities are often altered by captivity, which may hinder the success of species recovery programmes. However, these effects may be ameliorated by practices that minimize the most influential aspects of captivity and foster a more diverse microbiome. Natural exposure to environmental microbial reservoirs is thought to be important for maintaining microbial diversity, either through competition with immigrant microbiota or through regular colonization of specific bacteria [49]. Among captive animals, the inclusion of natural substrates (e.g. sand, water or access to natural habitat) has been shown to foster more abundant and diverse microbial communities [49,78]. It is important to note, however, that these approaches may pose health risks if microbial reservoirs contain pathogenic micro-organisms or chemical contaminants. Therefore, care must be taken in the selection of candidate microbial reservoirs for captive wildlife.
Hosts themselves can also serve as microbial reservoirs via contact with sympatric individuals [79], or even intra-individually across host tissues through self-inoculation [80]. Facilitating these interactions through conspecific co-habitation has been shown to increase microbial transmission between individuals and improve microbial community diversity [81], and thus could help to maintain natural microbiota and ameliorate the negative effects of captivity. Parental contact in early life may provide offspring with bacteria essential for major developmental events [82], and thus depriving captive individuals of natural interactions may disrupt necessary microbial transmission and be detrimental to the fitness of reintroduced wildlife. Indeed, early-life disruptions to microbial communities have been shown to influence the long-term trajectory of microbial communities [74], which may, in turn, affect the microbial community assembly of offspring via pre-natal transmission [83].
Because host diet largely determines the availability of energetic substrates that select for certain microbial taxa [84], care should be taken when designing captive diets. Greene et al. [78] demonstrated that the composition and function of gut microbiota among captive lemurs was rapidly altered by diet, and that more complex diets foster diverse gut microbial communities. It is thought that gut microbiota are also influenced by microbes contained on or within food items [85], suggesting that diets closely resembling those of wild conspecifics may help to maintain natural microbial communities among captive individuals. Furthermore, certain dietary nutrients have also been shown to foster abundant and taxonomically rich cutaneous bacterial communities in amphibians, possibly due to changes in dermal mucus production or immune function [67,86]. Therefore, breeding programmes should consider the potential impacts of industrially processed diets, and instead provide foods similar to those found in their native habitat to perhaps mitigate the loss of native microbiota.
Bioaugmentation, via probiotic therapy or microbiome transplantation, has emerged as a promising new strategy for mitigating disease risk and improving the health of both captive and wild animals [87]. For example, application of the probiotic Janthinobacterium lividum to the skin of amphibians decreases morbidity and mortality associated with Bd infections [51,88]. The positive effects of bioaugmentation appear to persist for several months after inoculation [88], and beneficial microbiota may even self-propagate through wildlife populations via inter-individual transmission [79]; however, the long-term stability of bioaugmented microbiota is unknown. Regardless, these results suggest that bioaugmentation of gut microbiota may also provide health benefits (figure 1), and thus conservation practitioners should consider implementing this technique in imperilled wildlife based on well-defined conservation objectives (box 2). While the potential benefits of bioaugmentation on wildlife management are exciting, less than 2% of all probiotic research has been directed at ecological applications [93]. For example, the success of bioaugmentation may be dependent on host [94] and environmental factors [95], thus some conditions may be more optimal for bioaugmentation than others. The effective transplantation of microbiota may sometimes require a reduction in resident microbes via antibiotic treatment [87], which may eliminate essential microbiota and lead to dysbioses. In any case, rigorous characterization and screening of candidate microbial taxa must be conducted to ensure that manipulation will not result in off-target effects on native microbial communities or adverse health outcomes. It is important to note that any manipulations of host gut microbial communities should be independently evaluated by veterinarians and institutional animal use committees to mitigate potential negative impacts on host health and to ensure that such procedures are acceptable under federal laws.
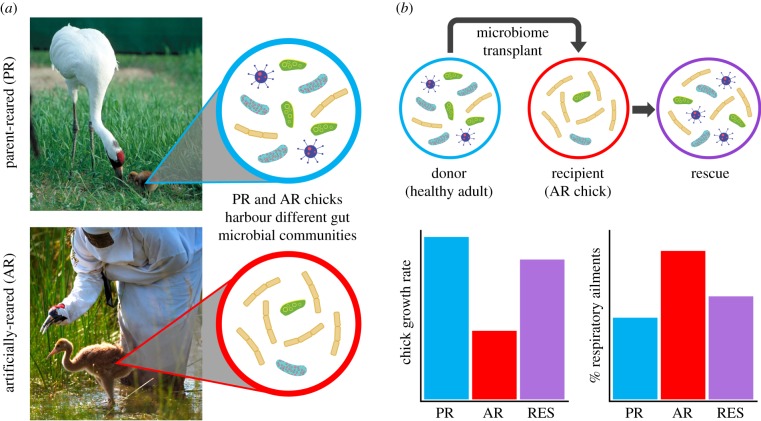
Figure 1.
Integrating microbiome and conservation research to enhance recovery of the endangered whooping crane. (a) The microbiomes of parent-reared and artificially reared crane chicks differ in gut microbiota composition. (b) Artificially reared crane chicks have reduced survival and increased rate of respiratory ailments, but transplanting the microbiome from healthy adult cranes into artificially reared chicks may ameliorate adverse health outcomes.
Box 2. Integrating microbiome and conservation research to enhance species recovery.
Many threatened and endangered species undergo captive breeding programmes to facilitate species recovery. For over 50 years, the captive rearing and reintroduction of the endangered whooping crane (Grus americana) has been necessary to prevent extinction and accelerate recovery of this once abundant migratory bird. While the reintroduction of captive reared chicks has helped to increase the number of whooping cranes in the wild, two on-going reintroductions are not yet self-sustaining, and one remnant reintroduced population in Florida is likely to be extinct within the next few years.
The recovery of whooping cranes is primarily limited by fledging success and hatch year survival [89]. Therefore, conservation scientists, often wearing crane costumes to avoid imprinting (figure 1a), have resorted to artificially rearing crane chicks to increase the likelihood of survival and enhance crane population recovery. However, artificially reared whooping crane chicks often exhibit high rates of respiratory ailments, slower growth rates, reduced survival and poor reductive output in the wild.
Can microbial transplants improve the health of artificially reared chicks?
Studies in other crane species have demonstrated that artificial rearing reduces the diversity of the gut microbiome in chicks [90]. This reduced gut microbial diversity during development may have life-long impacts on crane health (figure 1a). It is possible that adverse health outcomes may be ameliorated through microbial transfers. In general, the process is to collect donor material, dilute with saline, homogenize, filter to remove particulate matter and inoculate into a recipient via oral gavage [91]. Donor material can then be frozen and thawed for later administration [92]. For whooping cranes, faecal samples from healthy adults could be collected and inventoried using bacterial 16S rRNA sequencing, then gavaged to artificially reared chicks within the first few days of life. This approach could ‘rescue’ the microbiome of artificially reared chicks, thereby improving their health and fitness following reintroduction into the wild (figure 1b).
4. Working together: how conservation biologists and microbiome researchers can combine efforts to aid conservation
Bridging the fields of conservation biology and microbiome science will require communication and collaboration between experts in each field. However, this process involves numerous challenges, such as clarifying definitions that vary across fields, or communicating objectives and limitations for each side [7]. For example, if a captive individual of an endangered species is experiencing symptoms of a pathogenic bacterial infection, it may be treated with a broad-spectrum antibiotic in accordance with the standard procedures of the conservation institution. While this antibiotic treatment may complicate the interpretation of microbiome data, it is the opinion of the authors that conservation practices and objectives should take priority when designing experiments and collecting samples. That being said, such a treatment might also open up the opportunity for collaboration. For example, microbiome scientists can analyse how standard antibiotic treatment impacts the microbiome of captive animals [46], the results of which can be used by conservation practitioners to help weigh the costs and benefits of subsequent treatment.
The objective of this review is to highlight the relevance, possibilities and potential benefits of microbiome research for the field of conservation. Despite the challenges associated with integrating microbiome research into current wildlife management practices, conservation biologists and microbiome scientists have much to offer each other. Conservation biologists often have great familiarity with the problems, challenges and opportunities that exist within their systems, and may have specific research objectives already in mind that may relate to the microbiome (e.g. the effect of land use practices on the gut microbiome of wild animals). In return, microbiome scientists can provide technical expertise while relaying the limitations of microbiome science, thereby circumventing tendencies to overhype results [96]. Regardless of the study objective, experimental design is one of the most important factors influencing the downstream results and interpretations of microbiome studies [97]. Therefore, it is imperative that conservation biologists and microbiome scientists communicate early and often throughout collaborations, and be flexible and receptive to the needs of each side [7]. If implemented properly, the merging of ideas and techniques from these two fields can produce novel and meaningful results with the potential to increase our scientific understanding while advancing the field of wildlife conservation.
Supplementary Material
Data accessibility
This article has no additional data.
Authors' contributions
Co-first authors B.K.T. and S.S.F. conducted the literature review and wrote the majority of the paper. B.K.H. and K.D.K. assisted with writing, provided oversight and comments and edited the manuscript.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Cardinale BJ, et al. 2012. Biodiversity loss and its impact on humanity. Nature 486, 59 ( 10.1038/nature11148) [DOI] [PubMed] [Google Scholar]
- 2.Secretariat of the Convention on Biological Diversity (CBD). 2010. Global biodiversity outlook 3. Montreal, Canada: CBD Secretariat.
- 3.Wilson EO. 1992. The diversity of life. Cambridge, MA: The Belknap Press of Harvard University Press. [Google Scholar]
- 4.Soulé ME. 1985. What is conservation biology? BioScience 35, 727–734. ( 10.2307/1310054) [DOI] [Google Scholar]
- 5.Cooke SJ, Sack L, Franklin CE, Farrell AP, Beardall J, Wikelski M, Chown SL. 2013. What is conservation physiology? Perspectives on an increasingly integrated and essential science. Conserv. Physiol. 1, cot001 ( 10.1093/conphys/cot001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wikelski M, Cooke SJ. 2006. Conservation physiology. Trends Ecol. Evol. 21, 38–46. ( 10.1016/j.tree.2005.10.018) [DOI] [PubMed] [Google Scholar]
- 7.Campbell LM. 2005. Overcoming obstacles to interdisciplinary research. Conserv. Biol. 19, 574–577. ( 10.1111/j.1523-1739.2005.00058.x) [DOI] [Google Scholar]
- 8.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenney EA, Koelle K, Dunn RR, Yoder AD. 2018. The ecosystem services of animal microbiomes. Mol. Ecol. 27, 2164–2172. ( 10.1111/mec.14532) [DOI] [PubMed] [Google Scholar]
- 10.Kohl KD, Carey HV. 2016. A place for host–microbe symbiosis in the comparative physiologist's toolbox. J. Exp. Biol. 219, 3496–3504. ( 10.1242/jeb.136325) [DOI] [PubMed] [Google Scholar]
- 11.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260 ( 10.1038/nrg3182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. ( 10.1126/science.1224203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amato KR. 2013. Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microbiome Sci. Med. 1, 10–29. ( 10.2478/micsm-2013-0002) [DOI] [Google Scholar]
- 14.Antwis RE, et al. 2017. Fifty important research questions in microbial ecology. FEMS Microbiol. Ecol. 93, fix044. ( 10.1093/femsec/fix044) [DOI] [PubMed] [Google Scholar]
- 15.Hird SM. 2017. Evolutionary biology needs wild microbiomes. Front. Microbiol. 8, 725 ( 10.3389/fmicb.2017.00725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redford KH, Segre JA, Salafsky N, Martinez del Rio C, McAloose D. 2012. Conservation and the microbiome. Conserv. Biol. 26, 195–197. ( 10.1111/j.1523-1739.2012.01829.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahrndorff S, Alemu T, Alemneh T, Lund Nielsen J. 2016. The microbiome of animals: implications for conservation biology. Int. J. Genomics 2016, 5304028 ( 10.1155/2016/5304028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barelli C, et al. 2015. Habitat fragmentation is associated to gut microbiota diversity of an endangered primate: implications for conservation. Sci. Rep. 5, 14862 ( 10.1038/srep14862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amato KR, et al. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353. ( 10.1038/ismej.2013.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker C, Longo A, Haddad C, Zamudio K. 2017. Land cover and forest connectivity alter the interactions among host, pathogen and skin microbiome. Proc. R. Soc. B 284, 20170582 ( 10.1098/rspb.2017.0582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teyssier A, Rouffaer LO, Hudin NS, Strubbe D, Matthysen E, Lens L, White J. 2018. Inside the guts of the city: urban-induced alterations of the gut microbiota in a wild passerine. Sci. Total Environ. 612, 1276–1286. ( 10.1016/j.scitotenv.2017.09.035) [DOI] [PubMed] [Google Scholar]
- 22.Chang CW, Huang BH, Lin SM, Huang CL, Liao PC. 2016. Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol. 16, 33 ( 10.1186/s12866-016-0660-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J, Lu L, Jin C, Wang S, Zhou J, Ni Y, Fu Z, Jin Y. 2018. Effects of short term lead exposure on gut microbiota and hepatic metabolism in adult zebrafish. Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 209, 1–8. ( 10.1016/j.cbpc.2018.03.007) [DOI] [PubMed] [Google Scholar]
- 24.Kohl KD, Cary TL, Karasov WH, Dearing MD. 2015. Larval exposure to polychlorinated biphenyl 126 (PCB-126) causes persistent alteration of the amphibian gut microbiota. Environ. Toxicol. Chem. 34, 1113–1118. ( 10.1002/etc.2905) [DOI] [PubMed] [Google Scholar]
- 25.Kakumanu ML, Reeves AM, Anderson TD, Rodrigues RR, Williams MA. 2016. Honey bee gut microbiome is altered by in-hive pesticide exposures. Front. Microbiol. 7, 1255 ( 10.3389/fmicb.2016.01255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krynak KL, Burke DJ, Benard MF. 2017. Rodeo™ herbicide negatively affects Blanchard's Cricket Frogs (Acris blanchardi) survival and alters the skin-associated bacterial community. J. Herpetol. 51, 402–410. ( 10.1670/16-092) [DOI] [Google Scholar]
- 27.Bestion E, Jacob S, Zinger L, Di Gesu L, Richard M, White J, Cote J. 2017. Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat. Ecol. Evol. 1, 0161 ( 10.1038/s41559-017-0161) [DOI] [PubMed] [Google Scholar]
- 28.Fontaine SS, Novarro AJ, Kohl KD. 2018. Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J. Exp. Biol. 221, jeb. 187559 ( 10.1242/jeb.187559) [DOI] [PubMed] [Google Scholar]
- 29.Ramsby BD, Hoogenboom MO, Whalan S, Webster NS. 2018. Elevated seawater temperature disrupts the microbiome of an ecologically important bioeroding sponge. Mol. Ecol. 27, 2124–2137. ( 10.1111/mec.14544) [DOI] [PubMed] [Google Scholar]
- 30.Grottoli AG, et al. 2018. Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS ONE 13, e0191156 ( 10.1371/journal.pone.0191156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knutie SA. 2018. Relationships among introduced parasites, host defenses, and gut microbiota of Galapagos birds. Ecosphere 9, e02286 ( 10.1002/ecs2.2286) [DOI] [Google Scholar]
- 32.Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. 2015. Interactions between multiple helminths and the gut microbiota in wild rodents. Phil. Trans. R. Soc. B 370, 20140295 ( 10.1098/rstb.2014.0295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenney EA, Greene LK, Drea CM, Yoder AD. 2017. Down for the count: cryptosporidium infection depletes the gut microbiome in Coquerel's sifakas. Microb. Ecol. Health Dis. 28, 1335165 ( 10.1080/16512235.2017.1335165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walk ST, Blum AM, Ewing SA-S, Weinstock JV, Young VB. 2010. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm. Bowel Dis. 16, 1841–1849. ( 10.1002/ibd.21299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu Y, Hong P, Tang D, Qing H, Omondi Donde O, Wang H, Xiao B, Wu H. 2018. Comparison of intestinal microbes in female and male Chinese concave-eared frogs (Odorrana tormota) and effect of nematode infection on gut bacterial communities. MicrobiologyOpen e749 ( 10.1002/mbo3.749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganz HH, Doroud L, Firl AJ, Hird SM, Eisen JA, Boyce WM. 2017. Community-level differences in the microbiome of healthy wild mallards and those infected by Influenza A viruses. MSystems 2, e00188-e00116 ( 10.1128/mSystems.00188-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell LJ, Hammond SA, Price SJ, Sharma MD, Garner TW, Birol I, Helbing CC, Wilfert L, Griffiths AG. 2018. A novel approach to wildlife transcriptomics provides evidence of disease-mediated differential expression and changes to the microbiome of amphibian populations. Mol. Ecol. 27, 1413–1427. ( 10.1111/mec.14528) [DOI] [PubMed] [Google Scholar]
- 38.Rebollar EA, Hughey MC, Medina D, Harris RN, Ibáñez R, Belden LK. 2016. Skin bacterial diversity of Panamanian frogs is associated with host susceptibility and presence of Batrachochytrium dendrobatidis. ISME J. 10, 1682 ( 10.1038/ismej.2015.234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jani AJ, Briggs CJ. 2014. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl Acad. Sci. USA 111, E5049–E5058. ( 10.1073/pnas.1412752111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhanasiri AK, Brunvold L, Brinchmann MF, Korsnes K, Bergh Ø, Kiron V. 2011. Changes in the intestinal microbiota of wild Atlantic cod Gadus morhua L. upon captive rearing. Microb. Ecol. 61, 20–30. ( 10.1007/s00248-010-9673-y) [DOI] [PubMed] [Google Scholar]
- 41.Kohl KD, Brun A, Magallanes M, Brinkerhoff J, Laspiur A, Acosta JC, Caviedes-Vidal E, Bordenstein SR. 2017. Gut microbial ecology of lizards: insights into diversity in the wild, effects of captivity, variation across gut regions and transmission. Mol. Ecol. 26, 1175–1189. ( 10.1111/mec.13921) [DOI] [PubMed] [Google Scholar]
- 42.Wienemann T, Schmitt-Wagner D, Meuser K, Segelbacher G, Schink B, Brune A, Berthold P. 2011. The bacterial microbiota in the ceca of Capercaillie (Tetrao urogallus) differs between wild and captive birds. Syst. Appl. Microbiol. 34, 542–551. ( 10.1016/j.syapm.2011.06.003) [DOI] [PubMed] [Google Scholar]
- 43.Clayton JB, et al. 2016. Captivity humanizes the primate microbiome. Proc. Natl Acad. Sci. USA 113, 10 376–10 381. ( 10.1073/pnas.1521835113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohl KD, Skopec MM, Dearing MD. 2014. Captivity results in disparate loss of gut microbial diversity in closely related hosts. Conserv. Physiol. 2, cou009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menke S, Melzheimer J, Thalwitzer S, Heinrich S, Wachter B, Sommer S. 2017. Gut microbiomes of free-ranging and captive Namibian cheetahs: Diversity, putative functions and occurrence of potential pathogens. Mol. Ecol. 26, 5515–5527. ( 10.1111/mec.14278) [DOI] [PubMed] [Google Scholar]
- 46.Dahlhausen K, Doroud L, Firl A, Polkinghorne A, Eisen J. 2018. Characterization of shifts of koala (Phascolarctos cinereus) intestinal microbial communities associated with antibiotic treatment. PeerJ 6, e4452 ( 10.7717/peerj.4452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenzie V, et al. 2017. The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57, 690–704. ( 10.1093/icb/icx090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker MH, Richards-Zawacki CL, Gratwicke B, Belden LK. 2014. The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki). Biol. Conserv. 176, 199–206. ( 10.1016/j.biocon.2014.05.029) [DOI] [Google Scholar]
- 49.Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, Harris RN. 2014. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 8, 830 ( 10.1038/ismej.2013.200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyde ER, et al. 2016. The oral and skin microbiomes of captive komodo dragons are significantly shared with their habitat. MSystems 1, e00046–e00016 ( 10.1128/mSystems.00046-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kueneman JG, Woodhams DC, Harris R, Archer HM, Knight R, McKenzie VJ. 2016. Probiotic treatment restores protection against lethal fungal infection lost during amphibian captivity. Proc. R. Soc. B 283, 20161553 ( 10.1098/rspb.2016.1553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menke S, Meier M, Mfune JK, Melzheimer J, Wachter B, Sommer S. 2017. Effects of host traits and land-use changes on the gut microbiota of the Namibian black-backed jackal (Canis mesomelas). FEMS Microbiol. Ecol. 93, fix123 ( 10.1093/femsec/fix123) [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Marshall CW, Cheng M, Xu H, Li H, Yang X, Zheng T. 2017. Changes in land use driven by urbanization impact nitrogen cycling and the microbial community composition in soils. Sci. Rep. 7, 44049 ( 10.1038/srep44049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noguera JC, Aira M, Pérez-Losada M, Domínguez J, Velando A. 2018. Glucocorticoids modulate gastrointestinal microbiome in a wild bird. R. Soc. open sci. 5, 171743 ( 10.1098/rsos.171743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. 2011. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526. ( 10.1016/j.cmet.2011.02.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lapanje A, Zrimec A, Drobne D, Rupnik M. 2010. Long-term Hg pollution-induced structural shifts of bacterial community in the terrestrial isopod (Porcellio scaber) gut. Environ. Pollut. 158, 3186–3193. ( 10.1016/j.envpol.2010.07.001) [DOI] [PubMed] [Google Scholar]
- 57.Costa S, Lopes I, Proença DN, Ribeiro R, Morais PV. 2016. Diversity of cutaneous microbiome of Pelophylax perezi populations inhabiting different environments. Sci. Total Environ. 572, 995–1004. ( 10.1016/j.scitotenv.2016.07.230) [DOI] [PubMed] [Google Scholar]
- 58.Fields MW, Yan T, Rhee S-K, Carroll SL, Jardine PM, Watson DB, Criddle CS, Zhou J. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid–uranium waste. FEMS Microbiol. Ecol. 53, 417–428. ( 10.1016/j.femsec.2005.01.010) [DOI] [PubMed] [Google Scholar]
- 59.Claus SP, Guillou H, Ellero-Simatos S. 2016. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2, 16003 ( 10.1038/npjbiofilms.2016.3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinyayev TS, Kohan MJ, Herbin-Davis K, Creed JT, Thomas DJ. 2011. Preabsorptive metabolism of sodium arsenate by anaerobic microbiota of mouse cecum forms a variety of methylated and thiolated arsenicals. Chem. Res. Toxicol. 24, 475–477. ( 10.1021/tx200040w) [DOI] [PubMed] [Google Scholar]
- 61.Zogg GP, Zak DR, Ringelberg DB, White DC, MacDonald NW, Pregitzer KS. 1997. Compositional and functional shifts in microbial communities due to soil warming. Soil Sci. Soc. Am. J. 61, 475–481. ( 10.2136/sssaj1997.03615995006100020015x) [DOI] [Google Scholar]
- 62.Geffen Y, Rosenberg E. 2005. Stress-induced rapid release of antibacterials by scleractinian corals. Mar. Biol. 146, 931–935. ( 10.1007/s00227-004-1505-5) [DOI] [Google Scholar]
- 63.Meyer EA, Cramp RL, Hernando Bernal M, Franklin CE. 2012. Changes in cutaneous microbial abundance with sloughing: possible implications for infection and disease in amphibians. Dis. Aquat. Organ. 101, 235–242. ( 10.3354/dao02523) [DOI] [PubMed] [Google Scholar]
- 64.Kikuchi Y, Tada A, Musolin DL, Hari N, Hosokawa T, Fujisaki K, Fukatsu T. 2016. Collapse of insect gut symbiosis under simulated climate change. MBio 7, e01578-e01516 ( 10.1128/mBio.01578-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muletz-Wolz CR, Almario JG, Barnett SE, DiRenzo GV, Martel A, Pasmans F, Zamudio KR, Toledo LF, Lips KR. 2017. Inhibition of fungal pathogens across genotypes and temperatures by amphibian skin bacteria. Front. Microbiol. 8, 1551 ( 10.3389/fmicb.2017.01551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohl KD, Amaya J, Passement CA, Dearing MD, McCue MD. 2014. Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol. Ecol. 90, 883–894. ( 10.1111/1574-6941.12442) [DOI] [PubMed] [Google Scholar]
- 67.Antwis RE, Haworth RL, Engelmoer DJ, Ogilvy V, Fidgett AL, Preziosi RF. 2014. Ex situ diet influences the bacterial community associated with the skin of red-eyed tree frogs (Agalychnis callidryas). PLoS ONE 9, e85563 ( 10.1371/journal.pone.0085563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song SJ, et al. 2013. Cohabiting family members share microbiota with one another and with their dogs. elife 2, e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemieux-Labonté V, Tromas N, Shapiro BJ, Lapointe F-J. 2016. Environment and host species shape the skin microbiome of captive neotropical bats. PeerJ 4, e2430 ( 10.7717/peerj.2430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Terio K, Munson L, Marker L, Aldridge B, Solnick J. 2005. Comparison of Helicobacter spp. in cheetahs (Acinonyx jubatus) with and without gastritis. J. Clin. Microbiol. 43, 229–234. ( 10.1128/JCM.43.1.229-234.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pérez-Cobas AE, et al. 2012. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62, 1591–1601. ( 10.1136/gutjnl-2012-303184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP. 2016. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. ( 10.1016/j.tree.2016.06.008) [DOI] [PubMed] [Google Scholar]
- 73.Kohl KD, Varner J, Wilkening JL, Dearing MD. 2018. Gut microbial communities of American pikas (Ochotona princeps): evidence for phylosymbiosis and adaptations to novel diets. J. Anim. Ecol. 87, 323–330. ( 10.1111/1365-2656.12692) [DOI] [PubMed] [Google Scholar]
- 74.Knutie SA, Wilkinson CL, Kohl KD, Rohr JR. 2017. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 8, 86 ( 10.1038/s41467-017-00119-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. 2010. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 201009906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Apprill A, et al. 2014. Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals? PLoS ONE 9, e90785 ( 10.1371/journal.pone.0090785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shreiner AB, Kao JY, Young VB. 2015. The gut microbiome in health and in disease. Curr. Opin Gastroenterol. 31, 69 ( 10.1097/MOG.0000000000000139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greene LK, McKenney EA, O'Connell TM, Drea CM. 2018. The critical role of dietary foliage in maintaining the gut microbiome and metabolome of folivorous sifakas. Sci. Rep. 8, 14482 ( 10.1038/s41598-018-32759-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rebollar EA, Simonetti SJ, Shoemaker WR, Harris RN. 2016. Direct and indirect horizontal transmission of the antifungal probiotic bacterium Janthinobacterium lividum on green frog (Lithobates clamitans) tadpoles. Appl. Environ. Microbiol. 82, 2457–2466. ( 10.1128/AEM.04147-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiggins PJ, Smith JM, Harris RN, Minbiole KP. 2011. Gut of red-backed salamanders (Plethodon cinereus) may serve as a reservoir for an antifungal cutaneous bacterium. J. Herpetol. 45, 329–332. ( 10.1670/10-231.1) [DOI] [Google Scholar]
- 81.Nelson TM, Rogers TL, Carlini AR, Brown MV. 2013. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145. ( 10.1111/1462-2920.12022) [DOI] [PubMed] [Google Scholar]
- 82.Williams CL, Dill-McFarland KA, Sparks DL, Kouba AJ, Willard ST, Suen G, Brown AE. 2018. Dietary changes during weaning shape the gut microbiota of red pandas (Ailurus fulgens). Conserv. Physiol. 6, cox075 ( 10.1093/conphys/cox075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trevelline BK, MacLeod KJ, Knutie SA, Langkilde T, Kohl KD. 2018. In ovo microbial communities: a potential mechanism for the initial acquisition of gut microbiota among oviparous vertebrates. Biol. Lett. 14, 20180225 ( 10.1098/rsbl.2018.0225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Louis P, Scott KP, Duncan SH, Flint HJ. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102, 1197–1208. ( 10.1111/j.1365-2672.2007.03322.x) [DOI] [PubMed] [Google Scholar]
- 85.Kohl KD, Dearing MD. 2014. Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ. Microbiol. Rep. 6, 191–195. ( 10.1111/1758-2229.12118) [DOI] [PubMed] [Google Scholar]
- 86.Edwards CL, Byrne PG, Harlow P, Silla AJ. 2017. Dietary carotenoid supplementation enhances the cutaneous bacterial communities of the critically endangered southern corroboree frog (Pseudophryne corroboree). Microb. Ecol. 73, 435–444. ( 10.1007/s00248-016-0853-2) [DOI] [PubMed] [Google Scholar]
- 87.Bletz MC, Loudon AH, Becker MH, Bell SC, Woodhams DC, Minbiole KP, Harris RN. 2013. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820. ( 10.1111/ele.12099) [DOI] [PubMed] [Google Scholar]
- 88.Harris RN, et al. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3, 818 ( 10.1038/ismej.2009.27) [DOI] [PubMed] [Google Scholar]
- 89.Converse SJ, Strobel BN, Barzen JA. 2018. Reproductive failure in the eastern migratory population: the interaction of research and management. In Whooping cranes: biology and conservation (eds French JB, Converse SJ, Austin JA), pp. 161–178. Cambridge, MA: Academic Press. [Google Scholar]
- 90.Xie Y, Xia P, Wang H, Yu H, Giesy JP, Zhang Y, Mora MA, Zhang X. 2016. Effects of captivity and artificial breeding on microbiota in feces of the red-crowned crane (Grus japonensis). Sci. Rep. 6, 33350 ( 10.1038/srep33350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Borody TJ, Paramsothy S, Agrawal G. 2013. Fecal microbiota transplantation: indications, methods, evidence, and future directions. Curr. Gastroenterol. Rep. 15, 337 ( 10.1007/s11894-013-0337-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A. 2012. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 107, 761–767. ( 10.1038/ajg.2011.482) [DOI] [PubMed] [Google Scholar]
- 93.McKenzie VJ, Kueneman JG, Harris RN. 2018. Probiotics as a tool for disease mitigation in wildlife: insights from food production and medicine. Ann. NY Acad. Sci. 1429, 18–30. ( 10.1111/nyas.13617) [DOI] [PubMed] [Google Scholar]
- 94.Kohl KD, Cary TL, Karasov WH, Dearing MD. 2013. Restructuring of the amphibian gut microbiota through metamorphosis. Environ. Microbiol. Rep. 5, 899–903. ( 10.1111/1758-2229.12092) [DOI] [PubMed] [Google Scholar]
- 95.Williams CL, Willard S, Kouba A, Sparks D, Holmes W, Falcone J, Williams CH, Brown A. 2013. Dietary shifts affect the gastrointestinal microflora of the giant panda (Ailuropoda melanoleuca). J. Anim. Physiol. Anim. Nutr. (Berl) 97, 577–585. ( 10.1111/j.1439-0396.2012.01299.x) [DOI] [PubMed] [Google Scholar]
- 96.Hanage WP. 2014. Microbiome science needs a healthy dose of scepticism: to guard against hype, those interpreting research on the body's microscopic communities should ask five questions. Nature 512, 247–249. ( 10.1038/512247a) [DOI] [PubMed] [Google Scholar]
- 97.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. 2014. Conducting a microbiome study. Cell 158, 250–262. ( 10.1016/j.cell.2014.06.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.