Isolation of splenic macrophage subpopulations using an enzyme cocktail
Keywords: enzymatic dissociation, marginal zone macrophages, marginal zone metallophilic macrophages
Abstract
Tissue-resident macrophages in the spleen, including red pulp and white pulp macrophages, marginal zone macrophages (MZMs) and marginal zone metallophilic macrophages (MMMs), are highly heterogeneous as a consequence of adaptation to tissue-specific environments. Each macrophage sub-population in the spleen is usually identified based on the localization, morphology and membrane antigen expression by immunohistochemistry. However, their phenotypical and functional characteristics remain incompletely understood due to the difficulty of identification and isolation by flow cytometry. We used a cocktail of three enzymes (Collagenase D, Dispase I and DNase I), rather than traditional mechanical grinding, for isolation of each sub-population, which resulted in significant improvement of isolation of these macrophage sub-populations, particularly MZMs and MMMs, as determined by CD11bhiF4/80medTim4hi and CD11bhiF4/80medTim4med, respectively. This method should be helpful for molecular and functional characterization of each splenic resident macrophage sub-population.
Introduction
Tissue-resident macrophages play crucial roles in both tissue homeostasis and inflammation. Their functions and phenotypes are highly heterogeneous as a consequence of adaptation to tissue-specific environments (1, 2). Splenic macrophages show heterogeneity in their functions as well (3). The sub-populations of the macrophages in the spleen are defined by anatomical compartments of the red and white pulp regions separated by the marginal zone. Red pulp and white pulp (also named tingible body) macrophages mediate clearance of defective blood cells and external antigens, respectively (3, 4). On the other hand, the marginal zone area contains distinct sub-populations of sialic-acid binding immunoglobulin-like lectin 1 (CD169)+ macrophages and macrophage receptor with collagenous structure (MARCO)+ DC-Sign-related protein 1 (SIGNR1, CD209b)+ macrophages, which are called marginal zone metallophilic macrophages (MMMs) and marginal zone macrophages (MZMs), respectively. MMMs and MZMs share a close developmental and functional relationship. The development of both macrophages is regulated by the nuclear receptor LXR1α (5), and they play a key role in immune surveillance and adaptive immune responses by capturing blood borne antigens (3, 6) and apoptotic cell phagocytosis (efferocytosis) (7, 8). Nevertheless, mice deficient in marginal zone B cells also lack MZMs but not MMMs, implying that the developmental pathway of MZMs is different from that of MMMs (9, 10).
Each macrophage sub-population in the spleen is usually identified based on the localization in the spleen, morphology and membrane antigen expression by immunohistochemistry (6, 11). In addition, recent progress in the single cell RNA sequencing approach has allowed the elucidation of the transcriptional state of macrophages (12, 13). However, gene expression profiles and the related functions of those macrophage sub-populations remain incompletely understood, because those macrophage sub-populations in the spleen (except red pulp macrophages) are too few to be identified and too few to be isolated in sufficient numbers to be characterized by means of flow cytometry. We hypothesized that this was caused by inefficient isolation of the small populations among the large populations of various other types of hematopoietic and non-hematopoietic cells in the spleen that are obtained by the method that uses traditional mechanical grinding.
We have previously established an efficient method for isolation of non-hematopoietic cells, including follicular dendritic cells and marginal reticular cells, by digesting the spleen with a cocktail of three types of enzyme: DNase I, Collagenase D and Dispase I (14, 15). Although previous reports used either a single enzyme or combinations of two enzymes for isolation of cells from hematopoietic or non-hematopoietic tissues, the use of the three-enzyme mixture had not been tried. In the present study, we compared the traditional dissociation method of mechanical grinding with our method using the cocktail of three enzymes (Collagenase D, Dispase I and DNase I) for the isolation of splenic tissue macrophage sub-populations. We show that, while the sub-populations of MZMs and MMMs in the spleen after treatment with mechanical grinding were hard to detect by flow cytometry, use of the enzyme cocktail enabled us to efficiently isolate these two populations.
Methods
Mice
C57BL/6J mice were purchased from Clea Japan Inc. (Tokyo, Japan). Cd19-Cre mice were purchased from Jackson Laboratory and Cd19−/− mice were described previously (16). CD169-DTR mice were kindly provided by Masato Tanaka (Tokyo University of Pharmacy and Life Science, Japan) (8). Mice were maintained under specific pathogen-free conditions. All experiments were performed according to the guidelines of the Animal Ethics Committee of the University of Tsukuba Animal Research Center.
Antibodies
Fluorescein isothiocyanate (FITC)-conjugated anti-mouse CD19 (1D3), NK1.1 (PK136), Ly6g (1A8), phycoerythrin (PE)- conjugated anti-mouse NK1.1 (PK136), B220 (RA3-6B2), Ly6G(1A8), PE-CF594-conjugated anti-mouse SiglecF (E50-2440), allophycocyanin (APC)-Cy7-conjugated anti-mouse CD11b (M1/70), V500-conjugated anti-mouse I-A/I-E (M5/114.15.2), PE-conjugated mouse IgG1, κ Isotype Control (MOPC-21) and rat IgG2a, κ Isotype Control (R35-95) were purchased from BD Biosciences (San Jose, CA, USA). FITC- and PE-conjugated anti-mouse CD3 (145-2C11), FITC- and PE-conjugated anti-rat IgG2a isotype control (2A3) and anti-mouse CD16/32 (2.4G2) were purchased from Tombo Biosciences (San Diego, CA, USA). PE-Cy7-conjugated anti-mouse CD11c (N418), AlexaFluor700-conjugated anti-mouse Ly6C (HK1.4), PE- and PE-Cy7-conjugated anti-mouse Tim-4 (RMT4-54) and CD64 (X54-5/7.1) and Zombie Violet Fixable Viability Kit were purchased from BioLegend (San Diego, CA, USA). AlexaFluor647-conjugated anti-mouse F4/80 (Cl: A3-1) was purchased from Bio-Rad Laboratories (Berkeley, CA, USA). PE-conjugated anti-mouse SIGNR1 (REA125) and PE-conjugated isotype control (REA293) were purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). FITC-conjugated anti-mouse metallophilic macrophages (CD169) was purchased from Abcam (Cambridge, UK).
Isolation of splenic macrophages
Spleen cells were isolated by using a protocol that we have previously reported for the isolation of follicular dendritic cells (14, 15). In brief, the spleen was perfused with 10 ml of phosphate-buffered saline (PBS) injected into left ventricular prior to dissection. The spleen was then minced into a homogenous paste with a scalpel on a dish plate and then treated with 1 ml of an enzyme cocktail containing 1 mg ml−1 Collagenase D (Roche, Indianapolis, IN, USA), 100 µg ml−1 DNase I (Sigma, St Louis, MO, USA) and 0.6 U ml−1 Dispase (Roche) in complete Dulbecco’s modified Eagle’s medium (cDMEM) containing 2% fetal bovine serum (FBS). For mechanical grinding, the spleen was smashed and ground between the rough sides of frosted glass slides, and cells were collected in cDMEM containing 2% FBS. After incubation in a 24-well plate for 30 min at 37°C in a humidified incubator, cell suspensions were passed through a 100 µm Falcon nylon cell strainer (Corning, NY, USA) and cells were re-suspended in cDMEM containing 10% FBS and 5 mM ethylenediaminetetraacetic acid (EDTA). For lysis of red blood cells, cells were incubated with 1 ml of ammonium-chloride-potassium lysing buffer for 2 min, washed once with cDMEM, and then once with the washing buffer (PBS containing 2% FBS and 5 mM EDTA). Cells were re-suspended in the washing buffer at 109 ml−1 and incubated with 2.5 µg purified anti-mouse CD16/32 mAb for 108 cells for 20 min on ice. For sorting, non-lymphoid cells were enriched by negative selection by magnetic separation with anti-mouse CD4, anti-mouse CD8 and anti-mouse B220 magnetic particles (BD Pharmingen, San Diego, CA, USA) in accordance with the manufacturer’s instructions. Then cells were stained with Zombie Violet viability dye and anti-mouse CD16/CD32, followed by a mixture of FITC-conjugated anti-mouse CD3, anti-mouse CD19, anti-mouse Ly6G and anti-mouse NK1.1, PE-CF594-conjugated anti-mouse SiglecF, APC-Cy7-conjugated anti-mouse CD11b, PE-conjugated anti-mouse Tim-4 and AlexaFluor647-conjugated anti-mouse F4/80 for 30 min on ice. Cells were analyzed and sorted by FACSFortessa and FACS SORP (BD Biosciences), respectively (17). CD169-DTR mice were injected intra-peritoneally with diphtheria toxin (DT) or PBS 4 days prior to dissection of the spleen. To observe the effect of the enzyme cocktail on surface marker expression, we incubated peritoneal macrophages in the enzyme cocktail or in control medium as above and observed the expression of CD11b, F4/80 and Tim-4 by flow cytometry.
In vivo phagocytosis assay
Engulfment of apoptotic cells was assayed as reported previously with modification (18). Briefly, thymocytes from C57BL/6J mice were treated with 0.1 µM dexamethasone in DMEM containing 10% FBS for 12 h at 37°C to induce apoptosis, washed with PBS and incubated with 0.1 µg ml−1 pHrodo-SE for 30 min at room temperature. Reaction was stopped by adding FBS to a final concentration of 10% and washed with PBS containing 10% FBS. Cells were then washed with PBS and suspended in 200 ul PBS (5 × 108 ml–1) for use as prey. Apoptotic cells (1 × 108) were intravenously injected into the tails of 8-week-old C57BL/6J mice. Spleens were collected 2 h after injection and spleen cells were isolated by the enzyme digestion and analyzed by flow cytometry as described above.
Quantitative PCR
Total RNA was isolated by using Isogen reagent (Nippon Gene, Tokyo, Japan). For reverse transcription, we used a high-capacity complementary DNA reverse-transcription kit (Applied Biosystems, Carlsbad, CA, USA). Real-time PCR analysis of Cd169, Cd68, Marco and Cd209b was performed by using an ABI 7500 sequence detector (Applied Biosystems), Power SYBR Green PCR master mix (Applied Biosystems) and the appropriate primers. The Actb expression level was measured as the internal control to normalize the data. The primer sequences for the target genes were as follows: Cd169: forward, 5′-CAGGGCATCCTCGACTGTC-3′, reverse, 5′-GGAGCATCGTGAAGTTGGTTG-3′; Cd68: forward, 5′-TG TCTGATCTTGCTAGGACCG-3′, reverse, 5′-GAGAGTAACG GCCTTTTTGTGA-3′; Marco: forward, 5′-ACAGAGCCGAT TTTGACCAAG-3′, reverse, 5′-CAGCAGTGCAGTACCTG CC-3′; Cd209b: forward, 5′-CTGACAGATGAGCTTAC GTCCA-3′, reverse, 5′-CACAGGCGGAAGAGTTCAGTC-3′; Actb: forward, 5′-ACTGTCGAGTCGCGT CCA-3′, reverse, 5′-GCAGCGATATCGTCATCCAT-3′. The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The mRNA level was determined relative to that in whole splenic cells isolated by enzyme digestion.
Statistical analyses
Statistical analyses were performed by using the unpaired two-tailed Student’s t-test in GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA, USA). P <0.05 was considered statistically significant.
Results and discussion
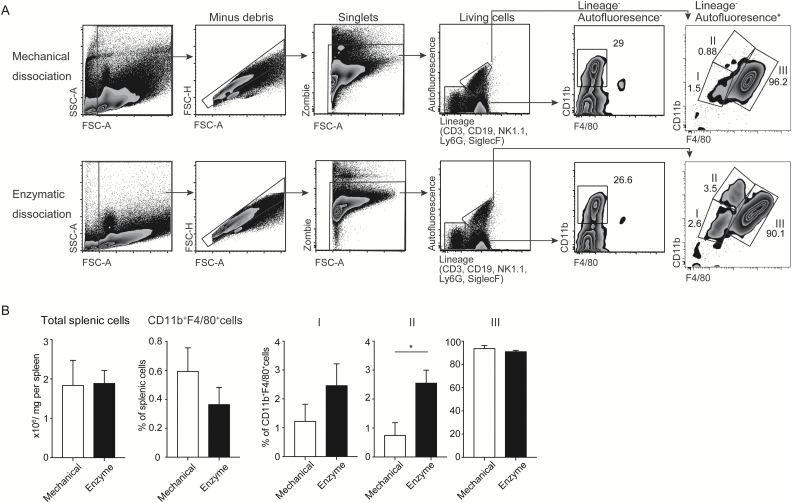
We first confirmed that the surface expression of CD11b and F4/80 on peritoneal macrophages was not affected by the treatment with the enzyme cocktail (Supplementary Figure 1). Therefore, we compared macrophage populations (CD11b+F4/80+) in the spleen after treatment with the enzyme cocktail with those with mechanical grinding by flow cytometry. The macrophage population was divided into three sub-populations: I (CD11bloF4/80lo), II (CD11bhiF4/80med) and III (CD11bloF4/80hi) (Fig. 1A). The total number of splenic cells and the macrophage population after the treatment with the enzyme cocktail were comparable to those after the treatment with mechanical grinding (Fig. 1B). However, sub-population II, but not I or III, was significantly increased after the treatment with the enzyme cocktail compared with that with mechanical grinding (Fig. 1A and B), suggesting that the mechanical grinding affects isolation of certain sub-populations of macrophages in the spleen.
Fig. 1.
Comparison of spleen dissociation by the enzyme cocktail with that by mechanical grinding. (A) Gating strategy of mechanically and enzymatically isolated splenic cells by flow cytometry. Cells were separated into three populations according to CD11b and F4/80 expression: I, CD11bdullF4/80dull; II, CD11bhiF4/80med; III, CD11bmedF4/80hi. A representative profile of flow cytometry analysis is shown. (B) Number and proportion of cells obtained from whole spleen and I–III sub-populations by mechanically and enzymatically isolated spleen macrophages (n = 3). Error bars indicate 1 SD; *P < 0.05. Data are representative of three independent experiments.
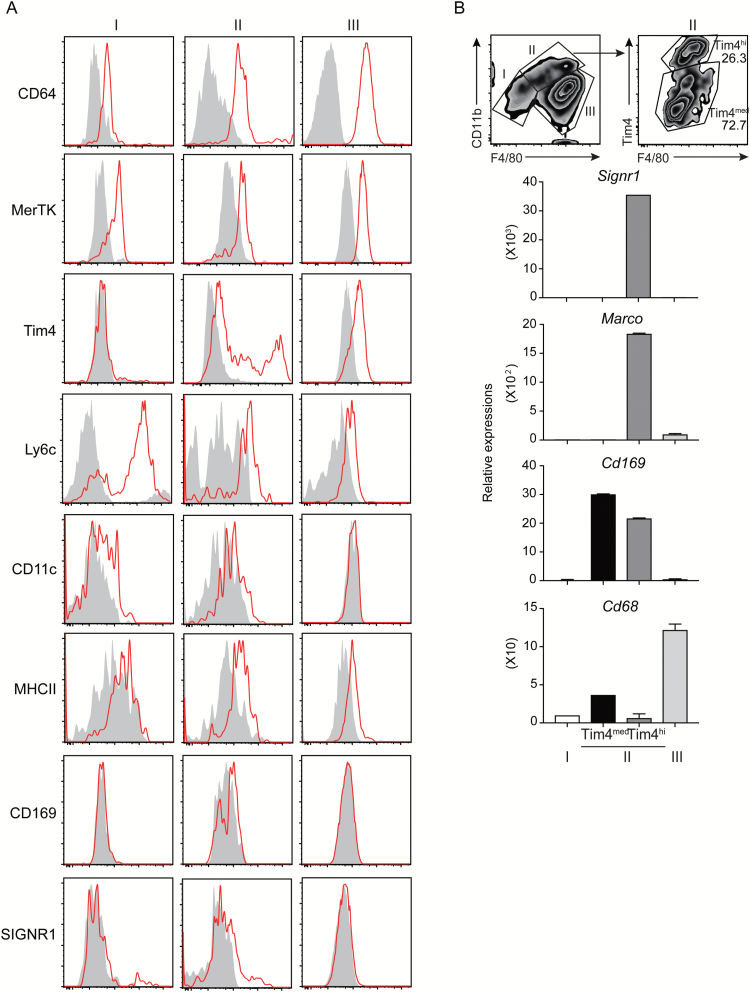
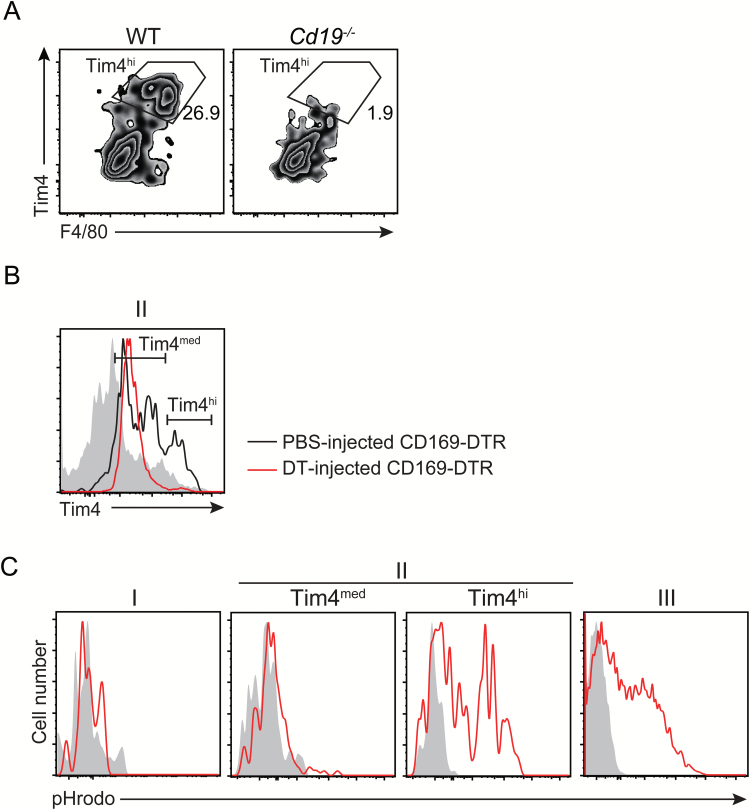
We then analyzed the cell surface antigen expression on each enzyme-dissociated macrophage sub-population by means of flow cytometry. Although the macrophage markers CD64 and MerTK were similarly expressed on all the sub-populations, the expression profile of Tim4, Ly6C, CD11c and MHC class II was different among these sub-populations (Fig. 2A). Sub-population I included the population that expressed Ly6C highly, suggesting that this population contains monocytes. We found that sub-population II was further divided into Tim4hi (II-Tim4hi) and Tim4med (II-Tim4med) populations (Fig. 2A and B). CD169 and SIGNR1, which are known as the specific antigens for MZMs and MMMs (11), respectively, were detected at very low levels on sub-population II isolated by the treatment with the enzymes or with mechanical grinding (Fig. 2A; Supplementary Figure 1B). Therefore, to further characterize the sub-populations, we sorted macrophages from the sub-populations of I, II-Tim4med, II-Tim4hi and III and analyzed mRNA expression of macrophage markers Signr1, Marco, Cd169 and Cd68. Signr1 and Marco were only expressed on sub-population II-Tim4hi, which was identical to the characteristics of MZMs (Fig. 2B) (3, 6). Indeed, this sub-population was significantly reduced in Cd19–/– mice (Fig. 3A), which developmentally lack MZMs as well as marginal zone B cells in the spleen (10), demonstrating that the sub-population II-Tim4hi corresponds to MZMs. On the other hand, sub-population II-Tim4med highly expressed Cd169, which is identical to the characteristics of MMMs (Fig. 2B) (3, 6). In addition, we observed that Cd169 was also expressed in sub-population II-Tim4hi. Treatment with DT significantly and partially decreased the size of sub-populations II-Tim4hi and II-Tim4med, respectively, in CD169-DTR mice, consistent with a previous report that used immunohistochemical analysis (Fig. 3B) (8). The major sub-population was III, which expressed Cd68 (Fig. 2B), and was identical to the characteristics of red pulp macrophages (2).
Fig. 2.
Characterization of macrophage sub-populations isolated by the enzyme cocktail. (A) Spleen cells were stained with monoclonal antibodies against CD64, MerTK, Tim4, Ly6C, CD11c, MHC class II, CD169 and SIGNR1; each sub-population of macrophages gated according to the strategy in Fig. 1 was analyzed for their expression. Red histograms indicate staining by the antibodies, and filled histograms indicate staining by the isotype control. (B) Each sub-population of macrophage was sorted from the spleen of mice (n = 5) and analyzed for relative gene expression to whole spleen cells by quantitative PCR.
Fig. 3.
Developmental and functional analyses of macrophage sub-populations. (A, B) Splenic cells from wild-type and Cd19−/− mice (A) and CD169-DTR mice injected with DT or PBS (B) were isolated by the enzyme cocktail and sub-population II of macrophages gated according to the strategy in Fig. 1 was analyzed by flow cytometry. (C) Apoptotic thymocytes were labeled with pHrodo and intravenously injected into mice. Splenic cells were isolated by enzyme digestion 2 h after injection and analyzed for pHrodo signals indicated by red histograms in each sub-population of macrophages by flow cytometry. Experiments were independently performed twice. Representative flow cytometry profiles are shown.
MZMs and red pulp macrophages are known to interact with apoptotic cells entering the spleen from the circulation and mediate efferocytosis (7, 8, 19, 20). Indeed, we observed that the sub-populations II-Tim4hi and III showed phagocytosis of apoptotic cells (Fig. 3C). Together, these results indicate that, while splenic tissue macrophage sub-populations, particularly MZMs and MMMs, have been difficult to dissociate by traditional mechanical grinding, the treatment with the enzyme cocktail has significantly improved the isolation of these macrophage sub-populations, which should be helpful for molecular and functional characterization of each sub-population.
Funding
This work was supported in part by grants provided by Japan Society for the Promotion of Science (KAKENHI) (grant numbers 16H06387 and 15H01365 to A.S. and 16H05350 and 17H05495 to C.N.-O.).
Supplementary Material
Acknowledgements
We thank M. Tanaka from the Tokyo University of Pharmacy and Life Science, Japan for providing us with the CD169-DTR mice. We thank S. Tochihara for secretarial assistance, and R. Hirochika for technical assistance.
Conflicts of interest statement: The authors declared no conflicts of interest.
References
- 1. Amit I. Winter D. R. and Jung S. 2016. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat. Immunol. 17:18. [DOI] [PubMed] [Google Scholar]
- 2. Davies L. C. Jenkins S. J. Allen J. E. and Taylor P. R. 2013. Tissue-resident macrophages. Nat. Immunol. 14:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mebius R. E. and Kraal G. 2005. Structure and function of the spleen. Nat. Rev. Immunol. 5:606. [DOI] [PubMed] [Google Scholar]
- 4. den Haan J. M. and Kraal G. 2012. Innate immune functions of macrophage subpopulations in the spleen. J. Innate Immun. 4:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. A-Gonzalez N., Quintana J. A., García-Silva S., et al. 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gordon S. and Plüddemann A. 2017. Tissue macrophages: heterogeneity and functions. BMC Biol. 15:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ravishankar B., Shinde R., Liu H. et al. 2014. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc. Natl Acad. Sci. USA 111:4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miyake Y. Asano K. Kaise H. Uemura M. Nakayama M. and Tanaka M. 2007. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J. Clin. Invest. 117:2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karlsson M. C. Guinamard R. Bolland S. Sankala M. Steinman R. M. and Ravetch J. V. 2003. Macrophages control the retention and trafficking of B lymphocytes in the splenic marginal zone. J. Exp. Med. 198:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. You Y. Zhao H. Wang Y. and Carter R. H. 2009. Cutting edge: primary and secondary effects of CD19 deficiency on cells of the marginal zone. J. Immunol. 182:7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gordon S. Plüddemann A. and Martinez Estrada F. 2014. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 262:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jaitin D. A., Kenigsberg E., Keren-Shaul H., et al. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gautier E. L., Shay T., Miller J., et al. ; Immunological Genome Consortium 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sato K. Honda S. I. Shibuya A. and Shibuya K. 2016. Improved protocol for the isolation of naïve follicular dendritic cells. Mol. Immunol. 78:140. [DOI] [PubMed] [Google Scholar]
- 15. Sato K. Honda S. I. Shibuya A. and Shibuya K. 2018. Cutting edge: identification of marginal reticular cells as phagocytes of apoptotic B cells in germinal centers. J. Immunol. 200:3691. [DOI] [PubMed] [Google Scholar]
- 16. Honda S., Sato K., Totsuka N., et al. 2016. Marginal zone B cells exacerbate endotoxic shock via interleukin-6 secretion induced by Fcα/μR-coupled TLR4 signalling. Nat. Commun. 7:11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mitchell A. J., Pradel L. C., Chasson L., et al. 2010. Technical advance: autofluorescence as a tool for myeloid cell analysis. J. Leukoc. Biol. 88:597. [DOI] [PubMed] [Google Scholar]
- 18. Miksa M. Komura H. Wu R. Shah K. G. and Wang P. 2009. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods 342:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGaha T. L. and Karlsson M. C. 2016. Apoptotic cell responses in the splenic marginal zone: a paradigm for immunologic reactions to apoptotic antigens with implications for autoimmunity. Immunol. Rev. 269:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGaha T. L. Chen Y. Ravishankar B. van Rooijen N. and Karlsson M. C. 2011. Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood 117:5403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.