Abstract
Sirt1 is an NAD-dependent, class III deacetylase that functions as a cellular energy sensor. In addition to its well-characterized effects in peripheral tissues, emerging evidence suggests that neuronal Sirt1 activity plays a role in the central regulation of energy balance and glucose metabolism. In this study, we generated mice expressing an enzymatically inactive form (N-MUT) or wild-type (WT) SIRT1 (N-OX) in mature neurons. N-OX male and female mice had impaired glucose tolerance, and N-MUT female, but not male, mice had improved glucose tolerance compared with that of WT littermates. Furthermore, glucose tolerance was improved in all mice with caloric restriction (CR) but was greater in the N-OX mice, who had better glucose tolerance than their littermates. At the reproductive level, N-OX females had impaired estrous cycles, with increased cycle length and more time in estrus. LH and progesterone surges were absent on the evening of proestrus in the N-OX mice, suggesting a defect in spontaneous ovulation, which was confirmed by the ovarian histology revealing fewer corpora lutea. Despite this defect, the mice were still fertile when mated to WT mice on the day of proestrus, indicating that the mice could respond to normal pheromonal or environmental cues. When subjected to CR, the N-OX mice went into diestrus arrest earlier than their littermates. Together, these results suggested that the overexpression of SIRT1 rendered the mice more sensitive to the metabolic improvements and suppression of reproductive cycles by CR, which was independent of circadian rhythms.
Keywords: neurons, fertility, caloric restriction, glucose intolerance
Sirtuin 1 (SIRT1) was first identified as a mammalian homolog of the yeast silent information regulator 2 (Sir2) protein, which is essential for lifespan extension due to caloric restriction(CR). The sirtuins are class III histone deacetylases that require NAD+ for enzyme activity [1] and these enzymes function as cellular energy sensors sensitive to the intracellular NAD+/NADH ratio.
SIRT1 is activated during fasting to increase fatty acid oxidation and gluconeogenesis, and suppress insulin secretion, insulin action, and adipogenesis [2–4]. Conversely, SIRT1 is reduced in adipose tissue by overnutrition in mouse models, human obesity, and in many brain regions [5–8]. Consequently, global transgenic overexpression improves glucose tolerance and insulin sensitivity even on a low-fat diet, because of increased brown adipose tissue activity, enhanced β3 adrenergic stimulation, and reduced inflammation [9–11]. The global knockout (KO) is difficult to interpret because most mice die in utero or perinatally [12–14], and those that survive on an outbred background are runted [14]. Study findings showed the global inactivation of the acetylase activity by deletion of exon 4 led to a similar phenotype with embryonic lethality, but the few surviving animals had higher triglyceride levels, higher levels of fatty acid release from adipose tissue, and greater hepatic steatosis on a high-fat diet [15, 16]. Similarly, introduction of a point mutation (H355Y) that inactivates the enzyme activity caused runting, elevated oxygen consumption, and male infertility [17], and increased the susceptibility to high-fat diet–induced metabolic dysfunction [18].
SIRT1 has also been reported to have tissue-specific effects. Forced reduction of Sirt1 in white adipose tissue by antisense oligonucleotides or conditional KO reduced body weight and white adipose tissue weight but increased macrophage infiltration, whereas overexpression prevents these effects [19]. Loss of SIRT1 in skeletal muscle reduces insulin sensitivity during CR [20]. Deletion in hepatocytes causes increased hepatic glucose production, fasting hyperglycemia, greater fasting-induced steatosis, and the development of late-onset obesity [16, 21–23]. A transgenic overexpressing line that uses the entire Sirt1 gene did not mimic intermittent fasting, despite enhancing brown adipose tissue function and protecting against high-fat diet–induced glucose intolerance [10, 11, 24–26].
Conflicting results have been obtained at the reproductive level. One study reported that the few surviving male mice from the global KO studies were infertile, as are the majority of female mice [14], but another study found that mice could be fertile [27, 28]. SIRT1 is expressed in spermatogonia and has been implicated in germ cell development in males [29, 30]. In the ovary, SIRT1 is expressed in large follicles, particularly in the oocyte and granulosa cells. Loss of SIRT1 causes small ovaries with early-stage follicular development but no evidence of ovulation [14, 31]. SIRT1 modulates androgen receptor and estrogen receptor-α function that may underlie some of the reproductive effects [32–35]. SIRT1 thus has beneficial effects in multiple tissues that ameliorate insulin resistance and support reproduction.
SIRT1 also pays a role in the central nervous system, but the results are less clear. Many studies have focused on the role of SIRT1 to prevent neuronal injury in ischemic stroke, Alzheimer’s disease, brain injury, and other models of neurodegeneration, potentially by reducing neuroinflammation [36–38]. SIRT1 also modulates cognitive function, synaptic plasticity, and learning [39]. Brain-specific deletion of Sirt1 using Nestin-cre decreases monoamine oxidase activity, increases serotonin levels, and decreases anxiety and depressive-like behavior [40, 41]. As in the periphery, SIRT1 expression and activity increase in the hypothalamus under fasting conditions, to regulate food intake and energy expenditure [42–44]. Some of these effects may be mediated by different neuronal populations, because deletion of Sirt1 in Pomc neurons decreases energy expenditure and increases susceptibility to diet-induced obesity in mice [45], but deletion of Sirt1 in Agouti-related peptide (AgRP) neurons has the opposite effect and decreases food intake and body weight [46, 47]. Mice lacking Sirt1 in Sf1-expressing neurons have increased susceptibility to diet-induced obesity and insulin resistance similar to the Pomc neuron KO, primarily due to decreased energy expenditure and skeletal muscle insulin sensitivity, but the mice are fertile, and transgenic overexpression prevents these effects [48].
Thus, it appears that SIRT1 may play distinctive and sometimes even opposing roles in energy balance and reproduction through different neuronal populations. Interpretation of the studies is complicated by the different approaches used to delete the gene. Some studies use deletion of exon 4, which encodes the deacetylase domain, to create a mutant protein lacking enzyme activity, but other studies delete exons 2 and 3, resulting in the introduction of a premature termination codon so no protein is produced. The widespread expression of SIRT1 in the periphery and central nervous system makes it difficult to assign a function for SIRT1 in a particular process; therefore, we have chosen to create neuron-specific genetic modifications using transgenic cre recombinase that is expressed in a wide range of mature neurons. Using this approach, we recently reported that loss of SIRT1 enzymatic activity in neurons in male mice reduced fasting and glucose-stimulated insulin levels while maintaining normoglycemia, suggesting increased insulin sensitivity [37]. In the current study, we characterized male and female mice expressing this enzymatically dead SIRT1 mutant, or conversely, conditionally overexpressing the wild-type (WT) SIRT1 protein, in mature neurons using synapsin-cre to distinguish peripheral from central effects on metabolism and reproduction.
1. Methods
A. Animals
Mice with cre recombinase expression under control of the neuron-specific Syntaxin1 promoter (Syn-cre) [49] were mated with mice with an exon 4 floxed Sirt1 allele or a floxed-STOP Sirt1 allele [50]. Expression of cre deletes exon 4 of Sirt1 that encodes the acetylase domain, thus creating a functionally inactive mutant (N-MUT; i.e., an enzymatically inactive form of SIRT1) from the floxed allele, or causes overexpression of wild-type SIRT1 by deletion of the transcriptional STOP sequence from the flox-STOP allele (N-OX; i.e., WT SIRT1). The cre driver was maintained on the female side, because the Syn-cre transgene is expressed in the testis [51]. Syn-cre mice express the cre transgene as early as embryonic day 12.5 in differentiated neurons throughout the brain [49].
Homozygous flox/cre- and heterozygous flox/cre- littermates from either the N-MUT or N-OX breeding pairs were used as WT control mice. In most experiments, no statistical difference was observed between the WT control mice from the N-MUT or N-OX breeding pairs, so the data were combined into a single WT group. Occasionally, statistically significant differences were observed between the control groups, in which case the data were analyzed against each separate control group. Mice were housed under a 12-hour light, 12-hour dark cycle (12:12 LD). Body weights were recorded weekly out to 90 days of age for both sexes. Males and females had access to standard chow and water ad libitum. Mouse procedures conformed to the Guide for Care and Use of Laboratory Animals of the US National Institutes of Health and were approved by the University of California, San Diego, Animal Subjects Committee.
B. Caloric Restriction
At 12 weeks of age, food intake by cohorts of female and male mice was measured for 2 weeks to assess ad libitum food intake. For the graded CR, mice were provided with 90% of prior food intake for week 1, then 80% for week 2, 70% for week 3, and then maintained at 60% of original intake for weeks 4 through 6. Body weight was measured weekly. The study was divided into four phases: before, stage 1 (weeks 1 and 2: 10% to 20% CR), stage 2 (weeks 3 and 4: 30% to 40% CR), and stage 3 (weeks 5 and 6: 40% CR). Estrous cycles were monitored in female mice by vaginal cytology for the entire 8 weeks. A cycle was defined as a day of diestrus or metestrus, followed by a day of proestrus, then a day of estrus. The length of the cycle was defined as the number of days between successive days of proestrus in normal diestrus-proestrus-estrus cycles
C. Puberty Onset and Fertility Assessment
Male and female pups were weaned at 21 days of age. Female pups were checked for vaginal opening as a sign of onset of puberty and monitored for first estrous. For fertility assessment, N-MUT, N-OX, or WT female mice were paired on the day of proestrus with known WT male breeders. The number of days to observable vaginal plugs, the number days to the first litter, and average number of pups born were recorded.
D. Tissue Collection and Histology
Testes, ovaries, brain, pituitary glands, and other tissues were collected after euthanasia for histology and for RNA extraction. Testes were fixed in Bouin’s solution for 6 hours and ovaries in formalin for 24 hours, followed by washing in 70% ethanol. Paraffin-embedded sections (5 μm) were cut, dewaxed, and stained with hematoxylin and eosin. Follicle number and stage, and corpora lutea number were counted in three to five sections from ovaries from four to five mice per group; counts are presented as the mean number per ovary [52]. Follicle stages were defined as follows: primary: having a single layer of cuboidal granulosa cells (GCs); secondary: having two or more layers of cuboidal GCs but no antrum; early antral: having small patches of clear space between GCs; antral: having clearly defined antrum; and atretic: having irregular oocyte morphology. Ovarian sections examined were separated by 50 µm. Images were scanned using an Aperio ImageScope and analyzed using the Imagescope software (Leica, Buffalo Grove, IL).
E. Gene Expression
Total RNA was extracted from the tissues using RNAbee (Tel-Test, Friendswood, TX) following the manufacturer’s instructions. First-strand cDNA was synthesized using a high capacity cDNA synthesis kit (Applied Biosystems, Waltham, MA). Targeted quantitative PCR (qPCR) assays were run in 20-µL triplicate reactions on a MJ Research Chromo4 instrument using iTaq SYBR Green Supermix (Bio-Rad, Hercules, CA). For gene expression assays using 7 nL microfluidic arrays (Fluidigm, San Francisco, CA), mRNA was extracted using RNAbee as before, but it was further purified using RNA purification kits from QIAGEN (Germantown, MD) or Macherey-Nagel (Bethlehem, PA), according to manufacturer’s instructions. Custom qPCR primers for a panel of targets were designed and synthesized by Fluidigm for their BioMark HD System. Gene expression levels were calculated after normalization to the housekeeping genes m36B4 or Gapdh using the comparative threshold cycle method and expressed as relative mRNA levels compared with the control. Primers are listed in an online repository [53].
F. Gonadotropin Measurements
Blood was collected from the tail vein of males and from females at diestrus and proestrus, and plasma was prepared. Plasma LH and FSH levels were measured by Luminex assay (catalog no. RPT86K; Millipore, Bedford, MA). Sensitivity of the assay for LH is 4.9 pg/mL and for FSH, 47.7 pg/mL, with an intra-assay coefficient of variation of 15%. For the GnRH stimulation test, tail-vein blood was collected before and 10 minutes after IP injection of 1 μg/kg GnRH (Sigma-Aldrich, St. Louis, MO), and gonadotropins were measured.
G. Intraperitoneal Glucose Tolerance and Insulin Tolerance Tests
Mice were subjected to IP glucose and insulin tolerance tests. Mice were fasted for 6 hours and then injected IP with glucose (1 g/kg body weight) or insulin (0.4 U/kg body weight). Tail-vein blood glucose level was measured at 0, 15, 30, 45, 60, 90, and 120 minutes after injection using a glucose meter (OneTouch Ultra; Bayer Healthcare, Tarrytown, NY). Glucose and insulin tolerance assays were performed for mice receiving normal chow and during the last week of 40% food restriction.
H. Insulin, Leptin, and Steroid Measurements
Animals were fasted for 6 hours. Glucose level was measured with a glucose meter, blood was collected from the tail vein, and plasma was obtained. Fasting insulin and leptin levels were measured using the Mouse Metabolic Kit (catalog no. K15124C-2; Meso Scale Discovery, Rockville, MD). Sensitivity of the assay was as follows: leptin: 43 pg/mL; insulin: 15 pg/mL; coefficients of variance: 6% and 12%, respectively. Estrogen, progesterone, and testosterone levels were measured using a Custom Steroid Hormone Panel Kit (Meso Scale Discovery) Sensitivity of the assay was as follows: estradiol 5 pg/mL; progesterone 70 pg/mL; testosterone 20 pg/mL; coefficients of variance: 7%, 15%, and 22% respectively.
I. Metabolic Assessment
Six mice per group (i.e., WT and N-OX), matched for body weight, were individually housed in a 12-chamber Clinical Laboratory Animal Monitoring System (Columbus Instruments, Columbus, OH) with controlled temperature, light, and feeding. Oxygen uptake, carbon dioxide output, respiratory exchange ratio, horizontal and vertical ambulatory movement, and feeding and drinking per 13-minute intervals were measured over 3 days. Data from the first 12 hours during day-1 acclimation were excluded from the analysis.
J. Circadian Activity
Female and male mice were raised under a 12:12 LD photoperiod until 3 months of age. Thereafter, mice were singly housed in standard shoebox cages equipped with running wheels (13-cm diameter). Cages were maintained in light-tight secondary enclosures with programmable fluorescent lighting for a three-phase assessment of circadian entrainment and free-running behavior conducted over ∼6 weeks. Initially, entrained behavior of each mouse was assessed over 2 weeks under the 12:12 LD. Thereafter, monitoring continued for an additional 2 weeks in a skeleton photoperiod consisting of two 1-hour light pulses coinciding with the first and last hours of the 12-hour light phase. This protocol removes potential masking effects of bright light on locomotor activity. Finally, endogenous circadian rhythmicity was assessed under continuous darkness over 7 to 10 days. A total of 48 mice were assessed in four cohorts.
Wheel turns were recorded and compiled into 6-minute bins. Data were analyzed offline using the Clocklab (Actimetrics, Wilmette, IL) suite of Matlab plugins (Mathworks, Natick, MA). For each animal, the following variables were calculated in light/dark and skeleton photoperiods over final 7-day spans of each photoperiod condition. The proper phasing of activity was assessed by calculating the timing of activity onset and offset, defined as the first and last time that the 7-day average activity profile rose above and below the daily mean, respectively. From these data were derived the phase angle of entrainment and the duration of elevated nighttime activity. Disruption of circadian timing was additionally assessed by calculating the proportion of activity intruding into the daytime (i.e., the central 8 hours of the light phase or skeleton daytime). Rhythm robustness was assessed by calculating the maximum amplitude of the χ2 periodogram. In the first 7 days of continuous darkness, the free-running period was estimated as the value with the highest amplitude in the χ2 periodogram and as the best-fit linear regression line through identified activity onsets. These methods yielded comparable estimates and only the former is reported here. Individually identified activity onsets were additionally used to determine the cycle-to-cycle variability in free-running activity onset. In all phases, the total amount of wheel-running activity without respect to its timing was also assessed.
All data were analyzed with four-way ANOVA with cohort, strain (OX vs MUT), Cre genotype (Cre+ or Cre-), and sex as between-subject factors. For phenotype screening, α was set initially at 0.05 without correction for multiple comparisons.
K. Statistical Analysis
Data were analyzed by ANOVA or Student t test, as appropriate, using GraphPad Prism (GraphPad, La Jolla, CA) or in R (https://www.r-project.org/). Pairwise comparisons were performed using Tukey post hoc tests after the ANOVA. Results are expressed as mean ± SE and considered significant at P < 0.05. Supplemental data are provided in an online repository [53].
2. Results
A. Neuronal SIRT1 Overexpression Caused Glucose Intolerance
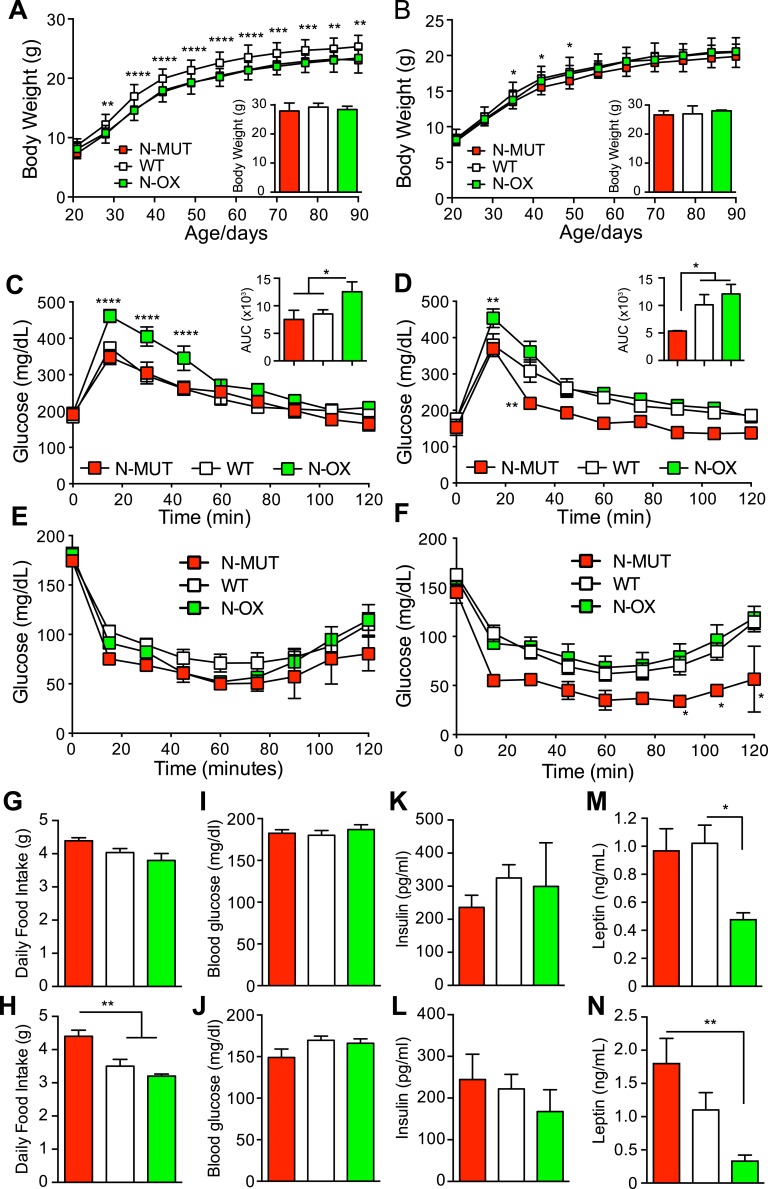
To test the role of neuronal SIRT1 in metabolic and reproductive regulation, we generated N-MUT [37] and N-OX mice. Assessment of Sirt1 mRNA expression in whole brains from N-OX mice indicated that the gene is overexpressed approximately 2.5-fold [53]. The body weights of male mice indicated a significant difference with genotype over the first 3 months, although no significant difference was observed at 8 to 9 months of age (Fig. 1A), and the female mice did not differ appreciably in body weight (Fig. 1B). Food intake did not differ in male mice, but the female N-MUT mice ate significantly more (Fig. 1G and 1H) despite having equivalent body weight. The male N-OX mice were significantly glucose intolerant (Fig. 1C), unlike the N-MUT mice, as we have previously reported [37]. Consistent with the male mice, the female N-OX mice also were glucose intolerant, but the N-MUT mice had significantly improved glucose tolerance compared with WT controls (Fig. 1D). Neither the N-MUT nor N-OX groups showed altered insulin tolerance in male mice, but the female N-MUT mice were significantly more insulin sensitive than WT or N-OX littermates (Fig. 1E and 1F). Fasting blood glucose and insulin levels and homeostatic model of assessment of insulin resistance were comparable between the groups and sexes (Fig. 1I–1L; and in a supplemental figure provided in an online repository [53]), but leptin levels were significantly lower in N-OX mice for both sexes (Fig. 1M and 1N).
Figure 1.
Neuronal SIRT1 caused glucose intolerance in mice. Data for N-MUT mice are shown in red, in white for WT littermates, and green for N-OX mice in green. For (A–D), Two-way ANOVA indicated significant genotype and time effects (P < 0.0001) but no interaction. (A) Body weights for male mice from 3 to 13 wk of age (n = 13 for N-MUT; n = 48 for WT; n = 24 for N-OX). Inset shows final body weight at 8 to 9 mo. (B) Body weights for female mice from 3 to 13 wk of age (n = 10 for N-MUT; n = 44 for WT; n = 20 for N-OX). Inset shows final body weight at 8 to 9 mo. (C) Glucose tolerance testing in male mice at 3 mo of age (n = 8 for N-OX; n = 16 for WT; n = 4 for N-MUT). Two-way ANOVA indicated significant interaction effect (P = 0.005). Inset shows AUC data. (D) Glucose tolerance testing in female mice at 3 mo of age (n = 9 for N-OX; n = 16 for WT; n = 6 for N-MUT). Inset shows AUC data. (E) Insulin tolerance testing in male mice at 3 mo of age (n = 9 for N-OX; n = 16 for WT; n = 6 for N-MUT). (F) Insulin tolerance testing in female mice at 3 mo of age (n = 10 for N-OX; n = 16 for WT; n = 6 for N-MUT). (E, F) Two-way ANOVA indicated significant genotype and time effects (P < 0.001 and P < 0.0001, respectively) but no interaction. (G) Daily food intake over 10 wk for male mice (n = 3 for N-MUT; n = 12 for WT; n = 4 for N-OX). (H) Daily food intake over 10 wk for female mice (n = 6 for N-MUT; n = 11 for WT; n = 5 for N-OX). (I) Fasting blood glucose data of male mice (n = 17 for N-OX; n = 32 for WT; n = 8 for N-MUT). (J) Fasting blood glucose data of female mice (n = 20 for N-OX; n = 32 for WT; n = 4 for N-MUT). (K) Fasting insulin data of male mice (n = 6 for N-MUT; n = 14 for WT; n = 6 for N-OX). (L) Fasting insulin data of female mice (n = 6 for N-MUT; n = 12 for WT; n = 6 for N-OX). (M) Fasting leptin data of male mice (n = 6 for N-MUT; n = 14 for WT; n = 6 for N-OX). (N) Fasting leptin data of female mice (n = 6 for N-MUT; n = 12 for WT; n = 6 for N-OX). Data are reported as mean ± SEM. *P < 0.05, **P < 0.01, *P < 0.001, and ****P < 0.0001 vs WT or as indicated. AUC, area under the curve.
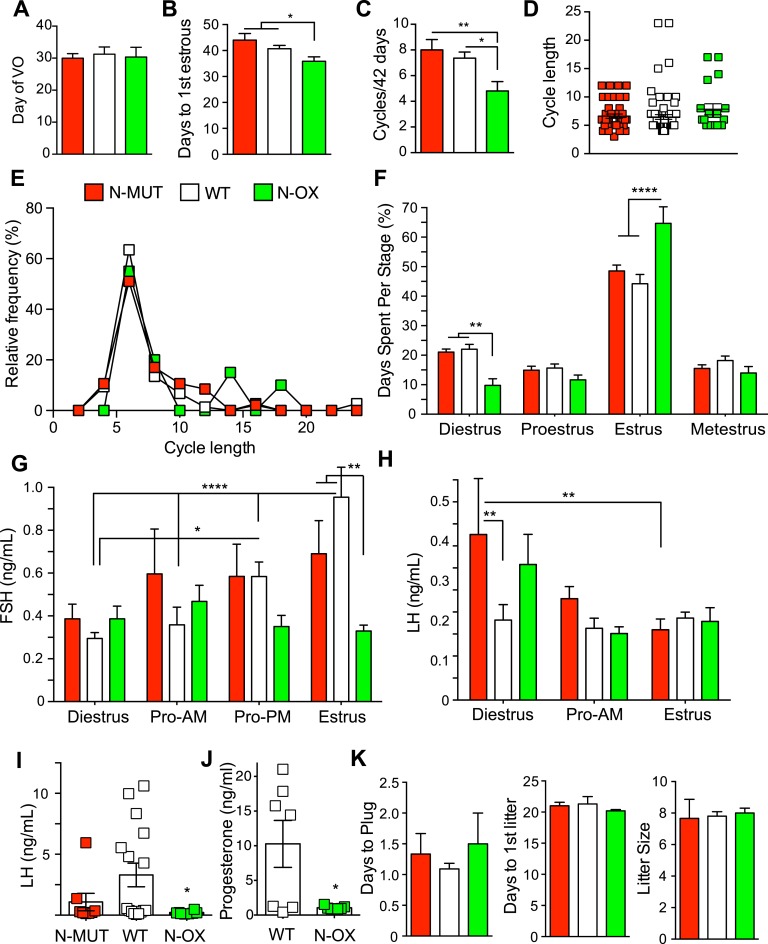
B. SIRT1 Overexpression Impaired Estrous Cycles
Neither N-MUT nor N-OX female mice differed in the day of vaginal opening (Fig. 2A), but the N-OX mice reached first estrous slightly earlier (Fig. 2B), indicating that all genotypes went through puberty normally. Assessment of the number of estrous cycles over 6 weeks after the onset of estrous cycles showed that N-OX mice had fewer cycles (Fig. 2C), and analysis of cycle length indicated that N-OX mice tended to have a slightly longer mean cycle length (6.6 days for N-MUT, 6.5 days for WT, and 7.8 days for N-OX; Fig. 2D) with more extended cycles (Fig. 2E). The N-OX mice had significantly more days of estrus and fewer days of diestrus (Fig. 2F). Representative cyclegrams are provided in an online repository [53]. No differences in FSH level were observed in N-OX mice at diestrus, the morning or evening of proestrus, or on estrus, but the WT mice had the expected higher FSH levels during late proestrus and estrus (Fig. 2G). LH levels were elevated at diestrus in N-MUT mice, but no differences were observed in early proestrus or on estrus (Fig. 2H). LH levels during late proestrus showed LH surges in N-MUT (n = 2 of 8) and WT (n = 8 of 16) mice, but no surges were observed in N-OX mice (Fig. 2I).
Figure 2.
Neuronal SIRT1 regulated reproduction in female mice. Data for N-MUT mice are shown in red, in white for WT littermates, and green for N-OX mice in green. (A) Day of vaginal opening (n = 8 for N-MUT; n = 40 for WT; n = 18 for N-OX). (B) Day of first estrous (n = 8 for N-MUT; n = 41 for WT; n = 21 for N-OX). (C) Number of estrous cycles by vaginal lavage over 6 wk (n = 6 for N-MUT; n = 11 for WT; n = 5 for N-OX). (D) Cycle length during the 6 wk (n = 44 for N-MUT; n = 74 for WT; n = 19 for N-OX). (E) Relative-frequency histogram of cycle lengths. (F) Percentage of d spent in each stage of the estrous cycle during the 6-wk cycling (n = 6 for N-MUT; n = 11 for WT; n = 5 for N-OX). Two-way ANOVA indicated significant stage effect (P < 0.0001) and significant interaction of stage and genotype (P < 0.0001). (G) FSH levels during diestrus, the morning of proestrus (Pro-AM), the afternoon of proestrus (Pro-PM), and the morning of estrus (n = 8 for N-MUT; n = 16 for WT; n = 8 for N-OX). Two-way ANOVA indicated significant stage effect (P = 0.015) and significant interaction of stage and genotype (P = 0.013). (H) LH levels during diestrus, the morning of proestrus, and the morning of estrus (n = 8 for N-MUT; n = 16 for WT; n = 8 for N-OX). Two-way ANOVA indicated significant stage effect (P = 0.013) but no interaction of stage and genotype. (I) LH levels during the afternoon of proestrus (n = 8 for N-MUT; n = 16 for WT; n = 8 for N-OX). Proestrus LH values did not follow a normal distribution, so N-OX mice had significantly lower LH levels than did WT mice by Kruskal-Wallis test (P = 0.033). (J) Progesterone levels during the afternoon of proestrus (n = 7 for WT; n = 7 for N-OX). (K) Days to plug, days to first litter, and litter size during fertility test (n = 3 for N-MUT; n = 11 for WT; n = 6 for N-OX). Data are reported as mean ± SEM. *P < 0.05, **P < 0.01, *P < 0.001, ****P < 0.0001 vs WT or as indicated.
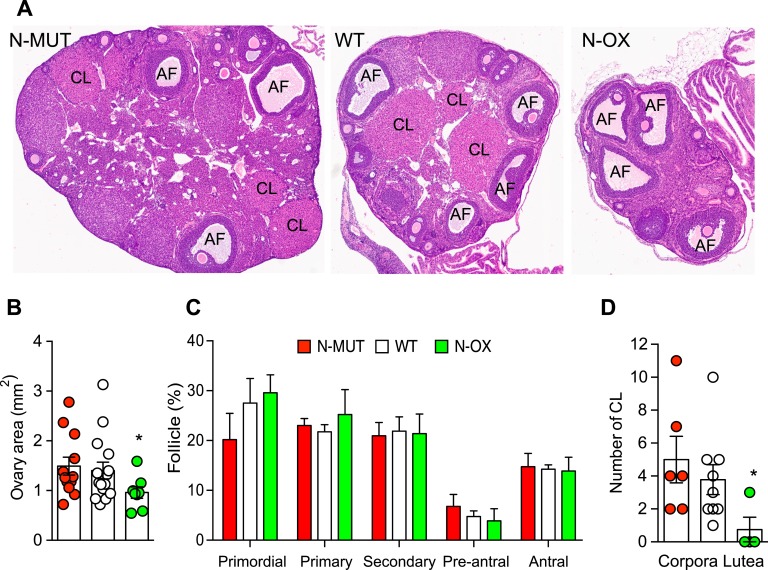
Measurement of progesterone levels confirmed that N-OX mice had not ovulated, compared with WT mice (Fig. 2J). These results suggested that spontaneous LH surges might be impaired in the N-OX mice, so we tested fertility directly by pairing female mice on proestrus with known male WT breeders. The number of days until the observation of vaginal plugs, the number of days until first litter, and the litter size did not show a difference associated with genotype (Fig. 1K), indicating the mice were fertile and could ovulate given normal environmental cues. Comparison of ovarian histology (Fig. 3A) showed that N-OX ovaries were smaller (Fig. 3B) and did not show a difference in follicle development (Fig. 3C), but corpora lutea were significantly reduced in the N-OX mice, consistent with the decreased LH surges and progesterone levels (Fig. 3D). Male mice were not studied reproductively, because the synapsin-cre is expressed in the testis [51].
Figure 3.
Overexpression of SIRT1 impaired ovulation. Data for N-MUT mice are shown in red, in white for WT littermates, and green for N-OX mice in green.. (A) Ovarian morphology. Representative hematoxylin- and eosin-stained sections of ovaries obtained at euthanasia. (B) Quantification of ovarian cross-sectional area for N-MUT (n = 12), WT (n = 15), and N-OX (n = 8) ovaries. *P < 0.05 vs N-MUT. (C) Quantification of follicle stage. Percentage of follicles at each stage for N-MUT (n = 6), WT (n = 9), and N-OX (n = 4) mice. (D) Quantification of CL for N-MUT (n = 6), WT (n = 9), and N-OX (n = 4) mice. *P < 0.05 vs WT. Data are reported as mean ± SEM. *Significant difference by post hoc testing.
C. Mice With Neuronal SIRT1 Overexpression Were More Sensitive to Improvements in Glucose Tolerance and Estrous Cycle Arrest, Due to CR
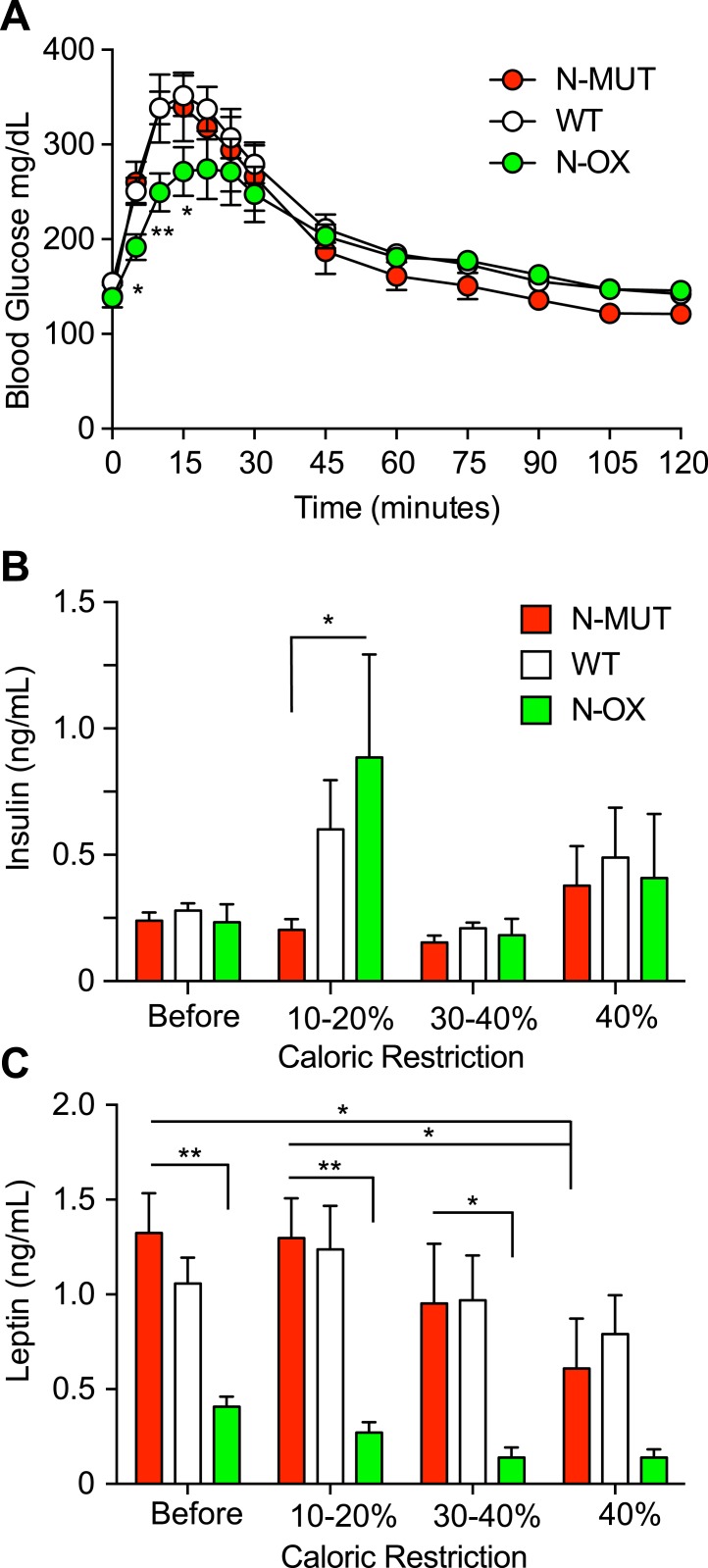
We then tested whether neuronal SIRT1 might alter the response to CR. To avoid the induction of an adrenal stress response due to a rapid reduction in food intake, we used a gradual 10% reduction of food intake over 4 weeks, then maintained the mice for an additional 2 weeks at 40% CR. Assessment of glucose tolerance during the last week showed that the N-OX mice had improved glucose tolerance (Fig. 4A). Insulin levels were higher in WT and N-OX mice during the initial stage of CR, consistent with fasting-induced insulin resistance, but then levels returned to normal (Fig. 4B). In contrast, leptin levels decreased progressively during CR (Fig. 4C) and the N-OX mice had consistently lower leptin levels, as we had seen earlier (Fig. 1M and 1N).
Figure 4.
Overexpression of SIRT1 enhanced the metabolic response to CR. Data for N-MUT mice are shown in red, in white for WT littermates, and green for N-OX mice in green. (A) Glucose tolerance test conducted after CR (n = 8 for N-OX; n = 17 for WT; n = 6 for N-MUT; n = 20 males; n = 11 females). Two-way ANOVA indicated a significant time effect (P < 0.0001) and time-genotype interaction (P = 0.0025). (B) Fasting insulin levels during CR (n = 14 for N-MUT; n = 26 for WT; n = 12 for N-OX; n = 28 males; n = 24 females). (C) Fasting leptin levels during CR (n = 14 for N-MUT; n = 26 for WT; n = 12 for N-OX; n = 28 males; n = 24 females). Two-way ANOVA indicated a significant genotype effect (P < 0.0001). Data are reported as mean ± SEM. *P < 0.05, **P < 0.01 among N-OX and WT and N-MUT, or as indicated, from post hoc testing. AF, antral follicle; CL, corpus luteum.
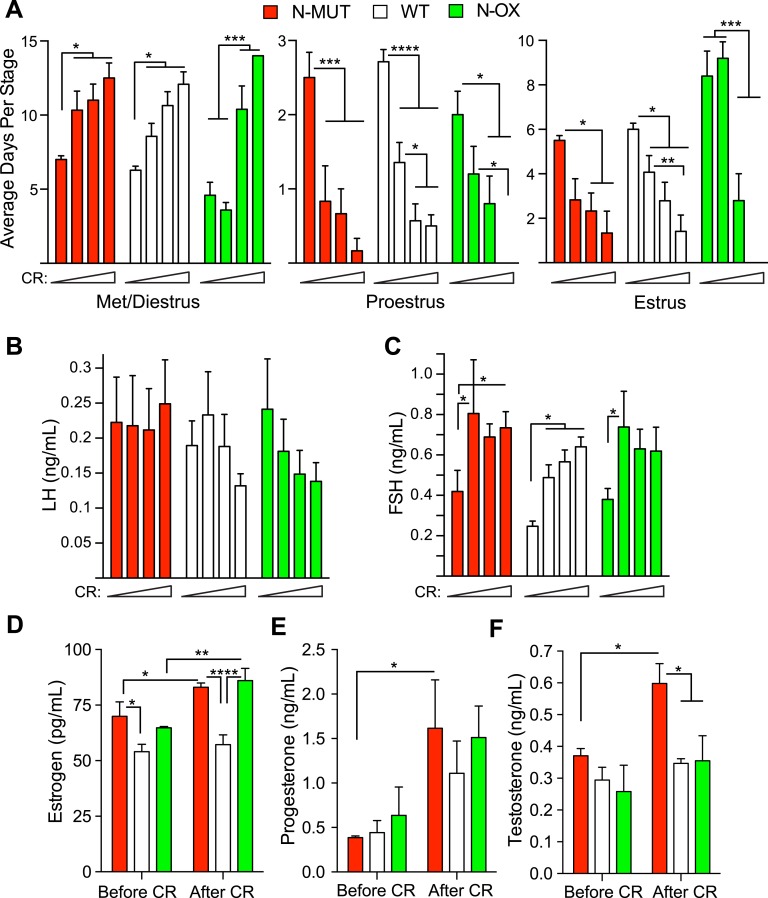
Estrous cycles were assessed in the female mice during the entire period of CR. N-MUT mice had the same response as WT mice: a gradual increase in the days of diestrus and a decrease in the days at proestrus and estrus (Fig. 5A). In contrast, the N-OX mice had fewer days at diestrus before CR, did not respond to the 10% to 20% CR, but showed a steep increase in the days of diestrus during the 30% to 40% CR such that all mice were in permanent diestrus by 40% CR. This was confirmed by determining the relative frequency of diestrus arrest at each CR stage for each genotype [53]. The WT and N-MUT mice had the highest frequency of arrest at 40% CR, but the N-OX mice had the highest frequency of arrest at 20% to 30% CR. Unexpectedly, we did not observe a significant decrease in LH levels during CR in any genotype (Fig. 5B), but we did observed an increase in FSH levels even at the lowest level of CR (Fig. 5C). Interestingly, this response was sexually dimorphic, because FSH levels decreased in male mice on CR, but LH levels did not change [53]. Inspection of ovarian histology at euthanasia showed that all mice had ceased cycling, because no corpora lutea were observed [53]. At time of euthanasia, estrogen levels were slightly higher in mice under CR but were significantly lower in WT mice than N-MUT and N-OX mice (Fig. 5D). Progesterone levels increased during CR (Fig. 5E); testosterone levels increased with CR, but there was also a genotype effect, with N-MUT mice having significantly higher levels of testosterone (Fig. 5F).
Figure 5.
Overexpression of SIRT1 enhanced the reproductive response to CR. Data for N-MUT mice are shown in red, in white for WT littermates, and green for N-OX mice in green. Estrous cycles were assessed by vaginal lavage in female mice during the 8 wk of CR. Data are reported as mean ± SEM. (A) Average number of d spent in met/diestrus, proestrus, or estrus during the four 2-wk stages of increasing CR for each genotype of the estrous cycle during the 6-wk cycling (n = 6 for N-MUT; n = 14 for WT; n = 5 for N-OX). Two-way ANOVA indicated a significant CR effect (P < 0.0001) and significant interaction of CR and genotype (P = 0.003) for met/diestrus, a CR effect (P < 0.0001) for proestrus; and a genotype effect (P = 0.017), a CR effect (P < 0.0001), and a significant interaction of CR and genotype (P = 0.011) for estrus. (B) LH levels during the four stages of CR (n = 6 for N-MUT; n = 12 for WT; n = 5 for N-OX). No significant differences were observed. (C) FSH levels during the four stages of CR (n = 6 for N-MUT; n = 12 for WT; n = 5 for N-OX). Two-way ANOVA indicated significant genotype (P = 0.022) and CR (P = 0.002) effects but no significant interaction. (D–F) Estradiol, progesterone, and testosterone levels before and after CR (n = 3 for N-MUT; n = 3 for WT; n = 3 for N-OX. Each sample was pooled from two animals). Two-way ANOVA indicated significant genotype (P = 0.0004) and CR (P = 0.0033) effects for estradiol, a CR effect (P = 0.0055) for progesterone, and genotype (P = 0.015) and CR (P = 0.019) effects for testosterone. No significant interactions of CR and genotype were observed. *P < 0.05, **P < 0.01, *P < 0.001, ****P < 0.0001 (post hoc testing). Met, metestrus.
D. Altered Hypothalamic Gene Expression in CR Mice
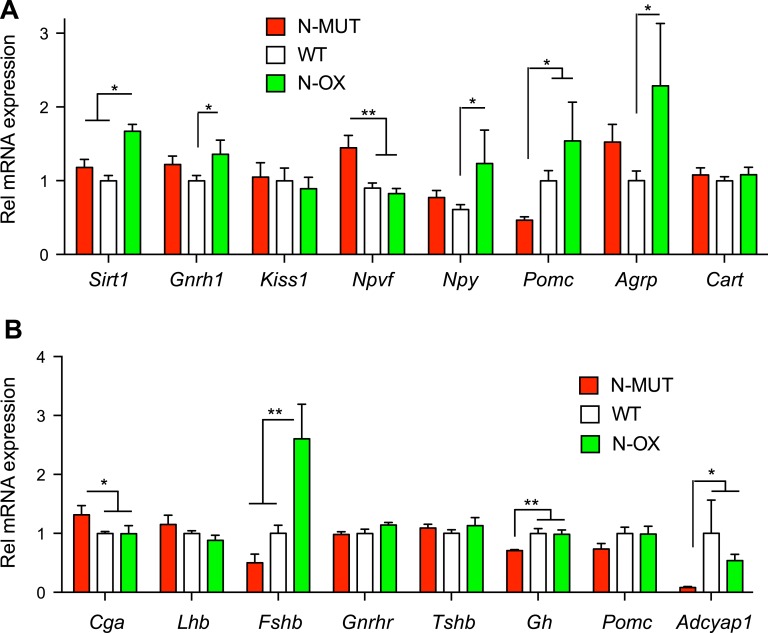
RNA was extracted from hypothalami and pituitaries of female mice euthanized at the end of CR. The N-OX mice had approximately twofold overexpression of Sirt1 by qPCR (Fig. 6A), as observed before (Fig. 1). Expression of neuropeptide Y (Npy), Pomc, and Agrp that control food intake and energy expenditure was increased in the N-OX mice, but the cocaine and amphetamine-regulated transcript (Cart) was unchanged (Fig. 6A). Expression of the reproductive neuropeptides gonadotropin-releasing hormone 1 (Gnrh1), and the RF-amide related peptide 3 precursor gene (Npvf) were significantly altered, but expression of the kisspeptin gene (Kiss1) was unchanged (Fig. 6A). Neuropeptide receptor gene expression did not change with genotype [53]. The same alterations in hypothalamic gene expression were not seen in male mice [53]. Assessment of pituitary gene expression in female mice indicated a threefold increase in Fshb gene expression in N-OX mice, an increase in the glycoprotein hormone α gene (Cga) in the N-MUT mice, and decreases in growth hormone (Gh) and pituitary adenylate-cyclase activating peptide (Adcyap1) gene expression in N-MUT mice (Fig. 6B).
Figure 6.
Hypothalamic and pituitary gene expression in N-MUT and N-OX mice. Data for N-MUT mice are shown in red, in white for WT littermates, and green for N-OX mice in green. (A) Gene expression in the hypothalami from female mice, determined by qPCR. (B) Gene expression in the pituitaries from female mice, determined by qPCR. Data are reported as mean ± SEM; n = 5 for N-MUT; n = 10 for WT, and n = 5 for N-OX. *P < 0.05, **P < 0.01. Rel, relative.
E. Altered Dark-Phase Activity in Mice With SIRT1 Overexpression
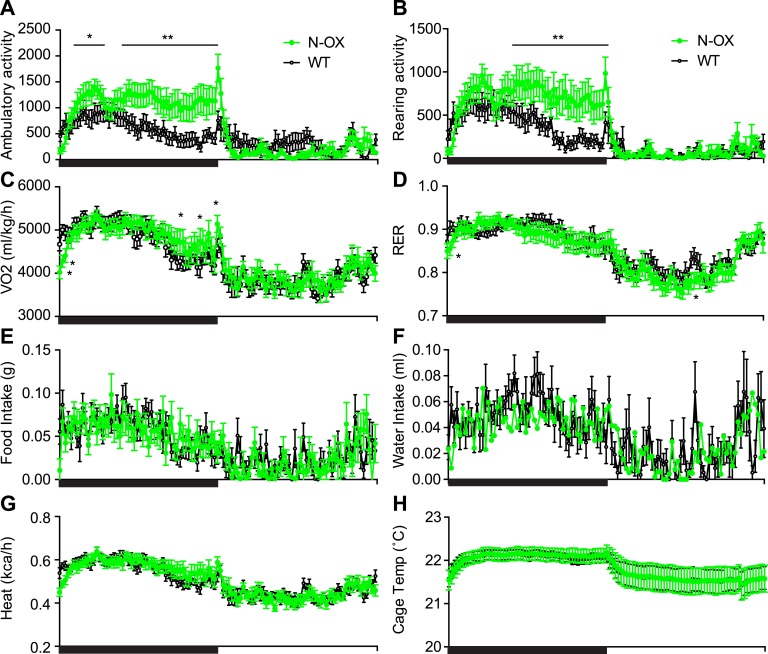
The N-OX mice were glucose intolerant; thus, we subjected N-OX and WT mice to a metabolic assessment with continuous monitoring over 3 days. The N-OX mice had greater diurnal changes in ambulatory and rearing activity (Fig. 7A and 7B). The onset of activity at the start of the dark phase was similar, but the N-OX mice were more active and sustained that activity throughout the entire dark phase, whereas the WT mice reduced activity in anticipation of the light phase. The difference in average activity level between the dark and light phases was also increased [53].
Figure 7.
Metabolic cage assessment of N-OX mice. Data of WT mice (n = 6) are shown in black and of N-OX mice (n = 6) in green. Graphs show diurnal patterns over 24 h measured per 13-min interval for the measures indicated by the y-axis labels. Horizontal black bar indicates period of lights off (6:00 pm to 6:00 am). (A) Ambulatory activity counts. (B) Rearing activity counts. (C) VO2 consumption. (D) Respiratory exchange ratio. (E) Food intake. (F) Water intake. (G) Heat produced. (H) Cage temperature. Data are reported as mean ± SEM. In all cases, repeated measures ANOVA indicated a significant time effect (P < 0.0001). Ambulatory activity, rearing activity, VO2, RER, and water intake were associated with significant genotype effects (P < 0.0001, P < 0.0001, P < 0.0001, P = 0.0004, and P = 0.04, respectively). *P < 0.05, **P < 0.01, by post hoc testing. RER, respiratory exchange ratio; Temp, temperature; VO2, oxygen consumption.
There was no significant difference in body weight between genotypes at the end of the assessment. Oxygen consumption was also altered, with lower consumption at the start of dark phase but higher consumption at the end of the dark phase (Fig. 7C), and there was a genotype effect in the respiratory exchange ratio, with greater lipid oxidative metabolism during the light phase in the N-OX mice, consistent with greater sensitivity to fasting (Fig. 7D). There was diurnal variation with food intake but no genotype effect, whereas there was a weak genotype effect with water intake (Fig. 7E and 7F). The expected diurnal variation occurred with heat production and cage temperature, but there was no genotype effect (Fig. 7G and 7H).
F. Unaltered Circadian Activity in Mice With Lack or Overexpression of Neuronal SIRT1
SIRT1 has been implicated in the regulation of the central circadian clock via deacetylation of PGC-1α [54]. In 12:12 LD, female mice had more robust circadian rhythms (P < 0.05) than did male mice [53] and had higher total and average activity counts (P < 0.001). Mice initiated wheel running shortly after lights off, according to the expected nocturnal pattern, and had the same period and activity onset and offset. The sex difference in rhythm robustness, average counts, and total counts persisted in the skeleton photoperiods (P < 0.05, 0.01, and 0.01 respectively). There were small genotype differences in rhythm robustness, 8-hour daytime counts, activity onset, and activity period, but none reached significance. In continuous darkness, a strain effect was associated with period (P < 0.05), with all MUT mice having a slightly shorter period, and a genotype effect was associated with rhythm robustness (P < 0.05), with the MUT genotype having greater robustness. In summary, SIRT1-overexpressing and mutant mice generated species-typical circadian locomotor activity rhythms, with the major difference being female mice were more active and had more robust rhythms.
3. Discussion
Previously, loss of SIRT1 enzyme activity in neurons did not alter glucose metabolism in male mice but increased insulin sensitivity in the brain and periphery, and partially protected mice from high-fat diet–induced obesity [37]. Here, we studied the same N-MUT mice but also studied the effect of overexpression of wild-type SIRT1 in neurons. In male mice, we observed that the N-MUT did not alter glucose tolerance as before, but we did observe decreased glucose tolerance in N-OX mice, which is consistent with the previous report on the N-MUT mice [37]. In contrast, N-MUT female mice had an improvement in glucose tolerance, whereas glucose intolerance was impaired in female N-OX mice. Unlike in the male mice, insulin sensitivity was slightly increased in the female N-MUT mice by insulin tolerance tests, but fasting blood glucose and insulin levels were not changed; consequently, the homeostatic model of assessment of insulin resistance was not significantly different. SIRT1 has been implicated in the response to food restriction; therefore, the mice were challenged with a gradual but progressive CR down to 60% of their normal food intake over 6 weeks. All mice had improved glucose tolerance, and the N-OX mice had better glucose tolerance than their littermates after CR, instead of worse glucose tolerance as had been seen for mice receiving ad libitum normal chow. This suggested that the overexpression of SIRT1 rendered the mice more sensitive to the metabolic effects of CR.
In a previous study, deletion of SIRT1 enzyme activity (Δexon 4) in neurons, astrocytes, glia, and the pituitary, using nestin-cre, disrupted the GH-IGF1 axis, causing runting [43]. Young mice had normal glucose tolerance but older mice were glucose intolerant. CR improved glucose tolerance in WT and the total-brain KO mice, indicating that SIRT1 enzyme activity was not required for the metabolic response to CR43. These KO mice had increased physical activity in a wheel-running experiment but did not respond to CR. A subsequent study showed this activity difference correlated with lower anxiety and increased exploratory drive due to decreased monoamine oxidase activity and increased serotonin and norepinephrine levels [41]. These studies were confounded, however, by the observation that SIRT1 is essential for neural fate determination in neural stem cells and could alter the final balance of neuronal, astrocyte, and oligodendrocyte populations, including Npy/AgRP and Pomc neurons [55–58]. More-focused deletion in specific neuronal populations has produced conflicting results. Deletion of Sirt1 in either Pomc or Sf1 neurons decreases energy expenditure and increases susceptibility to diet-induced obesity but has no effect in mice receiving normal amounts of chow [45, 48]. However, deletion in Agrp neurons has the opposite effect and decreases food intake and body weight [46, 47]. Transgenic SIRT1 overexpression in Pomc or Sf1 neurons increased energy expenditure, increased insulin sensitivity, and reduced body weight. The opposite effect was observed when SIRT1 was overexpressed in the mouse forebrain, using the CMKIIα promoter, which caused decreased energy expenditure and physical activity, resulting in obesity and glucose intolerance [59]. We did not see an effect on body weight in either the female N-OX or N-MUT mice receiving normal amounts of chow, similar to the Pomc or Sf1 neuron KOs, but we observed an increase in physical activity and oxygen consumption in female N-OX mice, and our mice were glucose intolerant. In addition, overexpression of SIRT1 using the mouse prion Prp promoter caused an enhanced response to CR, as measured by physical activity counts [44], in agreement with our study findings. These findings confirm the ability of SIRT1 to mediate the central metabolic effects of CR, but overexpression of SIRT1 alone may not be sufficient to mimic CR.
At the reproductive level, complete loss of Sirt1 results in hypogonadotropic hypogonadism, due to failure of Gnrh neurons to migrate [28]. Female pubertal development was normal in the N-OX and N-MUT mice. Overexpression of SIRT1 impaired estrous cycles and increased the number of days in estrus, perhaps because the N-OX mice did not have the normal increase in FSH levels during estrus to start the next follicular cycle. LH and progesterone surges were absent on the evening of proestrus in the N-OX mice, consistent with a defect in ovulation, which was confirmed by the ovarian histology that showed fewer corpora lutea. These findings are in agreement with those of a study reporting that the SIRT1 activator SRT1720 reduced ovary size and the number of corpora lutea [60]. Despite these defects, the mice were still fertile when mated to WT mice on the day of proestrus, indicating that the mice can respond to normal pheromonal or environmental cues. When subjected to CR, the N-OX mice went into diestrus arrest earlier than their littermates, suggesting that the overexpression of SIRT1 also renders mice more sensitive to the suppression of reproductive cycles. Unlike other fasting studies, LH levels did not decrease with the gradual CR, indicating that we avoided an adrenal stress response, which is known to suppress LH. We noticed a consistent increase in plasma FSH levels during CR in female, but not male, mice, independent of genotype. It is possible that this increase with CR allows the N-OX mice to leave estrus and reenter the estrous cycle before arresting in the next diestrus. The origin of this increase in FSH levels is not known, but we observed a significant increase in pituitary Fshb mRNA in the N-OX mice with CR. Interestingly, loss of SIRT1 has been implicated in increased Fshb production by pituitary gonadotropes [61]. We also observed increases in hypothalamic Npy and Agrp mRNAs in the N-OX mice on CR, which may underlie the effects of CR, because they inhibit GnRH neuronal suppression of reproductive cycles.
Last, SIRT1 has also been implicated in circadian rhythms in the suprachiasmatic nucleus (SCN) [54] and in relaying nutritional inputs to the clock via Sf1 neurons in the VMH [62]. The total-brain SIRT1 KO mice have increased photoperiod, and the SIRT1 overexpressors have decreased photoperiod, compared with WT controls [54]. In our study, neither N-OX nor N-MUT mice demonstrated a circadian phenotype, but it is possible that the synapsin-cre is not expressed in SCN neurons, although synapsin 1 is a reliable marker for synapse formation in the SCN [63]. Additional studies are needed to resolve this discrepancy.
Acknowledgments
We acknowledge the assistance of the Histology Core at the Moores’ Cancer Center, and the Genomics Core at the Sanford Consortium for Regenerative Medicine.
Financial Support: This work was funded in part by the National Institutes of Health (NIH) (Grants HD012303, CA155435, CA196853 to N.J.G.W.) and a VA Merit Review award (I01BX000130) to N.J.G.W. The Histology Core at the Moores’ Cancer Center is supported by NIH Grant CA023100.
Current Affiliation: E. Rickert’s current affiliation is BioMendics, 4209 State Route 44, Rootstown, Ohio 44272. M.O. Fernandez’s current affiliation is Laboratory of Neuroendocrinology, Instituto de Biología y Medicina Experimental, CONICET, Vuelta de Obligado 2490, C1428ADN, Buenos Aires, Argentina.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- 12:12 LD
cycle of 12 hours of light and 12 hours of dark
- AgRP
Agouti-related peptide
- CR
caloric restriction
- GC
granulosa cell
- KO
knockout
- N-OX
wild-type SIRT1
- N-MUT
enzymatically inactive form of SIRT1
- Npy
neuropeptide Y
- POMC
pro-opiomelanocortin
- qPCR
quantitative PCR
- SCN
suprachiasmatic nucleus
- Sirt1
sirtuin 1
- Syn-cre
Syntaxin1 promoter–controlled cre recombinase expression
- WT
wild type
References and Notes
- 1. Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5(1):253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McBurney MW, Clark-Knowles KV, Caron AZ, Gray DA. SIRT1 is a highly networked protein that mediates the adaptation to chronic physiological stress. Genes Cancer. 2013;4(3-4):125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boutant M, Cantó C. SIRT1 metabolic actions: integrating recent advances from mouse models. Mol Metab. 2013;3(1):5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chalkiadaki A, Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012;16(2):180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seyssel K, Alligier M, Meugnier E, Chanseaume E, Loizon E, Canto C, Disse E, Lambert-Porcheron S, Brozek J, Blond E, Rieusset J, Morio B, Laville M, Vidal H. Regulation of energy metabolism and mitochondrial function in skeletal muscle during lipid overfeeding in healthy men. J Clin Endocrinol Metab. 2014;99(7):E1254–E1262. [DOI] [PubMed] [Google Scholar]
- 7. Lomb DJ, Laurent G, Haigis MC. Sirtuins regulate key aspects of lipid metabolism. Biochim Biophys Acta. 2010;1804(8):1652–1657. [DOI] [PubMed] [Google Scholar]
- 8. Heyward FD, Walton RG, Carle MS, Coleman MA, Garvey WT, Sweatt JD. Adult mice maintained on a high-fat diet exhibit object location memory deficits and reduced hippocampal SIRT1 gene expression. Neurobiol Learn Mem. 2012;98(1):25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150(3):620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105(28):9793–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boutant M, Joffraud M, Kulkarni SS, García-Casarrubios E, García-Roves PM, Ratajczak J, Fernández-Marcos PJ, Valverde AM, Serrano M, Cantó C. SIRT1 enhances glucose tolerance by potentiating brown adipose tissue function. Mol Metab. 2014;4(2):118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100(19):10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23(1):38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/- mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151(6):2504–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ding RB, Bao J, Deng CX. Emerging roles of SIRT1 in fatty liver diseases. Int J Biol Sci. 2017;13(7):852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seifert EL, Caron AZ, Morin K, Coulombe J, He XH, Jardine K, Dewar-Darch D, Boekelheide K, Harper ME, McBurney MW. SirT1 catalytic activity is required for male fertility and metabolic homeostasis in mice. FASEB J. 2012;26(2):555–566. [DOI] [PubMed] [Google Scholar]
- 18. Caron AZ, He X, Mottawea W, Seifert EL, Jardine K, Dewar-Darch D, Cron GO, Harper ME, Stintzi A, McBurney MW. The SIRT1 deacetylase protects mice against the symptoms of metabolic syndrome. FASEB J. 2014;28(3):1306–1316. [DOI] [PubMed] [Google Scholar]
- 19. Gillum MP, Kotas ME, Erion DM, Kursawe R, Chatterjee P, Nead KT, Muise ES, Hsiao JJ, Frederick DW, Yonemitsu S, Banks AS, Qiang L, Bhanot S, Olefsky JM, Sears DD, Caprio S, Shulman GI. SirT1 regulates adipose tissue inflammation. Diabetes. 2011;60(12):3235–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schenk S, McCurdy CE, Philp A, Chen MZ, Holliday MJ, Bandyopadhyay GK, Osborn O, Baar K, Olefsky JM. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J Clin Invest. 2011;121(11):4281–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang RH, Li C, Deng CX. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int J Biol Sci. 2010;6(7):682–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, Wong K, Giles A, et al. Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology. 2014;146(2):539–549.e537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9(4):327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6(6):759–767. [DOI] [PubMed] [Google Scholar]
- 25. Boutant M, Kulkarni SS, Joffraud M, Raymond F, Métairon S, Descombes P, Cantó C. SIRT1 gain of function does not mimic or enhance the adaptations to intermittent fasting. Cell Reports. 2016;14(9):2068–2075. [DOI] [PubMed] [Google Scholar]
- 26. Boutant M, Cantó C. SIRT1: a novel guardian of brown fat against metabolic damage. Obesity (Silver Spring). 2016;24(3):554. [DOI] [PubMed] [Google Scholar]
- 27. Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Sante G, Wang L, Wang C, Jiao X, Casimiro MC, Chen K, Pestell TG, Yaman I, Di Rocco A, Sun X, Horio Y, Powell MJ, He X, McBurney MW, Pestell RG. Sirt1-deficient mice have hypogonadotropic hypogonadism due to defective GnRH neuronal migration. Mol Endocrinol. 2015;29(2):200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bell EL, Nagamori I, Williams EO, Del Rosario AM, Bryson BD, Watson N, White FM, Sassone-Corsi P, Guarente L. SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development. 2014;141(18):3495–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolthur-Seetharam U, Teerds K, de Rooij DG, Wendling O, McBurney M, Sassone-Corsi P, Davidson I. The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod. 2009;80(2):384–391. [DOI] [PubMed] [Google Scholar]
- 31. Tatone C, Di Emidio G, Vitti M, Di Carlo M, Santini S Jr, D’Alessandro AM, Falone S, Amicarelli F. Sirtuin functions in female fertility: possible role in oxidative stress and aging. Oxid Med Cell Longev. 2015;2015:659687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21(8):1807–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26(21):8122–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J Neurosci. 2011;31(48):17425–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore RL, Faller DV. SIRT1 represses estrogen-signaling, ligand-independent ERα-mediated transcription, and cell proliferation in estrogen-responsive breast cells. J Endocrinol. 2013;216(3):273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herskovits AZ, Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81(3):471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu M, Sarruf DA, Li P, Osborn O, Sanchez-Alavez M, Talukdar S, Chen A, Bandyopadhyay G, Xu J, Morinaga H, Dines K, Watkins S, Kaiyala K, Schwartz MW, Olefsky JM. Neuronal Sirt1 deficiency increases insulin sensitivity in both brain and peripheral tissues. J Biol Chem. 2013;288(15):10722–10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Albani D, Polito L, Forloni G. Sirtuins as novel targets for Alzheimer’s disease and other neurodegenerative disorders: experimental and genetic evidence. J Alzheimers Dis. 2010;19(1):11–26. [DOI] [PubMed] [Google Scholar]
- 39. Michán S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W, Mervis RF, Chen J, Guerin KI, Smith LE, McBurney MW, Sinclair DA, Baudry M, de Cabo R, Longo VD. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30(29):9695–9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abe-Higuchi N, Uchida S, Yamagata H, Higuchi F, Hobara T, Hara K, Kobayashi A, Watanabe Y. Hippocampal Sirtuin 1 signaling mediates depression-like behavior. Biol Psychiatry. 2016;80(11):815–826. [DOI] [PubMed] [Google Scholar]
- 41. Libert S, Pointer K, Bell EL, Das A, Cohen DE, Asara JM, Kapur K, Bergmann S, Preisig M, Otowa T, Kendler KS, Chen X, Hettema JM, van den Oord EJ, Rubio JP, Guarente L. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147(7):1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3(3):e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23(24):2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, Imai S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30(30):10220–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, Stuart RC, Perello M, Vianna CR, Nillni EA, Rahmouni K, Coppari R. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, Xu AW, Souza DO, Gao Q, Diano S, Gao XB, Horvath TL. Agrp neurons mediate Sirt1's action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J Neurosci. 2010;30(35):11815–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sasaki T, Kikuchi O, Shimpuku M, Susanti VY, Yokota-Hashimoto H, Taguchi R, Shibusawa N, Sato T, Tang L, Amano K, Kitazumi T, Kuroko M, Fujita Y, Maruyama J, Lee YS, Kobayashi M, Nakagawa T, Minokoshi Y, Harada A, Yamada M, Kitamura T. Hypothalamic SIRT1 prevents age-associated weight gain by improving leptin sensitivity in mice. Diabetologia. 2014;57(4):819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramadori G, Fujikawa T, Anderson J, Berglund ED, Frazao R, Michán S, Vianna CR, Sinclair DA, Elias CF, Coppari R. SIRT1 deacetylase in SF1 neurons protects against metabolic imbalance. Cell Metab. 2011;14(3):301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15(7):859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rempe D, Vangeison G, Hamilton J, Li Y, Jepson M, Federoff HJ. Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis. 2006;44(1):44–49. [DOI] [PubMed] [Google Scholar]
- 52. Myers M, Britt KL, Wreford NG, Ebling FJ, Kerr JB. Methods for quantifying follicular numbers within the mouse ovary. Reproduction. 2004;127(5):569–580. [DOI] [PubMed] [Google Scholar]
- 53.Rickert E, Fernandez M, Choi I, Gorman M, Olefsky J, Webster N. Supplementary data repository. 2018. 10.6084/m9.figshare.7396940.v1
- 54. Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153(7):1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cai Y, Xu L, Xu H, Fan X. SIRT1 and neural cell fate determination. Mol Neurobiol. 2016;53(5):2815–2825. [DOI] [PubMed] [Google Scholar]
- 56. Desai M, Li T, Han G, Ross MG. Programmed hyperphagia secondary to increased hypothalamic SIRT1. Brain Res. 2014;1589:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rafalski VA, Ho PP, Brett JO, Ucar D, Dugas JC, Pollina EA, Chow LM, Ibrahim A, Baker SJ, Barres BA, Steinman L, Brunet A. Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat Cell Biol. 2013;15(6):614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saharan S, Jhaveri DJ, Bartlett PF. SIRT1 regulates the neurogenic potential of neural precursors in the adult subventricular zone and hippocampus. J Neurosci Res. 2013;91(5):642–659. [DOI] [PubMed] [Google Scholar]
- 59. Wu D, Qiu Y, Gao X, Yuan XB, Zhai Q. Overexpression of SIRT1 in mouse forebrain impairs lipid/glucose metabolism and motor function. PLoS One. 2011;6(6):e21759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou XL, Xu JJ, Ni YH, Chen XC, Zhang HX, Zhang XM, Liu WJ, Luo LL, Fu YC. SIRT1 activator (SRT1720) improves the follicle reserve and prolongs the ovarian lifespan of diet-induced obesity in female mice via activating SIRT1 and suppressing mTOR signaling. J Ovarian Res. 2014;7(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lannes J, L’Hôte D, Garrel G, Laverrière JN, Cohen-Tannoudji J, Quérat B. Rapid communication: A microRNA-132/212 pathway mediates GnRH activation of FSH expression. Mol Endocrinol. 2015;29(3):364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Orozco-Solis R, Ramadori G, Coppari R, Sassone-Corsi P. SIRT1 relays nutritional inputs to the circadian clock through the Sf1 neurons of the ventromedial hypothalamus. Endocrinology. 2015;156(6):2174–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moore RY, Bernstein ME. Synaptogenesis in the rat suprachiasmatic nucleus demonstrated by electron microscopy and synapsin I immunoreactivity. J Neurosci. 1989;9(6):2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]