Abstract
Precision oncology applies genomic and other molecular analyses of tumor biopsies to improve the diagnosis and treatment of cancers. In addition to identifying therapeutic options, precision oncology tracks the response of a tumor to an intervention at the molecular level and detects drug resistance and the mechanisms by which it occurs. Integrative genomics can include sequencing specific panels of genes, exomes, or the entire triad of the patient’s germline and tumor exome plus tumor transcriptome. Although the capabilities of sequencing technologies continue to improve, widespread adoption of genomics-driven precision oncology in the clinic has been held back by logistical, regulatory, financial, and ethical considerations. Nevertheless, integrative clinical sequencing programs applied at the point of care have the potential to improve the clinical management of cancer patients.
The earliest documented examples of targeting the underlying mechanisms driving tumor growth to treat cancer might be George Beatson’s treatment of breast cancer patients by oophorectomy in 18961, and Charles Huggins use of castration to treat prostate cancer half a century later2. Although the mechanisms that underlie cancer have been investigated for more than a hundred years, clinical management remains rooted in morphological and histopathological methods to diagnose and estimate prognosis, while treatments rely on surgery to remove tumors followed by chemo- and/or radiation therapy to stop uncontrolled cell proliferation3,4.
Insights gained from the molecular characterization of aberrant genes, cell surface markers, hormonal/endocrine mediators, and signaling pathways associated with cancer have been incorporated into diagnostic and treatment strategies (Figure 1). The application of targeted therapies matched to specific aberrations for some cancers5–15, synthetic lethal targeting of DNA repair machinery in BRCA-deficient ovarian cancers5, and recent progress with immune checkpoint inhibitors in cancers with hypermutation/neo-antigen signatures6 have collectively fueled optimism that identification of molecular targets in individual cancers to enable targeted therapeutics could represent a general paradigm for cancer care. This optimism has been tempered by inconsistent responses to targeted therapies and emergence of drug resistance in many patients.
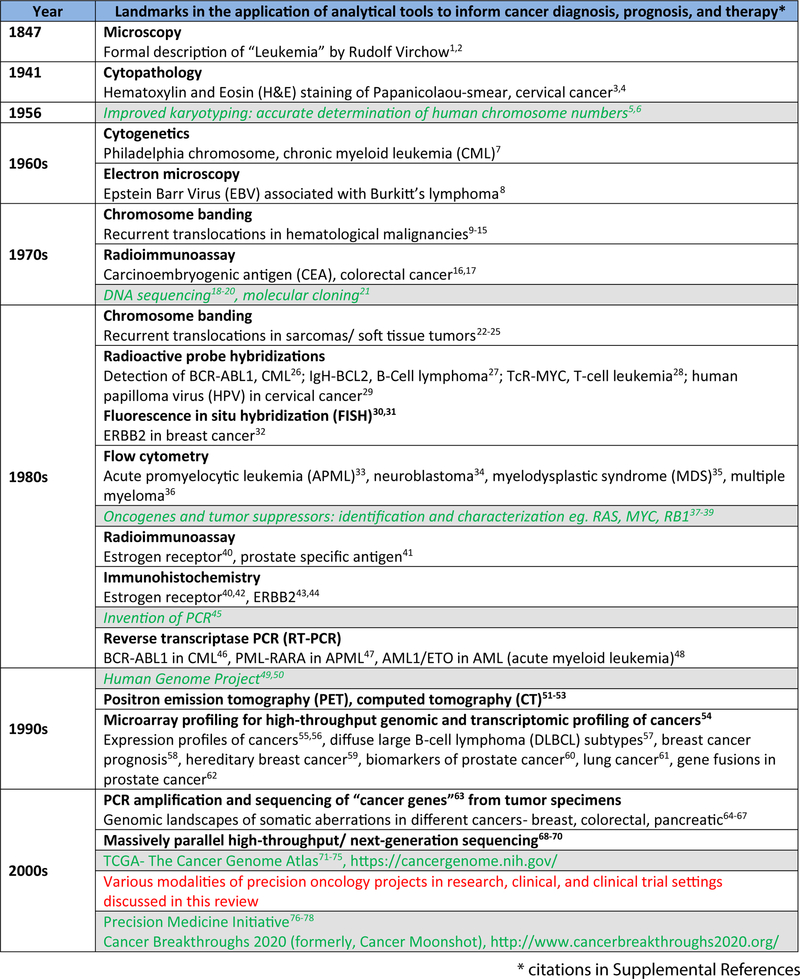
Figure 1. The progression of analytical tools in oncology.
The historical timeline highlights key developments in the assessment of cancer cells/tumor tissue, starting from the microscopic description of leukemia cells by Rudolf Virchow, up to the futuristic Cancer Breakthroughs 2020 project. It may be noted that the modern tool-kit for cancer analyses includes a range of old and new tools, and the high-throughput sequencing approaches add a highly informative component, complementary to other methods that include imaging, histopathology, and biochemical analyses.
Over the past decade, multiple large-scale genomic studies have identified genomic, transcriptomic, and proteomic aberrations that are specific to one cancer type or common among different cancers. These findings have suggested to many researchers that clinical management of individual cancer patients should be routinely informed by comprehensive molecular analyses of their tumors. Fortunately, policy and funding have kept pace with science, as exemplified by the Precision Medicine7,8, 9 and National Cancer Moonshot Initiatives10.
Precision medicine initiatives are poised to transform the paradigm of population-based clinical studies to define treatments for average patients into biomarker-driven clinical trials to identify the best treatments for individual patients 24,11. Initial clinical efforts have mainly focused on sequencing panels of well-validated therapeutic target genes, and have gradually expanded to include broader panels of cancer-associated genes. Less frequently, whole exome sequencing or comprehensive, integrative sequencing encompassing germline, genomic, and transcriptomic sequencing have been performed (Figure 2).
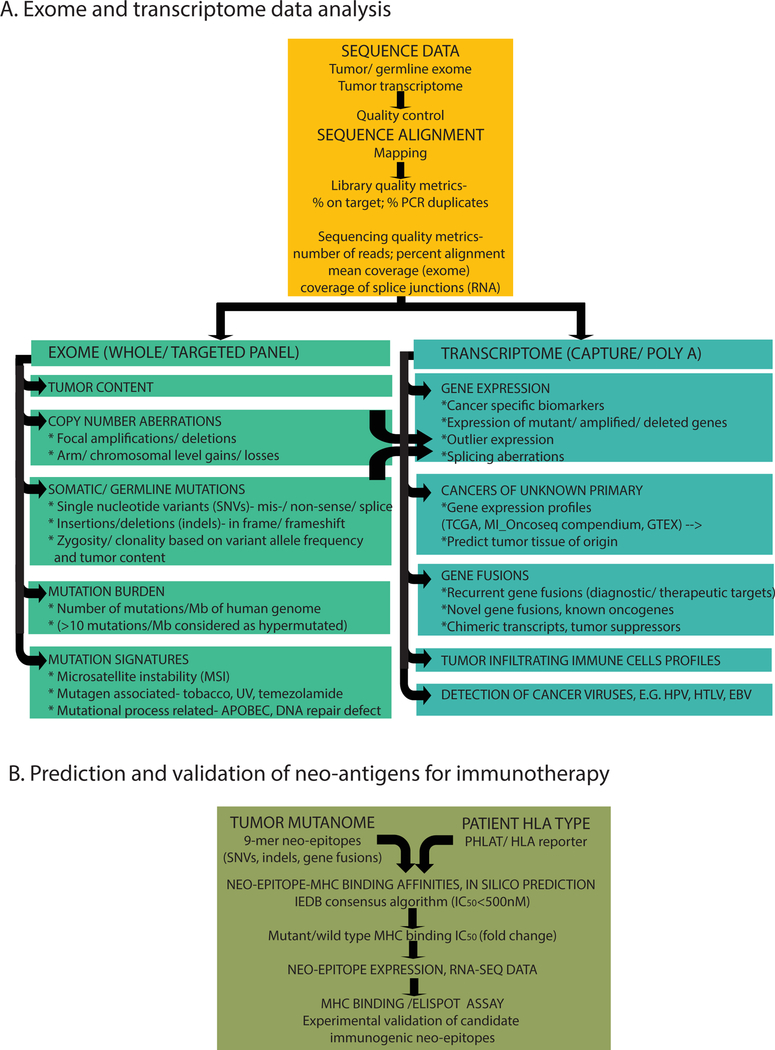
Figure 2. Integrative sequencing analysis to define the spectrum of cancer aberrations.
A. Bioinformatic workflow and classes of cancer aberrations identified. The primary sequencing data is subjected to different quality control metrics and aligned to the reference genome to define the indicated genomic and transcriptomic aberrations. B. Prediction of neo-antigens for immunotherapy. The flow chart indicates primary steps involved in in silico prediction of immunogenic cancer specific neo-antigens in tumor samples, for potential use in developing personalized peptide vaccines.
We review the application of precision oncology by sequencing gene panels, the use of whole exome capture or genome sequencing, as well as RNA sequencing, in clinical trials and routine clinical practice. We also review analytical setup and operational workflow of current integrative clinical sequencing programs. This is followed by a brief discussion of salient issues and directions for future developments.
Sequencing of gene panels for precision therapeutics
Specific aberrations in approximately 40 different cancer genes are represented in FDA (Food and Drug Administration)-approved targeted therapeutics and detected by FDA-mandated diagnostic assays that use cytogenetics, PCR, microarrays, or Sanger sequencing to detect mutations. High-throughput sequencing can, in principle, detect all of these mutations with sufficient sensitivity, and, thus, sequencing of panels of therapeutically targetable genes has emerged as an entry point for precision oncology 12, 13. For example, in a clinical trial that featured sequencing of 10 cancer genes in lung adenocarcinoma tissues from 733 patients, one or more oncogenic mutations were identified in 466 patients (64%), and 24 patients (3%) had mutations in two or more genes14. 260 patients received targeted therapy and achieved median survival of 3.5 years, compared with 2.4 years for 318 patients that did not receive matched therapy15.
Seizing the opportunity for immediate impact on patient care, the National Cancer Institute (NCI) initiated several clinical trials to test the application of biomarker-driven therapeutic approaches16. These trials include the Lung Master Protocol (Lung-MAP, S1400), which aims to target therapies for lung squamous cell carcinoma (SCC). SCC has no approved targeted therapies, and the frequency of actionable somatic aberrations in these cancers is so low (5%–20%) that traditional clinical trials are impractical17. In the Lung-MAP, NGS (next-generation sequencing) is used to identify actionable molecular abnormalities, and patients are randomized to targeted therapy or standard of care. Another example is the Adjuvant Lung Cancer Enrichment Marker Identification and Sequencing Trial (ALCHEMIST) in which early-stage lung cancer patients are screened for EGFR or ALK mutations by sequencing18, 19. In the Molecular Analysis for Therapy Choice (NCI-MATCH) clinical trial, biopsies of adult solid tumors and lymphomas are sequenced to screen for mutations in a panel of defined actionable genes, and patients are matched with either approved or investigational (Phase II) drugs20–23. The NCI-MATCH trial received such an enthusiastic response upon launch that enrollment had to be paused from January to April/May 2016 to allow for expansion of lab capacity, as well as addition of more than a dozen new treatment arms24. Similar trials were initiated for advanced solid tumors (NCI-MPACT; NCT01827384), pancreatic cancer (IMPaCT trial in Australia25), and thoracic malignancies (CUSTOM trial, NCT0130604540, 41), all involving sequencing of select target genes to be matched with precision therapies.
Sequencing extended panels of cancer genes
Following promising studies to identify hotspot mutations and single-nucleotide variants (SNVs) in specific genes, extended panels have been incorporated into recent analyses to detect SNVs, copy number variants (CNVs), structural rearrangements, and gene fusions. For example, the Memorial Sloan Kettering (MSK)-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) project used targeted sequencing of exons and selected introns of 341 cancer genes26 and was expanded to 410 genes in a follow-up study27. Likewise, Foundation Medicine sequenced exomes of 287 cancer-related genes plus intronic sequences from 19 genes involved in rearrangements or other aberrations28; later, this was expanded to 315 exomes, plus intronic sequences from 28 genes. Perhaps one of the largest such efforts is the University of Michigan’s MI_Oncoseq program, where exomes for a panel of 1700 cancer-related genes are captured for parallel sequencing of tumor and germline DNA29. Sequencing gene panels helps identify a broad range of cancer-associated aberrations but retains the advantages of cost effectiveness, fast output, and the use of limiting amounts of starting material, all preferred attributes for the application of NGS in clinical practice. Hand-in-hand with increasing participation of patients and cancer centers, more patients are being matched with therapeutics that are targeted for specific molecular aberrations but not yet approved for cancer treatment by the FDA. The American Society of Clinical Oncology (ASCO) has capitalized on these findings by launching a Targeted Agent and Profiling Utilization Registry (TAPUR) study (https://www.tapur.org/), which is a non-randomized clinical trial that will formally test the potential utility for off-label targeted therapies in cancer.
Exome or genome sequencing for individuals
The data from gene panel sequencing are indeed limited by the selection of genes in the panel but may also be limiting in chromosomal ploidy aberrations, arm level gains/losses, and unknown/new “cancer genes”. Whole exome capture sequencing analyzes the complete coding portion of the genome and provides a comprehensive genomic profile of aberrations in protein coding genes, arguably at relatively reduced coverage at individual loci; it is also costlier, takes longer, and is more resource and analysis intensive than using a limited panel of sequences.
Whole exome or genome sequencing has been particularly informative for analyzing exceptional therapeutic response or resistance. For example, a metastatic bladder cancer patient who showed an exceptional response to the mTOR inhibitor everolimus in a clinical trial categorized as “failed”, NCT00805129, showed loss of function mutations in TSC1 and NF2. These genes are associated with mTOR pathway activity but were not previously associated with therapeutic response30. Similar mutations were identified in additional bladder cancer cases, who may also potentially respond to everolimus. Similarly, activating mutations in mTOR were identified in an exceptional responder to everolimus and pazopanib31; mutation in RAD50 associated with loss of ATM signaling32 was found in an exceptional responder to treatment with CHK1 inhibitor in combination with DNA-damaging agent irinotecan; and an exceptional response to an IGF-1R-specific antibody was observed in a patient with ALK fusion-positive lung cancer33.
The underlying mechanisms for drug sensitivity or resistance are not always straightforward. For example, a pre-treatment tumor sample from a patient with stage IVA head and neck squamous cell carcinoma who showed a near complete histologic response to erlotinib revealed no EGFR alterations as expected; instead, the tumor harbored an activating mutation in MAPK1 (p.E322K) that enhanced EGFR phosphorylation, resulting in erlotinib sensitivity34. Further, sequencing of BRAF-mutant colorectal cancer biopsies pre- and post-treatment with RAF inhibitors, identified KRAS amplification and overexpression in one patient, BRAF amplification and overexpression in another case, and a putatively activating mutation in the RAF family protein, ARAF p.Q489L, plus a resistance mutation in MAP2K1 p.F53L in a third patient35. In a study to investigate markers associated with resistance to PD-1 immune checkpoint blockade in metastatic melanoma, whole exome sequencing identified loss of function mutations in JAK1, JAK2, or B2M36.
Remarkably, a review of 10 years of unpublished data from phase II clinical trials by the NCI Cancer Therapy Evaluation Program has estimated that as many as 10% of patients were “exceptional responders” in phase II clinical trials of therapies that failed to receive FDA approval37. NCI launched the Exceptional Responder Program in 2014 to systematically re-analyze these trials with the aim of identifying new combinations of aberrations and therapeutics37–39. These analyses will require comprehensive whole exome/genome analyses.
Despite evidence of utility, clinical sequencing programs have been deterred from application of whole exome or whole genome sequencing in routine clinical settings owing to the extra time, cost, resources, data storage, and analysis requirements. Instead, most sequencing centers sequence targeted panels that can be gradually expanded to incorporate additional genes/sequences of interest.
Precision oncology in routine clinical practice
Sequencing-based clinical precision oncology programs have only recently been implemented and only a few have reported results so far. Among these representative programs, the University of California San Diego (UCSD) Moores Cancer Center reported findings from the Profile Related Evidence Determining Individualized Cancer Therapy trial (PREDICT-UCSD; NCT02478931), wherein 347 patients with advanced solid malignancies were analyzed using Foundation Medicine exon capture panels; of these, 87 patients (25%) were treated with a matched therapy and had a slightly longer median progression-free survival compared with unmatched patients40. Mentioned above, the MSK-IMPACT27 project described an exome capture-based 410 gene panel assay for solid cancers (with matched germline samples), wherein more than 10,000 patients with advanced cancer have been analyzed so far, with up to 11% of patients enrolled in genomically matched clinical trials29. The MSKCC study prioritizes the use of targeted gene panels over whole exome sequencing to maximize throughput and depth of coverage, and, additionally, to reduce costs. The precision oncology program at the MD Anderson Cancer Center41 reported sequencing results from 1200 patients with advanced cancer, in which targeted sequencing of 201 genes or hotspot mutation analysis of 11 to 50 genes were performed. At least one alteration in a potentially actionable gene was noted in 945 patients (79%) using the larger panel, compared to only 527 patients (44%) with hotspot testing, supporting the use of large panels in routine clinical tests42. Weill Cornell Medical College–New York applied whole exome sequencing in routine clinical practice, analyzing tumor-normal pairs of 97 metastatic cancer cases, with informative aberrations observed in 91 patients, of whom 5 went on to receive targeted therapies43. These early reports are indicative of the immense interest in the community to deploy precision oncology in routine cancer care, even as questions of cost, choice of optimal analytical platforms, standardization of the assays and reporting metrics, and potential efficacy of these efforts continue to be deliberated.
Integrative clinical sequencing in precision oncology
Combining exome sequencing of germline and tumor tissue DNA with RNA sequencing can interrogate a wide array of somatic and germline aberrations in parallel (Figure 2) and has been effective in discovering actionable aberrations in osteosarcoma44, urothelial carcinoma45, non-small cell lung cancer (NSCLC)46, endometrioid endometrial carcinoma (EEC)47, melanoma48, 49, and a case of Sézary syndrome50.
The University of Michigan has implemented an integrative clinical sequencing program called MI_Oncoseq51 for all-comer advanced cancer patients with diverse tumor types in a hospital/academic setting. Briefly, as shown in Figure 3, participation in clinical sequencing is initiated by the attending physician. Following written informed consent from the patient, tumor biopsy and blood or buccal swab are used to extract DNA and RNA, which are sequenced and analyzed. These data are analyzed for potential clinical relevance and actionability through extensive literature survey of the disease, tumor specific aberrations, and potential therapeutic matches (BOX 1). Finally, the integrative molecular analyses are summarized and any therapeutic insights discussed at a multidisciplinary tumor board meeting attended by the referring physician and various key personnel involved in the entire MI_Oncoseq process. A final summary clinical report with clinical recommendations is provided for the referring physician. From start to finish, this process takes three weeks in most cases.
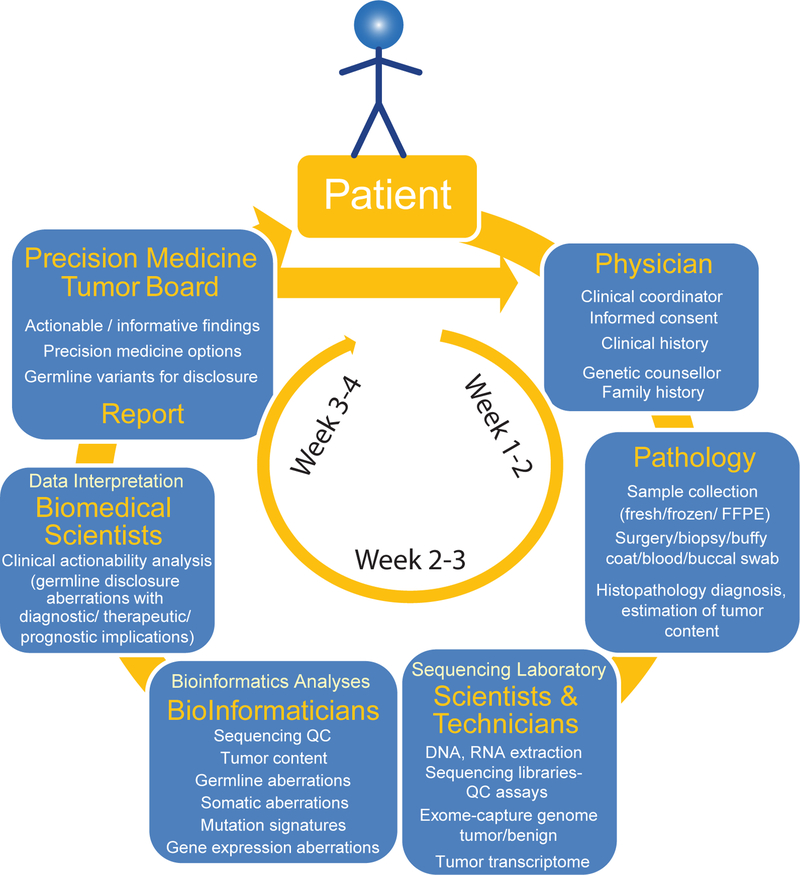
Figure 3. Workflow of integrative clinical sequencing for precision oncology.
The patient, in consultation with the cancer physician, enters the clinical sequencing program upon signing the informed consent. Along with documentation of detailed clinical history, a genetic counselor obtains family history of the patient to assess likely hereditary predisposition to cancer. Patient’s tumor biopsy is flash frozen in OCT blocks, and, along with blood or buccal swab, the samples are sent to the CLIA-certified sequencing laboratory. Histology sections of the tumor biopsy blocks are evaluated by a clinical pathologist for diagnosis and tumor content. DNA and RNA from tissue blocks with the highest tumor content and DNA from blood/buccal samples are used to generate sequencing libraries. Exome capture libraries from germline and tumor samples and the transcriptome library from tumor RNA are analyzed for germline and somatic aberrations. Potentially actionable molecular observations are identified and discussed at the multidisciplinary precision medicine tumor board (see also BOX 1), and a summary report of clinical recommendations is provided to the attending physician.
BOX 1. Precision Medicine Tumor Board.
The actionable germline and somatic findings along with clinical implications and therapeutic options for individual cases are discussed in the context of the following background information. Please refer to the main text of this article for detailed discussion of the clinical data analyses.
Clinical History:
Brief clinical histories of the patients, from the time of cancer diagnosis, key clinical signposts, prior therapies, responses, status of imaging, histological and/or molecular markers, history of response to therapies, disease progression etc. leading up to the details of biopsy of the clinical sequencing, are noted.
Family History:
Incidences of cancer in the family of patients are documented by a clinical geneticist to assess each case as likely familial or sporadic. Detailed in the main text, in a subset of cases, pathogenic germline variants are noted irrespective of a family history of cancer.
Pathology:
Hematoxylin and eosin stained sections of the tumor biopsy specimens to be used for sequencing are assessed by a pathologist for histopathology of the tumor specimen as well as estimation of tumor content. Tissue blocks adjudged to represent the highest tumor content are used for making sequencing libraries.
Samples for Sequencing Libraries:
DNA and RNA are simultaneously isolated from the same tumor tissue sections to ensure concordance of the samples used for genomic and transcriptomic analyses. DNA for germline sequencing is typically derived from blood samples in the case of solid cancers and from buccal swabs for hematological malignancies. Although frozen tumor samples are preferred starting material for sequencing DNA or RNA, recent technical and analytical improvements have facilitated routine use of formalin fixed-paraffin embedded (FFPE) samples for clinical sequencing28, 54, 66, 188–190. Quality of RNA sequencing is particularly sensitive to the integrity of the starting RNA material. To improve the information content of RNA-seq using suboptimal quality RNA samples, we have developed a transcriptome capture methodology using standard exome capture probes191.
QC of Sequencing Data:
Based on the guidelines proposed by the Standardization of Clinical Testing (Nex-StoCT) workgroup192, quality metrics of the sequence data are assessed before launching into mutation/gene expression analyses. Exome capture data from tumor samples with approximately 400X average coverage, matched with normal (blood or buccal) samples with approximately 300X average coverage, are considered optimal for analyses. The tumor content is estimated using a set of high quality SNV candidates on 2-copy genomic regions54. Sequencing quality is determined by a number of standardized criteria29, and sequencing libraries failing any of the quality metrics are flagged and factored during biological analysis and interpretation of the data.
Similar to the multidisciplinary cancer conferences (“tumor boards”) that are currently mandated by the American College of Surgeons to facilitate access to expertise in evolving technologies in accredited cancer programs52, 53, integrative precision oncology programs have implemented multidisciplinary molecular tumor boards to discuss molecular findings and make clinical recommendations66,77–81. MI_Oncoseq has implemented a multidisciplinary precision medicine tumor board (PMTB) comprising oncologists, cancer geneticists, genetic counsellors, pathologists, biologists, bioinformaticians, bioethicists, clinical study coordinators, and ad hoc expertise51, 54. Underscoring the vital importance of tumor boards in driving clinical sequencing efforts, and the need for standardized practices, ASCO is considering the development of a web-based, interactive molecular tumor board for educational purposes55.
At MI_Oncoseq PMTB meetings, individual cases are presented with clinical history, family history, tumor pathology and histopathology of specimens used for sequencing, summary details of sequencing libraries, quality control metrics of sequencing data, and estimation of tumor content based on the proportion of copy neutral heterozygous SNVs in the data [see BOX 1 for details of these components of MI_Oncoseq PMTB meetings]. This is followed by a detailed assessment of germline and cancer aberrations, potential clinical implications thereof, and proposed follow-up action items, all topics expanded upon in the next sections. Clinical coordinators then continue to track the clinical course of patients in consultation with the clinicians.
Germline DNA sequencing of cancer patients
Sequencing of the cancer patient’s germline DNA in parallel with tumor DNA has typically been undertaken to filter out germline polymorphisms from the somatic mutation data. However, identification of germline mutations in cancer predisposing genes has critical implications for the patient and their families, prompting active screening, surveillance, prophylactic actions, and preventative lifestyle adjustments, as well as can be informative with regard to treatment plans (Table 1). For example, germline aberrations in DNA repair pathway genes BRCA1/2, as well as ATM, CHEK2, and PALB2, have been associated with responsiveness to PARP inhibitor therapies in ovarian, breast, and prostate cancers56–60. Similarly, germline mutations in mismatch repair pathways have been associated with responsiveness to immune blockade therapy61.
Table 1. Summary of actionable germline aberrations in cancer predisposition genes.
Genes associated with predisposition or susceptibility to cancer included in NCCN guidelines, ACMG recommendations for reporting (Green et al, Genet. Med. 2013;15(7):565–574), currently being tested in clinical trials (see Supplementary Table 1A for details), or considered for reporting in MI_Oncoseq study.
| Gene MIM# | Gene | Cytoband | Chr coordinates (GRCh38) | Gene name | Phenotype* | NCCN guidelines | ACMG recommendation | Ongoing clinical trials** | MI_Oncoseq |
|---|---|---|---|---|---|---|---|---|---|
| 611731 | APC | 5q22.2 | 5:112707504–112846238 | Adenomatous polyposis coli | Colorectal cancer, somatic, 114500; Brain tumor-polyposis syndrome 2, AD, 175100; Hepatoblastoma, somatic, 114550; Adenomatous polyposis coli, AD, 175100; Gastric cancer, somatic, 613659; Adenoma, periampullary, somatic | X | X | X | X |
| 607585 | ATM | 11q22.3 | 11:108222483–108369101 | Ataxia-telangiectasia mutated (includes complementation groups A, C, D, and E) | Lymphoma, B-cell non-Hodgkin, somatic; Breast cancer, susceptibility to, AD, 114480; T-cell prolymphocytic leukemia, somatic; Lymphoma, mantle cell, somatic | X | X | ||
| 601215 | ATR | 3q23 | 3:142449234–142578825 | Ataxia-telangiectasia and Rad3-related (FRAP-related protein-1) | Cutaneous telangiectasia and cancer syndrome, familial, AD, 614564 | X | |||
| 601593 | BARD1 | 2q35 | 2:214725644–214809710 | BRCA1-associated RING domain 1 | Breast cancer, susceptibility to, AD, 114480 | X | |||
| 601299 | BMPR1A | 10q23.2 | 10:86755785–86927968 | Bone morphogenetic protein receptor, type IA | Polyposis, juvenile intestinal, AD, 174900; Polyposis syndrome, hereditary mixed, 2, 610069; Juvenile polyposis syndrome, infantile form, AD, 174900 | X | |||
| 113705 | BRCA1 | 17q21.31 | 17:43044294–43125482 | Breast cancer-1 gene | Pancreatic cancer, susceptibility to, 4, 614320; Breast-ovarian cancer, familial, 1, AD; Multifactorial), 604370 | X | X | X | X |
| 600185 | BRCA2 | 13q13.1 | 13:32315479–32399671 | BRCA2 gene | Breast cancer, male, susceptibility to, AD, 114480; Prostate cancer, AD, 176807; Wilms tumor, SM; AD, 194070; Pancreatic cancer 2, 613347; Medulloblastoma, AD, 155255; Glioblastoma 3, AR, 613029; Breast-ovarian cancer, familial, 2, AD, 612555 | X | X | X | |
| 605882 | BRIP1 | 17q23.2 | 17:61679185–61864119 | BRCA1-associated C-terminal helicase 1 | Breast cancer, early-onset, AD, 114480 | X | X | ||
| 192090 | CDH1 | 16q22.1 | 16:68737289–68835541 | Cadherin-1 (E-cadherin; uvomorulin) | Ovarian carcinoma, somatic, 167000; Gastric cancer, familial diffuse, with or without cleft lip and/or palate, AD, 137215; Endometrial carcinoma, somatic, 608089; Prostate cancer, susceptibility to, AD, 176807; Breast cancer, lobular, AD, 114480 | X | X | ||
| 604373 | CHEK2 | 22q12.1 | 22:28687742–28741865 | Checkpoint kinase 2, S. pombe, homolog of (RAD53, S. cerevisiae, homolog of) | Prostate cancer, familial, susceptibility to, AD, 176807; Breast cancer, susceptibility to, AD, 114480; Osteosarcoma, somatic, 259500 | X | X | X | |
| 606241 | DICER1 | 14q32.13 | 14:95086227–95158009 | Dicer, Drosophila, homolog of, 1 | Rhabdomyosarcoma, embryonal, 2, 180295; Pleuropulmonary blastoma, AD, 601200 | X | |||
| 131550 | EGFR | 7p11.2 | 7:55019031–55207337 | Epidermal growth factor receptor | Non-small cell lung cancer, susceptibility to, AR, 211980; Non-small cell lung cancer, response to tyrosine kinase inhibitor in, AR, 211980; Adenocarcinoma of lung, response to tyrosine kinase inhibitor in, AR, 211980 | X | |||
| 185535 | EPCAM | 2p21 | 2:47369147–47387027 | Epithelial cellular adhesion molecule | Colorectal cancer, hereditary nonpolyposis, type 8, 613244 | X | |||
| 607139 | FANCA | 16q24.3 | 16:89737550–89816657 | Fanconi anemia, complementation group A | Fanconi anemia, complementation group A, AR, 227650 | X | X | ||
| 136850 | FH | 1q43 | 1:241497556–241519784 | Fumarate Hydratase | Leiomyomatosis and renal cell cancer, AD, 150800 | X | X | X | |
| 610290 | GALNT12 | 9q22.33 | 9:98807698–98850080 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 | Colorectal cancer, susceptibility to, 1, 608812 | X | |||
| 604607 | HOXB13 | 17q21.32 | 17:48724762–48728748 | Homeobox B13 | Breast cancer, 114480; Prostate cancer, 610997 | X | |||
| 613733 | MEN1 | 11q13.1 | 11:64803513–64811293 | Menin | Adrenal adenoma, somatic; Parathyroid adenoma, somatic; Multiple endocrine neoplasia 1, AD, 131100; Lipoma, somatic; Carcinoid tumor of lung; Angiofibroma, somatic | X | |||
| 164860 | MET | 7q31.2 | 7:116672358–116798385 | Oncogene MET | Renal cell carcinoma, papillary, 1, familial and somatic, 605074; Hepatocellular carcinoma, childhood type, somatic, 114550 | X | |||
| 120436 | MLH1 | 3p22.2 | 3:36993349–37050845 | mutL, E. coli, homolog of, 1 | Colorectal cancer, hereditary nonpolyposis, type 2, 609310; Mismatch repair cancer syndrome, AR, 276300 | X | X | X | X |
| 600814 | MRE11A | 11q21 | 11:94415569–94512700 | Meiotic recombination 11, S. cerevisiae, homolog A of | Ataxia-telangiectasia-like disorder, AR, 604391 | X | X | X | |
| 609309 | MSH2 | 2p21-p16 | 2:47403066–47634500 | mutS, E. coli, homolog of, 2 | Mismatch repair cancer syndrome, AR, 276300; Colorectal cancer, hereditary nonpolyposis, type 1, AD, 120435 | X | X | X | |
| 600678 | MSH6 | 2p16.3 | 2:47783081–47806952 | MutS, E. coli, homolog of, 6 | Endometrial cancer, familial, 608089; Colorectal cancer, hereditary nonpolyposis, type 5, AD, 614350; Mismatch repair cancer syndrome, AR, 276300 | X | X | X | |
| 604933 | MUTYH | 1p34.1 | 1:45329241–45340924 | MutY, E. coli, homolog of | Adenomas, multiple colorectal, 608456; Gastric cancer, somatic, 613659; Colorectal adenomatous polyposis, AR, with pilomatricomas, SM, 132600 | X | X | X | |
| 602667 | NBN | 8q21.3 | 8:89933335–89984732 | Nibrin | Nijmegen breakage syndrome, AR, 251260; Leukemia, acute lymphoblastic, 613065; Aplastic anemia, 609135 | X | X | ||
| 607379 | NF2 | 22q12.2 | 22:29603555–29698599 | Merlin | Neurofibromatosis, type 2, AD, 101000; Meningioma, NF2-related, somatic, 607174; Schwannomatosis, AD, 162091 | X | X | ||
| 191315 | NTRK1 | 1q23.1 | 1:156815749–156881849 | Neurotrophic tyrosine kinase, receptor, type 1 | Medullary thyroid carcinoma, familial, AD, 155240 | X | |||
| 610355 | PALB2 | 16p12.2 | 16:23603161–23641356 | Partner and localizer of BRCA2 | Pancreatic cancer, susceptibility to, 3, 613348; Breast cancer, susceptibility to, AD, 114480; Fanconi anemia, complementation group N, 610832 | X | X | ||
| 600259 | PMS2 | 7p22.1 | 7:5970924–6009105 | Postmeiotic segregation increased, S. cerevisiae, 2, homolog of | Mismatch repair cancer syndrome, AR, 276300; Colorectal cancer, hereditary nonpolyposis, type 4, 614337 | X | X | X | |
| 601728 | PTEN | 10q23.31 | 10:87863437–87971929 | Phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | Glioma susceptibility 2, 613028; Endometrial carcinoma, somatic, 608089; Squamous cell carcinoma, head and neck, somatic, 275355; PTEN hamartoma tumor syndrome; Prostate cancer, somatic, 176807; Malignant melanoma, somatic, 155600; Meningioma, AD, 607174 | X | X | X | |
| 604040 | RAD50 | 5q31.1 | 5:132556923–132644620 | RAD50, S. cerevisiae, homolog of | Nijmegen breakage syndrome-like disorder, 613078 | X | |||
| 602774 | RAD51C | 17q22 | 17:58692537–58734341 | RAD51, S. cerevisiae, homolog of, C | Breast-ovarian cancer, familial, susceptibility to, 3, 613399; Fanconi anemia, complementation group O, AR, 613390 | X | X | ||
| 602954 | RAD51D | 17q12 | 17:35099791–35119868 | RAD51, S. cerevisiae, homolog of, D | Breast-ovarian cancer, familial, susceptibility to, 4, 614291 | X | |||
| 614041 | RB1 | 13q14.2 | 13:48303746–48481889 | Retinoblastoma-1 | Small-cell cancer of the lung, somatic, 182280; Retinoblastoma, trilateral, SM; AD, 180200; Retinoblastoma, SM; AD, 180200; Osteosarcoma, somatic, 259500; Bladder cancer, somatic, 109800 | X | X | ||
| 604610 | RECQL3 | 15q26.1 | 15:90717326–90815461 | DNA helicase, RecQ-like 3 | Bloom syndrome, AR, 210900 | X | |||
| 164761 | RET | 10q11.21 | 10:43077068–43130350 | RET transforming sequence; oncogene RET | Multiple endocrine neoplasia IIB, AD, 162300; Multiple endocrine neoplasia IIA, AD, 171400; Medullary thyroid carcinoma, AD, 155240; Pheochromocytoma, AD, 171300 | X | |||
| 613019 | SDHAF2 | 11q12.2 | 11:61430124–61446766 | Succinate dehydrogenase complex assembly factor 2 | Paragangliomas 2, AD, 601650 | X | |||
| 185470 | SDHB | 1p36.13 | 1:17018721–17054169 | Succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | Pheochromocytoma, AD, 171300; Paragangliomas 4, AD, 115310; Paraganglioma and gastric stromal sarcoma, 606864; Gastrointestinal stromal tumor, AD; Isolated cases, 606764 | X | |||
| 602413 | SDHC | 1q23.3 | 1:161314375–161364750 | Succinate dehydrogenase complex, subunit C, integral membrane protein, 15kD | Paragangliomas 3, AD, 605373; Paraganglioma and gastric stromal sarcoma, 606864; Gastrointestinal stromal tumor, AD; isolated cases, 606764 | X | |||
| 602690 | SDHD | 11q23.1 | 11:112086823–112095800 | Succinate dehydrogenase complex, subunit D, integral membrane protein | Carcinoid tumors, intestinal, AD, 114900; Paragangliomas 1, with or without deafness, AD, 168000; Paraganglioma and gastric stromal sarcoma, 606864; Merkel cell carcinoma, somatic; Cowden syndrome 3, 615106; Pheochromocytoma, AD, 171300 | X | |||
| 600993 | SMAD4 | 18q21.2 | 18:51030212–51085041 | Mothers against decapentaplegic, Drosophila, homolog of, 4 | Polyposis, juvenile intestinal, AD, 174900; Pancreatic cancer, somatic, 260350; Juvenile polyposis/hereditary hemorrhagic telangiectasia syndrome, AD, 175050 | X | |||
| 602216 | STK11 | 19p13.3 | 19:1205798–1228434 | Serine/threonine protein kinase-11 | Pancreatic cancer, (SM; AD; Multifactorial), 260350; Melanoma, malignant, somatic; Testicular tumor, somatic, 273300 | X | X | ||
| 191170 | TP53 | 17p13.1 | 17:7668401–7687549 | Tumor protein p53 | Glioma susceptibility 1, (SM; AD), 137800; Hepatocellular carcinoma, SM, 114550; Basal cell carcinoma 7, 614740; Colorectal cancer, AD, 114500; Pancreatic cancer, (SM; AD; Multifactorial), 260350; Choroid plexus papilloma, AD, 260500; Osteosarcoma, AR, 259500; Breast cancer, AD, 114480; Nasopharyngeal carcinoma, 607107; Adrenal cortical carcinoma, AR, 202300 | X | X | ||
| 605284 | TSC1 | 9q34.13 | 9:132891347–132945268 | Hamartin (tuberous sclerosis 1 gene) | Tuberous sclerosis-1, AD, 191100; Lymphangioleiomyomatosis, 606690 | X | |||
| 191092 | TSC2 | 16p13.3 | 16:2047803–2088719 | Tuberin (tuberous sclerosis 2 gene) | Tuberous sclerosis-2, AD, 613254; Lymphangioleiomyomatosis, somatic, 606690 | X | |||
| 608537 | VHL | 3p25.3 | 3:10141634–10153669 | VHL gene | Renal cell carcinoma, somatic, 144700; Pheochromocytoma, AD, 171300; Hemangioblastoma, cerebellar, somatic | X | |||
| 607102 | WT1 | 11p13 | 11:32387774–32435534 | Wilms tumor-1 | Mesothelioma, somatic, 156240; Wilms tumor, type 1, (SM; AD), 194070 | X |
AD: Autosomal dominant; AR: Autosomal recessive; SM: somatic mutation
See Supplementary Table 1A for details
Furthermore, several recent cancer sequencing studies have observed a high frequency of germline mutations in cancer pre-disposition genes among sporadic cases with no family history of cancer, as suggested by a recent analysis of SNP (single-nucleotide polymorphism) array-based GWAS (genome-wide association studies)62. Also, among 4,034 TCGA (The Cancer Genome Atlas) cancer cases representing 12 cancer types, rare germline truncations were noted in 114 cancer-susceptibility-associated genes, spanning 4% of acute myeloid leukemia (AML), 11% of stomach cancer, and up to 19% of ovarian cancer cases63. Not surprisingly, the germline mutation burden is high among pediatric cancer cases, where about 10% of the cases had notable germline findings in two studies54, 64. Incidentally, pathogenic germline mutations have been noted in a significant proportion of sporadic pediatric cancer cases64. Focusing on germline variants, the LCCC1108/UNCseq_ (NCT01457196) study involving 439 pediatric and adult cancer patients unselected for hereditary cancer predisposition identified 4.3% of the patients with pathogenic germline variants65. In addition to these pan-cancer studies, up to 11.8% of advanced prostate cancer cases were found to harbor pathogenic germline alterations66,67, with a significantly higher rate of germline mutations observed in metastatic cases compared to patients with localized tumors101. The frequent germline mutations observed in sporadic cancer patients argues for germline sequencing to be included as an integral part of routine clinical sequencing workflows, not restricted to patients with family history of cancer.
A list of clinically actionable germline variants commonly identified can be found in Table 1, and Supplementary Table 1A details current clinical trials centered on germline aberrations. In MI_Oncoseq workflow, germline variants referenced as pathogenic in ClinVar are reviewed by a clinical geneticist for implications for disclosure to the patient/family. Additionally, integration of the germline and somatic sequencing data helps define mutations showing loss of heterozygosity (LOH) in the tumor that may be missed if only the tumor was sequenced.
Copy number aberrations (CNAs):
Exome capture data from paired tumor and normal DNA is used to determine exome-wide somatic copy number aberrations by comparing the depth of coverage at all of the individual exons analyzed followed by segmentation analysis along the lines used for array cGH data, creating high resolution copy number profiles, while circumventing technical variations68. The genome-wide copy number profile at exon level resolution is plotted in a visually intuitive, color-coded linear chromogram to evaluate a variety of copy number aberrations, including focal or wider amplifications/deletions/copy losses or gains. Interestingly, in addition to identifying susceptibilities to classical therapies, aneuploidy and the burden of copy number loss have been associated with responsiveness to immunotherapy69, 70. These findings add another layer of potentially actionable information available from CNV analyses, and one that might be missed in highly-selective targeted gene panels. Analysis of data from cancer samples with low tumor content and extreme ploidy changes, however, continues to present analytical challenges still awaiting satisfactory resolution.
Somatic SNVs/indels:
Analysis of somatic mutations in cancers, many of which define canonical driver aberrations and therapeutic targets, likely represents the most emphasized output of clinical sequencing (Supplementary Table 1B). Pairwise analysis of tumor DNA samples compared with germline sequencing data helps distinguish germline polymorphisms from somatic mutation calls. At MI_Oncoseq, we also determine the variant allele fraction (the ratio of variant/reference reads), zygosity mutations, total number of somatic mutations, and mutation burden (number of mutations/Mb) (considered in the context of the published range of cohort specific mutation numbers in TCGA data71–73 and similar data from our compendium of more than 1,700 advanced cancer cases29).
To identify functionally relevant variants, hotspot, activating, or loss of function mutations based on recurrence in the COSMIC (Catalogue of Somatic Mutations in Cancer) database, as well as stop/gain SNVs or frameshifting insertions/deletions, are highlighted. Mutations close to hot-spots or involving functionally critical domains are also noted. Published literature on key mutations is reviewed manually with special attention given to therapeutic, prognostic, diagnostic, or mechanistic associations.
The level of expression of mutant genes often provides additional supportive evidence for the likely effect of the mutations. For example, splicing mutations show intron retention (e.g. CBL, NF1, ATM, TP53 etc.) or exon skipping (e.g. MET), and in-frame expression of large indels (e.g. NOTCH1, FOXA1, EGFRvIII or its variants) can occur. Additionally, a locus sometimes shows chromosomal gain or amplification but no corresponding increase in expression levels of resident genes.
Mutational signatures:
Analysis of the patterns of somatic aberrations in cancers has emerged as a source of clinically-actionable insights. Distinct patterns of genome-wide mutations in tri-nucleotide units observed in genome/exome sequencing data from diverse cancers have helped define signatures of somatic mutations characteristic of different tumor types, defective DNA recombination/repair pathways, and those that provide insights into the mechanism of carcinogenesis through external exposures such as UV radiation, tobacco, or alkylating chemotherapeutics like temozolomide71–76 (http://cancer.sanger.ac.uk/cosmic/signatures). Some hyper-mutated cancers, such as UV-induced malignant melanoma, have shown dramatic responsiveness to immunotherapies, associated with expression of neo-antigens by the cancer cells as a result of their increased mutational load77–82. Additionally, cases with a microsatellite instability (MSI) signature, typically but not always accompanied with loss of function mutations in mismatch repair (MMR) genes83, have been associated with responsiveness to immune checkpoint inhibitor therapy61, 84. Along similar lines, the signature of homologous repair deficiency (HRD)72 typically associated with mutations in BRCA1, BRCA2, and other fanconi anemia pathway genes85, is also observed in some sporadic cancers said to display “BRCAness”86, 87. Similar to BRCA mutation carriers, cancers displaying BRCAness have been associated with responsiveness to platinum-based therapies as well as PARP inhibitors88–93.
In addition to informing therapeutic avenues, mutational signatures of individual cases also help corroborate or qualify challenging diagnoses (e.g. a cancer of unknown primary showing distinct smoking signature characteristic of lung cancer71) or glean insights into specific mechanisms of tumor progression (APOBEC signature94, 95, signature of temozolomide treatment, etc.). Notably, whole exome or larger exome capture panels are better suited for mutation signature analyses; for more selective targeted panels, it may be useful to incorporate targeted probes to query for therapeutically informative signatures such as MSI and BRCAness.
Precision immunotherapy:
In recent years, a number of different immunotherapy approaches have shown promise in the clinic. Immune checkpoint blockade targeting CD28/CTLA4 or PD-1/PD-L1 has emerged as a promising therapeutic approach across diverse cancers71, 102–105. However, as only small subsets of patients benefit from the treatment, genomic or transcriptomic markers to predict response in genomes or transcriptomes are highly sought after (Figure 2B).
Neo-antigen peptide vaccines based on individual cancer mutanomes (all mutant protein coding sequences identified by high-throughput sequencing) have shown efficacy in protecting and treating the tumor in xenograft models78, 96 and are being tested in several ongoing clinical trials (for example, NCT02287428, NCT02950766, NCT01970358).
Adoptive cell therapy uses ex vivo expanded tumor-infiltrating lymphocytes (TILs), based on identification of an immunogenic neo-antigen showing high affinity binding to the patient’s MHC antigen97, 98. Unfortunately, the excitement of promising responses to immunotherapy across several cancer types is tempered by a relatively small percentage of patients achieving dramatic, durable responses and multiple modes of primary or acquired resistance99.
Several markers of sensitivity, response, and resistance to the various immunotherapies have been identified, including the level of tumor neoantigens17,140,141, 142, tumor genomic aberrations, gene expressions, profiles of TILs, and T-cell receptor (TCR) diversity100. Gene expression analysis tools like CIBERSORT101 and TIMER102 help define the profile of TILs from tumor RNA-seq data.
Transcriptome sequencing in the clinic
Many clinical sequencing workflows currently do not involve RNA sequencing of the tumor samples, possibly due to additional requirements of technical and analytical bandwidth as well as cost and time constraints. However, we and others have observed that a parallel analysis of genomic and RNA-seq data helps to identify expressed gene-fusions (including inactivating rearrangements involving tumor suppressors) and splicing aberrations and enables the detection of pathogenic viruses. Expression signatures of tumor biomarkers can help confirm/corroborate tumor diagnoses, and in cases of tumors of unknown primary origin, help predict the likely tissue/lineage of origin. Gene expression profiles also help assess the functional status of critical pathways. For example, expression levels of androgen receptor (AR) pathway genes, like ACPP, KLK2/3, SLC45A3, and TMPRSS2, help assess the status of AR pathway regulation in prostate cancer samples, irrespective of the status of AR gene or level of AR transcript. Similarly, specific mutations in cancer-associated pathway genes, like those of the NOTCH, WNT-beta-catenin, SHH, and HIPPO pathways, can be assessed for functional consequences in terms of expression levels of their downstream target genes. As mentioned in the previous section, RNA sequencing data has also found application in defining the expressed mutanome of cancer samples to nominate candidate neo-antigens for immunotherapy78, 81, 103. In an interesting analysis, Newman et al. 101 have defined gene expression patterns corresponding to various cell types comprising cancer tissues, including tumor infiltrating immune cells, providing a powerful tool to assess the immune reactive status of different tumors. A fortuitous application of RNA-seq is also in highly sensitive and specific detection of cancer virus/pathogens in tumor tissues, such as human papilloma virus (HPV16/18), human herpesvirus-4 (EBV), human T-lymphotropic virus (HTLV), and merkel cell polyoma virus, which is important for the application of immunotherapy and cancer virus vaccines. These diverse observations afforded by RNA-seq provide critical diagnostic and therapeutic insights that are not available with DNA sequencing alone104, 105.
Gene fusions:
A wide variety of gene fusions serve as diagnostic and prognostic biomarkers, as well as therapeutic targets, for several types of cancer106, 107. RNA-seq data is particularly useful in not only identifying gene fusions, but also providing an assessment of expression levels of the fusion transcripts. In MI_Oncoseq, we have identified ETV6-ABL154, NAB2-STAT6108, and various FGFR gene fusions109 using RNA-seq. Detection of chimeric RNAs involving tumor suppressor genes, showing loss of open reading frame/functional domains, although a relatively underexplored area of investigation, is another clinically-informative application of RNA sequencing data analyses107. Apart from gene fusions, RNA-seq can provide evidence of alternative splicing aberrations (AR-V7 in prostate cancer110, 111), novel isoforms with therapeutic implications (ALK alternative transcription initiation, ATI112), or exon skipping events, including exon 14 skipping in MET reported in subsets of lung cancer113, 114.
Gene expression analyses:
The RNA-seq data from tumor samples are assessed for expression of tumor type specific biomarkers/cell surface biomarkers, as well as additional biomarkers that are often part of the routine clinical work-up of patient samples (tested by immunohistochemistry, qRT-PCR, etc.). For instance, a readout of ESR1/PGR and ERBB2 expression data can confirm or qualify immunohistochemistry status of ER/PR/HER2 in breast cancer samples; similarly, RNA-seq expression of AR, KLK3, SLC45A3, ACPP, AMACR, TMPRSS2, and ERG provides an informative readout of the status of AR signaling and/or ERG fusion status in prostate cancers.
The biomarker analysis is particularly useful in cases of diagnostically-challenging specimens, as well as advanced cancer cases with unknown primary tissue of origin115–119. Besides expression of tissue specific biomarkers, we nominate the tissue type of tumors of unknown origin using a machine learning algorithm29, using a bootstrap aggregation of six different prediction models trained on RNA-seq data from 33 primary tumor types in TCGA and normal tissue expression data obtained from GTEX, TCGA, and the Human Proteome Atlas, based on a modification of a method by Vincent et. al120.
Finally, some therapeutic target genes show exceedingly high outlier expression in certain samples, with or without an observed genomic aberration, and may represent therapeutic avenues not derived from obvious genomic aberrations. For example, outlier expressions of MET in a case of esophageal carcinoma, ROS1 in a non-small cell lung cancer, and RET in a neuroendocrine carcinoma of the larynx were found to represent potential therapeutic targets in our study29.
Clinical report enables actionable recommendations
All of the potentially actionable or informative molecular aberrations in a patient’s tumor or germline discussed in the PMTB are summarized and submitted to the attending physician with specific recommendations relating to the individual cases. While the essential report format is similar across different tumor sequencing programs, specific details vary based on types of analyses. Collectively, all of the different aspects of the germline, somatic, and/or expression data are represented among the different cases analyzed (examples in Table 3), highlighting the critical importance of integrative analyses, as singular focus on exome sequencing would likely miss many of the actionable observations revealed through integration. A formal comparison of the different modalities may be moot to consider for future programs.
Table 3. Integrative sequencing provides an array of actionable observations.
Potentially actionable observations in a representative set of cases following integrative sequencing analyses highlights the spectrum of different kinds of aberrations including germline, somatic, and gene expression changes.
| Case | MO_1311 | TP_2132 | MO_1329 | MO_1233 | TP_2130 | MO_1102 | MO_1315 | MO_1331 | MO_1547 | MO_1177 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer | Renal Cell Carcinoma | Glioblastoma Multiforme | Chondrosarcoma | Adenoid Cystic Carcinoma | Sphenoid Sinus Squamous Cell Carcinoma | Thymic Carcinoma | Non-Small Cell Lung Cancer | Tongue Squamous Cell Carcinoma | Parotid Gland Cancer | Non-Small Cell Lung Carcinoma | |
| Gender/Age | M65 | M53 | F59 | F44 | M70 | M34 | F54 | F68 | F66 | M63 | |
| Germline | Germline_Pathogenic SNV/indels | FH p.K477 dup non-frameshift insertion | PMS2 p.R802* | - | - | - | MLH1 splicing mut. | - | - | - | - |
| Tumor exome/genome | Copy number_Amp | - | PDGFRA, KIT | - | - | - | - | - | MCL1, ARNT | MDM2, CDK4, CCND1 | PDGFA, BRD4 |
| Copy number_Del | - | PTEN | - | copy loss: CDKN1C, CDKN2A, CDKN2B, ARID1A, ARID1B, SMARCA2 | - | MLH1 copy loss | copy loss: CDKN2A, CDKN2B, SMARCA2, PTEN, TP53 | - | - | CDKN2A/2B | |
| SNV_gain of fn | - | - | IDH p.R132G | - | - | - | - | - | NOTCH1 p.L1678P, p.Q2444* (PEST domain) | - | |
| SNV_loss of fn | - | - | - | KDM6A pQ1311* (homozygous) | TP53 p.R273C, p.T125M | TP53 p.R282Q | TP53 p.S260T, KEAP1 p.R470H | IQGAP2 (G833*) | TP53 p.R175H | TP53 p.E285K; BAP1 p.W52* (homozygous) | |
| Indel_gain of fn | - | - | - | *NOTCH1 non-frameshift deletion (hotspot) activating | EGFR p.D770delinsDGF non-frameshift insertion | - | - | - | - | - | |
| Indel_loss of fn | - | PTEN | - | - | FAT1 p.T1818fs deletion | CDKN2A fs insertion | - | - | - | - | |
| Mutation signature | - | - | - | - | - | MSI Signature | - | - | MSI Signature | - | |
| Transcriptome | Gene Fusion | - | - | - | - | - | - | KIF5B-RET | - | - | NF2-OSBP2 (loss of NF2) |
| Gene Expression_Outlier | - | - | - | NOTCH1, FGFR2, ERBB3 | - | PD-L1 | - | MCL1, ARNT | - | PDGFA, BRD4 | |
| Gene Expression_Biomarkers | - | - | - | - | - | - | - | - | - | UPK3B, LRRN4, CALB2, WT-1, MSLN, PDPN | |
| Pathogens | Cancer Virus | - | - | - | - | - | - | HPV16 | - | - | |
| Clinical Action | FH p.K477dup is associated with Hereditary Leiomyomatosis and Renal Cell Carcinoma (HLRCC). | TKIs such as Nilotinib/Ponatinib; Genetic Counselling | IDH inhibitors | Clinical trial with Notch inhibitor | Eligible for FDA-approved Cetuximab | PD-1/ PD-L1 targeting immunotherapy | Cabozantinib in Patients With RET Fusion-Positive Advanced Non-Small Cell Lung Cancer; NCT01639508 | T Cell Receptor Immunotherapy Targeting HPV-16 E6 for HPV-Associated Cancers (NCT02280811) | Immunotherapy, NOTCH inhibitor, CDK4/6 inhibitor | Biomarkers supporting diagnosis of Mesothelioma. Clinical trial with BRD inhibitor (NCT01587703) | |
| Therapeutically actionable aberration | Diagnostically/prognostically actionable aberration | ||||||||||
Outlook for clinical sequencing in cancer
Currently, clinical sequencing programs largely focus on exome capture sequencing instead of sequencing the whole genome. However, it is increasingly apparent that recurrent aberrations in non-exonic regions of the genome, including promoters, enhancers, other regulatory elements, protein/RNA binding sites, intergenic loci of lncRNAs, and miRNA, need to be reconciled for a fuller assessment of cancer genomic aberrations. The recent discovery of hotspot mutations in the TERT promoter that lead to aberrant reactivation of telomerase was based on sequencing of a genomic locus defined by GWAS studies of familial melanoma130 as well as whole genome sequencing of melanoma samples131. This was followed by its detection in urothelial carcinoma132, brain cancer133, 134, and thyroid cancer135 using amplicon sequencing or targeted TERT promoter sequencing136. Fortuitously, the TERT promoter mutation hotspots happen to be located just upstream of the first exon and are, thus, captured by whole exome sequencing137–139. This example highlights the realm of somatic aberrations located outside the regions typically included in exon capture panels that are not being actively targeted by exome capture. Indeed, using whole genome sequencing data from TCGA and elsewhere, recurrent mutations in upstream regulatory elements have been described in DPH3, PLEKHS1, WDR74, and SDHD140, 141.
Further highlighting the importance of gene regulation in cancer, sequencing efforts have revealed numerous aberrations in chromatin-related genes142–148 across diverse tumor types. As an example, almost all cases of pediatric malignant rhabdoid tumors are characterized by the singular loss of SWI/SNF chromatic remodeling complex gene SMARCB1142, 144, 149–151. Similarly, almost 60% of bladder tumors show mutations in epigenetic modifiers152, 153, and a majority of pediatric diffuse intrinsic pontine gliomas harbor mutations in histone H3A/H3B154–157. Therapeutic approaches targeting epigenomic aberrations have primarily included DNA demethylation (DNMTase) inhibitors such as azacytidine, which is FDA-approved for use in myelodysplastic syndromes, and histone deacetylation (HDAC) inhibitors such as vorinostat (SAHA) and panobinostat, which are FDA-approved for cutaneous T-cell lymphoma and multiple myeloma, respectively. In this context, it is expected that high-throughput epigenomic profiling integrated with clinical sequencing will illuminate a mechanistic understanding of the molecular ramifications of aberrant DNA modification pathways in cancer to help inform wider application of precision therapeutic approaches targeting other epigenomic aberrations158, 159. Integration of methylome sequencing to profile epigenomic aberrations and proteomics will help further expand the field of actionable cancer aberrations.
Another exciting area of development with potential for immediate clinical impact is sequencing of minimally invasive “liquid biopsies”, including blood, cerebrospinal fluid, or urine from cancer patients. Analysis of circulating tumor cells (CTCs), exosomes, or cell free DNA/RNA (ctDNA/RNA) transcends the issues of sampling bias, tumor heterogeneity, and metastases not amenable to biopsy, and can help assess disease progression, response to therapy, emergence of resistance, or new therapeutic targets160–162. Marking a tangible advance in this arena, the FDA has recently approved detection of EGFR mutations in the ctDNA from blood of lung cancer patients as a companion diagnostic assay for erlotinib treatment163. The next frontier may be sensitive and robust detection of panels of “hotspot” aberrations in liquid biopsies164, 165.
Apart from genomic analyses, integration of gene-expression signatures with genes and small molecules166, metabolomic assessments167, and proteomic interactome maps168 represent areas of future development. However, functional characterization and translation of these data to inform clinical decisions could be more challenging than matching somatic aberrations with therapies.
Evaluation of workflows for integrative precision oncology
Cancer is a long-term disease which means that sequencing a tumor once (current practice) provides only a snapshot of a dynamic process. As sequencing becomes routine, sequencing of tumor biopsies at diagnosis, resection, progression, and after therapy will help generate a more complete picture of cancer development. Common examples of treatment-emergent alterations that could be detected by sequencing include the acquisition of mutations in the ligand binding domain of ESR1 following aromatase inhibitor therapies in breast cancer, AR amplification and mutations in prostate cancers following endocrine deprivation therapy, and mutations in receptor tyrosine kinases following treatment with TKIs. It is also important to determine if multiple targeting avenues are potentially available at an early stage in cancer. Eventually, clinical sequencing could supplant individual gene centric assays. First, we need evidence that sequencing provides a more sensitive and reliable detection modality than FDA-approved diagnostics. It is feasible that sequencing could serve as a primary diagnostic modality, along with histopathology and radiographic imaging.
Currently, the reported turnaround time for clinical sequencing analyses ranges from two to six weeks. Turnaround time is two weeks at Foundation Medicine, a month or less at MSKCC for the MSK-IMPACT study27 and Clinical Genomics Program, Taussig Cancer Institute, and Cleveland Clinic169, and between ten days to six weeks for the University of Michigan MI_Oncoseq54 study. This time frame may need to be further shortened to one to two weeks for routine clinical application.
Determining the efficacy of integrative pipelines is confounded by the fact that typical patients availing of clinical sequencing, such as at MI_Oncoseq, present with late stage, advanced disease who have received, and often failed, multiple therapies, have maximal mutational burden including therapy resistance mutations, and more or less arrived at a therapeutic cul de sac. In this setting, despite identification of compelling therapeutic leads, the patient’s physical condition often makes them ineligible for trials or incapable of tolerating treatment. Unfortunately, in a number of cases, within a month of providing samples for analysis, patients moved to hospice care, were lost to follow up, or died.
Several unforeseen circumstantial contingencies can also mitigate potential benefits from the findings. Patients enrolled for a clinical trial following a specific therapeutic indication after sequencing analysis may get placed on the control arm of the study, denying them opportunity to benefit from the specific information about their cancer; a number of such cases occurred in the gene fusion study at the University of Michigan, UMCC 2012.022. Having varying eligibilities for clinical trials across different institutions is also problematic; for example, activating mutations in PIK3CA are common in breast cancer, but patients displaying hot-spot activating mutation in PIK3CA being treated at the University of Michigan cannot enroll in the ongoing PI3Ki SIGNATURE trial, as it excludes breast and prostate cancer. The need to negotiate with pharma and insurance companies to consider sequencing results as rationale for providing drugs for off-label use on compassionate grounds is also a constant hurdle.
Unlike the rigorous assessment that novel drugs or therapeutics are subjected to, precision oncology is fairly new and empirical evidence of its effectiveness remains equivocal. A systematic, multicenter randomized, controlled phase 2 trial (SHIVA; NCT01771458) directly comparing the efficacy of off-label molecularly-targeted therapies based on tumor molecular profiling with conventional therapy, observed no significant improvement in progression-free survival in the targeted therapy group in a cohort of heavily pre-treated cancer patients12, 170. Elsewhere, the NCI initiated comparative effectiveness research (CER) to systematically assess the efficacy of cancer genomics and precision medicine. Based on early findings of seven research studies and a follow-up workshop, they reported “insufficient evidence of clinical utility of precision medicine in translating genomic discoveries into clinical practice”171. Representing a skeptical position on the efficacy of precision oncology, the hematologist–oncologist Vinay Prasad at Oregon Health and Science University recently weighed the rather few reports of exceptional responses to targeted therapies against a preponderance of failed attempts. Given the paucity of randomized clinical trials formally testing the metrics of success, the very premise and promise of precision oncology was questioned172, 173. On a more positive note, in a recent prospective clinical trial to evaluate the clinical benefit of high-throughput genomic analyses (MOSCATO 01), actionable molecular alterations were identified in up to 48% of the cases analyzed (411 of 843 patients), of which 199 patients could be treated with a matched targeted therapy174. 7% of the successfully screened patients were assessed as having benefited from this approach in terms of progression-free survival on matched therapy as compared to prior therapy.
In our opinion, the metrics of the utility of precision oncology should be considered in the context of adding value to the standard of care, not apart from it. Much of what is the standard of care is already part of ‘precision oncology’, including all the diagnostic/prognostic markers and targeted therapies matched with specific aberrations currently in use. The latest high-throughput methodologies only help to scale up and expedite the assays over a broader range of cancers, providing access to molecular information that encompasses our collective knowledgebase. In this sense, the current forays in precision oncology would help generate an integrative knowledgebase of clinical, molecular, and therapeutic aspects of cancers that could usher in the next phase in the quest for a cancer cure.
Clinical sequencing data co-operatives
As numerous institutional efforts in precision oncology have grown, many initiatives to harness the information from collective datasets are underway. To formally test the suitability and efficacy of off-label use of targeted therapeutics, ASCO has launched a clinical trial (TAPUR) that will use genomic profiling data to match and test the utility of molecularly targeted cancer drugs outside the indications approved by the FDA and generate a registry of effective off-label usage20. In a different approach, AACR has launched the Project Genomics Evidence Neoplasia Information Exchange (GENIE), wherein seven independent clinical sequencing programs will pool their collective clinical, sequencing, treatment, and follow-up data to populate a public data repository reference. A joint research program undertaken by the National Human Genome Research Institute (NHGRI) and the NCI, called Clinical Sequencing Exploratory Research (CSER), is coordinating several research programs to help define optimal use and implementation of clinical sequencing tests175, 176. Under this conglomerate initiative, diverse issues such as considerations for validation of NGS variants177, reporting germline findings178–180, diagnostic yield of tumor sequencing data181, classification of variants182, incorporation of sequencing data in electronic health records183, genetic counselling184, and social and behavioral research185 are explored. Another co-operative effort is exemplified by the Oncology Research Information Exchange Network (ORIEN), comprising 11 US-based cancer centers sharing clinical, molecular, and therapy related data to help match patients with appropriate clinical trials based on their molecular profile. Industry is also participating; Medical Evidence Development Consortium (Med-C), a non-profit organization floated by Genentech, Roche, and Eli Lilly, plans to develop uniform, standardized work flows for matching cancer mutations with targeted therapies, intuitive to clinicians and insurance companies. Similar initiatives are mooted in the international setting through the Clinical Cancer Genome Task Team of the Global Alliance for Genomics and Health186, as well as European data centers187
Precision FDA was launched on December 15, 2015 to provide a private workspace in a public setting to make precision oncology studies available to users without access to big sequencing facilities (https://precision.fda.gov/). Users will have access to Genome in a Bottle, reference DNA for validating human genome sequences developed by the National Institute of Standards and Technology. Users will also be able to compare their results to previously validated reference results and share their results with other users, track changes, and obtain feedback. See BOX 2 and Table 2 for more details on these and other resources.
BOX 2. Community resources and data repositories serving precision oncology.
High-throughput, data-intensive applications of precision oncology have produced a plethora of databases, repositories, and online portals, catering to various distinct niches that are extensively utilized in clinical sequencing data analyses (Table 2). These include repositories of primary sequencing data, such as SRA (Short Reads Archive) and dbGAP, the database hosted by NCBI that stores and provides high-throughput genomic/transcriptomic/methylome and other data relating to genotype and phenotype in humans. Public databases of germline variations, such as 1000 Genomes Project and ExAC, provide very useful compendia of genetic polymorphism in the human population. Of these, the 1000 Genomes Project, concluded in 2015193, 194, provides a comprehensive reference of common human genomic variations compiled from 2,504 individuals representing 26 distinct populations worldwide. An even more expansive public resource called the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/) has aggregated exome sequencing data from multiple large-scale sequencing projects (including the 1000 Genome Study), spanning a total of 60,706 unrelated individuals from various disease-specific and population genetic studies. Reference databases with curated, annotated information on pathogenic germline aberrations associated with cancer include OMIM195, Leiden Open Variation Database (LoVD)196, 197, and NCBI ClinVar198. Primary cancer sequencing data repositories include The Cancer Genome Atlas (TCGA), International Cancer Genomics Consortium (ICGC), and University of California Santa Cruz (UCSC) Cancer Genomics Browser. These provide valuable references for assessing recurrence of rare somatic variants, estimation of tumor type specific mutation burden, mutation signature analyses, and comparisons of gene expression, among other applications. Data visualization portals include cBioportal for TCGA data and UCSC Xena Browser for data across multiple consortia. There is a compendium of somatic aberrations in cancer (COSMIC), as well as databases providing multidimensional assessment of somatic mutations, including the Turnkey Variant Analysis Project (TVAP) of National Human Genome Research Institute (NHGRI) that provides multiple popular open source bioinformatics tools for detection, interpretation, and visualization of high-throughput sequencing data, and database of curated mutations (DoCM). Finally, to explore models of “community” sharing of collective data repositories, the Genomic Data Commons (GDC) program of NCI and Project Genomics Evidence Neoplasia Information Exchange (GENIE) launched by AACR aim to foster unified data repositories that enable data sharing, analyses, and clinical interpretations across cancer genomic studies.
Table 2. Online resources for precision oncology studies.
A list of online portals and databases catering to different data analysis requirements for precision oncology.
| Data portals for mutations and/or germline variations | Website |
|---|---|
| 1000 Genomes Project, global reference for human genetic variation | http://www.1000genomes.org/ |
| Cancer genomics data portal_ICGC | https://dcc.icgc.org/ |
| Cancer genomics data portal_TCGA | https://tcga-data.nci.nih.gov/docs/publications/tcga/ |
| cBioPortal, visualization, analysis and download of cancer genomics data | http://www.cbioportal.org |
| ClinVar, Database of genomic variants related to human health | http://www.ncbi.nlm.nih.gov/clinvar |
| COSMIC, Catalog of somatic mutations in cancer | http://cancer.sanger.ac.uk/cosmic/ |
| dbGAP, Database of Genotypes and Phenotypes | http://www.ncbi.nlm.nih.gov/gap |
| dbNSFP, Database of Functional Predictions for SNVs | https://sites.google.com/site/jpopgen/dbNSFP |
| dbSNP, Database of Single Nucleotide Polymorphisms (SNPs) | http://www.ncbi.nlm.nih.gov/snp |
| dbVar, Database of genomic structural variations | http://www.ncbi.nlm.nih.gov/dbvar |
| DoCM, Database of Curated Mutations | http://docm.genome.wustl.edu/about |
| Ensemble Variant Effect Predictor | http://useast.ensembl.org/info/docs/tools/vep/index.html |
| ExAC, Exome Aggregation Consortium | http://exac.broadinstitute.org/ |
| Exome Variant Server | http://evs.gs.washington.edu/EVS |
| Genome Modeling Tools, Washington Univ. St. Louis | http://gmt.genome.wustl.edu |
| Human Gene Mutation Database | http://www.hgmd.org |
| IARC (WHO) TP53 mutation Database | http://p53.iarc.fr |
| Intogen mutational cancer drivers database | https://www.intogen.org/search |
| IGV, Integrative Genomics Viewer | http://software.broadinstitute.org/software/igv/ |
| LOVD, Leiden Open Variation Database | http://www.lovd.nl |
| MuSiC, Mutational significance in cancer genomes | http://tvap.genome.wustl.edu/tools/music/ |
| Pediatric Cancer Genome Project | http://explore.pediatriccancergenomeproject.org/ |
| SRA, Short Reads Archive | http://www.ncbi.nlm.nih.gov/sra |
| The Turnkey Variant Analysis Project | http://tvap.genome.wustl.edu/ |
| UCSC Cancer Genome Browser | https://genome-cancer.ucsc.edu/ |
| Xena, Integration/visualization of in-house data with public data | http://xena.ucsc.edu/ |
| Data portals for integration of -omics data with clinical interpretation/resources | |
| AACR_Project Genomics Evidence Neoplasia Information Exchange (GENIE) | http://www.aacr.org/Research/Research/Pages/aacr-project-genie.aspx#.WMs4uWfau70 |
| Cancer Commons knowledgebase | https://www.cancercommons.org/patients-caregivers/ |
| Cancer Resource | http://data-analysis.charite.de/care/ |
| CancerLinQ | http://cancerlinq.org/ |
| CIViC, Clinical interpretations of variants in cancer | https://civic.genome.wustl.edu |
| Clinical Trials | http://clinicaltrials.gov |
| CollabRx | http://www.collabrx.com/ |
| Electronic Medical Records and Genomics (eMERGE) | https://emerge.mc.vanderbilt.edu/ |
| Gene cards | http://www.genecards.org/ |
| GeneInsight | http://geneinsight.com/ |
| Genetic Testing Registry | https://www.ncbi.nlm.nih.gov/gtr/ |
| Genomic Data Commons | https://gdc.cancer.gov/ |
| GTEX, genotype-tissue expresssion data | http://www.gtexportal.org/home/ |
| Malacards | http://www.malacards.org/ |
| My Cancer Genome, Vanderbilt-Ingram Cancer Center | http://www.mycancergenome.org/ |
| N-of-One, clinical interpretation service | http://n-of-one.com/ |
| Personalized Cancer Therapy | https://pct.mdanderson.org/#/ |
| Pubmed Clinical | https://www.ncbi.nlm.nih.gov/pubmed/clinical |
| NIH Collaboratory | https://www.nihcollaboratory.org |
| Data portals providing gene-drug knowledgebase | |
| Cancer Driver Log | https://candl.osu.edu/ |
| DGIdb, The drug gene interaction database | http://dgidb.genome.wustl.edu/ |
| Drugs@FDA: FDA Approved Drug Products | http://www.accessdata.fda.gov/scripts/cder/daf/index.cfm |
| Drug Bank | http://www.drugbank.ca/about |
| Gene Drug Knowledge Database | https://www.synapse.org/#!Synapse:syn2370773/wiki/ |
| IUPHAR/BPS, guide to pharmacology | http://www.guidetopharmacology.org/download.jsp |
| NCI Drug Dictionary | https://www.cancer.gov/publications/dictionaries/cancer-drug |
| Personalized cancer Therapy, MD Anderson Cancer Center | https://pct.mdanderson.org |
| Pharmacogenomics Research Network sequence platform | http://www.pgrn.org/ |
| PharmaGKB | https://www.pharmgkb.org/ |
| SuperTarget | http://bioinf-apache.charite.de/supertarget_v2/ |
| TARGET, Tumor alterations relevant for genomics-driven therapy | http://www.broadinstitute.org/cancer/cga/target |
| TTD, Therapeutic Targets Database | http://xin.cz3.nus.edu.sg/group/ttd/ttd.asp |
Conclusion
The incorporation of clinical sequencing analyses in oncology represents the culmination of a long-standing quest to systematically link tumor specific molecular aberrations with mechanistically-targeted therapies to inform individual patient treatment. It is envisaged that widespread access to the high-resolution molecular data on individual cancer cases, along with attendant clinical data, therapy details, and follow-up information, should help close the gaps in our understanding of cancer progression and pave the way for improved cancer treatments, as well as anticipate and overcome resistance to drugs. A sobering disclaimer is due at this stage; we are not there yet.
Supplementary Material
Supplementary Table 1. A. Germline aberrations currently in clinical trials. An expansion from Table 1 in the main manuscript, current clinical trials are detailed that test therapeutics matched to the indicated germline variant. B. Summary of actionable somatic aberrations in cancer genes. Actionable somatic aberrations are listed along with other details such as therapeutics matched to the aberration and associated clinical trials or PubMed Identifier (PMID) references. Data are collated from the Cancer Genome Interpreter database (https://www.cancergenomeinterpreter.org/biomarkers), My Cancer Genome (https://www.mycancergenome.org/), and in-house MI_Oncoseq database.
Acknowledgements
This work was supported in part by the NIH Early Detection Research Network Award U01 CA214170, the NIH Clinical Sequencing Exploratory Research (CSER) Award NIH 1UM1HG006508, a Prostate SPORE Award P50 CA186786, and awards from the Prostate Cancer Foundation. A.M.C. is an American Cancer Society Research Professor, a Howard Hughes Medical Institute Investigator, and a Taubman Scholar of the University of Michigan. We thank Stephanie Ellison, scientific writer, for editorial help with the manuscript and Robin Kunkel for figure artwork. Helpful discussions with the members of the MI_Oncoseq team including Dan Robinson, Robert Lonigro, Marcin Cieslik, Yi-Mi Wu, Saravana Mohan Dhanasekaran, Pankaj Vats, and Xuhong Cao are gratefully acknowledged.
REFERENCES
- 1.Beatson G ON THE TREATMENT OF INOPERABLE CASES OF CARCINOMA OF THE MAMMA: SUGGESTIONS FOR A NEW METHOD OF TREATMENT, WITH ILLUSTRATIVE CASES. The Lancet 148, 162–165. [PMC free article] [PubMed] [Google Scholar]
- 2.Huggins C, Stevens RE Jr & Hodges CV Studies on prostatic cancer: Ii. the effects of castration on advanced carcinoma of the prostate gland. Archives of Surgery 43, 209–223 (1941). [Google Scholar]
- 3.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S The Emperor of All Maladies: A Biography of Cancer. Booklist 107, 25 (2010). [Google Scholar]
- 5.Ledermann J et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 366, 1382–1392 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 7.The Precision Medicine Initiative https://www.whitehouse.gov/precision-medicine. (2015).
- 8.Collins FS & Varmus H A new initiative on precision medicine. N Engl J Med 372, 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varmus H The Transformation of Oncology. Science 352, 1 (2016). [DOI] [PubMed] [Google Scholar]
- 10.McCarthy M US president endorses “moonshot” effort to cure cancer. Bmj 352, i213 (2016). [DOI] [PubMed] [Google Scholar]
- 11.de Bono JS & Ashworth A Translating cancer research into targeted therapeutics. Nature 467, 543–549 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Le Tourneau C et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol (2015). [DOI] [PubMed]
- 13.Mullard A Use of personalized cancer drugs runs ahead of the science. Nature (2015).
- 14.Kris MG et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311, 1998–2006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasche B & Grant SC Non-small cell lung cancer and precision medicine: a model for the incorporation of genomic features into clinical trial design. JAMA 311, 1975–1976 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Abrams J et al. National Cancer Institute’s Precision Medicine Initiatives for the new National Clinical Trials Network. Am Soc Clin Oncol Educ Book, 71–76 (2014). [DOI] [PubMed]
- 17.Herbst RS et al. Lung Master Protocol (Lung-MAP)-A Biomarker-Driven Protocol for Accelerating Development of Therapies for Squamous Cell Lung Cancer: SWOG S1400. Clin Cancer Res 21, 1514–1524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alden RS, Mandrekar SJ & Oxnard GR Designing a definitive trial for adjuvant targeted therapy in genotype defined lung cancer: the ALCHEMIST trials. Chin Clin Oncol 4, 37 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Govindan R et al. ALCHEMIST Trials: A Golden Opportunity to Transform Outcomes in Early-Stage Non-Small Cell Lung Cancer. Clin Cancer Res 21, 5439–5444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brower V NCI-MATCH pairs tumor mutations with matching drugs. Nat Biotechnol 33, 790–791 (2015). [DOI] [PubMed] [Google Scholar]
- 21.McNeil C NCI-MATCH launch highlights new trial design in precision-medicine era. J Natl Cancer Inst 107 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Mullard A NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov 14, 513–515 (2015). [DOI] [PubMed] [Google Scholar]
- 23.NCI Prepares to Launch MATCH Trial. Cancer Discov 5, 685 (2015). [Google Scholar]
- 24.Colwell J NCI-MATCH Trial Draws Strong Interest. Cancer Discov 6, 334 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Chantrill LA et al. Precision Medicine for Advanced Pancreas Cancer: The Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) Trial. Clinical Cancer Research (2015). [DOI] [PubMed]
- 26.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman DM et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today 20, 1422–1428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frampton GM et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31, 1023–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson DR et al. Integrative clinical genomics of metastatic cancer. Nature advance online publication (2017). [DOI] [PMC free article] [PubMed]
- 30.Iyer G et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 338, 221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagle N et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov 4, 546–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Ahmadie H et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov 4, 1014–1021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovly CM et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 20, 1027–1034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Allen EM et al. Genomic Correlate of Exceptional Erlotinib Response in Head and Neck Squamous Cell Carcinoma. JAMA Oncol 1, 238–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahronian LG et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov 5, 358–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaretsky JM et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. New England Journal of Medicine 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takebe N, McShane L & Conley B Biomarkers: exceptional responders-discovering predictive biomarkers. Nat Rev Clin Oncol 12, 132–134 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Chang DK, Grimmond SM, Evans TRJ & Biankin AV Mining the genomes of exceptional responders. Nature Reviews Cancer 14, 291–292 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Mehra N, Lorente D & de Bono JS What have we learned from exceptional tumour responses?: Review and perspectives. Curr Opin Oncol 27, 267–275 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Schwaederle M et al. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol Cancer Ther 15, 743–752 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Meric-Bernstam F et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arango NP et al. A feasibility study of returning clinically actionable somatic genomic alterations identified in a research laboratory. Oncotarget 8, 41806–41814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltran H et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol 1, 466–474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry JA et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A 111, E5564–5573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellmunt J et al. Somatic Copy Number Abnormalities and Mutations in PI3K/AKT/mTOR Pathway Have Prognostic Significance for Overall Survival in Platinum Treated Locally Advanced or Metastatic Urothelial Tumors. PLoS One 10, e0124711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol 32, 121–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y et al. Clinical significance of CTNNB1 mutation and Wnt pathway activation in endometrioid endometrial carcinoma. J Natl Cancer Inst 106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Network TCGA Genomic Classification of Cutaneous Melanoma. Cell 161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LoRusso PM et al. Pilot Trial of Selecting Molecularly Guided Therapy for Patients with Non-V600 BRAF-Mutant Metastatic Melanoma: Experience of the SU2C/MRA Melanoma Dream Team. Mol Cancer Ther (2015). [DOI] [PMC free article] [PubMed]
- 50.Sekulic A et al. Personalized treatment of Sezary syndrome by targeting a novel CTLA4:CD28 fusion. Mol Genet Genomic Med 3, 130–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roychowdhury S et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 3, 111ra121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.American College of Surgeons/Commission on Cancer. Cancer Program Standards 2012: Ensuring Patient-Centered Care V1.0 Chicago: American College of Surgeons; 2012; (2012). [Google Scholar]
- 53.Keating NL et al. Tumor boards and the quality of cancer care. J Natl Cancer Inst 105, 113–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mody RJ et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA 314, 913–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schilsky RL Implementing personalized cancer care. Nat Rev Clin Oncol 11, 432–438 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Farmer H et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Gelmon KA et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 12, 852–861 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Ledermann J et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 15, 852–861 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Mateo J et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Noll R et al. Long-term safety and anti-tumour activity of olaparib monotherapy after combination with carboplatin and paclitaxel in patients with advanced breast, ovarian or fallopian tube cancer. Br J Cancer 113, 396–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le DT et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y et al. Most common ‘sporadic’ cancers have a significant germline genetic component. Human molecular genetics 23, 6112–6118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu C et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun 6, 10086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 373, 2336–2346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seifert BA et al. Germline Analysis from Tumor-Germline Sequencing Dyads to Identify Clinically Actionable Secondary Findings. Clin Cancer Res (2016). [DOI] [PMC free article] [PubMed]
- 66.Robinson D et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pritchard CC et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 375, 443–453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lonigro RJ et al. Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia 13, 1019–1025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davoli T, Uno H, Wooten EC & Elledge SJ Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanetti M Chromosomal chaos silences immune surveillance. Science 355, 249–250 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Alexandrov LB et al. Mutational signatures associated with tobacco smoking in human cancer. Science 354, 618–622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexandrov LB & Stratton MR Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 24, 52–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Helleday T, Eshtad S & Nik-Zainal S Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 15, 585–598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Behjati S et al. Mutational signatures of ionizing radiation in second malignancies. Nat Commun 7, 12605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roberts SA & Gordenin DA Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer 14, 786–800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brahmer JR et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 366, 2455–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castle JC et al. Exploiting the mutanome for tumor vaccination. Cancer Res 72, 1081–1091 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Homet Moreno B & Ribas A Anti-programmed cell death protein-1/ligand-1 therapy in different cancers. Br J Cancer 112, 1421–1427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li GM Mechanisms and functions of DNA mismatch repair. Cell Res 18, 85–98 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Dudley JC, Lin MT, Le DT & Eshleman JR Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 22, 813–820 (2016). [DOI] [PubMed] [Google Scholar]
- 85.D’Andrea AD & Grompe M The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 3, 23–34 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Lord CJ & Ashworth A BRCAness revisited. Nat Rev Cancer 16, 110–120 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Turner N, Tutt A & Ashworth A Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4, 814–819 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY & Stratton MR A mutational signature in gastric cancer suggests therapeutic strategies. Nat Commun 6, 8683 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Akashi-Tanaka S et al. BRCAness predicts resistance to taxane-containing regimens in triple negative breast cancer during neoadjuvant chemotherapy. Clin Breast Cancer 15, 80–85 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Engert F, Kovac M, Baumhoer D, Nathrath M & Fulda S Osteosarcoma cells with genetic signatures of BRCAness are susceptible to the PARP inhibitor talazoparib alone or in combination with chemotherapeutics. Oncotarget (2016). [DOI] [PMC free article] [PubMed]
- 91.Hong S et al. Complete Durable Response From Carboplatin and Olaparib in a Heavily Pretreated Triple-Negative Metastatic Breast Cancer With Germline BRCA2 and “BRCAness” Mutations. J Oncol Pract 12, 270–272 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Lorusso D et al. Prospective phase II trial of trabectedin in BRCA-mutated and/or BRCAness phenotype recurrent ovarian cancer patients: the MITO 15 trial. Ann Oncol 27, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Waddell N et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henderson S, Chakravarthy A, Su X, Boshoff C & Fenton TR APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 7, 1833–1841 (2014). [DOI] [PubMed] [Google Scholar]
- 95.Kuong KJ & Loeb LA APOBEC3B mutagenesis in cancer. Nat Genet 45, 964–965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gubin MM et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tran E et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rosenberg SA et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharma P, Hu-Lieskovan S, Wargo JA & Ribas A Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu XS & Mardis ER Applications of Immunogenomics to Cancer. Cell 168, 600–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Newman AM et al. Robust enumeration of cell subsets from tissue expression profiles. Nature methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li B et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 17, 174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kreiter S et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.TCGA Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khoury JD et al. Landscape of DNA virus associations across human malignant cancers: analysis of 3,775 cases using RNA-Seq. Journal of virology 87, 8916–8926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rowley JD, Le Beau, Michelle M & Rabbitts TH Chromosomal Translocations and Genome Rearrangements in Cancer (Springer, 2015). [Google Scholar]
- 107.Kumar-Sinha C, Kalyana-Sundaram S & Chinnaiyan AM Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome medicine 7, 129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robinson DR et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet 45, 180–185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu YM et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3, 636–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Antonarakis ES et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371, 1028–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scher HI et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol (2016). [DOI] [PMC free article] [PubMed]
- 112.Wiesner T et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 526, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kong-Beltran M et al. Somatic Mutations Lead to an Oncogenic Deletion of Met in Lung Cancer. Cancer Research 66, 283–289 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Dhanasekaran SM et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Handorf CR Gene expression analysis and immunohistochemistry in evaluation of cancer of unknown primary: time for a patient-centered approach. Journal of the National Comprehensive Cancer Network : JNCCN 9, 1415–1420 (2011). [DOI] [PubMed] [Google Scholar]
- 116.Wei IH, Shi Y, Jiang H, Kumar-Sinha C & Chinnaiyan AM RNA-Seq accurately identifies cancer biomarker signatures to distinguish tissue of origin. Neoplasia 16, 918–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Massard C, Loriot Y & Fizazi K Carcinomas of an unknown primary origin--diagnosis and treatment. Nat Rev Clin Oncol 8, 701–710 (2011). [DOI] [PubMed] [Google Scholar]
- 118.Matthew EM et al. A multiplexed marker-based algorithm for diagnosis of carcinoma of unknown primary using circulating tumor cells. Oncotarget 7, 3662–3676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oien KA & Dennis JL Diagnostic work-up of carcinoma of unknown primary: from immunohistochemistry to molecular profiling. Ann Oncol 23 Suppl 10, x271–277 (2012). [DOI] [PubMed] [Google Scholar]
- 120.Vincent M, Perell K, Nielsen FC, Daugaard G & Hansen NR Modeling tissue contamination to improve molecular identification of the primary tumor site of metastases. Bioinformatics 30, 1417–1423 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Dietel M et al. A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance. Cancer Gene Ther 22, 417–430 (2015). [DOI] [PubMed] [Google Scholar]
- 122.Peters S, Michielin O & Zimmermann S Dramatic response induced by vemurafenib in a BRAF V600E-mutated lung adenocarcinoma. J Clin Oncol 31, e341–344 (2013). [DOI] [PubMed] [Google Scholar]
- 123.Haroche J et al. Dramatic efficacy of vemurafenib in both multisystemic and refractory Erdheim-Chester disease and Langerhans cell histiocytosis harboring the BRAF V600E mutation. Blood 121, 1495–1500 (2013). [DOI] [PubMed] [Google Scholar]
- 124.David Michael Hyman et al. in 2014 ASCO Annual Meeting, Vol. 32 (ed. Oncol JC) 5s(suppl; abstr 2533^) (2014). [Google Scholar]
- 125.Tiacci E et al. Targeting Mutant BRAF in Relapsed or Refractory Hairy-Cell Leukemia. N Engl J Med 373, 1733–1747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ross JS et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. International Journal of Cancer 138, 881–890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu J et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 64, 636–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Spratt DE, Zumsteg ZS, Feng FY & Tomlins SA Translational and clinical implications of the genetic landscape of prostate cancer. Nat Rev Clin Oncol 13, 597–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaji D, Miura Y & Takano T Olaparib in platinum-sensitive ovarian cancer. N Engl J Med 367, 179; author reply 179–180 (2012). [DOI] [PubMed] [Google Scholar]
- 130.Horn S et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013). [DOI] [PubMed] [Google Scholar]
- 131.Huang FW et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Borah S et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347, 1006–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reitman ZJ, Pirozzi CJ & Yan H Promoting a new brain tumor mutation: TERT promoter mutations in CNS tumors. Acta Neuropathol 126, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Killela PJ et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 110, 6021–6026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vinagre J et al. Frequency of TERT promoter mutations in human cancers. Nat Commun 4, 2185 (2013). [DOI] [PubMed] [Google Scholar]
- 136.Cancer Genome Atlas Research, N. et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372, 2481–2498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Piscuoglio S et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J Pathol 238, 508–518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abedalthagafi MS et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet 208, 345–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Assie G et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet 46, 607–612 (2014). [DOI] [PubMed] [Google Scholar]
- 140.Weinhold N, Jacobsen A, Schultz N, Sander C & Lee W Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 46, 1160–1165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fredriksson NJ, Ny L, Nilsson JA & Larsson E Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet 46, 1258–1263 (2014). [DOI] [PubMed] [Google Scholar]
- 142.Versteege I et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 143.Wilson BG & Roberts CW SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11, 481–492 (2011). [DOI] [PubMed] [Google Scholar]
- 144.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F & Delattre O SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 10, 145–171 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Ronan JL, Wu W & Crabtree GR From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet 14, 347–359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cancer Genome Atlas Research, N. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Varela I et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.The Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Biegel JA et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res 59, 74–79 (1999). [PubMed] [Google Scholar]
- 150.Jackson EM et al. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res 15, 1923–1930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sevenet N et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet 65, 1342–1348 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gui Y et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43, 875–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Guo G et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet 45, 1459–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khuong-Quang DA et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124, 439–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lewis PW et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schwartzentruber J et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 157.Wu G et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46, 444–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ahuja N, Sharma AR & Baylin SB Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu Rev Med 67, 73–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Baylin SB & Jones PA A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11, 726–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Krishnamurthy N, Spencer E, Torkamani A & Nicholson L Liquid Biopsies for Cancer: Coming to a Patient near You. J Clin Med 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Karachaliou N, Mayo-de-Las-Casas C, Molina-Vila MA & Rosell R Real-time liquid biopsies become a reality in cancer treatment. Ann Transl Med 3, 36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Aravanis AM, Lee M & Klausner RD Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 168, 571–574 (2017). [DOI] [PubMed] [Google Scholar]
- 163.Uchida J et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin Chem 61, 1191–1196 (2015). [DOI] [PubMed] [Google Scholar]
- 164.Guo N et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep 6, 33519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heitzer E, Ulz P, Geigl JB & Speicher MR Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies. Mol Oncol 10, 494–502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lamb J et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 167.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rolland T et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sohal DP et al. Prospective Clinical Study of Precision Oncology in Solid Tumors. J Natl Cancer Inst 108 (2015). [DOI] [PubMed] [Google Scholar]
- 170.Wang AZ Precision cancer medicine: Hype or hope? Science Translational Medicine 7, 306ec164–306ec164 (2015). [Google Scholar]
- 171.Simonds NI et al. Comparative effectiveness research in cancer genomics and precision medicine: current landscape and future prospects. J Natl Cancer Inst 105, 929–936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Prasad V, Fojo T & Brada M Precision oncology: origins, optimism, and potential. Lancet Oncol 17, e81–86 (2016). [DOI] [PubMed] [Google Scholar]
- 173.Prasad V Perspective: The precision-oncology illusion. Nature 537, S63 (2016). [DOI] [PubMed] [Google Scholar]
- 174.Massard C et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov (2017). [DOI] [PubMed]
- 175.Foster MW, Mulvihill JJ & Sharp RR Evaluating the utility of personal genomic information. Genet Med 11, 570–574 (2009). [DOI] [PubMed] [Google Scholar]
- 176.Green RC et al. Clinical Sequencing Exploratory Research Consortium: Accelerating Evidence-Based Practice of Genomic Medicine. Am J Hum Genet 99, 246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Beck TF, Mullikin JC, Program NCS & Biesecker LG Systematic Evaluation of Sanger Validation of Next-Generation Sequencing Variants. Clin Chem 62, 647–654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Raymond VM et al. Germline Findings in Tumor-Only Sequencing: Points to Consider for Clinicians and Laboratories. J Natl Cancer Inst 108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Amendola LM et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet 99, 247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jarvik GP & Browning BL Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am J Hum Genet 98, 1077–1081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Parsons DW et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol (2016). [DOI] [PMC free article] [PubMed]
- 182.Amendola LM et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 25, 305–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shirts BH et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc (2015). [DOI] [PMC free article] [PubMed]
- 184.Everett JN, Mody RJ, Stoffel EM & Chinnaiyan AM Incorporating genetic counseling into clinical care for children and adolescents with cancer. Future Oncol (2016). [DOI] [PMC free article] [PubMed]
- 185.Gray SW et al. Social and behavioral research in genomic sequencing: approaches from the Clinical Sequencing Exploratory Research Consortium Outcomes and Measures Working Group. Genet Med 16, 727–735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lawler M et al. Sharing Clinical and Genomic Data on Cancer - The Need for Global Solutions. N Engl J Med 376, 2006–2009 (2017). [DOI] [PubMed] [Google Scholar]
- 187.Auffray C et al. Making sense of big data in health research: Towards an EU action plan. Genome Medicine 8, 71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wagle N et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2, 82–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Beltran H et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol 63, 920–926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Van Allen EM et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 20, 682–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cieslik M et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 25, 1372–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gargis AS et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 30, 1033–1036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Abecasis GR et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF & Hamosh A OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43, D789–798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fokkema IF, den Dunnen JT & Taschner PE LOVD: easy creation of a locus-specific sequence variation database using an “LSDB-in-a-box” approach. Hum Mutat 26, 63–68 (2005). [DOI] [PubMed] [Google Scholar]
- 197.Fokkema IF et al. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 32, 557–563 (2011). [DOI] [PubMed] [Google Scholar]
- 198.Landrum MJ et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44, D862–868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. A. Germline aberrations currently in clinical trials. An expansion from Table 1 in the main manuscript, current clinical trials are detailed that test therapeutics matched to the indicated germline variant. B. Summary of actionable somatic aberrations in cancer genes. Actionable somatic aberrations are listed along with other details such as therapeutics matched to the aberration and associated clinical trials or PubMed Identifier (PMID) references. Data are collated from the Cancer Genome Interpreter database (https://www.cancergenomeinterpreter.org/biomarkers), My Cancer Genome (https://www.mycancergenome.org/), and in-house MI_Oncoseq database.