Abstract
The lysosome-mediated degradation pathway known as macroautophagy is the most versatile means through which cells can eliminate and recycle unwanted materials. Through both selective and non-selective means, macroautophagy can degrade a wide range of cargoes from bulk cytosol to organelles and aggregated proteins. Although studies of disorders such as Parkinson’s disease and Amyotrophic Lateral Sclerosis suggest that autophagic and lysosomal dysfunction directly contributes to disease, this had not been the case for the polyglutamine disorder Huntington’s disease (HD), for which there was little indication of a disruption in the autophagic-lysosomal system. This supported the possibility of targeting autophagy as a much needed therapeutic approach to combat this disease. Possibly challenging this view, however, are a recent set of studies suggesting that the protein affected in Huntington’s disease, huntingtin, might mechanistically contribute to macroautophagy. In this review, we will explore how autophagy might impact or be impacted by HD pathogenesis, and whether a therapeutic approach centering on autophagy may be possible for this yet incurable disease.
Introduction
Huntington’s disease is an autosomal-dominantly inherited neurodegenerative disease characterized by cognitive dysfunction, psychiatric disturbances and severe motor dysfunction (Sturrock and Leavitt 2010). The primary site of neurodegeneration is the medium spiny neurons of the caudate putamen, which worsens in a dorsomedial to ventrolateral direction, whereas the interneurons of the region are largely spared (Vonsattel, Keller et al. 2011)). As the severity of pathology worsens, the affected regions broaden to include regions of the cortex, cerebellum, thalamus and white matter (Vonsattel, Keller et al. 2011). The age of onset is typically midlife, and the duration of the disease can extend through decades (Sturrock and Leavitt 2010, Vonsattel, Keller et al. 2011).
In a landmark study of reverse genetics in 1998, the genetic cause of HD was identified as a trinucleotide repeat expansion mutation of cysteine, adenine, and guanine (CAG) in the coding region of the ubiquitously expressed gene now known as the HD gene (HDCRG 1993). At the protein level, the mutation leads to the production of abnormally long tracts of polyglutamine (polyQ) repeats near the N-terminus of the HD gene product, huntingtin (Htt) (Gusella, MacDonald et al. 1993, Hoogeveen, Willemsen et al. 1993, Li, Schilling et al. 1993). One of the most marked features of the mutant protein is its propensity to accumulate and aggregate (Perutz, Johnson et al. 1994, Scherzinger, Lurz et al. 1997, Bauerlein, Saha et al. 2017) and to form the intranuclear and intracytoplasmic neuronal inclusions which are now considered a pathological hallmark of the disease (Roizin, Stellar et al. 1979, Davies, Turmaine et al. 1997, DiFiglia, Sapp et al. 1997, Gutekunst, Li et al. 1999). Given that the rediscovery of protein aggregation in this disease was due to the creation of the first HD mouse model, the R6/2 model (Mangiarini, Sathasivam et al. 1996, Davies, Turmaine et al. 1997, DiFiglia, Sapp et al. 1997), much of what we understand about HD is based on experimental model systems, and validation in patient material is limited (Vonsattel 2008, Cepeda, Cummings et al. 2010). A recent study using neurons generated from human HD fibroblasts however suggests that mechanistic validation using endogenously expressed mutant Htt might soon be forth coming (Victor, Richner et al. 2018).
The presence of the polyQ expansion at the NH3-terminus of Htt, as well as the fact that Htt can be proteolyzed to shorter fragments (Goldberg, Nicholson et al. 1996, Kim, Yi et al. 2001, Mende-Mueller, Toneff et al. 2001, Lunkes, Lindenberg et al. 2002, Graham, Deng et al. 2006, Landles, Sathasivam et al. 2010, Bhat, Yan et al. 2014) raised question as to what form of the protein was preferentially accumulating: A short NH3-terminal fragment of the Htt protein (of which there are several possible lengths); the complete 348 kDa protein with the polyQ expansion; or a combination of both. Although it is clear from single cell-based systems to in vivo models such as flies and mice that the kinetics of aggregation is significantly affected by the length of the Htt protein itself, it is difficult to know how relevant these differences might be in a disease that onsets mid-life and lasts for decades. To further complicate matters, more recent studies using Htt fragment- and full length- models suggest that the trinucleotide repeat mutation might also be problematic at the level of RNA, leading to the production and accumulation of proteins containing dipeptide repeats (Banez-Coronel, Ayhan et al. 2015), alternative splicing (Neueder and Bates 2018), as well as to the creation of RNA foci (Rudnicki, Pletnikova et al. 2008, de Mezer, Wojciechowska et al. 2011, Krzyzosiak, Sobczak et al. 2012, Jain and Vale 2017).
HD and Htt
Expression of Htt is essential for life, and elimination of HD gene expression results in embryonic lethality between days embryonic (E) 8.5 to 9.5 in mice (Duyao, Ambrose et al. 1993, Nasir, Floresco et al. 1995, Zeitlin, Liu et al. 1995). Subsequent genetic studies eliminating HD gene expression postnatally found that the loss of Htt can have consequences both within and outside of the brain, even when expression is eliminated in the adult (Dragatsis, Levine et al. 2000, Wang, Liu et al. 2016, Dietrich, Johnson et al. 2017). Although the vast majority of patients with HD are heterozygous for the expansion mutation, rare cases in which patients are homozygous for the mutation revealed that solely a loss of Htt function is unlikely to be the cause of HD (Wexler, Young et al. 1987, Myers, Leavitt et al. 1989). Moreover, there is little indication that the symptoms of disease are dependent on dosage of either the mutant or the normal allele, and homozygous patients are not more affected than heterozygous individuals (Wexler, Young et al. 1987, Myers, Leavitt et al. 1989). Taken together, the genetic studies in mice and humans indicate that HD pathogenesis cannot be ascribed to a pure loss of Htt function.
Nonetheless, given how the expanded polyQ stretch can affect protein-protein interactions, and the solubility of Htt, a partial loss of function due to the polyQ mutation cannot be excluded (Liu and Zeitlin 2017), and might exacerbate, possibly in a cell type specific manner, mutant Htt toxicity. Htt is a large, neuronally enriched 350 kDa protein predominated by HEAT motifs, indicative of its role as a scaffold protein (Guo, Bin et al. 2018), and has been implicated in a broad range of functions from transcriptional regulation, nucleo-cytoplasmic shuttling, mitochondrial dynamics, vesicle trafficking, synaptic function, and anti-apoptotic activity (reviewed in (Harjes and Wanker 2003, Schulte and Littleton 2011, Reddy and Shirendeb 2012, Saudou and Humbert 2016)). Additionally, studies have shown Htt to be an integrator of transport along the cellular cytoskeleton through its roles in endocytosis, endosomal motility, and axonal transport and can regulate trafficking and transport dynamics within a cell (Gauthier, Charrin et al. 2004, Gunawardena and Goldstein 2005, Her and Goldstein 2008, Caviston and Holzbaur 2009, Power, Srinivasan et al. 2012, Liot, Zala et al. 2013, Zala, Hinckelmann et al. 2013, Wong and Holzbaur 2014). Importantly, reports have also shown huntingtin and the huntingtin-associated protein 1 (HAP1) to be involved in axonal transport through interactions with both the anterograde and retrograde transport machinery (Engelender, Sharp et al. 1997, Li, Gutekunst et al. 1998, McGuire, Rong et al. 2006, Caviston, Ross et al. 2007, Wong and Holzbaur 2014). Although Htt is reported to associate with components of both motor complexes the exact role of Htt in axonal transport is unclear, but recent studies in D. melanogaster suggests that dHtt acts locally at cargo interaction sites to regulate processivity (Weiss and Littleton 2016).
Htt is expressed ubiquitously across all cell types, but observed most highly in neurons especially in the neocortex, the cerebellar cortex, the striatum and the hippocampus (Li, Schilling et al. 1993, Fusco, Chen et al. 1999). It is distributed throughout the cell as a cytosolic protein as well as in the nucleus, axonal processes and at synapses, and in association with various organelles and structures including microtubules, clathrin-coated vesicles, caveolae, mitochondria and synaptosomes (DiFiglia, Sapp et al. 1995, Velier, Kim et al. 1998, BorrellPages, Zala et al. 2006, Reddy and Shirendeb 2012). Htt is expressed broadly early in development, and reaches its highest expression in the developing brain (Bhide, Day et al. 1996). Given its early expression and the breadth of functions in which it has been implicated, it is perhaps unsurprising that neurodevelopmental alterations have been reported recently in patients carrying this disease-causing mutation (Kerschbamer and Biagioli 2015, Lee, Conrad et al. 2018). Segregating whether these events are due to a partial loss-of-function of Htt or a mutation-dependent gain-of-function remains to be seen. Moreover, how and if these neurodevelopmental changes might influence HD onset and pathogenesis is unclear.
Selective degradation of aggregated protein by MA
The abnormal accumulation of the mutant htt protein (mHtt) gave rise to a question posed across all proteinopathies, which was whether the aggregated species of mHtt contributed to pathogenesis. Although large, visible intraneuronal inclusions are found in at most 10% of cortical, striatal and thalamic neurons in adult HD (Vonsattel, Keller et al. 2011), the distribution of smaller oligomeric species remains unclear. Interestingly, studies across HD mouse models suggest that abolishing expression of mutant Htt in symptomatic animals is sufficient to bring about therapeutic reversal, including significantly lowering levels of aggregation in the brain (Yamamoto, Lucas et al. 2000, Regulier, Trottier et al. 2003, Harper, Staber et al. 2005, DiFiglia, Sena-Esteves et al. 2007, Kordasiewicz, Stanek et al. 2012). These data indicated that adult neurons have the capacity to eliminate proteinaceous inclusions. Should the aggregated structures contribute to pathogenesis, then identification of how these inclusions are eliminated could lead to a therapeutic approach to combat HD and other proteinopathies.
Studies by several groups implicated the lysosome-mediated degradation pathway macroautophagy (MA) to play a critical role in aggregate-clearance (Johnston, Ward et al. 1998, Ravikumar, Duden et al. 2002, Iwata, Christianson et al. 2005, Iwata, Riley et al. 2005, Yamamoto, Cremona et al. 2006). MA is one of three autophagy pathways that traffics cytosolic cargoes for lysosomal degradation (review in (Kaushik and Cuervo 2012, Yamamoto and Yue 2014)). In MA, cargoes are captured into a transient organelle known as an autophagosome, a multilamellar structure formed through a coordinated effort of several proteins, including those known as the Autophagy-related genes (Atg) (Mizushima, Yoshimori et al. 2011). Although MA is not the exclusive pathway through which aggregated proteins might be eliminated (Juenemann, Schipper-Krom et al. 2013, Schipper-Krom, Juenemann et al. 2014, Hjerpe, Bett et al. 2016, Dikic 2017) its contribution is robust enough that the elimination of mutant Htt aggregates is often used as a primary screen to identify proteins involved in autophagic degradation (Bjorkoy, Lamark et al. 2005, Yamamoto, Cremona et al. 2006, Filimonenko, Stuffers et al. 2007, Filimonenko, Isakson et al. 2010, Korac, Schaeffer et al. 2013, Eenjes, Dragich et al. 2016, Wold, Lim et al. 2016).
MA can package cargoes in a selective manner through adaptor proteins that scaffold protein aggregates to the growing membrane of the autophagosome, a pathway known as aggrephagy (Yamamoto and Yue 2014, Dikic 2017, Gatica, Lahiri et al. 2018). There are several adaptor proteins including the autophagy receptor proteins p62/Sequestesome-1 (p62/sqstm1)(Shin 1998, Seibenhener, Babu et al. 2004, Bjorkoy, Lamark et al. 2005, Pankiv, Clausen et al. 2007), Near BRCA 1 (Nbr1) (Kirkin, Lamark et al. 2009, Lamark, Kirkin et al. 2009), and optineurin (Optn)(Korac, Schaeffer et al. 2013), in addition to the selectivity adaptor protein Autophagy Linked FYVE protein (Alfy)(Simonsen, Birkeland et al. 2004, Filimonenko, Isakson et al. 2010), that have been linked to autophagy-mediated clearance of aggregates. How or why multiple adaptor proteins are required for selective degradation is unclear. Genetic studies in mice, as well as a recent study in a recombinant system (Zaffagnini, Savova et al. 2018) indicate that the most well-studied adaptor protein, p62, acts to sequester and consolidate misfolded proteins and other cargoes preparing them for degradation through its ubiquitin binding domain and a self-interacting PB1 domain (Bjorkoy, Lamark et al. 2005, Pankiv, Clausen et al. 2007). p62, like all autophagy receptor proteins, contain a LIR domain giving it the ability to bind to the mammalian orthologues of the yeast protein, Atg8, which can be found in a cytosolic form or covalently attached to the autophagosome membrane (Pankiv, Clausen et al. 2007).
Although the ability to bind to mammalian Atg8 orthologues such as Microtubule associated protein light chain 3B (LC3B) is modeled to be sufficient to bring the nascent autophagic membrane to the aggregate-structure (Rogov, Dotsch et al. 2014), other adaptor proteins such as Alfy appear necessary for the clearance of protein aggregates, including those containing mutant Htt (Filimonenko, Isakson et al. 2010). Alfy is a 380 kDa protein that has a series of protein-protein domains and a protein-lipid interaction domain in the C-terminus (Simonsen, Birkeland et al. 2004). Through these domains, Alfy can bind to the p62-positive inclusion (Clausen, Lamark et al. 2010) and the Atg5–12-16L complex (Filimonenko, Isakson et al. 2010), which acts as an E3-like protein that helps conjugate Atg8 proteins to the autophagosome membrane. In addition, the FYVE domain binds to a key lipid on the autophagosome membrane, phosphatidylinositol 3-monophosphate, and is speculated to stabilize the growing autophagosome membrane to the cargo (Simonsen, Birkeland et al. 2004, Filimonenko, Isakson et al. 2010). Although Alfy also has a LIR domain, unlike p62, Alfy interacts with a subset of Atg8 homologs, specifically the GABA-A receptor associated protein (GABARAP) family of proteins but not the LC3 family (Lystad, Ichimura et al. 2014). The physiologic significance of the differential interaction with the two different Atg8 families remains unclear.
A role for Alfy in the clearance of aggregated proteins first came to light in flies, in which the loss of expression of the Drosophila homolog, Blue cheese (Bchs), led to neurodegeneration and the accumulation of ubiquitinated proteins (Finley, Edeen et al. 2003). Studies in stable mammalian cell lines (Filimonenko, Isakson et al. 2010) and primary cells derived from mice (Dragich, Kuwajima et al. 2016) similarly found that the depletion or loss of Alfy expression could accelerate aggregate-accumulation. In addition, increasing Alfy expression diminishes aggregate-load in primary neurons, and protects against polyQ toxicity in a fly eye model (Filimonenko, Isakson et al. 2010). Subsequent studies confirmed that the diminished aggregate load is due to the ability of Alfy to eliminate preformed inclusions, rather than interfering with aggregate-formation (Eenjes, Dragich et al. 2016). Given how Alfy may play a rate-limiting role in aggregate-clearance, establishing how Alfy expression impacts protein accumulation in mouse models of HD will provide much needed insight into how aggregation might influence disease pathogenesis.
Nonetheless, it should be noted that studies affecting cargo adaptor proteins rather than autophagy directly can have unexpected results. For example, Nukina and colleagues found that the loss of p62 led to partial neuroprotection in two mouse models of HD (Kurosawa, Matsumoto et al. 2015), rather than potentially enhancing toxicity by slowing the ability of inclusion to form and be eliminated. Instead, the loss of p62 significantly diminished the number of nuclear inclusions, and augmented the number of cytosolic inclusions. These unexpected findings might in part be due to a specific feature of the models used; the R6/2 and HD109QG models, are aggressive, early onset models of HD, that demonstrate predominantly intranuclear accumulations of a short mutant Htt fragment. Given the ability of this short mutant Htt fragment to aggregate in vitro and in cell-based systems, it was often assumed that the polyQ expansion was sufficient to drive aggregation and accumulation, regardless of whether it was in a nuclear or cytoplasmic compartment. However, the work by Kurosawa et al. brought to light how the nuclear aggregation observed in these mice requires p62, whereas the cytosolic aggregation does not. These data suggest that the intranuclear sequestration of the exon1 Htt fragment by p62 is maladaptive, potentially preventing the turnover of the mutant Htt fragment by MA, and aggravate the intranuclear toxicity of the fragment mutant Htt. Thus the loss of p62 was partially protective, but only partial because the mutant Htt was still present and aggregating in the cytosol. Given that this study also shows how wholesale disruption of MA in the same HD mice (in the presence of p62) increases toxicity, this strongly suggest that ongoing MA might be continuing to confer protection regardless of the presence of p62. The potential role of other adaptor proteins working in the absence of p62, or the ability of non-adaptor mediated capture of aggregates by the autophagosome might explain this effect. These observations highlight the importance of studying different HD models simultaneously that express different Htt fragments to full length protein to fully understand how pathogenesis might evolve when augmenting or diminishing autophagic degradation.
Another consideration when examining the role of MA in HD requires our better understanding of how MA is utilized by the different cells of the CNS. Although the contribution of polyQ toxicity in glial cells cannot be disregarded (Myers, Vonsattel et al. 1991, Huang, Wei et al. 2015, Jin, Peng et al. 2015, Phillips, Joshi et al. 2016, Teo, Hong et al. 2016), how autophagy in these cells might be relevant remains uncertain because virtually nothing is known about MA in astrocytes, microglia or oligodendrocytes, especially in context of a disease. Even our understanding of MA in neurons, which represent one of the first cell types in which MA was described, is still in its early stages. An early study in cultured neurons by Hollenbeck (Hollenbeck 1993) suggested that autophagosomes could form in the distal axon tip and mature as it trafficked retrograde towards the soma. Recently, this study was elegantly revisited and expanded upon by Holzbaur and colleagues (Maday and Holzbaur 2012, Maday, Wallace et al. 2012, Maday and Holzbaur 2014, Maday and Holzbaur 2016), extending the observations from dorsal root ganglia neurons to hippocampal and cortical neurons. These studies together with observations made by Yue, Burke and others, highlight how that axon health (Komatsu, Wang et al. 2007), and axonal die-back in the presence of toxins (Cheng, Kim et al. 2011), are MAdependent. Moreover, the Holzbaur lab indicate that MA is not limited to axons, and MA might spatially be defined in a cargo-dependent manner (Wong and Holzbaur 2015, Maday and Holzbaur 2016). Given that mutant Htt deposits are observed throughout the neuron, and differently across neuronal subtypes, modulating MA might impact discrete aspects of HD pathogenesis such as the age-of-onset or rate-of-progression.
Autophagy: A direct link to HD?
Recently, studies in Drosophilia and mice implicated Htt to play a significant role in selective autophagy. Zeitlin and colleagues first found that the loss of the polyQ stretch, even in normal Htt appeared to enhance autophagic capacity in neurons (Zheng, Clabough et al. 2010). Although it was unclear why a simple deletion of the polyQ domain would lead to these observation, Steffan proposed in 2010 that Htt showed structural similarities to three yeast selective autophagy proteins Atg11, Atg23, and Vac8 in tandem (Steffan 2010). In a follow up study, Steffan and colleagues found common Htt interactions with an Atg11 binding partners, as well as the identification of a conserved motif of Htt that mediates some of these interactions (Ochaba, Lukacsovich et al. 2014). Additionally, they reported that the loss of Htt function in Drosophila and mouse CNS led to protein accumulation, which corresponded with an independent study indicating that depletion of Htt or its interactor, HAP1, impaired retrograde transport of autophagosomes containing non-degraded selective cargo, thereby leading to accumulation (Wong and Holzbaur 2014). Soon after, a collaborative effort by Zheng, Cuervo and colleagues similarly found that Htt functions as a scaffold for selective autophagy by promoting cargo recognition and autophagy initiation in both fly and mammalian cells, mediating the binding between p62 and the autophagy-initiating kinase, ULK1 (Rui, Xu et al. 2015), suggesting that a loss of autophagy regulation might contribute to HD pathogenesis (Gelman, Rawet-Slobodkin et al. 2015). Given that Htt has already been implicated to play a role in endocytosis (DiFiglia, Sapp et al. 1995, Velier, Kim et al. 1998) and in trafficking of cargoes from mitochondria to different vesicle populations, (Reddy and Shirendeb 2012, White, Anderson et al. 2015), including autophagosomes (Wong and Holzbaur 2014), an involvement of Htt in autophagy might be unsurprising. Nonetheless the question remains as to what extent the polyQ mutation impacts Htt function in this regard in patients. The mutation lengths often used in the experimental paradigms far exceed the typical adult onset repeat length of 44, and the impact of these shorter repeat lengths are of little study. Moreover, even mice and cells with transgenic overexpression of mutant Htt with greater than one hundred polyQ, or homozygous knock-in mice with greater polyQ lengths do not exhibit gross alterations in autophagic function, and the clearance of aggregates by autophagy can still occur (Heng, Duong et al. 2010, Baldo, Soylu et al. 2013, Eenjes, Dragich et al. 2016). This would suggest, that although Htt function might be a critical player in autophagy and vesicle trafficking overall, it is likely that despite the presence of the polyQ mutation, there is still sufficient Htt function to drive these events. Moreover, these observations also suggest that therapeutic interventions overcoming the losses due to the HD mutation might be possible.
Autophagy and HD: A therapeutic approach?
The activation of autophagy has been a sought after approach for a wide array of disorders with adult onset neurodegenerative diseases only representing a small subset. However, this has been particularly challenging especially in cells of the CNS for different reasons. For example, although starvation and the subsequent inhibition of the serine/threonine mTOR is the first identified means to activate MA (and led to the discovery of autophagy), it is widely accepted that MA can occur despite mTOR activation, such as by modulating the Vps34-Beclin1 kinase complex(Yamamoto, Cremona et al. 2006, Johnson, Melia et al. 2012, Sarkar 2013, Manzoni, Mamais et al. 2016). In addition, to what extent autophagy induction is achieved via mTOR inhibition in neurons is unclear (Du, Hickey et al. 2009, Alirezaei, Kemball et al. 2010, Dubinsky, Dastidar et al. 2014, Tang, Gudsnuk et al. 2014, Maday and Holzbaur 2016). Taken together, how MA is most effectively activated in the adult brain remains uncertain.
Aside from the molecular basis of autophagy in the CNS, global activation of autophagy would by definition extend to organs beyond the brain and spinal cord. Thus, even though the focus of neurodegenerative diseases such as HD is the brain, one must consider the importance of other organs such as striated muscle. Muscle wasting is a significantly devastating aspect of the disease, and ultimately contributes to the demise of HD patients (Mielcarek and Isalan 2015). Sandri and colleagues have found that the role for MA in muscle wasting is complex, and that although MA is activated during catabolic conditions, it is also essential to maintain muscle mass (Masiero, Agatea et al. 2009, Bonaldo and Sandri 2013, Schiaffino, Dyar et al. 2013). Whereas there is still much to be learned about why patients lose muscle mass in HD, augmenting MA systemically by using approaches that mimic catabolic conditions might have devastating effects on patients.
Experimentally however, many of the mouse models of HD do not appear to manifest the cachexic symptoms, and thus the impact of mTOR inhibitors and other compounds that might lead to mTOR inhibition such as rapamycin, CC1–779, glucose and glucose-6-phosphate can be examined for their impact on the proteinopathy, and all have been shown to enhance the clearance of mutant huntingtin fragments, reduces formation of aggregate-prone proteins and is protective against cytotoxicity in cell, Drosophila and mouse models of HD (Ravikumar, Duden et al. 2002, Berger, Ravikumar et al. 2006). A key question remains, however, whether these observations are solely due to the activation of MA, or whether mTOR inhibition decreased translation of the exogenously expressed mutant Htt protein. Potential mTOR-independent activators of MA, primarily driven through modulators of intracellular calcium have also been identified to enhance mutant Htt clearance, primarily in cell- and Drosophila-based models of HD. (Sarkar, Floto et al. 2005, Criollo, Maiuri et al. 2007, Zhang, Yu et al. 2007, Williams, Sarkar et al. 2008). How calcium might influence MA is complex, as it can both increase and decrease MA, depending on the source of the intracellular calcium (Johnson, Melia et al. 2012). In mouse models, therapeutic benefit likely through the activation of AMPK has been shown to be evoked by trehalose and metformin in the R6/2 mouse model (Tanaka, Machida et al. 2004, Ma, Buescher et al. 2007, Sarkar, Davies et al. 2007), and in the N171–82Q mouse model (Jiang, Wei et al. 2015).
Although the number of studies have been limited, no obvious deficits in the expression of the core MA machinery has been reported in HD patient samples (Hodges, Strand et al. 2006, Kuhn, Thu et al. 2011), suggesting that if MA in brains can be selectively activated, the possibility for potential therapeutic intervention exists. Interestingly, Luthi-Carter and colleagues found that in the caudate nucleus of patient samples, levels of the adaptor proteins Alfy and Optn were significantly decreased, suggesting that the selective turnover of aggregated proteins and mitochondria might selectively diminished. Thus, enhancing cargo recognition by these adaptor proteins, possibly via posttranslational modifications, might be a way through which this can be overcome. Finally, studies in animal models suggest that MA regulation through the Beclin1Vps34 complex might be diminished (Mealer, Murray et al. 2014). Nonetheless, MA capacity might be positively impacted by increasing that activity of transcription factor EM (TFEB), which transcriptionally regulates lysosomal degradation(Sardiello 2016), which has shown promise in mouse models (Tsunemi, Ashe et al. 2012). Ultimately, targeting the upregulation of autophagy as a therapeutic strategy has some promise, but rather than global activation of a status of metabolic stress, other means such as through activation of cargo adaptors might be beneficial. However, it is clear that much research still must be performed before the development of beneficial therapies may be created.
Conclusion
Although a powerful means to establish whether aggregated proteins might contribute to pathogenesis, the recent implication that Htt itself might be necessary for selective MA brings to question how effective autophagy might be harnessed for therapeutic benefit. The uncovering of Htt as a potential modulator of autophagy specifically highlights how much we are still learning about this fascinating pathway. Given that the vast majority of patients are heterozygous for the mutation gives rise to a key question, to what extent are Htt levels rate limiting for autophagy. Moreover, Htt’s already implicated roles in vesicle trafficking, and how that relates to its proposed role in selective autophagy remains to be uncovered. On-going studies examining how modulating autophagic processes on HD pathogenesis in mouse models, might give insight into these question. Nonetheless, it is clear that working with experimental models alone will not suffice, and a back and forth between patient materials, with relevant polyQ lengths, will be essential to fully define how much a loss of function of Htt is contributing to disease. Taken together, there is still significant work to be done to establish the importance of MA as both a contributor and savior against this devastating disease.
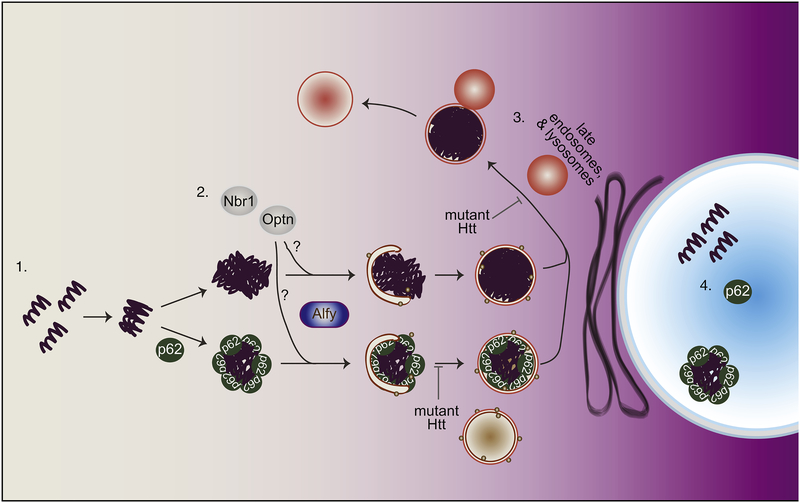
Figure 1.
Schematic summary of macroautophagy (MA) in relation to Huntington’s disease (HD).
1. In the cytosol, mutant Htt oligomerizes then form aggregates in p62-dependent and – independent manner. 2. Targeting p62-positive, and possibly p62-negative aggregated proteins are then targeted to the autophagosome-membrane, with the help of the selectivity adaptor protein Alfy. Other autophagy receptors implicated in the turnover of aggregates include Optn and Nbr1, but whether then are involved together or separately remains unclear. Mutant Htt may impede cargo capture leading to the formation of empty autophagosomes, although the mechanism through which this occurs is uncertain. 3. Upon autophagosome formation, the autophagosome matures and acidifies by fusing into the endolysosomal system. Mutant Htt may impede autophagosome maturation, by disrupting retrograde trafficking of the autophagosome back into the soma. 4. In the nucleus, mutant Htt can also accumulates. This accumulation and aggregation, at least of a Htt fragment encoded by exon 1 of the HD gene, appears to require the presence of p62.
Acknowledgments
This work was supported by the National Science Foundation Predoctoral Fellowship (KRC), Hereditary Disease Foundation (AY) and NINDS RO1 NS063973 (AY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alirezaei M, Kemball CC, Flynn CT, Wood MR, Whitton JL and Kiosses WB (2010). “Shortterm fasting induces profound neuronal autophagy.” Autophagy 6(6): 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo B, Soylu R and Petersen A (2013). “Maintenance of basal levels of autophagy in Huntington’s disease mouse models displaying metabolic dysfunction.” PLoS One 8(12): e83050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, Yachnis AT, Troncoso JC and Ranum LP (2015). “RAN Translation in Huntington Disease.” Neuron 88(4): 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W and Fernandez-Busnadiego R (2017). “In Situ Architecture and Cellular Interactions of PolyQ Inclusions.” Cell 171(1): 179–187 e110. [DOI] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ and Rubinsztein DC (2006). “Rapamycin alleviates toxicity of different aggregate-prone proteins.” Hum Mol Genet 15(3): 433–442. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Yan S, Wang CE, Li S and Li XJ (2014). “Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A.” Proc Natl Acad Sci U S A 111(15): 5706–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhide PG, Day M, Sapp E, Schwarz C, Sheth A, Kim J, Young AB, Penney J, Golden J, Aronin N and DiFiglia M (1996). “Expression of normal and mutant huntingtin in the developing brain.” J Neurosci 16(17): 5523–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H and Johansen T (2005). “p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death.” J Cell Biol 171(4): 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo P and Sandri M (2013). “Cellular and molecular mechanisms of muscle atrophy.” Dis Model Mech 6(1): 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell-Pages M, Zala D, Humbert S and Saudou F (2006). “Huntington’s disease: from huntingtin function and dysfunction to therapeutic strategies.” Cell Mol Life Sci 63(22): 2642–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP and Holzbaur EL (2009). “Huntingtin as an essential integrator of intracellular vesicular trafficking.” Trends Cell Biol 19(4): 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M and Holzbaur EL (2007). “Huntingtin facilitates dynein/dynactin-mediated vesicle transport.” Proc Natl Acad Sci U S A 104(24): 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda C, Cummings DM, Andre VM, Holley SM and Levine MS (2010). “Genetic mouse models of Huntington’s disease: focus on electrophysiological mechanisms.” ASN Neuro 2(2): e00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HC, Kim SR, Oo TF, Kareva T, Yarygina O, Rzhetskaya M, Wang C, During M, Talloczy Z, Tanaka K, Komatsu M, Kobayashi K, Okano H, Kholodilov N and Burke RE (2011). “Akt suppresses retrograde degeneration of dopaminergic axons by inhibition of macroautophagy.” J Neurosci 31(6): 2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A and Johansen T (2010). “p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy.” Autophagy 6(3): 330–344. [DOI] [PubMed] [Google Scholar]
- Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, Molgo J, Diaz J, Lavandero S, Harper F, Pierron G, di Stefano D, Rizzuto R, Szabadkai G and Kroemer G (2007). “Regulation of autophagy by the inositol trisphosphate receptor.” Cell Death Differ 14(5): 1029–1039. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L and Bates GP (1997). “Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation.” Cell 90(3): 537–548. [DOI] [PubMed] [Google Scholar]
- de Mezer M, Wojciechowska M, Napierala M, Sobczak K and Krzyzosiak WJ (2011). “Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference.” Nucleic Acids Res 39(9): 3852–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich P, Johnson IM, Alli S and Dragatsis I (2017). “Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis.” PLoS Genet 13(7): e1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, Davies S, Bates G, Vonsattel J and Aronin N (1997). “Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain.” Science 277: 1990–1993. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA and et al. (1995). “Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons.” Neuron 14(5): 1075–1081. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, Manoharan M, Sah DW, Zamore PD and Aronin N (2007). “Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits.” Proc Natl Acad Sci U S A 104(43): 17204–17209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I (2017). “Proteasomal and Autophagic Degradation Systems.” Annu Rev Biochem 86: 193–224. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS and Zeitlin S (2000). “Inactivation of hdh in the brain and testis results in progressive neurodegeneration and sterility in mice.” Nat Genet 26(3): 300–306. [DOI] [PubMed] [Google Scholar]
- Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon MS, Eenjes E, Bosco JR, Fox LM, Lystad AH, Oo TF, Yarygina O, Mita T, Waguri S, Ichimura Y, Komatsu M, Simonsen A, Burke RE, Mason CA and Yamamoto A (2016). “Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain.” Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Hickey RW, Bayir H, Watkins SC, Tyurin VA, Guo F, Kochanek PM, Jenkins LW, Ren J, Gibson G, Chu CT, Kagan VE and Clark RS (2009). “Starving neurons show sex difference in autophagy.” J Biol Chem 284(4): 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubinsky AN, Dastidar SG, Hsu CL, Zahra R, Djakovic SN, Duarte S, Esau CC, Spencer B, Ashe TD, Fischer KM, MacKenna DA, Sopher BL, Masliah E, Gaasterland T, Chau BN, Pereira de Almeida L, Morrison BE and La Spada AR (2014). “Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy.” Cell Metab 20(4): 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M and et al. (1993). “Trinucleotide repeat length instability and age of onset in Huntington’s disease.” Nat Genet 4(4): 387–392. [DOI] [PubMed] [Google Scholar]
- Eenjes E, Dragich JM, Kampinga HH and Yamamoto A (2016). “Distinguishing aggregate formation and aggregate clearance using cell based assays.” J Cell Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur EL and Ross CA (1997). “Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin.” Hum Mol Genet 6(13): 2205–2212. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A and Yamamoto A (2010). “The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy.” Mol Cell 38(2): 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H and Simonsen A (2007). “Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease.” J Cell Biol 179(3): 485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M and McKeown M (2003). “blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration.” J Neurosci 23(4): 1254–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco FR, Chen Q, Lamoreaux WJ, Figueredo-Cardenas G, Jiao Y, Coffman JA, Surmeier DJ, Honig MG, Carlock LR and Reiner A (1999). “Cellular localization of huntingtin in striatal and cortical neurons in rats: lack of correlation with neuronal vulnerability in Huntington’s disease.” J Neurosci 19(4): 1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V and Klionsky DJ (2018). “Cargo recognition and degradation by selective autophagy.” Nat Cell Biol 20(3): 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, De Mey J, MacDonald ME, Lessmann V, Humbert S and Saudou F (2004). “Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules.” Cell 118(1): 127–138. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rawet-Slobodkin M and Elazar Z (2015). “Huntingtin facilitates selective autophagy.” Nat Cell Biol 17(3): 214–215. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, Graham RK, Bromm M, Kazemi-Esfarjani P, Thornberry NA, Vaillancourt JP and Hayden MR (1996). “Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract.” Nat Genet 13(4): 442–449. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW and Hayden MR (2006). “Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin.” Cell 125(6): 1179–1191. [DOI] [PubMed] [Google Scholar]
- Gunawardena S and Goldstein LS (2005). “Polyglutamine diseases and transport problems: deadly traffic jams on neuronal highways.” Arch Neurol 62(1): 46–51. [DOI] [PubMed] [Google Scholar]
- Guo Q, Bin H, Cheng J, Seefelder M, Engler T, Pfeifer G, Oeckl P, Otto M, Moser F, Maurer M, Pautsch A, Baumeister W, Fernandez-Busnadiego R and Kochanek S (2018). “The cryo-electron microscopy structure of huntingtin.” Nature 555(7694): 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME, Ambrose CM and Duyao MP (1993). “Molecular genetics of Huntington’s disease.” Arch Neurol 50(11): 1157–1163. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM and Li XJ (1999). “Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology.” J Neurosci 19(7): 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes P and Wanker EE (2003). “The hunt for huntingtin function: interaction partners tell many different stories.” Trends Biochem Sci 28(8): 425–433. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL and Davidson BL (2005). “From the Cover: RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model.” Proc Natl Acad Sci U S A 102(16): 5820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HDCRG (1993). “A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes.” Cell 72(6): 971–983. [DOI] [PubMed] [Google Scholar]
- Heng MY, Duong DK, Albin RL, Tallaksen-Greene SJ, Hunter JM, Lesort MJ, Osmand A, Paulson HL and Detloff PJ (2010). “Early autophagic response in a novel knock-in model of Huntington disease.” Hum Mol Genet 19(19): 3702–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her LS and Goldstein LS (2008). “Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin.” J Neurosci 28(50): 13662–13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, Osborne G, Bates GP, Glickman MH, Trost M, Knebel A, Marchesi F and Kurz T (2016). “UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome.” Cell 166(4): 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L and LuthiCarter R (2006). “Regional and cellular gene expression changes in human Huntington’s disease brain.” Hum Mol Genet 15(6): 965–977. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ (1993). “Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport.” J Cell Biol 121(2): 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogeveen AT, Willemsen R, Meyer N, de Rooij KE, Roos RA, van Ommen GJ and Galjaard H (1993). “Characterization and localization of the Huntington disease gene product.” Hum Mol Genet 2(12): 2069–2073. [DOI] [PubMed] [Google Scholar]
- Huang B, Wei W, Wang G, Gaertig MA, Feng Y, Wang W, Li XJ and Li S (2015). “Mutant huntingtin downregulates myelin regulatory factor-mediated myelin gene expression and affects mature oligodendrocytes.” Neuron 85(6): 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS and Kopito RR (2005). “Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation.” Proc Natl Acad Sci U S A 102(37): 13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA and Kopito RR (2005). “HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin.” J Biol Chem 280(48): 40282–40292. [DOI] [PubMed] [Google Scholar]
- Jain A and Vale RD (2017). “RNA phase transitions in repeat expansion disorders.” Nature 546(7657): 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wei W, Gaertig MA, Li S and Li XJ (2015). “Therapeutic Effect of Berberine on Huntington’s Disease Transgenic Mouse Model.” PLoS One 10(7): e0134142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Peng Q, Hou Z, Jiang M, Wang X, Langseth AJ, Tao M, Barker PB, Mori S, Bergles DE, Ross CA, Detloff PJ, Zhang J and Duan W (2015). “Early white matter abnormalities, progressive brain pathology and motor deficits in a novel knock-in mouse model of Huntington’s disease.” Hum Mol Genet 24(9): 2508–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CW, Melia TJ and Yamamoto A (2012). “Modulating macroautophagy: a neuronal perspective.” Future Med Chem 4(13): 1715–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL and Kopito RR (1998). “Aggresomes: a cellular response to misfolded proteins.” J Cell Biol 143(7): 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenemann K, Schipper-Krom S, Wiemhoefer A, Kloss A, Sanz Sanz A and Reits EA (2013). “Expanded polyglutamine-containing N-terminal huntingtin fragments are entirely degraded by mammalian proteasomes.” J Biol Chem 288(38): 27068–27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S and Cuervo AM (2012). “Chaperone-mediated autophagy: a unique way to enter the lysosome world.” Trends Cell Biol 22(8): 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschbamer E and Biagioli M (2015). “Huntington’s Disease as Neurodevelopmental Disorder: Altered Chromatin Regulation, Coding, and Non-Coding RNA Transcription.” Front Neurosci 9: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N and DiFiglia M (2001). “Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis.” Proc Natl Acad Sci U S A 98(22): 12784–12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I and Johansen T (2009). “A role for NBR1 in autophagosomal degradation of ubiquitinated substrates.” Mol Cell 33(4): 505–516. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr., Iwata J, Kominami E, Chait BT, Tanaka K and Yue Z (2007). “Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration.” Proc Natl Acad Sci U S A 104(36): 1448914494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J and Dikic I (2013). “Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates.” J Cell Sci 126(Pt 2): 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, Hung G, Bennett CF and Cleveland DW (2012). “Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis.” Neuron 74(6): 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A and Kozlowski P (2012). “Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target.” Nucleic Acids Res 40(1): 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Thu D, Waldvogel HJ, Faull RL and Luthi-Carter R (2011). “Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain.” Nat Methods 8(11): 945–947. [DOI] [PubMed] [Google Scholar]
- Kurosawa M, Matsumoto G, Kino Y, Okuno M, Kurosawa-Yamada M, Washizu C, Taniguchi H, Nakaso K, Yanagawa T, Warabi E, Shimogori T, Sakurai T, Hattori N and Nukina N (2015). “Depletion of p62 reduces nuclear inclusions and paradoxically ameliorates disease phenotypes in Huntington’s model mice.” Hum Mol Genet 24(4): 1092–1105. [DOI] [PubMed] [Google Scholar]
- Lamark T, Kirkin V, Dikic I and Johansen T (2009). “NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets.” Cell Cycle 8(13): 1986–1990. [DOI] [PubMed] [Google Scholar]
- Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H, Finkbeiner S, Sun B, Gafni J, Ellerby LM, Trottier Y, Richards WG, Osmand A, Paganetti P and Bates GP (2010). “Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease.” J Biol Chem 285(12): 8808–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Conrad A, Epping E, Mathews K, Magnotta V, Dawson JD and Nopoulos P (2018). “Effect of Trinucleotide Repeats in the Huntington’s Gene on Intelligence.” EBioMedicine 31: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Gutekunst CA, Hersch SM and Li XJ (1998). “Interaction of huntingtin-associated protein with dynactin P150Glued.” J Neurosci 18(4): 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Schilling G, Young W. S. d., Li XJ, Margolis RL, Stine OC, Wagster MV, Abbott MH, Franz ML, Ranen NGand et al. (1993). “Huntington’s disease gene (IT15) is widely expressed in human and rat tissues.” Neuron 11(5): 985–993. [DOI] [PubMed] [Google Scholar]
- Liot G, Zala D, Pla P, Mottet G, Piel M and Saudou F (2013). “Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites.” J Neurosci 33(15): 6298–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP and Zeitlin SO (2017). “Is Huntingtin Dispensable in the Adult Brain?” J Huntingtons Dis 6(1): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunkes A, Lindenberg KS, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer GB, Mandel JL and Trottier Y (2002). “Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions.” Mol Cell 10(2): 259–269. [DOI] [PubMed] [Google Scholar]
- Lystad AH, Ichimura Y, Takagi K, Yang Y, Pankiv S, Kanegae Y, Kageyama S, Suzuki M, Saito I, Mizushima T, Komatsu M and Simonsen A (2014). “Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures.” EMBO Rep 15(5): 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma TC, Buescher JL, Oatis B, Funk JA, Nash AJ, Carrier RL and Hoyt KR (2007). “Metformin therapy in a transgenic mouse model of Huntington’s disease.” Neurosci Lett 411(2): 98–103. [DOI] [PubMed] [Google Scholar]
- Maday S and Holzbaur EL (2012). “Autophagosome assembly and cargo capture in the distal axon.” Autophagy 8(5): 858–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S and Holzbaur EL (2014). “Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway.” Dev Cell 30(1): 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S and Holzbaur EL (2016). “Compartment-Specific Regulation of Autophagy in Primary Neurons.” J Neurosci 36(22): 5933–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE and Holzbaur EL (2012). “Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons.” J Cell Biol 196(4): 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW and Bates GP (1996). “Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice.” Cell 87(3): 493–506. [DOI] [PubMed] [Google Scholar]
- Manzoni C, Mamais A, Roosen DA, Dihanich S, Soutar MP, Plun-Favreau H, Bandopadhyay R, Hardy J, Tooze SA, Cookson MR and Lewis PA (2016). “mTOR independent regulation of macroautophagy by Leucine Rich Repeat Kinase 2 via Beclin-1.” Sci Rep 6: 35106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S and Sandri M (2009). “Autophagy is required to maintain muscle mass.” Cell Metab 10(6): 507–515. [DOI] [PubMed] [Google Scholar]
- McGuire JR, Rong J, Li SH and Li XJ (2006). “Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons.” J Biol Chem 281(6): 3552–3559. [DOI] [PubMed] [Google Scholar]
- Mealer RG, Murray AJ, Shahani N, Subramaniam S and Snyder SH (2014). “Rhes, a striatalselective protein implicated in Huntington disease, binds beclin-1 and activates autophagy.” J Biol Chem 289(6): 3547–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF and Hook VY (2001). “Tissue-specific proteolysis of Huntingtin (htt) in human brain: evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum.” J Neurosci 21(6): 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M and Isalan M (2015). “A shared mechanism of muscle wasting in cancer and Huntington’s disease.” Clin Transl Med 4(1): 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T and Ohsumi Y (2011). “The role of Atg proteins in autophagosome formation.” Annu Rev Cell Dev Biol 27: 107–132. [DOI] [PubMed] [Google Scholar]
- Myers RH, Leavitt J, Farrer LA, Jagadeesh J, McFarlane H, Mastromauro CA, Mark RJ and Gusella JF (1989). “Homozygote for Huntington disease.” Am J Hum Genet 45(4): 615–618. [PMC free article] [PubMed] [Google Scholar]
- Myers RH, Vonsattel JP, Paskevich PA, Kiely DK, Stevens TJ, Cupples LA, Richardson EP Jr. and Bird ED (1991). “Decreased neuronal and increased oligodendroglial densities in Huntington’s disease caudate nucleus.” J Neuropathol Exp Neurol 50(6): 729–742. [DOI] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O’Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG and Hayden MR (1995). “Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes.” Cell 81(5): 811–823. [DOI] [PubMed] [Google Scholar]
- Neueder A and Bates GP (2018). “RNA Related Pathology in Huntington’s Disease.” Adv Exp Med Biol 1049: 85–101. [DOI] [PubMed] [Google Scholar]
- Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, Mao K, Lau AL, Yeung SY, Humbert S, Saudou F, Klionsky DJ, Finkbeiner S, Zeitlin SO, Marsh JL, Housman DE, Thompson LM and Steffan JS (2014). “Potential function for the Huntingtin protein as a scaffold for selective autophagy.” Proc Natl Acad Sci U S A 111(47): 16889–16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G and Johansen T (2007). “p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy.” J Biol Chem 282(33): 24131–24145. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M and Finch JT (1994). “Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases.” Proc Natl Acad Sci U S A 91(12): 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips OR, Joshi SH, Squitieri F, Sanchez-Castaneda C, Narr K, Shattuck DW, Caltagirone C, Sabatini U and Di Paola M (2016). “Major Superficial White Matter Abnormalities in Huntington’s Disease.” Front Neurosci 10: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power D, Srinivasan S and Gunawardena S (2012). “In-vivo evidence for the disruption of Rab11 vesicle transport by loss of huntingtin.” Neuroreport 23(16): 970–977. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R and Rubinsztein DC (2002). “Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy.” Hum Mol Genet 11(9): 1107–1117. [DOI] [PubMed] [Google Scholar]
- Reddy PH and Shirendeb UP (2012). “Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington’s disease.” Biochim Biophys Acta 1822(2): 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulier E, Trottier Y, Perrin V, Aebischer P and Deglon N (2003). “Early and reversible neuropathology induced by tetracycline-regulated lentiviral overexpression of mutant huntingtin in rat striatum.” Hum Mol Genet 12(21): 2827–2836. [DOI] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T and Kirkin V (2014). “Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy.” Mol Cell 53(2): 167–178. [DOI] [PubMed] [Google Scholar]
- Roizin L, Stellar S and Liu JC (1979). Neuronal nuclear-cytoplasmic changes in Huntington’s chorea: electron microscope investigations. Advances in Neurology, Huntington’s Disease. [Google Scholar]
- Chase TN, Wexler NS and Barbeau A New York, Raven Press; 23: 95–122. [Google Scholar]
- Rudnicki DD, Pletnikova O, Vonsattel JP, Ross CA and Margolis RL (2008). “A comparison of huntington disease and huntington disease-like 2 neuropathology.” J Neuropathol Exp Neurol 67(4): 366374. [DOI] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM and Zhang S (2015). “Huntingtin functions as a scaffold for selective macroautophagy.” Nat Cell Biol 17(3): 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M (2016). “Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases.” Ann N Y Acad Sci 1371(1): 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S (2013). “Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers.” Biochem Soc Trans 41(5): 1103–1130. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A and Rubinsztein DC (2007). “Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alphasynuclein.” Journal of Biological Chemistry 282(8): 5641–5652. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ and Rubinsztein DC (2005). “Lithium induces autophagy by inhibiting inositol monophosphatase.” J Cell Biol 170(7): 11011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F and Humbert S (2016). “The Biology of Huntingtin.” Neuron 89(5): 910–926. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H and Wanker EE (1997). “Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo.” Cell 90(3): 549–558. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Dyar KA, Ciciliot S, Blaauw B and Sandri M (2013). “Mechanisms regulating skeletal muscle growth and atrophy.” FEBS J 280(17): 4294–4314. [DOI] [PubMed] [Google Scholar]
- Schipper-Krom S, Juenemann K, Jansen AH, Wiemhoefer A, van den Nieuwendijk R, Smith DL, Hink MA, Bates GP, Overkleeft H, Ovaa H and Reits E (2014). “Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies.” FEBS Lett 588(1): 151–159. [DOI] [PubMed] [Google Scholar]
- Schulte J and Littleton JT (2011). “The biological function of the Huntingtin protein and its relevance to Huntington’s Disease pathology.” Curr Trends Neurol 5: 65–78. [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR and Wooten MW (2004). “Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation.” Mol Cell Biol 24(18): 8055–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J (1998). “P62 and the sequestosome, a novel mechanism for protein metabolism.” Arch Pharm Res 21(6): 629–633. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A and Stenmark H (2004). “Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes.” J Cell Sci 117(Pt 18): 4239–4251. [DOI] [PubMed] [Google Scholar]
- Steffan JS (2010). “Does Huntingtin play a role in selective macroautophagy?” Cell Cycle 9(17): 34013413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturrock A and Leavitt BR (2010). “The clinical and genetic features of Huntington disease.” J Geriatr Psychiatry Neurol 23(4): 243–259. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M and Nukina N (2004). “Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease.” Nat Med 10(2): 148–154. [DOI] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J and Sulzer D (2014). “Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits.” Neuron 83(5): 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo RT, Hong X, Yu-Taeger L, Huang Y, Tan LJ, Xie Y, To XV, Guo L, Rajendran R, Novati A, Calaminus C, Riess O, Hayden MR, Nguyen HP, Chuang KH and Pouladi MA (2016). “Structural and molecular myelination deficits occur prior to neuronal loss in the YAC128 and BACHD models of Huntington disease.” Hum Mol Genet 25(13): 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E and La Spada AR (2012). “PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function.” Sci Transl Med 4(142): 142ra197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N and DiFiglia M (1998). “Wildtype and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways.” Exp Neurol 152(1): 34–40. [DOI] [PubMed] [Google Scholar]
- Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, Huh CJ, Zhang B, Davidson BL, Yang XW and Yoo AS (2018). “Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes.” Nat Neurosci 21(3): 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP (2008). “Huntington disease models and human neuropathology: similarities and differences.” Acta Neuropathol 115(1): 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, Keller C and Cortes Ramirez EP (2011). “Huntington’s disease - neuropathology.” Handb Clin Neurol 100: 83–100. [DOI] [PubMed] [Google Scholar]
- Wang G, Liu X, Gaertig MA, Li S and Li XJ (2016). “Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis.” Proc Natl Acad Sci U S A 113(12): 3359–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KR and Littleton JT (2016). “Characterization of axonal transport defects in Drosophila Huntingtin mutants.” J Neurogenet 30(3–4): 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler NS, Young AB, Tanzi RE, Travers H, Starosta-Rubinstein S, Penney JB, Snodgrass SR, Shoulson I, Gomez F, Ramos Arroyo MA and et al. (1987). “Homozygotes for Huntington’s disease.” Nature 326(6109): 194–197. [DOI] [PubMed] [Google Scholar]
- White JA, 2nd, Anderson E, Zimmerman K, Zheng KH, Rouhani R and Gunawardena S(2015). “Huntingtin differentially regulates the axonal transport of a sub-set of Rab-containing vesicles in vivo.” Hum Mol Genet 24(25): 7182–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA and Rubinsztein DC (2008). “Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway.” Nat Chem Biol 4(5): 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS, Lim J, Lachance V, Deng Z and Yue Z (2016). “ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington’s disease models.” Mol Neurodegener 11(1): 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC and Holzbaur EL (2014). “The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation.” J Neurosci 34(4): 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC and Holzbaur EL (2015). “Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria.” Autophagy 11(2): 422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML and Rothman JE (2006). “Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway.” J Cell Biol 172(5): 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ and Hen R (2000). “Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease.” Cell 101(1): 57–66. [DOI] [PubMed] [Google Scholar]
- Yamamoto A and Yue Z (2014). “Autophagy and Its Normal and Pathogenic States in the Brain.” Annu Rev Neurosci. [DOI] [PubMed] [Google Scholar]
- Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, Ebner M, Peterbauer T, Sztacho M, Trapannone R, Tarafder AK, Sachse C and Martens S (2018). “p62 filaments capture and present ubiquitinated cargos for autophagy.” EMBO J 37(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelieres FP, Marco S and Saudou F (2013). “Vesicular glycolysis provides on-board energy for fast axonal transport.” Cell 152(3): 479–491. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE and Efstratiadis A (1995). “Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue.” Nat Genet 11(2): 155–163. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D and Yuan J (2007). “Small molecule regulators of autophagy identified by an image-based high-throughput screen.” Proc Natl Acad Sci U S A 104(48): 19023–19028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC and Zeitlin SO (2010). “Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice.” PLoS Genet 6(2): e1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]