Abstract
Yeast prions have become important models for the study of the basic mechanisms underlying human amyloid diseases. Yeast prions are pathogenic (unlike the [Het-s] prion of Podospora anserina), and most are amyloid-based with the same in-register parallel β-sheet architecture as most of the disease-causing human amyloids studied. Normal yeast cells eliminate the large majority of prion variants arising, and several anti-prion/anti-amyloid systems that eliminate them have been identified. It is likely that mammalian cells also have anti-amyloid systems, which may be useful in the same way humoral, cellular, and innate immune systems are used to treat or prevent bacterial and viral infections.
Keywords: amyloid, inositol phosphate, prion, chaperone, innate immunity, in-register parallel, Upf proteins, Siw14, Hsp104, Btn2, Cur1, [URE3], [PSI+]
Herb Tabor, my mentor
I began my scientific career after completing my medical internship when I became a postdoc with Herb and Celia Tabor in 1967 at the National Institutes of Health. I was fortunate to be working with someone who was such an exceptional model scientist. After 3 years as a postdoc with Jerry Hurwitz in New York, I returned to Herb's department in an independent position. Herb had built a department of diverse interests that was unified by daily literature and weekly work seminars, both featuring intense discussion and critique. It was an excellent environment for learning and developing one's own program. Although I became the Lab Chief (NIH's equivalent of the Department Chair) in 1996, my relationship with Herb has remained essentially unchanged for over 50 years (it took me ∼25 of those years before I switched from “Dr. Tabor” to “Herb”). Herb Tabor is totally devoted to science, has always done experiments himself, is far more concerned about getting the right answer to a problem than in making a splash, and is the least vain of any scientist I have ever met. His total honesty, sincerity, and scientific focus have been a model I have attempted to emulate (though largely without success). His devotion to science did not mean neglect of his family, but rather a total involvement of his family in science. Celia White Tabor worked together with Herb for many years, and all of his wonderful children have had science careers. A large part of Herb's life has been science in the lab, but an equal or maybe even larger part has been scientific publishing. His technologically forward-looking approach and his broadening the scope and enlarging the size of the JBC had an enormous impact.
Among the lessons I have learned from Herb are:
Any scientific area has the potential to be interesting, if studied with enough depth and skill.
“Depth is better than width.”
The word “critical” was used in a positive sense to describe someone who could thoughtfully, fairly, and accurately evaluate a piece of work.
“Kinetics are meaningless.”
“Some scientists may not want to be acknowledged because they do not think much of your work.”
“I've told you all I know, maybe more.”
Although my own work has taken a largely genetic direction, the lessons I learned from Herb are not specific to any area of science, and I know I have benefited greatly from his constant example.
The word “prion,” meaning “infectious protein,” without any nucleic acid being required for the transmission of the infection, originated in studies of the transmissible spongiform encephalopathies of mammals (1–3). However, the proof of the existence of a prion had to await the discovery that the nonchromosomal genetic elements, [URE3] and [PSI+] of Saccharomyces cerevisiae, are based on altered infectious forms of Ure2p and Sup35p, respectively (4). Both the mammalian transmissible spongiform encephalopathies and the yeast prions are self-propagating amyloids. Amyloid is a linear polymer of a particular protein, a sort of one-dimensional crystal in which the predominant structure is β-sheet, with the β-strands perpendicular to the long axis of the filaments. Of late, evidence has been found for prion-like behavior, or even frank infection, for such classical amyloidoses as amyloidosis A (5, 6), Alzheimer's disease (7–10), multiple system atrophy (11), amyotrophic lateral sclerosis (12), and type 2 diabetes (13). This developing unification of the human amyloidoses around the prion model enhances the importance of an in-depth study of the prions of S. cerevisiae, with their facile genetic manipulability.
Yeast prions
Two long-known nonchromosomal genetic elements of yeast, [URE3] (14) and [PSI+] (15), were identified as prions of Ure2p and Sup35p based on their outré genetic properties (4). Ure2p, a repressor of genes encoding transporters and assimilation enzymes for poor nitrogen sources, is active when a good nitrogen source is available (16), but in [URE3] strains, Ure2p is largely trapped in infectious amyloid filaments (17–20). The aggregated form is evidently largely inactive as [URE3] cells have a phenotype similar to ure2Δ cells, the inappropriate derepression of the controlled genes despite the presence of a good nitrogen source. Sup35p is a subunit (with Sup45) of the translational termination factor (21, 22). In [PSI+] cells, Sup35p is in infectious amyloid aggregates (23–29), and increased readthrough of stop codons ensues.
A wide array of other prions has now been found in S. cerevisiae and one particularly revealing prion in the filamentous fungus Podospora anserina (30). [PIN+] is an amyloid-based prion of Rnq1p, a protein of unknown function (31–33). [PIN+] is manifest only (so far) by its facilitation of the (nonetheless rare) generation of other prions, originally [PSI+] (31) and later [URE3] (34) and [SWI+] (35). Extensive evidence indicates that this stimulation of prion formation occurs by an inefficient form of the same seeding process that is involved in propagation of all of the amyloid-based yeast prions (36). There is clinical and experimental evidence that similar cross-seeding is an important feature of human amyloidosis (37, 38). Because nearly all known pathogenic amyloids have a similar architecture (see below), it is likely that this potentiation of formation of one prion/amyloid by another is a general phenomenon.
Prion variants are a feature of all pathogenic prions, whether of animals or yeast (39). A single prion protein with a single sequence can be the basis of a wide array of prion variants (or prion strains), with distinct biological properties and different amyloid conformations (29, 40). Each variant is relatively stably propagated, implying that there must be a mechanism by which the amyloid filaments act as a template to force monomers joining the end of the filaments to assume the same conformation as molecules already in the filament. In yeast, prion variants may differ in the intensity of their phenotype (strong versus weak), stability of propagation, ability to propagate in the face of overproduction or deficiency of various chaperones or other cell components, ability to cross interspecies or intraspecies barriers, and other properties. Using a nonselective system, it was shown that the [PSI+] prion exists as a “cloud” of prion variants that segregate from each other as cells grow and mutate at some frequency (41), thus establishing the “prion cloud” model (39, 42).
Prion domains are the part of the protein that actually forms the amyloid and is roughly the same as the part needed to transmit the prion (17, 19, 26, 43). Both the extent of amyloid structure and the region needed to faithfully propagate the prion vary with the prion variant (29, 44, 45). The “prion domains” have normal nonprion functions. The Ure2p prion domain is necessary for the stability of the whole molecule against degradation, and thus for the full nitrogen regulation function (46). The prion domain of Sup35p is necessary for general turnover of mRNAs (47), for cytoskeleton-associated translation (48), and for recovery from the stationary phase (49).
Structure of infectious yeast prion amyloids
In an attempt to show that there were sequences in the prion domains of Ure2p and Sup35p needed for prion formation, it was, surprisingly, found that randomly shuffling these domains produced sequences that in all cases were able to form prions (50–52)! This proved that it was not the sequence but rather the amino acid content of these domains that made them suitable for prion formation, and detailed analysis has revealed which residues favor or impair prion formation (53, 54). The sequence independence of prion formation, combined with the well-known barriers to prion propagation produced by even a single amino acid difference in some cases (55–57), indicated that the faithful propagation of prion variant/strain information was not based on “complementarity,” as for DNA or RNA, but rather a principle of “identity” (52). Any complementarity feature (self-complementarity in this case) would be destroyed by shuffling the sequence. It was realized that an anti-parallel β-sheet, a β-helix, or an out-of-register parallel β-sheet would rely on complementarity between neighboring amino acids in different molecules (52). However, a parallel, in-register β-sheet features rows of identical amino acid residues along the long axis of the filament, such as had already been shown for Aβ amyloid (58). Shuffling the sequence would not prevent identical residues from interacting in a parallel in-register structure, only their order would change. For this reason, we proposed that the prion amyloids of Ure2p and Sup35p had an in-register parallel β-sheet architecture (52).
Based on extensive studies of highly infectious amyloids of Sup35p, Ure2p, and Rnq1p, using solid-state NMR (59–63) and EM (particularly mass/length measurements) (64–66), it is clear that each of these prion amyloids in fact does have a folded, parallel, in-register, β-sheet architecture. Indeed, the amyloids formed by the shuffled prion domains also had this architecture (67).
Mechanism of conformational templating
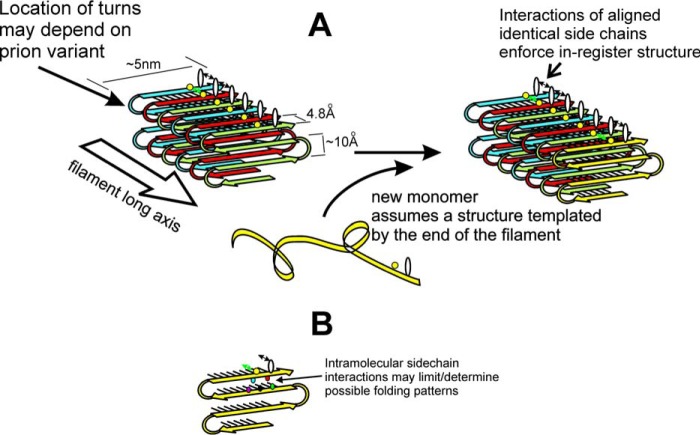
The fact that a single protein sequence can be the basis for an array of prion variants apparently differing only in protein conformation implies that there must be a mechanism for proteins to template their conformation and to transmit this conformation information to other molecules of the same sequence. This is the central mystery of prions. Based on the folded in-register parallel β-sheet architecture of yeast prion amyloids, we have suggested that the same forces of favorable interaction between identical side chains that keeps this architecture in-register also ensures that a new molecule joining the end of a filament will adopt the same structure as molecules already in the filament (Fig. 1) (68, 69). A line of glutamine residues along the long axis of a filament can form a line of hydrogen bonds, but only if the molecules are in-register. Thus, this interaction stabilizes the in-register architecture. The same is true of a line of asparagines, serines, or threonines. A line of identical hydrophobic residues can have a series of stabilizing hydrophobic interactions if the structure is in-register. Charged residues would of course destabilize an in-register structure, and charged residues are rare in the prion domains of Ure2p, Sup35p, and Rnq1p. Furthermore, mutations blocking prion transmission are often uncharged-to-charged amino acid changes (55, 56). A molecule of the prion protein joining the end of the filament, to have these favorable side-chain interactions, must have a turn between the β-strands at the same location as those molecules already in the filament. This results in the end of the filament acting as a template to transmit conformation information to the new molecule joining the end of the filament (68, 69). This templating explains how proteins can act as genes, templating conformation in analogy to the templating of sequence by replicating DNA or RNA (Fig. 1).
Figure 1.
A, folded parallel in-register β-sheet architecture of yeast prion amyloids Sup35p, Ure2p, and Rnq1p (59–61, 63). It is proposed that prion variants differ largely in the location of the turns in the strands/folds in the β-sheet (B), with favorable H-bonding or hydrophobic interactions among aligned side chains ensuring alignment of identical residues in adjacent molecules. These favorable interactions drive new molecules joining the end of the filament to have their turns in the same places as molecules already in the filament, and this constitutes templating of conformation (68, 69). Reprinted with permission from Ref. 69. This research was originally published in Biochemistry. Wickner, R. B., Edskes, H. K., Bateman, D. A., Kelly, A. C., Gorkovskiy, A., Dayani, Y., and Zhou, A. Amyloids and yeast prion biology. Biochemistry 2013;52:1514–1527. © the American Chemical Society.
Nearly all of the pathogenic amyloids associated with human diseases that have been examined have the same folded in-register parallel β-sheet architecture (58, 70–77) as observed for these yeast prions, with one exception being transthyretin (78–80). It is likely that templating of conformational information for the human pathologic amyloids proceeds by the same mechanism discussed here.
Yeast prions [PSI+], [URE3], [SWI+], and [PIN+] are pathogenic
Most variants of [PSI+] and [URE3] arising in WT laboratory strains severely slow growth or actually kill the host cells (81). A beneficial prion variant would be widespread in wild strains, particularly because they are infectious, but [PSI+], [URE3], [SWI+], and [PIN+] are known to be rare in wild isolates (82–84), more rare than the 2-μm DNA plasmid that is known to cause a >1% growth detriment (82, 85). These and other results show that no variants of these prions are beneficial on the net (86).
Anti-prion systems in yeast
There is a wide array of cellular components whose overproduction or deficiency leads to the loss of one or another prion (36, 87, 88). These studies have been important in establishing mechanisms affecting prions, but it is generally unclear which of these effects represent physiologic (or pathophysiologic) responses of cells to prions. It is of particular interest to know whether there are systems that cure yeast prions in a normal cell, without overproduction or deletion of anything, and it is these that we refer to as “anti-prion” systems. As expected for a pathogenic infectious agent, the cell indeed has defenses against yeast prions at several levels. Prion formation is partially blocked; having formed, several systems cure the nascent prions; and other systems prevent prion-based pathology (Table 1). As with DNA repair systems, these anti-prion systems are each only partially effective.
Table 1.
Yeast anti-prion systems active without overexpression or deficiency
5PP-IP5 is 5-diphosphoinositol pentakisphosphate.
| Anti-prion protein | Anti-prion protein activity | Prion affected | Effect | Ref. |
|---|---|---|---|---|
| Ssb1p, Ssb2p | Ribosome-associated Hsp70 | [PSI+] | Prevents [PSI+] formation | 92, 94, 142 |
| Btn2p | Collector of aggregates | [URE3] | Cures by sequestration of amyloid filaments | 104, 107 |
| Cur1p | ?? | [URE3] | Cures by unknown mechanism | 104, 107, 110 |
| Hsp104 | Disaggregating chaperone | [PSI+] | Cures–mechanism controversial: asymmetric segregation or blocking Hsp70 access for filament cleavage | 115, 125–127, 130 |
| Siw14p | 5-Pyrophosphate-inositol-hexakisphosphate pyrophosphatase | [PSI+] | Cures by lowering 5PP-IP5 levels | 131, 132 |
| Upf1p, Upf2p, Upf3p | Nonsense-mediated mRNA decay factors | [PSI+] | Cures, perhaps by either binding Sup35p monomers or binding to filaments blocking extension | 138 |
| Sis1p | Hsp40 chaperone | [PSI+] | Prevents lethality of normally mild variant | 95 |
| Lug1p/YRL352wp | F-box protein; substrate-specifying subunit of cullin ubiquitin ligase | [URE3] | Allows growth of [URE3] strains on glycerol | 96 |
Ribosome-associated Hsp70s inhibit [PSI+] formation
Ssb1 and Ssb2 are Hsp70 family members that are associated with the ribosomes and are involved in folding of nascent polypeptides (89, 90). For example, in the absence of the Ssb proteins, aggregated proteins accumulate in the cell (91). In ssb1Δ ssb2Δ cells, the frequency of [PSI+] arising is about 10-fold higher than in a normal cell (92, 93). Restoration of Ssb1 does not cure the [PSI+] variants arising in the double mutant, indicating that the Ssb proteins largely prevent [PSI+] generation, rather than blocking their propagation (92). However, dissociation of Ssb's from ribosomes in cells grown on minimal medium has a [PSI+]-curing effect (94).
Hsp40 family member Sis1p prevents toxicity of otherwise mild [PSI+] prion variants
Although a majority of [PSI+] prion variants are highly toxic to cells, the variants usually studied do not produce an obvious growth defect. The studies of wild strains (above) show that even these variants are detrimental in the wild. However, Masison's group (95) has shown that these “mild” [PSI+] variants have the potential to prevent cell growth except for the action of the Sis1p protein. Sis1p is essential for growth, but deletion of the C-terminal domain of the protein does not substantially affect growth of [psi−] cells. However, [PSI+] cells with the Sis1p C-terminal deletion grow poorly or not at all (95).
F-box protein Ylr352w/Lug1 prevents toxicity of mild [URE3]
A general screen for genes that are essential specifically in [URE3] cells was carried out using transposon mutagenesis with the Hermes transposon (96). Genes that could be mutated in a [ure-o] cell, but not in a [URE3−1] strain, were inferred to be either necessary for propagation of [URE3] or for cell growth/survival in the presence of the prion. The most dramatic hit from the screen was YLR352W (renamed LUG1 for “lets [URE3] grow”) (96), encoding a substrate-specifying subunit of a cullin-containing multisubunit E3 ubiquitin ligase (97). Other subunits of this complex and subunit-modifying proteins also were detected in the screen (96). Lug1p is one of 20 substrate-specifying subunits with a common sequence called the “F-box” (97–100), but the substrates specified by Lug1p are unknown. The lug1Δ [URE3] strains could not grow on glycerol media, whereas lug1Δ [ure-o] cells grew normally. The growth defect was suppressed by overproduction of Hap4p, a transcription factor that stimulates expression of mitochondria-related genes, consistent with the requirement for oxidation for utilization of glycerol (96). The growth defect of lug1Δ [URE3] strains was also suppressed by mutation of GLN1, encoding glutamine synthase. Like cells carrying [URE3], gln1 mutants are defective in nitrogen catabolite repression, the diminished expression of genes for using poor nitrogen sources when a good nitrogen source is available. This indicates that the inability to utilize glycerol of the lug1 [URE3] strains was not a consequence of failure to control nitrogen catabolism. Growth of lug1Δ strains on proline, a condition that is known to shut off Ure2p's activity in repressing NCR genes, does not affect cell growth, confirming the conclusion that Ure2p has another function independent of NCR (96). It remains unclear what is the relevant target of Lug1p, but it is clear that this protein prevents the pathologic effects of an otherwise relatively mild variant of [URE3].
The same screen also showed a substantial decrease in detection of mutations in many chaperone genes, including some that had not previously been implicated in [URE3] prion propagation. This effect was observed for HSP82 and HSC82 (encoding Hsp90 paralogs), HSP104 and SSA2 (known to be necessary for [URE3] propagation (101, 102)), YDJ1 and CAJ1 (Hsp40s), STI1 (co-chaperone of Hsp90s), FES1 and SSE1 (nucleotide exchange factors for Hsp70s known to affect [URE3] (103)), HSP26 and HSP42 (“small” heat-shock proteins), SSB1 and SSB2 (ribosome-associated Hsp70s), and SSA1 and SBA1. Whether these effects reflect specific toxicities of [URE3] prevented by the chaperones, or a synergistic toxicity of prion-induced stress and stress due to chaperone deficiency, remains to be determined. Nearly all of these chaperones have close human homologs (HSP104 is a notable exception), making understanding this phenomenon of particular interest.
Normal levels of Btn2p and Cur1p cure most [URE3] variants
Overproduction of either of two paralogs, Btn2p or Cur1p, cure [URE3] (104). In the process, Btn2p collects the aggregates of Ure2p at one site in the cell (instead of being scattered about the cytoplasm), and this collection spot co-localizes with a spot of Btn2p (104). Apparently, on cell division, the probability of daughter cells getting any filaments is thereby reduced, producing frequent cured cells (104). Consistent with this “sequestration model,” Btn2p can also collect nonprion aggregates of other proteins at a single cellular site (105, 106). However, Cur1p did not co-localize with Ure2p aggregates in the course of its curing of [URE3] (104).
To determine whether normal levels of Btn2p or Cur1p have an anti-prion action, a series of [URE3] prion variants was isolated in a btn2Δ cur1Δ strain, and the normal levels of each protein were restored by mating or by other means (107). Remarkably, nearly all (∼90%) of these [URE3] variants were cured in the WT environment, although most were quite stable in the btn2Δ cur1Δ strain. Indeed, the frequency of [URE3] arising spontaneously was elevated about 5-fold in btn2Δ cur1Δ strains (107). A propagon-measuring method (108) revealed that variants of [URE3] with high seed number (presumably many amyloid filaments per cell) could only be cured by overproduction of Btn2p or Cur1p, whereas those variants with low seed numbers were cured by normal levels of these proteins (107). Thus, there are far more prions arising than the few that manage to escape the cellular anti-prion systems.
Btn2p overproduction curing of [URE3] requires Hsp42 (107), a small chaperone also known to be involved in collecting nonprion aggregates of denatured proteins (109). Hsp42 overproduction can also cure [URE3] (107). Btn2p overproduction curing of [URE3] does not require Cur1p nor does Cur1p overproduction curing require Btn2p (107). The facts that Cur1p does not co-localize with Ure2p aggregates in the process of curing [URE3] (104) and that Cur1p overproduction curing does not require Hsp42 (107) suggest that Cur1p and Btn2p cure by different mechanisms, despite their sequence similarity (110).
Btn2p was previously known as a mediator of protein trafficking in the endosome pathway (111, 112), and Btn3p, an inhibitor of Btn2p's action in protein sorting, also inhibits its ability to sequester the [URE3] amyloid filaments and cure the prion (113). This is the only connection yet defined between the protein-sorting and aggregate-collecting activities of Btn2p.
Normal levels of the Hsp104 disaggregase cures most [PSI+] variants
Hsp104 is a disaggregating chaperone that, in concert with Hsp40s and Hsp70s, disaggregates denatured proteins allowing cells to recover from heat shock (114). Hsp104, working with Hsp70s, Hsp40s, and nucleotide exchange factors, cleaves prion amyloid filaments to generate new filament ends and thus new growing points (23, 87, 115–118, 121). However, overproduction of Hsp104 cures the [PSI+] prion (115) and [URE3] to a minor extent (104). The propagation-promoting activity of Hsp104 involves its removing monomers from the middle of an amyloid filament thereby breaking it into two pieces (119, 120). Inhibition of Hsp104 by millimolar concentrations of guanidine results in the arrest of new filament formation, and eventual curing of the prion in most progeny cells as the filaments are diluted by cell growth (108, 122–124).
The mechanism of the [PSI+]-curing effect of Hsp104 overproduction is still controversial, despite extensive study. One line of evidence shows that Hsp104 has a specific binding site in the M (middle) domain of Sup35p (125) and that excessive Hsp104 blocks access of Hsp70s to the filaments, thereby preventing filament cleavage (126). Another study shows that overproduced Hsp104 results in asymmetric segregation of prion seeds, resulting in frequent loss of the prion (127). A third group proposes that the overproduction curing activity of Hsp104 is a “trimming” activity, removing monomers from the ends (in contrast to cleavage of filaments in the middle) (128).
To determine whether normal levels of Hsp104 cure some [PSI+] variants as they arise, we used the same approach as discussed above for Btn2 and Cur1. We used the hsp104T160M mutant that propagates [PSI+] normally but cannot cure [PSI+] even if overproduced (129). A series of [PSI+] variants isolated in this strain was tested for the ability to propagate in an HSP104 host, and about half were unable to do so (130). The frequency of [PSI+] arising spontaneously was elevated over 10-fold in hsp104T160M hosts. These results indicate that the normal Hsp104 (without overexpression) cures many [PSI+] variants as they arise (130).
Inositol pyro-/polyphosphates and [PSI+] propagation
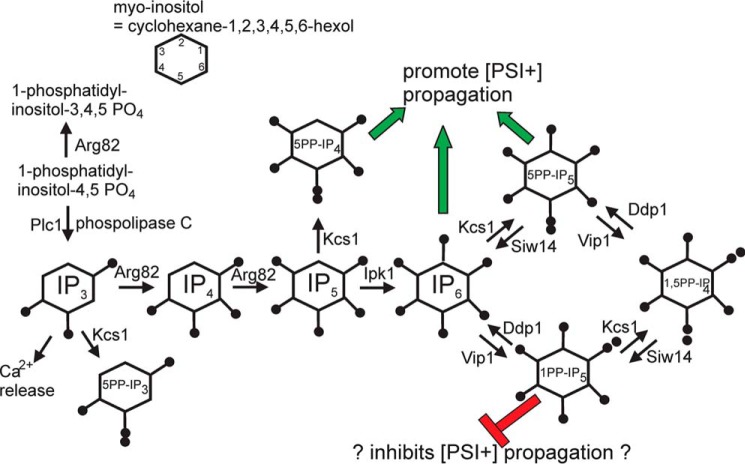
In a general screen of the knockout collection for mutants defective in anti-[PSI+] activity, two isolates were siw14::kanMX mutants (131), lacking a pyrophosphatase specifically active on 5-pyrophosphoinositol pentakisphosphate, and thus having elevated levels of this compound (Fig. 2) (132). We found that nearly all [PSI+] variants require some inositol poly/pyrophosphate and that 5-pyrophosphoinositol tetrakisphosphate, inositol hexakisphosphate, or 5-pyrophosphoinositol pentakisphosphate could support [PSI+] propagation (131). In the absence of the 5-pyrophosphoinositol polyphosphates, elevated levels of the 1-pyrophosphoinositol polyphosphates inhibited [PSI+] propagation (131). The target(s) of the inositol poly/pyrophosphates in exerting these effects is at this time unclear. The inositol poly/pyrophosphates have a wide array of activities (133, 134), making this a difficult task. Wu et al. (135) prepared affinity resins with the inositol poly/pyrophosphates that support [PSI+] and identified yeast proteins that adhere to columns of these resins. Among these were Hsp26p, Sse1p, and Ssb1,2, each of which has been reported to affect [PSI+] generation or propagation (92, 103, 136, 137), and they must be considered candidates for mediators of action of the inositol poly/pyrophosphates.
Figure 2.
Inositol pyro/polyphosphate pathways showing [PSI+]-supporting (green arrows) and [PSI+]-inhibiting (red symbol) species (modified from Ref. 131). IP3, inositol 1,4,5-trisphosphate; IP4, inositol tetrakisphosphate; 5PP-IP5, 5-diphosphoinositol pentakisphosphate; IP6, inositol 1,2,3,4,5,6-hexakisphosphate.
Upf proteins, mediators of nonsense-mediated mRNA decay, cure many [PSI+] variants
The screen described above also returned deletions of UPF1 and UPF3 (138), genes encoding components of the nonsense-mediated mRNA decay apparatus, responsible for elimination of mRNAs carrying a premature stop codon (139). Because these genes also affect the genetic test for [PSI+] (readthrough of a premature stop codon), it was shown in several different ways that the effect of the Upf proteins was on prion propagation and not on the assay. The upf mutants have a >10-fold increased frequency of [PSI+] generation, and nearly all of the [PSI+] variants produced in a upf mutant are cured by simply replacing the normal amount of the Upf protein (138). The Upf proteins normally form a complex with the translation termination factors Sup35p and Sup45p on the ribosome, and it is argued that this complex stabilizes Sup35p in its normal form preventing conversion to the amyloid form (138). Upf1p, at sub-stoichiometric levels, inhibits amyloid formation in vitro by Sup35p (but not by Ure2p), suggesting that a direct interaction is responsible for blocking prion propagation (138). Examination of a series of UPF1 and UPF2 mutants showed a poor correlation of [PSI+]-curing activity (138) with nonsense-mediated decay (NMD) efficiency, RNA helicase, ATPase, or RNA-binding activity, but a better correlation with their reported (140, 141) formation of the Upf1–2-3 complex and binding to Sup35p.
It is possible that the binding of Sup35p monomers by the Upf complex competes with their binding to amyloid filaments. Alternatively, the binding of the Upf1–2-3 complex to the ends of Sup35p amyloid filaments may block access of Sup35p monomers to these growth points. We infer that normal protein–protein interactions naturally compete with and may even reverse abnormal interactions. This may have general implications applicable to treatment of disease.
Study of anti-prion systems reveals an unexpected abundance of prion variants
At this time the various anti-prion systems appear to be specific for a particular prion, although only limited tests have been done. Several systems cure [PSI+] variants, and it is not yet known whether they work independently or in concert. It is evident in yeast that prions are arising at a dramatically higher rate than was previously appreciated and that the array of anti-prion systems cures most of them. There are also anti-prion systems that prevent prion formation, and others that make some of those prions that do succeed in arising less harmful than they would otherwise be. It is hoped that detailed study of the yeast anti-prion systems will facilitate discovery of homologous or analogous mammalian systems active against the many human amyloidoses.
Footnotes
This work was supported by the Intramural Program of the NIDDK, National Institutes of Health. This JBC Review is part of a collection honoring Herbert Tabor on the occasion of his 100th birthday. The author declares that he has no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Alper T., Cramp W. A., Haig D. A., and Clarke M. C. (1967) Does the agent of scrapie replicate without nucleic acid? Nature 214, 764–766 10.1038/214764a0 [DOI] [PubMed] [Google Scholar]
- 2. Griffith J. S. (1967) Self-replication and scrapie. Nature 215, 1043–1044 10.1038/2151043a0 [DOI] [PubMed] [Google Scholar]
- 3. Prusiner S. B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 4. Wickner R. B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science 264, 566–569 10.1126/science.7909170 [DOI] [PubMed] [Google Scholar]
- 5. Kisilevsky R., and Boudreau L. (1983) The kinetics of amyloid deposition. I. The effects of amyloid-enhancing factor and splenectomy. Lab. Invest. 48, 53–59 [PubMed] [Google Scholar]
- 6. Kisilevsky R., Raimondi S., and Bellotti V. (2016) Historical and current concepts of fibrillogenesis and in vivo amyloidogenesis: implications of amyloid tissue targeting. Front. Mol. Biosci. 3, 17 10.3389/fmolb.2016.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jucker M., and Walker L. C. (2013) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 10.1038/nature12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watts J. C., Condello C., Stöhr J., Oehler A., Lee J., DeArmond S. J., Lannfelt L., Ingelsson M., Giles K., and Prusiner S. B. (2014) Serial propagation of distinct strains of Aβ prions from Alzheimer's disease patients. Proc. Natl. Acad. Sci. U.S.A. 111, 10323–10328 10.1073/pnas.1408900111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaunmuktane Z., Mead S., Ellis M., Wadsworth J. D., Nicoll A. J., Kenny J., Launchbury F., Linehan J., Richard-Loendt A., Walker A. S., Rudge P., Collinge J., and Brandner S. (2015) Evidence for human transmission of amyloid-β pathology and cerebral amyloid angiopathy. Nature 525, 247–250 10.1038/nature15369 [DOI] [PubMed] [Google Scholar]
- 10. Lu J. X., Qiang W., Yau W. M., Schwieters C. D., Meredith S. C., and Tycko R. (2013) Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell 154, 1257–1268 10.1016/j.cell.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prusiner S. B., Woerman A. L., Mordes D. A., Watts J. C., Rampersaud R., Berry D. B., Patel S., Oehler A., Lowe J. K., Kravitz S. N., Geschwind D. H., Glidden D. V., Halliday G. M., Middleton L. T., Gentleman S. M., et al. (2015) Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 112, E5308–E5317 10.1073/pnas.1514475112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ayers J. I., Fromholt S. E., O'Neal V. M., Diamond J. H., and Borchelt D. R. (2016) Prion-like propagation of mutant SOD1 misfolding and motor neuron disease. Acta Neuropathol. 131, 103–114 10.1007/s00401-015-1514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mukherjee A., Morales-Scheihing D., Salvadores N., Moreno-Gonzalez I., Gonzalez C., Taylor-Presse K., Mendez N., Shahnawaz M., Gaber A. O., Sabek O. M., Fraga D. W., and Soto C. (2017) Induction of IAPP amyloid deposition and associated diabetic abnormalities by a prion-like mechanism. J. Exp. Med. 214, 2591–2610 10.1084/jem.20161134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lacroute F. (1971) Non-mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106, 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox B. S. (1965) PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20, 505–521 10.1038/hdy.1965.65 [DOI] [Google Scholar]
- 16. Cooper T. G. (2002) Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: connecting the dots. FEMS Microbiol. Rev. 26, 223–238 10.1111/j.1574-6976.2002.tb00612.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masison D. C., and Wickner R. B. (1995) Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93–95 10.1126/science.270.5233.93 [DOI] [PubMed] [Google Scholar]
- 18. Edskes H. K., Gray V. T., and Wickner R. B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. U.S.A. 96, 1498–1503 10.1073/pnas.96.4.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor K. L., Cheng N., Williams R. W., Steven A. C., and Wickner R. B. (1999) Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283, 1339–1343 10.1126/science.283.5406.1339 [DOI] [PubMed] [Google Scholar]
- 20. Brachmann A., Baxa U., and Wickner R. B. (2005) Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24, 3082–3092 10.1038/sj.emboj.7600772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., and Philippe M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14, 4065–4072 10.1002/j.1460-2075.1995.tb00078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stansfield I., Jones K. M., Kushnirov V. V., Dagkesamanskaya A. R., Poznyakovski A. I., Paushkin S. V., Nierras C. R., Cox B. S., Ter-Avanesyan M. D., and Tuite M. F. (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14, 4365–4373 10.1002/j.1460-2075.1995.tb00111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paushkin S. V., Kushnirov V. V., Smirnov V. N., and Ter-Avanesyan M. D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 10.1002/j.1460-2075.1996.tb00675.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paushkin S. V., Kushnirov V. V., Smirnov V. N., and Ter-Avanesyan M. D. (1997) In vitro propagation of the prion-like state of yeast Sup35 protein. Science 277, 381–383 10.1126/science.277.5324.381 [DOI] [PubMed] [Google Scholar]
- 25. Patino M. M., Liu J.-J., Glover J. R., and Lindquist S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273, 622–626 10.1126/science.273.5275.622 [DOI] [PubMed] [Google Scholar]
- 26. King C.-Y., Tittmann P., Gross H., Gebert R., Aebi M., and Wüthrich K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. U.S.A. 94, 6618–6622 10.1073/pnas.94.13.6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glover J. R., Kowal A. S., Schirmer E. C., Patino M. M., Liu J.-J., and Lindquist S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 10.1016/S0092-8674(00)80264-0 [DOI] [PubMed] [Google Scholar]
- 28. King C.-Y., and Diaz-Avalos R. (2004) Protein-only transmission of three yeast prion strains. Nature 428, 319–323 10.1038/nature02391 [DOI] [PubMed] [Google Scholar]
- 29. Tanaka M., Chien P., Naber N., Cooke R., and Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 10.1038/nature02392 [DOI] [PubMed] [Google Scholar]
- 30. Coustou V., Deleu C., Saupe S., and Begueret J. (1997) The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. U.S.A. 94, 9773–9778 10.1073/pnas.94.18.9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., and Liebman S. W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sondheimer N., and Lindquist S. (2000) Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172 10.1016/S1097-2765(00)80412-8 [DOI] [PubMed] [Google Scholar]
- 33. Derkatch I. L., Bradley M. E., Hong J. Y., and Liebman S. W. (2001) Prions affect the appearance of other prions: the story of [PIN]. Cell 106, 171–182 10.1016/S0092-8674(01)00427-5 [DOI] [PubMed] [Google Scholar]
- 34. Bradley M. E., Edskes H. K., Hong J. Y., Wickner R. B., and Liebman S. W. (2002) Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. U.S.A. 99, Suppl. 4, 16392–16399 10.1073/pnas.152330699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du Z., Goncharoff D. K., Cheng X., and Li L. (2017) Analysis of [SWI+] formation and propagation events. Mol. Microbiol. 104, 105–124 10.1111/mmi.13616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liebman S. W., and Chernoff Y. O. (2012) Prions in yeast. Genetics 191, 1041–1072 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Westermark G. T., Fändrich M., Lundmark K., and Westermark P. (2018) Non-cerebral amyloidoses: aspects on seeding, cross-seeding and transmission. Cold Spring Harb. Perspect. Med. 8, a024323 10.1101/cshperspect.a024323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moreno-Gonzalez I., Edwards Iii G., Salvadores N., Shahnawaz M., Diaz-Espinoza R., and Soto C. (2017) Molecular interaction between type 2 diabetes and Alzheimer's disease through cross-seeing of protein misfolding. Mol. Psychiatry 22, 1327–1334 10.1038/mp.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collinge J., and Clarke A. R. (2007) A general model of prion strains and their pathogenicity. Science 318, 930–936 10.1126/science.1138718 [DOI] [PubMed] [Google Scholar]
- 40. Bessen R. A., and Marsh R. F. (1994) Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68, 7859–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bateman D. A., and Wickner R. B. (2013) The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet. 9, e1003257 10.1371/journal.pgen.1003257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanaka M., Chien P., Yonekura K., and Weissman J. S. (2005) Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121, 49–62 10.1016/j.cell.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 43. Ter-Avanesyan M. D., Dagkesamanskaya A. R., Kushnirov V. V., and Smirnov V. N. (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137, 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bradley M. E., and Liebman S. W. (2004) The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol. Microbiol. 51, 1649–1659 10.1111/j.1365-2958.2003.03955.x [DOI] [PubMed] [Google Scholar]
- 45. Chang H.-Y., Lin J.-Y., Lee H.-C., Wang H.-L., and King C.-Y. (2008) Strain-specific sequences required for yeast prion [PSI+] propagation. Proc. Natl. Acad. Sci. U.S.A. 105, 13345–13350 10.1073/pnas.0802215105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shewmaker F., Mull L., Nakayashiki T., Masison D. C., and Wickner R. B. (2007) Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176, 1557–1565 10.1534/genetics.107.074153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hosoda N., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., and Katada T. (2003) Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278, 38287–38291 10.1074/jbc.C300300200 [DOI] [PubMed] [Google Scholar]
- 48. Li X., Rayman J. B., Kandel E. R., and Derkatch I. L. (2014) Functional role of Tia1/Pub1 and Sup35 prion domains: directing protein synthesis machinery to the tubulin cytoskeleton. Mol. Cell 55, 305–318 10.1016/j.molcel.2014.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franzmann T. M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A. S., Nüske E., Richter D., Baumeister W., Grill S. W., Pappu R. V., Hyman A. A., and Alberti S. (2018) Phase separation of a yeast protein promotes cellular fitness. Science 359, eaao5654 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- 50. Ross E. D., Baxa U., and Wickner R. B. (2004) Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24, 7206–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ross E. D., Edskes H. K., Terry M. J., and Wickner R. B. (2005) Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. U.S.A. 102, 12825–12830 10.1073/pnas.0506136102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ross E. D., Minton A., and Wickner R. B. (2005) Prion domains: sequences, structures and interactions. Nat. Cell Biol. 7, 1039–1044 10.1038/ncb1105-1039 [DOI] [PubMed] [Google Scholar]
- 53. Toombs J. A., Liss N. M., Cobble K. R., Ben-Musa Z., and Ross E. D. (2011) [PSI+] maintenance is dependent on the composition, not the primary sequence, of the oligopeptide repeat domain. PLoS ONE 6, e21953 10.1371/journal.pone.0021953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toombs J. A., McCarty B. R., and Ross E. D. (2010) Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30, 319–332 10.1128/MCB.01140-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doel S. M., McCready S. J., Nierras C. R., and Cox B. S. (1994) The dominant PNM2− mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DePace A. H., Santoso A., Hillner P., and Weissman J. S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252 10.1016/S0092-8674(00)81467-1 [DOI] [PubMed] [Google Scholar]
- 57. King C. Y. (2001) Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol. 307, 1247–1260 10.1006/jmbi.2001.4542 [DOI] [PubMed] [Google Scholar]
- 58. Antzutkin O. N., Balbach J. J., Leapman R. D., Rizzo N. W., Reed J., and Tycko R. (2000) Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 97, 13045–13050 10.1073/pnas.230315097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shewmaker F., Wickner R. B., and Tycko R. (2006) Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. U.S.A. 103, 19754–19759 10.1073/pnas.0609638103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baxa U., Wickner R. B., Steven A. C., Anderson D. E., Marekov L. N., Yau W.-M., and Tycko R. (2007) Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 46, 13149–13162 10.1021/bi700826b [DOI] [PubMed] [Google Scholar]
- 61. Wickner R. B., Dyda F., and Tycko R. (2008) Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. U.S.A. 105, 2403–2408 10.1073/pnas.0712032105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shewmaker F., Kryndushkin D., Chen B., Tycko R., and Wickner R. B. (2009) Two prion variants of Sup35p have in-register β-sheet structures, independent of hydration. Biochemistry 48, 5074–5082 10.1021/bi900345q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gorkovskiy A., Thurber K. R., Tycko R., and Wickner R. B. (2014) Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc. Natl. Acad. Sci. U.S.A. 111, E4615–E4622 10.1073/pnas.1417974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Diaz-Avalos R., King C. Y., Wall J., Simon M., and Caspar D. L. (2005) Strain-specific morphologies of yeast prion amyloids. Proc. Natl. Acad. Sci. U.S.A. 102, 10165–10170 10.1073/pnas.0504599102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baxa U., Taylor K. L., Wall J. S., Simon M. N., Cheng N., Wickner R. B., and Steven A. (2003) Architecture of Ure2p prion filaments: the N-terminal domain forms a central core fiber. J. Biol. Chem. 278, 43717–43727 10.1074/jbc.M306004200 [DOI] [PubMed] [Google Scholar]
- 66. Chen B., Thurber K. R., Shewmaker F., Wickner R. B., and Tycko R. (2009) Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 14339–14344 10.1073/pnas.0907821106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shewmaker F., Ross E. D., Tycko R., and Wickner R. B. (2008) Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry 47, 4000–4007 10.1021/bi7024589 [DOI] [PubMed] [Google Scholar]
- 68. Wickner R. B., Shewmaker F., Kryndushkin D., and Edskes H. K. (2008) Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. Bioessays 30, 955–964 10.1002/bies.20821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wickner R. B., Edskes H. K., Bateman D. A., Kelly A. C., Gorkovskiy A., Dayani Y., and Zhou A. (2013) Amyloids and yeast prion biology. Biochemistry 52, 1514–1527 10.1021/bi301686a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Der-Sarkissian A., Jao C. C., Chen J., and Langen R. (2003) Structural organization of α-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 278, 37530–37535 10.1074/jbc.M305266200 [DOI] [PubMed] [Google Scholar]
- 71. Margittai M., and Langen R. (2004) Template-assisted filament growth by parallel stacking of tau. Proc. Natl. Acad. Sci. U.S.A. 101, 10278–10283 10.1073/pnas.0401911101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cobb N. J., Sönnichsen F. D., McHaourab H., and Surewicz W. K. (2007) Molecular architecture of human prion protein amyloid: a parallel, in-register β-structure. Proc. Natl. Acad. Sci. U.S.A. 104, 18946–18951 10.1073/pnas.0706522104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Luca S., Yau W.-M., Leapman R., and Tycko R. (2007) Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry 46, 13505–13522 10.1021/bi701427q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tycko R., Savtchenko R., Ostapchenko V. G., Makarava N., and Baskakov I. V. (2010) The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry 49, 9488–9497 10.1021/bi1013134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ladner C. L., Chen M., Smith D. P., Platt G. W., Radford S. E., and Langen R. (2010) Stacked sets of parallel, in-register β-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J. Biol. Chem. 285, 17137–17147 10.1074/jbc.M110.117234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Groveman B. R., Dolan M. A., Taubner L. M., Kraus A., Wickner R. B., and Caughey B. (2014) Parallel in-register intermolecular β sheet architecture for prion seeded PrP amyloids. J. Biol. Chem. 289, 24129–24142 10.1074/jbc.M114.578344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tuttle M. D., Comellas G., Nieuwkoop A. J., Covell D. J., Berthold D. A., Kloepper K. D., Courtney J. M., Kim J. K., Barclay A. M., Kendall A., Wan W., Stubbs G., Schwieters C. D., Lee V. M., George J. M., and Rienstra C. M. (2016) Solid-state NMR structure of a pathogenic fibril of a full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 10.1038/nsmb.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Blake C., and Serpell L. (1996) Synchrotron x-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous β-sheet helix. Structure 4, 989–998 10.1016/S0969-2126(96)00104-9 [DOI] [PubMed] [Google Scholar]
- 79. Serag A. A., Altenbach C., Gingery M., Hubbell W. L., and Yeates T. O. (2002) Arrangement of subunits and ordering of β-strands in an amyloid sheet. Nat. Struct. Biol. 9, 734–739 10.1038/nsb838 [DOI] [PubMed] [Google Scholar]
- 80. Bateman D. A., Tycko R., and Wickner R. B. (2011) Experimentally derived structural constraints for amyloid fibrils of wild-type transthyretin. Biophys. J. 101, 2485–2492 10.1016/j.bpj.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McGlinchey R. P., Kryndushkin D., and Wickner R. B. (2011) Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. U.S.A. 108, 5337–5341 10.1073/pnas.1102762108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakayashiki T., Kurtzman C. P., Edskes H. K., and Wickner R. B. (2005) Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. U.S.A. 102, 10575–10580 10.1073/pnas.0504882102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bateman D. A., and Wickner R. B. (2012) [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: intraspecies 'species barriers'. Genetics 190, 569–579 10.1534/genetics.111.136655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Halfmann R., Jarosz D. F., Jones S. K., Chang A., Lancaster A. K., and Lindquist S. (2012) Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368 10.1038/nature10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kelly A. C., Shewmaker F. P., Kryndushkin D., and Wickner R. B. (2012) Sex, prions and plasmids in yeast. Proc. Natl. Acad. Sci. U.S.A. 109, E2683–E2690 10.1073/pnas.1213449109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wickner R. B., Shewmaker F. P., Bateman D. A., Edskes H. K., Gorkovskiy A., Dayani Y., and Bezsonov E. E. (2015) Yeast prions: structure, biology and prion-handling systems. Microbiol. Mol. Biol. Rev. 79, 1–17 10.1128/MMBR.00041-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Masison D. C., and Reidy M. (2015) Yeast prions are useful for studying protein chaperones and protein quality control. Prion 9, 174–183 10.1080/19336896.2015.1027856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wickner R. B., Bezsonov E. E., Son M., Ducatez M., DeWilde M., and Edskes H. K. (2018) Anti-prion systems in yeast and inositol polyphosphates. Biochemistry 57, 1285–1292 10.1021/acs.biochem.7b01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pfund C., Lopez-Hoyo N., Ziegelhoffer T., Schilke B. A., Lopez-Buesa P., Walter W. A., Wiedmann M., and Craig E. A. (1998) The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17, 3981–3989 10.1093/emboj/17.14.3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chernoff Y. O., and Kiktev D. A. (2016) Dual role of ribosome-associated chaperones in prion formation and propagation. Curr. Genet. 62, 677–685 10.1007/s00294-016-0586-2 [DOI] [PubMed] [Google Scholar]
- 91. Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., and Deuerling E. (2010) A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189, 57–68 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chernoff Y. O., Newnam G. P., Kumar J., Allen K., and Zink A. D. (1999) Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI] prion. Mol. Cell. Biol. 19, 8103–8112 10.1128/MCB.19.12.8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Amor A. J., Castanzo D. T., Delany S. P., Selechnik D. M., van Ooy A., and Cameron D. M. (2015) The ribosome-associated complex antagonizes prion formation in yeast. Prion 9, 144–164 10.1080/19336896.2015.1022022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kiktev D. A., Melomed M. M., Lu C. D., Newnam G. P., and Chernoff Y. O. (2015) Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 96, 621–632 10.1111/mmi.12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kirkland P. A., Reidy M., and Masison D. C. (2011) Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 188, 565–577 10.1534/genetics.111.129460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Edskes H. K., Mukhamedova M., Edskes B. K., and Wickner R. B. (2018) Hermes transposon mutagenesis shows [URE3] prion pathology prevented by a ubiquitin-targeting protein: evidence for carbon/nitrogen assimilation cross-talk and a second function for Ure2p in Saccharomyces cerevisiae. Genetics 209, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Seol J. H., Shevchenko A., Shevchenko A., and Deshaies R. J. (2001) Skip1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3, 384–391 10.1038/35070067 [DOI] [PubMed] [Google Scholar]
- 98. Sarikas A., Hartmann T., and Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220 10.1186/gb-2011-12-4-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hua Z., and Vierstra R. D. (2011) The cullin-ring ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334 10.1146/annurev-arplant-042809-112256 [DOI] [PubMed] [Google Scholar]
- 100. Jonkers W., and Rep M. (2009) Lessons from fungal F-box proteins. Eukaryot. Cell 8, 677–695 10.1128/EC.00386-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Moriyama H., Edskes H. K., and Wickner R. B. (2000) [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20, 8916–8922 10.1128/MCB.20.23.8916-8922.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Roberts B. T., Moriyama H., and Wickner R. B. (2004) [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast 21, 107–117 10.1002/yea.1062 [DOI] [PubMed] [Google Scholar]
- 103. Kryndushkin D., and Wickner R. B. (2007) Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2149–2154 10.1091/mbc.e07-02-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kryndushkin D. S., Shewmaker F., and Wickner R. B. (2008) Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 27, 2725–2735 10.1038/emboj.2008.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kryndushkin D., Ihrke G., Piermartiri T. C., and Shewmaker F. (2012) A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol. Microbiol. 86, 1531–1547 10.1111/mmi.12075 [DOI] [PubMed] [Google Scholar]
- 106. Malinovska L., Kroschwald S., Munder M. C., Richter D., and Alberti S. (2012) Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 23, 3041–3056 10.1091/mbc.e12-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wickner R. B., Bezsonov E., and Bateman D. A. (2014) Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. U.S.A. 111, E2711–E2720 10.1073/pnas.1409582111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Cox B., Ness F., and Tuite M. (2003) Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165, 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Specht S., Miller S. B., Mogk A., and Bukau B. (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 195, 617–629 10.1083/jcb.201106037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Barbitoff Y. A., Matveenko A. G., Moskalenko S. E., Zemlyanko O. M., Newnam G. P., Patel A., Chernova T. A., Chernoff Y. O., and Zhouravleva G. A. (2017) To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 105, 242–257 10.1111/mmi.13697 [DOI] [PubMed] [Google Scholar]
- 111. Chattopadhyay S., and Pearce D. A. (2002) Interaction with Btn2p is required for localization of Rsg1p: Btn2p-mediated changes in arginine uptake in Saccharomyces cerevisiae. Eukaryot. Cell 1, 606–612 10.1128/EC.1.4.606-612.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kama R., Robinson M., and Gerst J. E. (2007) Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol. 27, 605–621 10.1128/MCB.00699-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kanneganti V., Kama R., and Gerst J. E. (2011) Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol. Biol. Cell 22, 1648–1663 10.1091/mbc.e10-11-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Glover J. R., and Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- 115. Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., and Liebman S. W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- 116. Wegrzyn R. D., Bapat K., Newnam G. P., Zink A. D., and Chernoff Y. O. (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21, 4656–4669 10.1128/MCB.21.14.4656-4669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Winkler J., Tyedmers J., Bukau B., and Mogk A. (2012) Chaperone networks in protein disaggregation and prion propagation. J. Struct. Biol. 179, 152–160 10.1016/j.jsb.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 118. Matveenko A. G., Barbitoff Y. A., Jay-Garcia L. M., Chernoff Y. O., and Zhouravleva G. A. (2018) Differential effects of chaperones on yeast prions: CURrent views. Curr. Genet. 64, 317–325 10.1007/s00294-017-0750-3 [DOI] [PubMed] [Google Scholar]
- 119. Ness F., Ferreira P., Cox B. S., and Tuite M. F. (2002) Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22, 5593–5605 10.1128/MCB.22.15.5593-5605.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mogk A., Kummer E., and Bukau B. (2015) Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Reidy M., Miot M., and Masison D. C. (2012) Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics 192, 185–193 10.1534/genetics.112.142307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Jung G., and Masison D. C. (2001) Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43, 7–10 10.1007/s002840010251 [DOI] [PubMed] [Google Scholar]
- 123. Ferreira P. C., Ness F., Edwards S. R., Cox B. S., and Tuite M. F. (2001) The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357–1369 10.1046/j.1365-2958.2001.02478.x [DOI] [PubMed] [Google Scholar]
- 124. Jung G., Jones G., and Masison D. C. (2002) Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. U.S.A. 99, 9936–9941 10.1073/pnas.152333299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Helsen C. W., and Glover J. R. (2012) Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J. Biol. Chem. 287, 542–556 10.1074/jbc.M111.302869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Winkler J., Tyedmers J., Bukau B., and Mogk A. (2012) Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 198, 387–404 10.1083/jcb.201201074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ness F., Cox B. S., Wonwigkarm J., Naeimi W. R., and Tuite M. F. (2017) Over-expression of the molecular chaperone Hsp104 in Saccharomyces cerevisiae results in the malpartition of [PSI+] propagons Mol. Microbiol. 104, 125–143 10.1111/mmi.13617 [DOI] [PubMed] [Google Scholar]
- 128. Greene L. E., Zhao X., and Eisenberg E. (2018) Curing of [PSI+] by Hsp104 overexpression: clues to solving the puzzle. Prion 12, 9–15 10.1080/19336896.2017.1412911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hung G. C., and Masison D. C. (2006) N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173, 611–620 10.1534/genetics.106.056820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gorkovskiy A., Reidy M., Masison D. C., and Wickner R. B. (2017) Hsp104 at normal levels cures many [PSI+] variants in a process promoted by Sti1p, Hsp90 and Sis1p. Proc. Natl. Acad. Sci. U.S.A. 114, E4193–E4202 10.1073/pnas.1704016114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wickner R. B., Kelly A. C., Bezsonov E. E., and Edskes H. K. (2017) Prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. U.S.A. 114, E8402–E8410 10.1073/pnas.1714361114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Steidle E. A., Chong L. S., Wu M., Crooke E., Fiedler D., Resnick A. C., and Rolfes R. J. (2016) A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 protein selectively cleaves the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J. Biol. Chem. 291, 6772–6783 10.1074/jbc.M116.714907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shears S. B. (2018) Intimate connections: inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell. Physiol. 233, 1897–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hatch A. J., Odom A. R., and York J. D. (2017) Inositol phosphate multikinase dependent transcriptional control. Adv. Biol. Regul. 64, 9–19 10.1016/j.jbior.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wu M., Chong L. S., Perlman D. H., Resnick A. C., and Fiedler D. (2016) Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 113, E6757–E6765 10.1073/pnas.1606853113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Fan Q., Park K.-W., Du Z., Morano K. A., and Li L. (2007) The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177, 1583–1593 10.1534/genetics.107.077982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Duennwald M. L., Echeverria A., and Shorter J. (2012) Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 10, e1001346 10.1371/journal.pbio.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Son M., and Wickner R. B. (2018) Nonsense-mediated mRNA decay factors cure most [PSI+] prion variants. Proc. Natl. Acad. Sci. U.S.A. 115, E1184–E1193 10.1073/pnas.1717495115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. He F., and Jacobson A. (2015) Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 49, 339–366 10.1146/annurev-genet-112414-054639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Weng Y., Czaplinski K., and Peltz S. W. (1996) Genetic and biochemical characterization of mutations in the ATPase and helicase region of the Upf1 protein. Mol. Cell. Biol. 16, 5477–5490 10.1128/MCB.16.10.5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. He F., Ganesan R., and Jacobson A. (2013) Intra- and intermolecular regulatory interactions in Upf1p, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol. Cell. Biol. 33, 4672–4684 10.1128/MCB.01136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Chacinska A., Szczesniak B., Kochneva-Pervukhova N. V., Kushnirov V. V., Ter-Avanesyan M. D., and Boguta M. (2001) Ssb1 chaperone is a [PSI+] prion-curing factor. Curr. Genet. 39, 62–67 10.1007/s002940000180 [DOI] [PubMed] [Google Scholar]