Abstract
Toxoplasma gondii is a ubiquitous, obligate intracellular eukaryotic parasite that causes congenital birth defects, disease in immunocompromised individuals, and blindness. Protein glycosylation plays an important role in the infectivity and evasion of immune responses of many eukaryotic parasites and is also of great relevance to vaccine design. Here we demonstrate that micronemal protein 2 (MIC2), a motility-associated adhesin of T. gondii, has highly glycosylated thrombospondin repeat (TSR) domains. Using affinity-purified MIC2 and MS/MS analysis along with enzymatic digestion assays, we observed that at least seven C-linked and three O-linked glycosylation sites exist within MIC2, with >95% occupancy at these O-glycosylation sites. We found that addition of O-glycans to MIC2 is mediated by a protein O-fucosyltransferase 2 homolog (TgPOFUT2) encoded by the TGGT1_273550 gene. Even though POFUT2 homologs are important for stabilizing motility-associated adhesins and for host infection in other apicomplexan parasites, loss of TgPOFUT2 in T. gondii had only a modest impact on MIC2 levels and the wider parasite proteome. Consistent with this, both plaque formation and tachyzoite invasion were broadly similar in the presence or absence of TgPOFUT2. These findings indicate that TgPOFUT2 O-glycosylates MIC2 and that this glycan, in contrast to previous findings in another study, is dispensable in T. gondii tachyzoites and for T. gondii infectivity.
Keywords: glycosylation, Toxoplasma gondii, fucosyltransferase, proteomics, MS
Introduction
The phylum Apicomplexa is comprised of a large group of obligate intracellular eukaryotic parasites, many of which have medical and agricultural significance. Pathogenic apicomplexan species include Plasmodium spp. (malaria), Cryptosporidium spp. (cryptosporidiosis), Theileria spp. (theileriosis), Babesia spp. (babesiosis), and the most ubiquitous of all, Toxoplasma gondii (toxoplasmosis). T. gondii infects 30–80% of the human population, and, although largely self-limiting in healthy individuals, it can cause major problems in the immunosuppressed and lead to congenital birth defects when contracted while pregnant (1). Some countries have extremely high rates of progressive blindness caused by toxoplasmic retinopathy, which has no curative treatment (1–3).
All apicomplexan parasites must migrate through host tissues and invade cells to survive and to cause disease; thus, this process is considered an excellent therapeutic target. Central to dissemination and invasion is a unique form of cellular locomotion termed “gliding motility.” Gliding motility requires the apical release of transmembrane adhesins from microneme organelles onto the parasite surface (4), which provides an anchor to the extracellular environment and/or host cells. The current model posits that motility is initiated when an actomyosin-based “glideosome,” which lies just underneath the plasma membrane, binds to the cytoplasmic tails of adhesins and drags them to the rear of the parasite, exerting a forward-acting force, thus driving forward motion (5–7).
Motility-associated adhesins in Apicomplexa vary between species and differ in expression across the various life cycle stages, reflecting the diversity of host cells that are targeted by this group of parasites. For example, Plasmodium spp. use a range of adhesins that specifically bind to erythrocyte receptors in asexual blood stages and other adhesins in stages that infect mosquitoes or the human liver (8, 9). Although little is known about the host cell receptors of T. gondii, a diverse array of putative adhesins is contained within their micronemes, which goes some way toward explaining the remarkably diverse range of host species and cell types that can be infected by this zoonotic parasite. Common features exist among these motility and invasion-associated adhesin proteins, including the recurrence of one or more thrombospondin repeat (TSR)5 domains (10).
Apicomplexan TSR-containing adhesin proteins are the only examples of TSRs found outside metazoa, but their function in mediating infection remains poorly understood. Metazoan TSRs are glycosylated in the endoplasmic reticulum with the O-linked glycan α-d-glucopyranosyl-1,3-β-l-fucopyranoside (GlcFuc) and C-linked α-d-mannopyranosides (C-Man) (11). Protein O-fucosyltransferase 2 (POFUT2) initiates metazoan TSR O-glycosylation and plays an important role in the folding and stabilization of these proteins (12–16). Recently, TSR O-glycosylation has also been observed in Plasmodium falciparum and Plasmodium vivax (17, 18); this O-glycan is initiated by a homolog of POFUT2 and is important for stabilizing proteins with TSR domains and efficient host infection (19). O-glycosylation of TSR domains may also be an important consideration in vaccine design (20).
Endogenous TSR O-glycosylation has yet to be observed in T. gondii; however, it does encode a putative POFUT2 (TGGT1_273550), and recombinant expression of T. gondii proteins in Chinese hamster ovary cells results in modification of parasite protein with the GlcFuc and C-Man glycans (21). Here we generate a T. gondii line to facilitate bulk purification of micronemal protein 2 (MIC2) directly from tachyzoites to map endogenous glycosylation sites on this motility-associated adhesin that possesses six TSR domains. We go on to demonstrate that a T. gondii POFUT2 is encoded by TGGT1_273550 and that loss of O-glycosylation has only a modest impact on MIC2 levels, the wider tachyzoite proteome, and infectivity.
Results
MIC2 is glycosylated at multiple sites to high occupancy
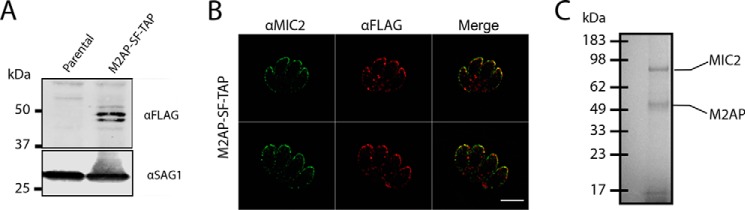
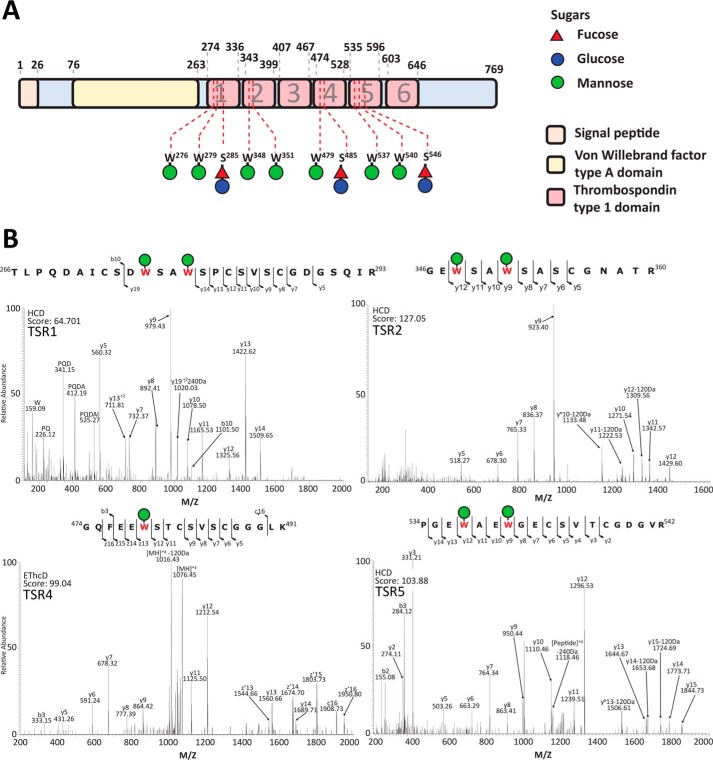
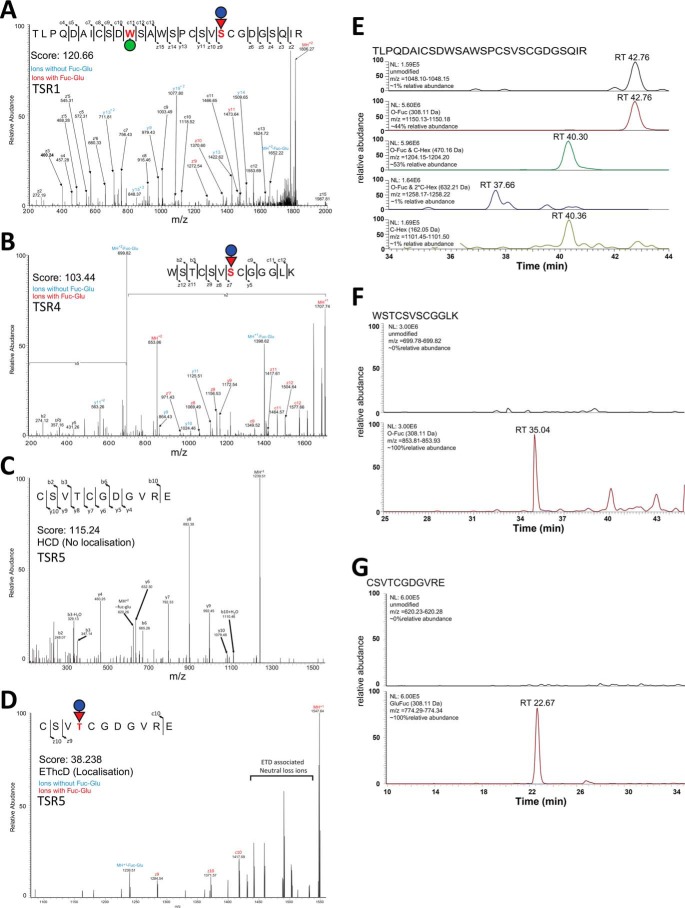
To explore the glycosylation of MIC2, we generated a T. gondii line in which the MIC2 associated protein (M2AP) possessed a C-terminal strep-FLAG tandem affinity purification (SF-TAP) tag to allow convenient purification of the M2AP–MIC2 complex. Western blotting using FLAG antibodies confirmed that M2AP was tagged (Fig. 1A). Further, we showed, using immunofluorescence, that M2AP–SF-TAP largely co-localizes with MIC2, as expected (Fig. 1B). The SF-TAP–tagged M2AP enabled simple high-yielding batch purification of the M2AP–MIC2 complex from bulk tachyzoite cultures (Fig. 1C). We then identified glycosylation events using MS/MS analysis in combination with multiple enzymatic digestions approaches. Enzymatic digestions comprised of trypsin, gluC, and sequential gluC and then trypsin, which was analyzed using higher-energy collision dissociation (HCD) and electron transfer and higher-energy collision dissociation (EThcD) fragmentation approaches (Ref. 22 and Table S1). Using these approaches, we detected multiple C-glycosylation and O-glycosylation events in five of the six TSR domains of MIC2 (Fig. 2, A and B, and Fig. S1). Within these TSR domains a total of five previously unreported C-glycosylation events were observed (Fig. 2B), corresponding to Trp276, Trp279, Trp348, Trp351, and Trp479, all lying within the C-mannosyltransferase consensus motifs (WXXW/C (23)). Because of the stable nature of C-glycosylation, these sites were readily assigned with rich fragmentation around Trp residues, enabling confident identification. Electron transfer–based fragmentation was used to localize the labile O-glycosylation events to three sites within MIC2: Ser285 (Fig. 3A), Ser485 (Fig. 3B), and Thr546 (Fig. 3, C and D). Using EThcD, fragmentation ions decorated with and without GlcFuc were observed (Fig. 3, A and D), both supporting the presence of the labile GlcFuc modification and the sites of modification. An additional O-glycosylation event was also observed within TSR2 on the semitryptic peptide 346GEWSAWSASCGNATR360, but the exact site of modification was unable to be precisely assigned (Table S1). We compared the abundance of unmodified and modified peptides covering these sites to better understand O-glycan occupancy (Table S2), and all of the O-glycosylated peptides were found to be the dominant observable species (Fig. 3, E–G). To summarize, MIC2 is endogenously modified with at least four O-glycans (GlcFuc) and seven C-glycans (C-Man) (Figs. 2 and 3) within TSR1, 2, 4, and 5, with the O-glycan occupancy greater than 95% for sites Ser285, Ser485, and Thr546.
Figure 1.
Establishment and validation of the M2AP SF-TAP T. gondii line. A, addition of a C-terminally appended SF-TAP tag enables detection of M2AP within the T. gondii line M2AP SF-TAP compared with the parental line. B, M2AP, detected using α-FLAG, co-localizes with MIC2 within intracellular tachyzoites. Scale bar corresponds to 5 μm. C, enrichment of M2AP SF-TAP tagged protein enables the isolation of the MIC2-M2AP complex to high purity, as determined by Coomassie-stained gels (image reproduced from Fig. 4C).
Figure 2.
Characterization of MIC2 glycosylation events. A, within MIC2, ten glycosylation sites were identified, corresponding to seven C-glycosylation events and three GlcFuc sites of modification. The residues modified and the corresponding carbohydrate modification are mapped to the MIC2 sequence. All observed glycosylation events lie within the TSR domains. B, peptides containing observed the C-glycosylation events Trp276, Trp279, Trp348, Trp351, Trp479, Trp537 and Trp540.
Figure 3.
Sites of O-fucosylations of MIC2 are modified at a high occupancy. A, analysis of tryptic digested MIC2 with EThcD fragmentation enabled identification of the C-glycosylated and GlcFuc within 266TLPQDAICSDWSAWSPCSVSCGDGSQIR293, with the site of GlcFuc localized to S285. B, analysis of trypsin- and GluC-digested MIC2 with EThcD fragmentation enabled identification of glycosylated 479WSTCSVSCGGGLK491, with the site of GlcFuc localized to Ser485. Analysis of GluC-digested MIC2 using a combination of HCD (C) and EThcD (D) fragmentation enabled localization of GlcFuc to Thr546 on the glycopeptide 543CSVTCGDGVRE553. The GlcFuc-containing glycoforms of these peptides were the most abundant forms observed (E–G), supporting the hypothesis that these sites are occupied at high occupancy.
TGGT1_273550 encodes the T. gondii POFUT2
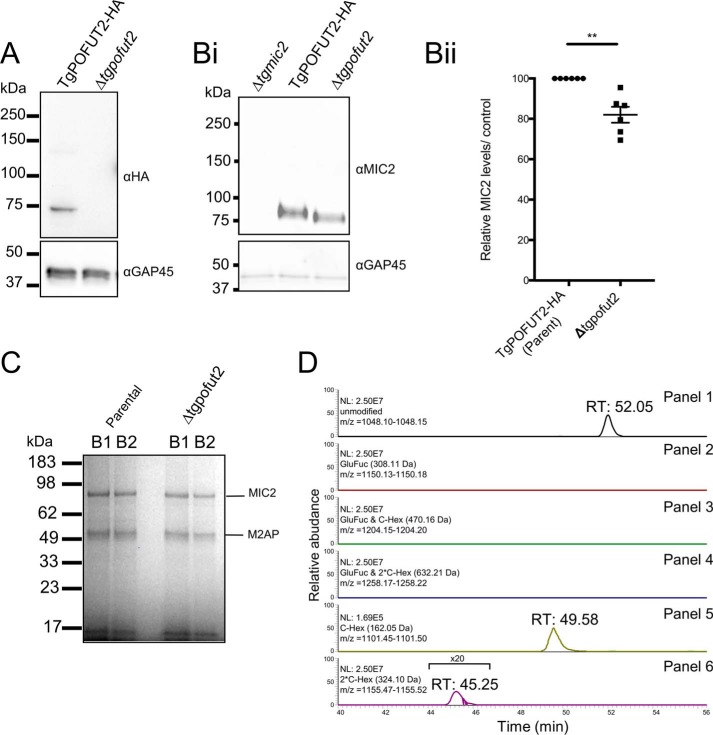
We were interested in determining the enzyme responsible for deposition of O-glycans on MIC2. A BLAST search of the T. gondii GT1 genome using Homo sapiens POFUT2 (CAC24557.1 (24)) as the query sequence suggested that the protein encoded by TGGT1_273550 was most likely the T. gondii POFUT2 homolog. To investigate the function of TgPOFUT2, we first introduced a C-terminal triple HA (HA3) epitope tag using single cross-over recombination (Fig. 4A). To ensure that HA tagging did not interfere with function, we also monitored MIC2 glycosylation in TgPOFUT2-HA and could show that this was indistinguishable from the parent M2AP-SF line (Fig. S3). This then allowed us to monitor disruption of TgPOFUT2 using CRISPR/Cas9-based gene knockout (Fig. 4A and Fig. S2, A and B), which was all performed in the M2AP–SF-TAP genetic background to provide a convenient means to purify and analyze MIC2 in the absence of TgPOFUT2 (Δtgpofut2). We first looked to see whether loss of TgPOFUT2 impacted MIC2 protein levels, as observed for thrombospondin repeat anonymous protein (TRAP) in P. falciparum (19). Using quantitative western blots, we found only a small but consistent reduction in abundance compared with parental lines (Fig. 4B, i and ii). Interestingly, we also observed reproducible faster migration of MIC2 by SDS-PAGE, perhaps reflecting the small mass change and/or an increase in polypeptide hydrophobicity that results from loss of multiple O-glycans (Fig. 4B, i). We then used the SF-TAP handle to purify the MIC2–M2AP complex from both parental and Δtgpofut2 lines as before (Fig. 4C) and subjected these samples to trypsin digestion followed by MS analysis. In doing so, we could observe no difference in the ability of M2AP to precipitate MIC2, as observed by peptide abundance between both components, suggesting no difference in interaction between these two proteins. Monitoring the MIC2 glycopeptide 266TLPQDAICSDWSAWSPCSVSCGDGSQIR293, which can be both C- and O-glycosylated, provided insights into glycosylation changes within the Δtgpofut2 line (Fig. 4D, panels 2–4). We could not observe any GlcFuc-containing forms of this peptide, but we were readily able to identify the unmodified form of this peptide (retention time, 53.06 min) and both the singly and doubly C-glycosylated forms of this peptide (retention time, 49.58 and 45.25 min, respectively) (Fig. 4D, panels 5 and 6, Tables S3 and S4). Collectively, these data suggests that the T. gondii POFUT2 is encoded by TGGT1_273550 and that TSR O-glycosylation plays only a minor role in regulating MIC2 abundance.
Figure 4.
TGGT1_273550 encodes T. gondii POFUT2. A, endogenously tagging TgPOFUT2 with HA enabled detection in tachyzoites. This band is absent in a Δtgpofut2 line. B, loss of POFUT2 leads to a change in the migration of MIC2 and a small, ∼20% decrease in MIC2. Data points refer to individual biological replicates, showing mean ± S.E. p values were calculated using a paired t test; **, p <0.01. C, Coomassie-stained gel of isolation of M2AP enables co-isolation of MIC2 in the Δtgpofut2 line at comparable levels, with B1 and B2 corresponding to two independent isolations from biological replicates. D, for the peptide 266TLPQDAICSDWSAWSPCSVSCGDGSQIR293, unmodified, single, and doubled C-glycosylated peptides from MIC2 were identified, but no GlcFuc-containing species were observed. Extracted ion chromatograms demonstrate that these identified forms are clearly detectible in panels 1, 5, and 6, whereas even at the MS1 level, no ions corresponding to the GlcFuc glycoforms were observed in panels 2–4.
Disruption of TgPOFUT2 results in modest alterations across the proteome
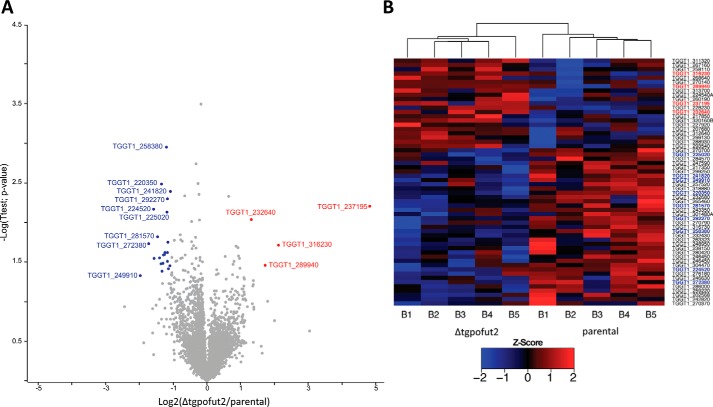
Because glycosylation systems can target multiple protein substrates, and because this modification is a known stabilizing factor, we hypothesized that the loss of TgPOFUT2 might disrupt a range of proteins beyond MIC2 (25, 26). To gain a better understanding of underlying changes that result from the loss of O-glycosylation in T. gondii, we conducted a global proteomics analysis comparing the parental strain with Δtgpofut2 using label-free quantitative (LFQ) proteomics (Table S5) (27). A total of 3839 unique T. gondii proteins were identified across biological replicates, with >3000 quantified proteins within each of the five parental and Δtgpofut2 replicates. We observed only modest changes in the proteome in response to the loss of TgPOFUT2 (Fig. 5A). These modest changes are reflected in the Pearson correlation (>0.95) observed between samples (Fig. S4). Using conventional thresholds (fold change more than ±1-fold and p >0.05), we observed a total of 26 proteins that decreased in abundance, whereas four increased in abundance within Δtgpofut2 compared with the parent (Fig. 5A). Even with less stringent thresholds (fold change more than ±1-fold and p >0.075), few additional alterations are observed across the proteome of the Δtgpofut2 strain (Fig. 5B, 37 decreasing and 21 increasing respectively). Although modest, these alterations are consistent across biological replicates (Fig. 5B) and suggest that loss of TSR O-glycosylation leads to small but real changes in the T. gondii proteome. Importantly, no TSR-containing proteins were observed to undergo significant changes in abundance in response to loss of TgPOFUT2.
Figure 5.
Quantitative proteomic analysis of Δtgpofut2 compared with the parental line. Label-free quantification of isolated tachyzoites was undertaken to compare Δtgpofut2 with the parental line. A, identified proteins are presented as a volcano plot depicting mean LFQ intensity ratios of Δtgpofut2 versus the parental line plotted against logarithmic t test p values from five biological experiments of each line. B, heat map of z-scored values of the proteins observed to change between the Δtgpofut2 and the parental line, demonstrating the consistency of these changes across experiments.
TSR O-glycosylation is not important for lytic stage growth, MIC2 trafficking, or host cell invasion
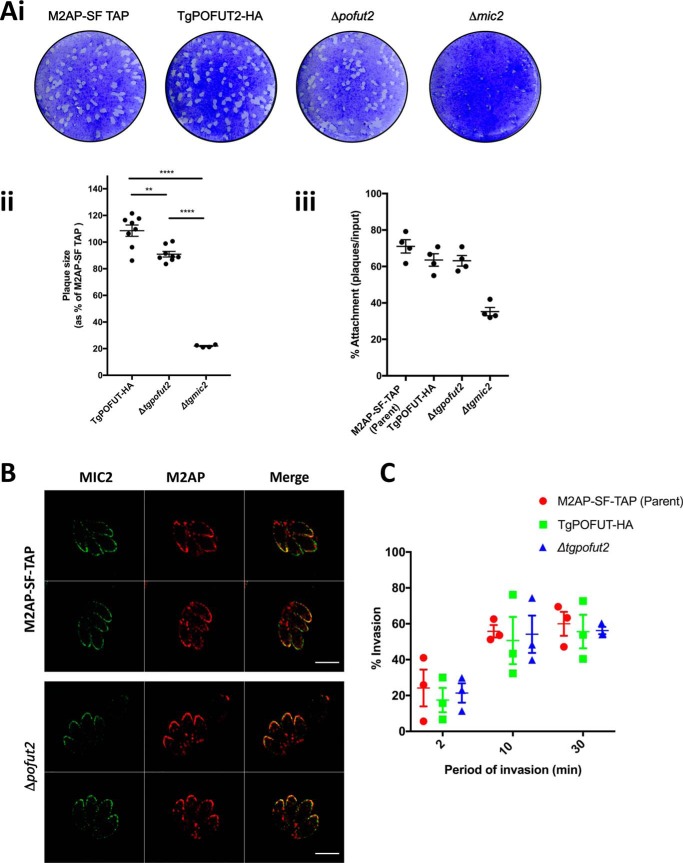
We furthered our phenotypic analysis to reveal whether TgPOFUT2 contributes to lytic stage growth in relation to what is known about MIC2 in T. gondii. To do this, we first performed plaque assays where parental and Δtgpofut2 parasites were left to grow on human foreskin fibroblast (HFF) monolayers over 7–8 days, and then zones of lysis (plaques) were quantitated by size and number. Here we confirmed, as described previously, that parasites lacking MIC2 cause a reduction in plaque size compared with the parental line (Fig. 6A) (28–30). Δtgpofut2 tachyzoites, on the other hand, had mildly smaller plaques than the parental TgPOFUT2-HA line, but not nearly as severe as complete loss of MIC2 (Fig. 6A, i and ii), which is consistent with the mild CRISPR fitness score (−0.34 log2). We could not detect any significant difference in host cell attachment of Δtgpofut2 compared with the dramatic defect upon complete loss of MIC2 (Fig. 6A, iii).
Figure 6.
Phenotypic analysis of Δtgpofut2 compared with the parental line. A, i, morphological assessment of plaque assays comparing the parental line (TgM2AP-SF TAP), TgPOFUT2-HA, Δtgpofut2 and Δmic2). ii, numerical assessment of plaque size compared with the TgM2AP-SF TAP parental line and Δmic2. iii, numerical assessment of plaque capacity, as a surrogate of host cell attachment, compared across all lines. B, IFA assessment of localization of MIC2, detected using αMIC2, and M2AP, detected using αFLAG, suggests that O-glycosylation plays no detectable role in protein trafficking. Scale bars correspond to 5 μm. C, invasion assays at 2, 10, and 30 min demonstrate no detectable difference in invasion capacity. Each data point represents the average value across a biological replicate and are collectively represented using mean ± S.E. p values were calculated using a one-way analysis of variance for A, ii and iii, and two-way analysis of variance for C; **, p <0.01; ****, p <0.0001.
Posttranslational modifications can affect protein trafficking, and we therefore assessed the localization of M2AP and MIC2 in Δtgpofut2 by IFA. Here we could observe no difference in localization of either MIC2 or M2AP, suggesting that O-glycosylation plays no detectable role in protein trafficking (Fig. 6B). We also specifically monitored for defects in host cell invasion after 2-, 10-, and 30-min incubation on HFFs (Fig. 6C). Again, we could see no difference in invasion capacity at any time point when using this assay, suggesting that TgPOFUT2 is not important for MIC2 function in vitro.
Discussion
The recent recognition of the importance of glycosylation at the host–pathogen interface in parasites such as P. falciparum (19) and Trypanosoma brucei (31) has demonstrated the need to better understand glycosylation within phylogenetically distinct eukaryotic parasites. Here we demonstrate that T. gondii, the causative agent of toxoplasmosis, has a functional POFUT2 homologue that is responsible for initiating O-glycosylation of TSR domains on MIC2. We confirm that at least three sites in MIC2 (Ser285, Ser485, and Thr546; Fig. 3) are modified by GlcFuc in addition to seven sites of C-glycosylation. These O-glycosylation sites lie within the previously proposed POFUT2 consensus sequon CXX(S/T)C (11, 24, 32) and suggest that TgPOFUT2 has a similar substrate preference as metazoan POFUT2 enzymes. Our data demonstrates that these sites are modified to a high occupancy, with few peptide species lacking the O-glycans observed in parental MIC2 preparations. The loss of O-glycans in the Δtgpofut2 line did not appear to impact C-glycosylation, as previously observed sites were readily detected within Δtgpofut2, suggesting that, in T. gondii MIC2, these two modifications operate independently of each other.
Previous work on mammalian POFUT2 has revealed that O-glycosylation is essential for mice with homozygous disruption of POFUT2 being embryonic lethal after implantation (15). In humans, loss of O-glycan elongation because of mutation in the β-1,3-glucosyltransferase (B3GLCT) (33, 34) leads to Peter plus syndrome (35). These defects are driven by the requirement for the complete GlcFuc disaccharide for endoplasmic reticulum quality control, where this glycan influences the folding and stability of proteins with TSR domains (36, 37). Similarly, we have observed that loss of O-fucosylation in P. falciparum sporozoites leads to destabilization of the key adhesin TRAP (19, 38). However, within T. gondii, loss of TgPOFUT2 appears not to be the case. We show that loss of TgPOFUT2 only leads to a small change in MIC2 levels and has no detectable difference in its ability to bind to its accessory protein M2AP or affect its protein trafficking. The small change in MIC2 levels is accompanied by a modest reduction (∼20%) in plaque size, which is consistent with the recent CRISPR-based fitness score assigned by Sidik et al. (39) to TgPOFUT2 (−0.34 log2), which, compared with loss of MIC2 (−1.17 log2), is very mild. However, we cannot discount that TgPOFUT2 may be more important for in vivo (mouse) infection models, where optimal tissue dissemination and invasion are critical for parasite survival. It may also be that TgPOFUT2 plays an important role in other stages of the parasite's lifecycle (e.g. bradyzoites and enteric feline stages), which was not assessed in this study. Indeed, the P. falciparum POFUT2 ortholog does not appear to have a detectable role in blood stages but does during transmissible stages (19).
Consistent with the nonessential nature of TgPOFUT2 in tachyzoites, we observed few changes across the proteome (Fig. 4). The changes we observed were modest in magnitude but consistent across replicates, suggesting that TgPOFUT2 targets a limited repertoire of substrates, in line with other POFUT2 enzymes (24, 32). Interestingly, the absence of any dramatic decreases in abundance suggests that few proteins undergo destabilization, as observed with TRAP in P. falciparum (19). A number of proteins were observed to decreased within Δtgpofut2, which may contribute to the observed phenotypes, including TGGT1_270700, now known as MYR2, which was recently shown to be essential for effector translocation into the host cell (40). We did not assess effector translocation in this study, and therefore this is worthy of more attention in the future. Additionally, we note that multiple members of the glycosylphosphatidylinositol-anchored SRS superfamily (41) are altered in response to Δtgpofut2, with an observed decrease in TGGT1_292270 (SRS36C) and an increase in TGGT1_292280 (SRS36D). As these surface antigens have been shown to modulate multiple aspects of infection (42), the function of these proteins may be worth following up. Surprisingly the most profound change we observed within the proteome of Δtgpofut2 was an increase in the hypothetical protein TGGT1_237195. This protein is not associated with T. gondii parasite fitness in vitro (39), and it has yet to be characterized how or why the loss of Δtgpofut2 results in an increase in abundance. It is important to note that none of these proteins contain the TgPOFUT2 CXX(S/T)C sequon, suggesting that they are not direct targets of TgPOFUT2 but may be influenced indirectly by the loss of O-fucosylation. We did not see any difference in MIC2 levels in our global proteome analysis as we saw with quantitate Western blotting, suggesting that this technique may not be sensitive enough to detect milder changes in protein levels.
The fact that TSR O-glycosylation does not appear to be important for the stability or trafficking of MIC2, in contrast to observations from TRAP in P. falciparum (19), may be due to the intimate association of MIC2 and M2AP. M2AP contributes to trafficking of this important adhesin to the micronemes and assists with its function (43, 44). Other coccidian parasites, including Neospora caninum and Eimeria tenella also express orthologs of T. gondii M2AP (45, 46). However, TRAP and related proteins in P. falciparum are not known to associate with an analogous protein, and therefore it is plausible that this makes O-glycosylation more important for protein folding and trafficking in these species. We have also shown here that MIC2 is heavily C-mannosylated, which alludes to the possibility that this modification could be important for the function of MIC2. This possibility is reflected by the fitness score assigned by Sidik et al. 39 to the putative C-mannosyltransferase in T. gondii (TGGT1_280400, −2.37 log2), which is suggestive of severe defects in tachyzoite function, although this remains to be further investigated.
During compilation of this manuscript, we were made aware of another body of work that also functionally characterizes POFUT2 in T. gondii. Bandini et al. (62) largely come to the same conclusion as reported here, including identification of similar glycosylation sites and demonstrating that TgPOFUT2 is responsible for their deposition. Further, Bandini et al. (62) also demonstrate that loss of TgPOFUT2 results in an apparent mild decrease in MIC2 levels (although they do not quantify this). Their study does diverge from this one in concluding that Δtgpofut2 has a defect in host cell attachment and invasion, which appears to be similar to complete loss of MIC2. From careful comparison of experimental protocols, the only difference we have identified is the method by which invasion assays were performed. We used [K+] shift to promote synchronous invasion whereas the Bandini et al. study (62) did not. [K+] shift strongly promotes cytosolic Ca2+ signaling and microneme secretion, resulting in synchronous motility and invasion (47, 48). It could be that these differences in conditions are enough to subtly effect invasion efficiency and thus explain the differences in the two studies. However, it is important to note that if, indeed, Δtgpofut2 has an invasion defect, then this would be the first mutant to do so without having an appreciable change in plaquing capacity. Regardless, dissecting out the reasons for the differences between the two studies might reveal novel and interesting biology regarding the role of fucosylation in MIC2 and invasion biology more generally.
In summary, TgPOFUT2 is responsible for O-glycosylation of TSR domains in MIC2. Loss of this modification leads to only small changes in MIC2 abundance, in contrast to the fate of TRAP in P. falciparum (19) and little to no impact on parasite invasiveness in vitro. Loss of TgPOFUT2 also provides no profound changes in the tachyzoite proteome. Taken together, this demonstrates that TgPOFUT2 is dispensable for replication of T. gondii tachyzoites.
Materials and methods
Plasmid construction and transfection
The 3′ portion of the tgm2ap (TGGT1_214940) ORF was PCR-amplified using the primers 5′-TACTTCCAATCCAATTTAATGCTGCTTGAGCCGTGACAACAGATTAC-3′ and 5′-TCCTCCACTTCCAATTTTAGCCGCCTCATCGTCACTCGGCAGACGGC-3′ and LIC cloned into vector pSF-TAP.LIC.DHFR-TS as described previously (49). The pM2AP.SF-TAP construct was linearized within the tgm2ap homology region with PflMI prior to transfection into RHΔku80:HXGPRT tachyzoites (49). The 3′ portion of the tgpofut2 (TGGT1_273550) ORF was PCR-amplified using the following primers: 5′-TAGTAGATCTAGCGATTAGCACTTTTTGG-3′ and 5′-AGCCCTAGGCAGTGTCGAACTGGGGTC-3′ and ligated into the BglII/AvrII sites of pgCH (Gra 1 5′-CAT-BglII/AvrII 3xHA). The pgCH-POFUT2–3xHA construct was linearized within the tgpofut2 homology region with MfeI prior to transfection into RHΔku80:HXG tachyzoites (49). This construct was linearized with Mfe1 (New England Biolabs) prior to transfection.
Tgpofut2 was knocked out using a unique guide selected by EuPaGDT (http://grna.ctegd.uga.edu/batch_tagging.html;6 61) combined with a homologous repair template containing the BLE cassette. The CRISPR target plasmid (50) was constructed by Q5 mutagenesis (New England Biolabs) with the common reverse primer 5′-AACTTGACATCCCCATTTAC-3′ (51) and the forward primer 5′-GAGACGGTAAGAACTGAGACGGTTTTAGAGCTAGAAATAGCAAG-3′. The BLE cassette was then amplified using Primestar Max (Takara) containing homologous flanking regions on either side of the Cas9 cut site with primers 5′-TGTCTGCTCAACCACCGTCGCTTGCTGTTAGGCCTCGTCGTCTAGAAGGTGGATGCGGGA-3′ and 5′-TGAATGGGAGACACGAGAGGAAGACGGTAAGAACTGAGCGATGTGGAGTCGTCTCAAGCG-3′. 10 μg of the Cas9 plasmid was combined with 20 μg of PCR product and then precipitated using ethanol/sodium acetate prior to transfection. The dried DNA was resuspended in 3 μl of elution buffer (Qiagen), followed by 20 μl pf P3 solution (Lonza). A washed parasite pellet containing ∼106 tachyzoites was then resuspended in this solution and transfected using the code FI-115 in a 16-well nucleocuvette strip in an Amaxa 4D nucleofector (Lonza). Parasites containing the knockout construct were then selected as normal following addition of phleomycin and subsequently subcloned until a stable population was obtained. All transfections proceeded using either a Gene Pulser II (Bio-Rad) or an Amaxa 4D nucleofector (Lonza). Gene Pulser II transfection took place at 1.5 kV and 25 μF, as is standard with 15 μg of purified linearized DNA when seeking homologous integration.
Parasite culture
Transfection and in vitro culture T. gondii tachyzoites were cultured under standard conditions. Briefly, HFFs (ATCC SCRC-1041) were grown in Dulbecco's Modified Eagle's Medium (DME) supplemented with 10% heat-inactivated cosmic calf serum (Hyclone) until confluency was reached. Upon T. gondii infection, HFFs with were refreshed with DME supplemented with 1% fetal calf serum. All cells were grown in humidified incubators at 37 °C/10% CO2.
Purification of the M2AP–MIC2 complex
Parasites obtained from four T150 flasks confluent with HFFs were purified by filtration and collected by centrifugation (1000 × g, 10 min). The parasite pellet was washed once on ice with PBS (2 ml). The parasite pellet was resuspended in 1 ml of lysis buffer (50 mm Tris (pH 8), 150 mm NaCl, 1% Triton X-100, Roche Mini Protease Inhibitor Mixture, DNase) and lysed at room temperature for 20 min. The lysate was centrifuged (10,000 × g, 10 min, 4 °C) to remove cellular debris, and the supernatant was incubated with Streptactin II resin (200 μl of 50% slurry) at 4 °C for 1–2 h with nutation. The resin was collected in microspin columns (2000 × g, 2 min, 4 °C) and washed three times with 50 mm Tris (pH 8), 150 mm NaCl, 0.1% Triton X-100. The resin was then incubated with 2.5 mm desthiobiotin, 50 mm Tris (pH 8), 150 mm NaCl (200 μl per column, 20 min, 4 °C) and the eluate collected by centrifugation (2000 × g, 2 min, 4 °C) to provide the purified MIC2–M2AP complex.
Tryptic digest of gel-separated proteins
Affinity-purified MIC2–M2AP was separated using SDS-PAGE, fixed, and visualized with Coomassie G-250 according to the protocol of Kang et al. (52). Bands of interest were excised and destained in a 50:50 solution of 50 mm NH4HCO3/100% ethanol for 20 min at room temperature with shaking at 750 rpm. Destained samples were then washed with 100% ethanol, vacuum-dried for 20 min, and rehydrated in 50 mm NH4HCO3 plus 10 mm DTT. Reduction was carried out for 60 min at 56 °C with shaking. The reducing buffer was then removed, and the gel bands were washed twice in 100% ethanol for 10 min to remove residual DTT. Reduced ethanol-washed samples were sequentially alkylated with 55 mm iodoacetamide in 50 mm NH4HCO3 in the dark for 45 min at room temperature. Alkylated samples were then washed with two rounds of 100% ethanol and vacuum-dried. Alkylated samples were then rehydrated with 12 ng/μl trypsin (Promega) in 40 mm NH4HCO3 at 4 °C for 1 h. Excess trypsin was removed, gel pieces were covered in 40 mm NH4HCO3 and incubated overnight at 37 °C. Peptides were concentrated and desalted using C18 stage tips (53) before analysis by LC-MS.
In-solution Glu-C/trypsin digestion and double digestion
Affinity-purified MIC2–M2AP was resuspend in 50 μl of 20% trifluoroethanol and a diluted equal volume of reduction/alkylation buffer (40 mm tris(2-carboxyethyl)phosphine, 80 mm chloroacetamide, and 100 mm NH4HCO3). Samples were then heated at 40 °C for 30 min to aid denaturation and reduction/alkylation in the dark. Glu-C or trypsin was added (1/50 w/w) and allowed to incubate overnight at 37 °C. For double digestion after the initial Glu-C digestion, trypsin (1/50 w/w) was added and allowed to incubate overnight at 37 °C. Digested samples were acidified to a final concentration of 0.5% formic acid and desalted using C18 stage tips (53) before analysis by LC-MS.
Characterization of MIC2 using reverse-phase LC-MS
Purified peptides were resuspend in buffer A and separated using a two-column chromatography set up composed of a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μl/min for 5 min and infused into an Orbitrap FusionTM LumosTM TribridTM mass spectrometer (Thermo Fisher Scientific) or OrbitrapTM Q-exactiveTM HF (Thermo Fisher Scientific) at 300 nl/min via the analytical column using a Dionex Ultimate 3000 Ultra-Performance Liquid Chromatography (UPLC) instrument (Thermo Fisher Scientific). 75-min gradients were run, altering the buffer composition from 1% buffer B to 28% B over 45 min, then from 28% buffer B to 40% buffer B over 10 min, and then from 40% buffer B to 100% buffer B over 2 min. The composition was held at 100% buffer B for 3 min and then dropped to 3% buffer B over 5 min and held at 3% buffer B for another 10 min. The LumosTM mass spectrometer was operated in a data-dependent mode automatically switching between acquisition of a single Orbitrap MS scan (120,000 resolution) every 3 s and MS-MS scan. For each ion selected for fragmentation, Orbitrap HCD (maximum fill time, 100 ms; automatic gain control (AGC) 2 × 105 with a resolution of 30,000), Orbitrap EThcD (maximum fill time, 100 ms; AGC 5 × 104 with a resolution of 30,000), and ion trap collision-induced dissociation (for each selected precursor; maximum fill time, 100 ms; AGC 2 × 104) was performed. The Q-exactiveTM HF mass spectrometer was operated in a data-dependent mode automatically switching between acquisition of a single Orbitrap MS scan (120,000 resolution) and 20 MS-MS scans (Orbitrap HCD; maximum fill time, 100 ms; AGC 2 × 105).
Digestion of complex protein lysates for quantitative proteome
Parasites obtained from four T150 flasks confluent with HFFs were purified by filtration and collected by centrifugation (1000 × g, 10 min). The parasite pellet was washed three times on ice with ice-cold PBS (2 ml). Parasites were lysed in ice-cold guanidine hydrochloride lysis buffer (6 m GdnHCl, 100 mm Tris (pH 8.5), 10 mm tris(2-carboxyethyl)phosphine, and 40 mm 2-chloroacetamide) and boiled at 95 °C for 10 min with shaking at 2000 rpm to shear DNA and inactivate protease activity, according to the protocol of Humphrey et al. (54). Lysates were then cooled for 10 min on ice and then boiled again at 95 °C for 10 min with shaking at 2000 rpm. Lysates were cooled, and protein concentration was determined using a BCA assay. 100 μg of protein from each sample was acetone-precipitated by mixing 4 volumes of ice-cold acetone with 1 volume of sample. Samples were precipitated overnight at −20 °C and then spun down at 4000 × g for 10 min at 4 °C. The precipitated protein pellets were resuspended with 80% ice-cold acetone and precipitated for an additional 4 h at −20 °C. Samples were spun down at 17,000 × g for 10 min at 4 °C to collect precipitated protein, the supernatant was discarded, and excess acetone was driven off at 65 °C for 5 min. Dried protein pellets were resuspended in 6 m urea, 2 m thiourea, 40 mm NH4HCO3, and reduced/alkylated prior to digestion with Lys-C (1/200 w/w) and then trypsin (1/50 w/w) overnight, as described previously (55). Digested samples were acidified to a final concentration of 0.5% formic acid and desalted using C18 stage tips (53) before analysis by LC-MS.
Quantitative proteome of Δtgpofut2 and parental lines using reverse-phase LC-MS
Purified peptides were resuspend in buffer A and separated using a two-column chromatography setup composed of a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μl/min for 5 min and infused into an Orbitrap FusionTM LumosTM TribridTM mass spectrometer (Thermo Fisher Scientific) at 300 nl/min via the analytical column using a Dionex Ultimate 3000 UPLC (Thermo Fisher Scientific). 180-min gradients were run, altering the buffer composition from 1% buffer B to 28% buffer B over 150 min, then from 28% buffer B to 40% buffer B over 10 min, and then from 40% buffer B to 100% buffer B over 2 min. The composition was held at 100% buffer B for 3 min and then dropped to 3% buffer B over 5 min and held at 3% buffer B for another 10 min. The LumosTM mass spectrometer was operated in a data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan (120,000 resolution) every 3 s and MS-MS scans (Orbitrap HCD; maximum fill time, 60 ms; AGC 2 × 105 with a resolution of 15,000).
Mass spectrometry data analysis
Identification of modification events within MIC2 and LFQ analysis was accomplished using MaxQuant (v1.5.3.1) (56). For characterization of MIC2, searches were performed against the T. gondii (strain ATCC 50853/GT1) proteome (Uniprot proteome ID UP000005641, downloaded February 4, 2017; 8,450 entries). For LFQ analysis, searches were performed against the T. gondii (strain ATCC 50853/GT1) proteome as well as the human (Uniprot proteome ID UP000005640; H. sapiens, downloaded October 24, 2013; 84,843 entries). For MIC2 searches, carbamidomethylation of cysteine was set as a fixed modification and the variable modifications of oxidation of methionine, C-glycosylation (+162.05 Da to W, allowing the loss of 120 Da because of the characteristic cross-ring fragmentation of the C-glycoside), and O-fucosylation (+308.11 Da to Ser or Thr, allowing a neutral loss of 308.11). For LFQ searches, carbamidomethylation of cysteine was set as a fixed modification and the variable modifications of oxidation of methionine and acetylation of protein N termini. Searches were performed with semitrypsin cleavage specificity for MIC2 analysis and trypsin cleavage specificity, allowing two miscleavage events for LFQ experiments with a maximum false discovery rate of 1.0% set for protein and peptide identifications. To enhance the identification of peptides between samples, the Match Between Runs option was enabled, with a precursor match window set to 2 min and an alignment window of 10 min. For label-free quantitation, the MaxLFQ option within Maxquant (27) was enabled in addition to the requantification module. The resulting protein group output was processed within the Perseus (v1.4.0.6) (57) analysis environment to remove prior reverse matches and common protein contaminates. For LFQ comparisons, missing values were imputed using Perseus and Pearson correlations visualized using R. All MS proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE (58) partner repository with the dataset identifiers PXD010714 and PXD011440.
Quantitative Western blotting
Parasite samples were lysed for 30 min at 4 °C in 1% (v/v) Triton X-100 and 1 mm MgCl2 in PBS (Gibco), supplemented with final 1× cOmplete protease inhibitors (Sigma) and 0.2% (v/v) benzonase (Merck). Cleared samples were then mixed with an equivalent volume of 2× sample buffer and run on gradient gels. Proteins were then transferred to nitrocellulose and blocked with 5% (w/v) skim milk. Primary and secondary antibodies were diluted in milk/PBS solution. Membranes were probed with mouse anti-MIC2 6D10 and rabbit anti-GAP45 (52) and immunodecorated with LI-COR IRDye 800CW– and IRDye 680RD–conjugated secondary antibodies. Quantitation of the signal proceeded using ImageJ and normalizing the MIC2 signal against GAP45 as a loading control.
Immunofluorescence assay
IFA analysis was performed as described here and elsewhere (59). Briefly, tachyzoite-infected HFFs were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100 (v/v), and blocked in 3% BSA/PBS (w/v). Primary and secondary antibodies were added sequentially, washing with PBS in between. Secondary antibodies were conjugated with Alexa Fluor 488 and 594 (Invitrogen). Samples were then imaged on an AP DeltaVision Elite microscope equipped with a CoolSnap2 charge-coupled device (CCD) detector and captured with SoftWorx software (GE Healthcare). Samples were then processed in ImageJ and assembled in Adobe Photoshop.
Invasion assay
The invasion assay proceeded as described previously (60). Briefly, parasites were resuspended in a high [K+] buffer (142 mm) to suppress motility, added to host cells, and allowed to settle. The buffer was then exchanged with DME supplemented with 1% fetal bovine serum, and parasites were allowed to invade for 2, 10, and 30 min at 37 °C. Then they were chemically fixed in 2.5% formaldehyde and 0.02% glutaraldehyde for 10 min each. The wells were then washed three times with PBS (pH 7.4) and were blocked with 3% BSA in PBS (pH 7.4) overnight at 4 °C. IFA proceeded as standard using antibodies against SAG1 to detect extracellular parasites and then permeabilization with 0.1% Triton X-100. GAP45 antibodies were then used to mark all parasites. The invasion rate was determined by counting the number of SAG1+ parasites compared with total (GAP45+).
Plaque assay
Plaque assays were carried out by inoculating 120 parasites into each well of a 6-well plate in D1 medium and allowed to grow undisturbed for 7–8 days. Cultures were fixed in 4% paraformaldehyde and incubated for 1 h at room temperature. Host cells were then stained using 2% crystal violet and washed.
Author contributions
S. K., M. J. C., A. J., A. D. U., R. J. S., and N. E. S. investigation; S. K., M. J. C., A. J., A. D. U., R. J. S., and N. E. S. methodology; S. K., M. J. C., A. J., A. D. U., M.-H. H., R. J. S., V. C., C. J. T., E. D. G.-B., and N. E. S. writing-review and editing; V. C. and N. E. S. resources; C. J. T. and E. D. G.-B. conceptualization; C. J. T. and E. D. G.-B. supervision; C. J. T., E. D. G.-B., and N. E. S. funding acquisition; E. D. G.-B. project administration; N. E. S. visualization; N. E. S. writing-original draft.
Supplementary Material
Acknowledgments
We thank L. David Sibley for the kind gift of the pCas9-GFP plasmid and anti-MIC2 mAb 6D10 as well as Giel Van Dooren for the kind gift of the pgCH plasmid. We would also like to thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for support, maintenance, and access to MS infrastructure. We are also grateful for institutional support from the Victorian State Government Operational Infrastructure Support and the Australian Government NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS).
Note added in proof
Fig. 1 contained an inadvertently duplicated image in the version of this article published as a Paper in Press on December 4, 2018. This error has now been corrected and does not affect the results or conclusions of this work.
This work was supported by National Health and Medical Research Council of Australia (NHMRC) Project Grant APP1100164 (to N. E. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1–S5.
The MS proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD010714 and PXD011440.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- TSR
- thrombospondin repeat
- GlcFuc
- β-d-glucopyranosyl-1,3-α-l-fucopyranoside
- C-Man
- C-linked α-d-mannopyranoside
- SF-TAP
- strep-FLAG tandem affinity purification
- HCD
- higher-energy collision dissociation
- EThcD
- electron transfer and higher-energy collision dissociation
- HA
- hemagglutinin
- LFQ
- label-free quantitative
- HFF
- human foreskin fibroblast
- TRAP
- thrombospondin repeat anonymous protein
- IFA
- immunofluorescence assay
- LIC
- ligation-independent cloning
- DME
- Dulbecco's Modified Eagle's Medium
- AGC
- automatic gain control.
References
- 1. Furtado J. M., Smith J. R., Belfort R. Jr., Gattey D., and Winthrop K. L. (2011) Toxoplasmosis: a global threat. J. Glob. Infect. Dis. 3, 281–284 10.4103/0974-777X.83536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Montoya J. G., and Liesenfeld O. (2004) Toxoplasmosis. Lancet 363, 1965–1976 10.1016/S0140-6736(04)16412-X [DOI] [PubMed] [Google Scholar]
- 3. Jones J., Lopez A., and Wilson M. (2003) Congenital toxoplasmosis. Am. Fam. Physician 67, 2131–2138 [PubMed] [Google Scholar]
- 4. Frénal K., Dubremetz J. F., Lebrun M., and Soldati-Favre D. (2017) Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 15, 645–660 10.1038/nrmicro.2017.86 [DOI] [PubMed] [Google Scholar]
- 5. Carruthers V. B., Giddings O. K., and Sibley L. D. (1999) Secretion of micronemal proteins is associated with toxoplasma invasion of host cells. Cell. Microbiol. 1, 225–235 10.1046/j.1462-5822.1999.00023.x [DOI] [PubMed] [Google Scholar]
- 6. Boucher L. E., and Bosch J. (2015) The apicomplexan glideosome and adhesins: structures and function. J. Struct. Biol. 190, 93–114 10.1016/j.jsb.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tardieux I., and Baum J. (2016) Reassessing the mechanics of parasite motility and host-cell invasion. J. Cell Biol. 214, 507–515 10.1083/jcb.201605100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horuk R., Chitnis C. E., Darbonne W. C., Colby T. J., Rybicki A., Hadley T. J., and Miller L. H. (1993) A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science 261, 1182–1184 10.1126/science.7689250 [DOI] [PubMed] [Google Scholar]
- 9. Cowman A. F., Tonkin C. J., Tham W. H., and Duraisingh M. T. (2017) The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 22, 232–245 10.1016/j.chom.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huynh M. H., and Carruthers V. B. (2006) Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog. 2, e84 10.1371/journal.ppat.0020084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofsteenge J., Huwiler K. G., Macek B., Hess D., Lawler J., Mosher D. F., and Peter-Katalinic J. (2001) C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 276, 6485–6498 10.1074/jbc.M008073200 [DOI] [PubMed] [Google Scholar]
- 12. Ricketts L. M., Dlugosz M., Luther K. B., Haltiwanger R. S., and Majerus E. M. (2007) O-fucosylation is required for ADAMTS13 secretion. J. Biol. Chem. 282, 17014–17023 10.1074/jbc.M700317200 [DOI] [PubMed] [Google Scholar]
- 13. Wang L. W., Dlugosz M., Somerville R. P., Raed M., Haltiwanger R. S., and Apte S. S. (2007) O-fucosylation of thrombospondin type 1 repeats in ADAMTS-like-1/punctin-1 regulates secretion: implications for the ADAMTS superfamily. J. Biol. Chem. 282, 17024–17031 10.1074/jbc.M701065200 [DOI] [PubMed] [Google Scholar]
- 14. Benz B. A., Nandadasa S., Takeuchi M., Grady R. C., Takeuchi H., LoPilato R. K., Kakuda S., Somerville R. P. T., Apte S. S., Haltiwanger R. S., and Holdener B. C. (2016) Genetic and biochemical evidence that gastrulation defects in Pofut2 mutants result from defects in ADAMTS9 secretion. Dev. Biol. 416, 111–122 10.1016/j.ydbio.2016.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du J., Takeuchi H., Leonhard-Melief C., Shroyer K. R., Dlugosz M., Haltiwanger R. S., and Holdener B. C. (2010) O-fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev. Biol. 346, 25–38 10.1016/j.ydbio.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niwa Y., Suzuki T., Dohmae N., and Simizu S. (2015) O-Fucosylation of CCN1 is required for its secretion. FEBS Lett. 589, 3287–3293 10.1016/j.febslet.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 17. Swearingen K. E., Lindner S. E., Flannery E. L., Vaughan A. M., Morrison R. D., Patrapuvich R., Koepfli C., Muller I., Jex A., Moritz R. L., Kappe S. H. I., Sattabongkot J., and Mikolajczak S. A. (2017) Proteogenomic analysis of the total and surface-exposed proteomes of Plasmodium vivax salivary gland sporozoites. PLoS Negl. Trop. Dis. 11, e0005791 10.1371/journal.pntd.0005791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swearingen K. E., Lindner S. E., Shi L., Shears M. J., Harupa A., Hopp C. S., Vaughan A. M., Springer T. A., Moritz R. L., Kappe S. H., and Sinnis P. (2016) Interrogating the Plasmodium sporozoite surface: identification of surface-exposed proteins and demonstration of glycosylation on CSP and TRAP by mass spectrometry-based proteomics. PLoS Pathog. 12, e1005606 10.1371/journal.ppat.1005606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopaticki S., Yang A. S. P., John A., Scott N. E., Lingford J. P., O'Neill M. T., Erickson S. M., McKenzie N. C., Jennison C., Whitehead L. W., Douglas D. N., Kneteman N. M., Goddard-Borger E. D., and Boddey J. A. (2017) Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat. Commun. 8, 561 10.1038/s41467-017-00571-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goddard-Borger E. D., and Boddey J. A. (2018) Implications of Plasmodium glycosylation on vaccine efficacy and design. Future Microbiol. 13, 609–612 10.2217/fmb-2017-0284 [DOI] [PubMed] [Google Scholar]
- 21. Hoppe C. M., Albuquerque-Wendt A., Bandini G., Leon D. R., Shcherbakova A., Buettner F. F. R., Izquierdo L., Costello C. E., Bakker H., and Routier F. H. (2018) Apicomplexan C-mannosyltransferases modify thrombospondin type I-containing adhesins of the TRAP family. Glycobiology 28, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frese C. K., Altelaar A. F., van den Toorn H., Nolting D., Griep-Raming J., Heck A. J., and Mohammed S. (2012) Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84, 9668–9673 10.1021/ac3025366 [DOI] [PubMed] [Google Scholar]
- 23. Hartmann S., and Hofsteenge J. (2000) Properdin, the positive regulator of complement, is highly C-mannosylated. J. Biol. Chem. 275, 28569–28574 10.1074/jbc.M001732200 [DOI] [PubMed] [Google Scholar]
- 24. Chen C. I., Keusch J. J., Klein D., Hess D., Hofsteenge J., and Gut H. (2012) Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 31, 3183–3197 10.1038/emboj.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nothaft H., and Szymanski C. M. (2013) Bacterial protein N-glycosylation: new perspectives and applications. J. Biol. Chem. 288, 6912–6920 10.1074/jbc.R112.417857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X., and Qian K. (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 10.1038/nrm.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andenmatten N., Egarter S., Jackson A. J., Jullien N., Herman J. P., and Meissner M. (2013) Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat. Methods 10, 125–127 10.1038/nmeth.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitelaw J. A., Latorre-Barragan F., Gras S., Pall G. S., Leung J. M., Heaslip A., Egarter S., Andenmatten N., Nelson S. R., Warshaw D. M., Ward G. E., and Meissner M. (2017) Surface attachment, promoted by the actomyosin system of Toxoplasma gondii is important for efficient gliding motility and invasion. BMC Biol. 15, 1 10.1186/s12915-016-0343-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gras S., Jackson A., Woods S., Pall G., Whitelaw J., Leung J. M., Ward G. E., Roberts C. W., and Meissner M. (2017) Parasites lacking the micronemal protein MIC2 are deficient in surface attachment and host cell egress, but remain virulent in vivo. Wellcome Open Res. 2, 32 10.12688/wellcomeopenres.11594.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinger J., Nešić D., Ali L., Aresta-Branco F., Lilic M., Chowdhury S., Kim H. S., Verdi J., Raper J., Ferguson M. A. J., Papavasiliou F. N., and Stebbins C. E. (2018) African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat. Nat. Microbiol. 3, 932–938 10.1038/s41564-018-0187-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valero-González J., Leonhard-Melief C., Lira-Navarrete E., Jiménez-Osés G., Hernández-Ruiz C., Pallarés M. C., Yruela I., Vasudevan D., Lostao A., Corzana F., Takeuchi H., Haltiwanger R. S., and Hurtado-Guerrero R. (2016) A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat. Chem. Biol. 12, 240–246 10.1038/nchembio.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kozma K., Keusch J. J., Hegemann B., Luther K. B., Klein D., Hess D., Haltiwanger R. S., and Hofsteenge J. (2006) Identification and characterization of a β1,3-glucosyltransferase that synthesizes the Glc-β1,3-Fuc disaccharide on thrombospondin type 1 repeats. J. Biol. Chem. 281, 36742–36751 10.1074/jbc.M605912200 [DOI] [PubMed] [Google Scholar]
- 34. Sato T., Sato M., Kiyohara K., Sogabe M., Shikanai T., Kikuchi N., Togayachi A., Ishida H., Ito H., Kameyama A., Gotoh M., and Narimatsu H. (2006) Molecular cloning and characterization of a novel human β1,3-glucosyltransferase, which is localized at the endoplasmic reticulum and glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domain. Glycobiology 16, 1194–1206 10.1093/glycob/cwl035 [DOI] [PubMed] [Google Scholar]
- 35. Lesnik Oberstein S. A., Kriek M., White S. J., Kalf M. E., Szuhai K., den Dunnen J. T., Breuning M. H., and Hennekam R. C. (2006) Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am. J. Hum. Genet. 79, 562–566 10.1086/507567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vasudevan D., Takeuchi H., Johar S. S., Majerus E., and Haltiwanger R. S. (2015) Peters plus syndrome mutations disrupt a noncanonical ER quality-control mechanism. Curr. Biol. 25, 286–295 10.1016/j.cub.2014.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vasudevan D., and Haltiwanger R. S. (2014) Novel roles for O-linked glycans in protein folding. Glycoconj. J. 31, 417–426 10.1007/s10719-014-9556-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sultan A. A., Thathy V., Frevert U., Robson K. J., Crisanti A., Nussenzweig V., Nussenzweig R. S., and Ménard R. (1997) TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell 90, 511–522 10.1016/S0092-8674(00)80511-5 [DOI] [PubMed] [Google Scholar]
- 39. Sidik S. M., Huet D., Ganesan S. M., Huynh M. H., Wang T., Nasamu A. S., Thiru P., Saeij J. P. J., Carruthers V. B., Niles J. C., and Lourido S. (2016) A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell 166, 1423–1435.e12 10.1016/j.cell.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marino N. D., Panas M. W., Franco M., Theisen T. C., Naor A., Rastogi S., Buchholz K. R., Lorenzi H. A., and Boothroyd J. C. (2018) Identification of a novel protein complex essential for effector translocation across the parasitophorous vacuole membrane of Toxoplasma gondii. PLoS Pathog. 14, e1006828 10.1371/journal.ppat.1006828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jung C., Lee C. Y., and Grigg M. E. (2004) The SRS superfamily of Toxoplasma surface proteins. Int. J. Parasitol. 34, 285–296 10.1016/j.ijpara.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 42. Wasmuth J. D., Pszenny V., Haile S., Jansen E. M., Gast A. T., Sher A., Boyle J. P., Boulanger M. J., Parkinson J., and Grigg M. E. (2012) Integrated bioinformatic and targeted deletion analyses of the SRS gene superfamily identify SRS29C as a negative regulator of Toxoplasma virulence. MBio 3, e00321–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huynh M. H., Liu B., Henry M., Liew L., Matthews S. J., and Carruthers V. B. (2015) Structural basis of Toxoplasma gondii MIC2-associated protein interaction with MIC2. J. Biol. Chem. 290, 1432–1441 10.1074/jbc.M114.613646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huynh M. H., Rabenau K. E., Harper J. M., Beatty W. L., Sibley L. D., and Carruthers V. B. (2003) Rapid invasion of host cells by Toxoplasma requires secretion of the MIC2-M2AP adhesive protein complex. EMBO J. 22, 2082–2090 10.1093/emboj/cdg217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rabenau K. E., Sohrabi A., Tripathy A., Reitter C., Ajioka J. W., Tomley F. M., and Carruthers V. B. (2001) TgM2AP participates in Toxoplasma gondii invasion of host cells and is tightly associated with the adhesive protein TgMIC2. Mol. Microbiol. 41, 537–547 10.1046/j.1365-2958.2001.02513.x [DOI] [PubMed] [Google Scholar]
- 46. Huynh M. H., Opitz C., Kwok L. Y., Tomley F. M., Carruthers V. B., and Soldati D. (2004) Trans-genera reconstitution and complementation of an adhesion complex in Toxoplasma gondii. Cell. Microbiol. 6, 771–782 10.1111/j.1462-5822.2004.00403.x [DOI] [PubMed] [Google Scholar]
- 47. Moudy R., Manning T. J., and Beckers C. J. (2001) The loss of cytoplasmic potassium upon host cell breakdown triggers egress of Toxoplasma gondii. J. Biol. Chem. 276, 41492–41501 10.1074/jbc.M106154200 [DOI] [PubMed] [Google Scholar]
- 48. Uboldi A. D., Wilde M. L., McRae E. A., Stewart R. J., Dagley L. F., Yang L., Katris N. J., Hapuarachchi S. V., Coffey M. J., Lehane A. M., Botte C. Y., Waller R. F., Webb A. I., McConville M. J., and Tonkin C. J. (2018) Protein kinase A negatively regulates Ca2+ signalling in Toxoplasma gondii. PLoS Biol. 16, e2005642 10.1371/journal.pbio.2005642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huynh M. H., and Carruthers V. B. (2009) Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 10.1128/EC.00358-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shen B., Brown K. M., Lee T. D., and Sibley L. D. (2014) Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5, e01114–e01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Long S., Wang Q., and Sibley L. D. (2016) Analysis of noncanonical calcium-dependent protein kinases in Toxoplasma gondii by targeted gene deletion using CRISPR/Cas9. Infect. Immun. 84, 1262–1273 10.1128/IAI.01173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kang D., Gho Y. S., Suh M., and Kang C. (2002) Highly sensitive and fast protein detection with Coomassie Brilliant Blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bull. Korean Chem. Soc. 23, 2 [Google Scholar]
- 53. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 54. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 10.1038/nbt.3327 [DOI] [PubMed] [Google Scholar]
- 55. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., and Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 57. Tyanova S., Temu T., Carlson A., Sinitcyn P., Mann M., and Cox J. (2015) Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 15, 1453–1456 10.1002/pmic.201400449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Coffey M. J., Dagley L. F., Seizova S., Kapp E. A., Infusini G., Roos D. S., Boddey J. A., Webb A. I., and Tonkin C. J. (2018) Aspartyl protease 5 matures dense granule proteins that reside at the host-parasite interface in Toxoplasma gondii. MBio 9, e01796–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kafsack B. F., Beckers C., and Carruthers V. B. (2004) Synchronous invasion of host cells by Toxoplasma gondii. Mol. Biochem. Parasitol. 136, 309–311 10.1016/j.molbiopara.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 61. Peng D., and Tarleton R. (2015) EuPaGDT: A web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. Genom. 1, e000033 10.1099/mgen.0.000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bandini G., Leon D. R., Hoppe C. M., Zhang Y., Agop-Nersesian C., Shears M. J., Mahal L. K, Routier F. H., Costello C. E., and Samuelson J. (2019) O-Fucosylation of thrombospondin-like repeats is required for processing of microneme protein 2 and for efficient host cell invasion by Toxoplasma gondii tachyzoites. J. Biol. Chem. 294, 1967–1983 10.1074/jbc.RA118.005179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.