Abstract
Drosophila Nedd4 (dNedd4) is a HECT E3 ubiquitin ligase present in two major isoforms: short (dNedd4S) and long (dNedd4Lo), with the latter containing two unique regions (N terminus and Middle). Although dNedd4S promotes neuromuscular synaptogenesis (NMS), dNedd4Lo inhibits it and impairs larval locomotion. To explain how dNedd4Lo inhibits NMS, MS analysis was performed to find its binding partners and identified SH3PX1, which binds dNedd4Lo unique Middle region. SH3PX1 contains SH3, PX, and BAR domains and is present at neuromuscular junctions, where it regulates active zone ultrastructure and presynaptic neurotransmitter release. Here, we demonstrate direct binding of SH3PX1 to the dNedd4Lo Middle region (which contains a Pro-rich sequence) in vitro and in cells, via the SH3PX1-SH3 domain. In Drosophila S2 cells, dNedd4Lo overexpression reduces SH3PX1 levels at the cell periphery. In vivo overexpression of dNedd4Lo post-synaptically, but not pre-synaptically, reduces SH3PX1 levels at the subsynaptic reticulum and impairs neurotransmitter release. Unexpectedly, larvae that overexpress dNedd4Lo post-synaptically and are heterozygous for a null mutation in SH3PX1 display increased neurotransmission compared with dNedd4Lo or SH3PX1 mutant larvae alone, suggesting a compensatory effect from the remaining SH3PX1 allele. These results suggest a post-synaptic–specific regulation of SH3PX1 by dNedd4Lo.
Keywords: E3 ubiquitin ligase, sorting nexin (SNX), synapse, ubiquitin, protein degradation
Introduction
Neuromuscular synaptogenesis (NMS)3 is the process by which axons innervate muscles during development. In Drosophila, NMS begins with motor axon filopodia exploring the muscle surface, releasing presynaptic neurotransmitters that stimulate an evoked postsynaptic response on the specified target developing myotube surface (1). Upon myotube differentiation, the mature muscle pattern and synaptic zones are formed, and neurotransmitter receptors are transported to the postsynaptic membrane (2). Growth cone differentiation into presynaptic terminals then occurs, forming fully functional neuromuscular junctions (NMJs) that can receive signals from the central nervous system and elicit postsynaptic responses, resulting in larval locomotor activity (2). In Drosophila, the larval body wall muscles are present in two identical hemisegments, each containing 30 unique body wall muscles. These body wall muscles are innervated by 32 glutamatergic neurons in a stereotypic pattern (3, 4). The regions of connection between axons and target muscles at NMJs are called synaptic boutons. There are a variety of proteins that play roles in regulating the development and function of Drosophila NMJs, including some that belong to the ubiquitin system (5, 6).
The ubiquitin-proteasome system regulates protein turnover, trafficking/sorting, and localization in cells (7). Ubiquitination is carried out by E1, E2, and E3 enzymes, with the latter (E3 ubiquitin ligase) responsible for substrate recognition and attachment of ubiquitin. We previously identified the HECT E3 ubiquitin ligase Drosophila (d) Nedd4 as a regulator of NMS (8, 9). Like other Nedd4 proteins, dNedd4 contains an N-terminal C2 domain, which in its mammalian orthologue Nedd4, is responsible for plasma membrane localization (10, 11) and intramolecular inhibitory interactions with the catalytic HECT domain (12, 13), 3 WW domains that usually recognize substrates by binding to PY motifs (L/PPXY) (8, 14, 15), and a C-terminal catalytic HECT domain (16, 17).
In Drosophila, dNedd4 is present as two major splice isoforms: dNedd4-short (dNedd4S) and dNedd4-long (dNedd4Lo). These isoforms differ only in the presence of two unique regions in dNedd4Lo: a N-terminal region generated by an alternate start codon, and a Middle (Mid) region generated by an additional exon between the WW1 and WW2 domains (9). We previously showed that dNedd4S acts as a positive regulator of NMS in Drosophila, where it functions in the muscle to ubiquitinate and internalize Commissureless (Comm) from the muscle surface (8). In contrast, dNedd4Lo inhibits NMS, and when dNedd4Lo is overexpressed in the muscle, this causes abnormal innervation along the SNb branch from muscle 13 to 12, impaired larval locomotion, and enhanced larval lethality (9). Interestingly, whereas dNedd4S expression remains high throughout the embryonic stage when NMS is taking place, dNedd4Lo transcript levels decrease significantly at the onset of NMS (at embryonic stage 11 of Drosophila development) and remain reduced until NMS is complete (9). This prompts us to investigate the biological consequences of disrupting this critical reduction in dNedd4Lo levels. Additionally, further examination of dNedd4Lo function also showed that the inhibition of NMS by dNedd4Lo requires the catalytic HECT domain, as well as both the unique N-terminal and Mid regions (9). We thus hypothesized that this regulation may be the result of the unique regions of dNedd4Lo interacting with other proteins.
To identify interacting partners to the unique regions of dNedd4Lo, Drosophila embryo lysates were incubated with purified, GST-immobilized N-terminal and Mid regions and analyzed by MS. Two of the high-confidence interacting partners were identified as binding partners to dNedd4Lo were Drosophila amphiphysin (dAmph) and SH3PX1. dAmph, which binds to the unique N-terminal region of dNedd4Lo, was shown to be degraded by dNedd4Lo in the muscle and to regulate T-tubule formation and larval locomotion (18). Our screen also identified SH3PX1 as a binding partner to the dNedd4Lo unique Mid region.
SH3PX1 is the Drosophila orthologue of mammalian sorting nexins 9, 18, and 33 (SNX9, SNX18, and SNX33), which function in protein and endosomal vesicle sorting, cargo adaptors, endocytosis membrane trafficking, and cytoskeletal remodeling (19–22). SH3PX1 contains a N-terminal SH3 domain that binds proline-rich motifs, a PHOX homology (PX) domain that functions in membrane recruitment and phosphoinositide binding, and a C-terminal BAR domain that senses and induces plasma membrane curvature and also provides a dimerization interface (23). Mammalian SNX9 is expressed in the pre-synaptic compartment of cultured hippocampal neurons, where it regulates synaptic vesicle endocytosis (24). In Drosophila, SH3PX1 is localized both pre- and post-synaptically at the NMJ as well as throughout the muscle (25). At the pre-synapse, SH3PX1 regulates the active zone ultrastructure and promotes pre-synaptic neurotransmission through an interaction with Nervous wreck (25). A role for SH3PX1 at the post-synapse has not been reported.
Here, we show that the SH3 domain of SH3PX1 binds the dNedd4Lo (Mid) region, which contains a proline-rich sequence. In Drosophila S2 cells, overexpression of dNedd4Lo leads to reduced levels of SH3PX1 at the cell periphery. In vivo, SH3PX1 co-localizes post-synaptically with dNedd4Lo at the subsynaptic reticulum (SSR). Overexpression of dNedd4Lo in the post-synaptic muscle, but not pre-synaptically, leads to reduced SH3PX1 levels at the SSR, but not throughout the muscle. Although homozygous null SH3PX1 mutant larvae and larvae expressing dNedd4Lo in muscle show reduced locomotor activity, larvae that overexpress dNedd4Lo and are heterozygous for a SH3PX1 null mutation do not display further impairment of larval locomotion, suggesting a lack of genetic interaction between dNedd4Lo and SH3PX1 in regulating locomotion. Interestingly, whereas SH3PX1 heterozygous mutants and dNedd4Lo-expressing larvae have impaired evoked excitatory junction potential amplitude, larvae expressing dNedd4Lo and heterozygous null for SH3PX1 display rescued EJP amplitudes.
Results
SH3PX1 interacts with dNedd4Lo
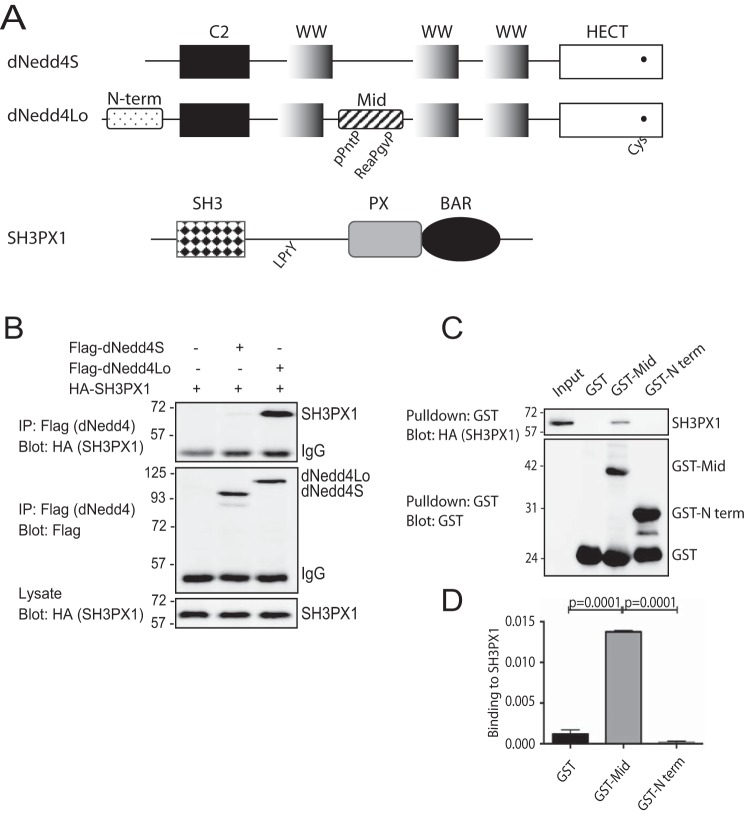
We previously identified an interaction between dNedd4Lo-Mid and SH3PX1 using mass spectrometry (MS) (Fig. 1A) (18). To validate the MS results, we first tested whether SH3PX1 co-immunoprecipitates (co-IP) with Nedd4Lo in cells. Drosophila Schneider 2 (S2) cells were transiently co-transfected with HA-SH3PX1 and FLAG-dNedd4Lo or -dNedd4S, FLAG-dNedd4 immunoprecipitated (IP) and the IP immunoblotted for HA-SH3PX1. The results show co-IP of HA-SH3PX1 with dNedd4Lo, but not dNedd4S (Fig. 1B).
Figure 1.
SH3PX1 interacts with dNedd4Lo. A, schematic representation of dNedd4Lo, dNedd4S, and SH3PX1 (not to scale). The unique N terminus and middle regions and proline-rich sequences of dNedd4Lo are indicated. B, co-IP: Drosophila Schneider 2 (S2) cells were co-transfected with equal amounts of HA-SH3PX1 and FLAG (Flag)-dNedd4S or FLAG-dNedd4Lo. Cells were lysed and the FLAG-dNedd4 proteins were IP using anti-FLAG beads, and immunoblotted with anti-HA antibodies to detect co-IP of SH3PX1 (top panel). C, SH3PX1 interacts with the dNedd4Lo Middle unique region in vitro: purified dNedd4Lo unique N-terminal (N-term) and Middle (Mid) regions or GST alone were immobilized on GST beads and incubated with lysates from Drosophila S2 cells expressing HA-SH3PX1, and subjected to GST pulldown. The samples were resolved by SDS-PAGE and immunoblotted with HA antibodies to determine binding of SH3PX1. D, quantitation of in vitro binding experiments in C (Student's t test, n = 3 independent experiments).
Next, we tested binding between the dNedd4Lo-Middle region (dNedd4Lo-Mid) that was identified in the MS analysis and SH3PX1 using an in vitro pulldown assay. Only dNedd4Lo was tested here because it was the only isoform that bound SH3PX1. GST-immobilized unique Mid- or N-terminal (control) regions of dNedd4Lo, or GST alone (control), were incubated with lysates from Drosophila S2 cells that were transiently expressing HA-tagged SH3PX1. Immunoblotting with anti-HA antibody showed that SH3PX1 binds only to the Mid region of dNedd4Lo in vitro and not the N-terminal region or GST alone (Fig. 1, C and D). Together, these binding experiments validate the MS results.
SH3PX1 binds via its SH3 domain to a Pro-rich motif in the unique middle region of dNedd4Lo
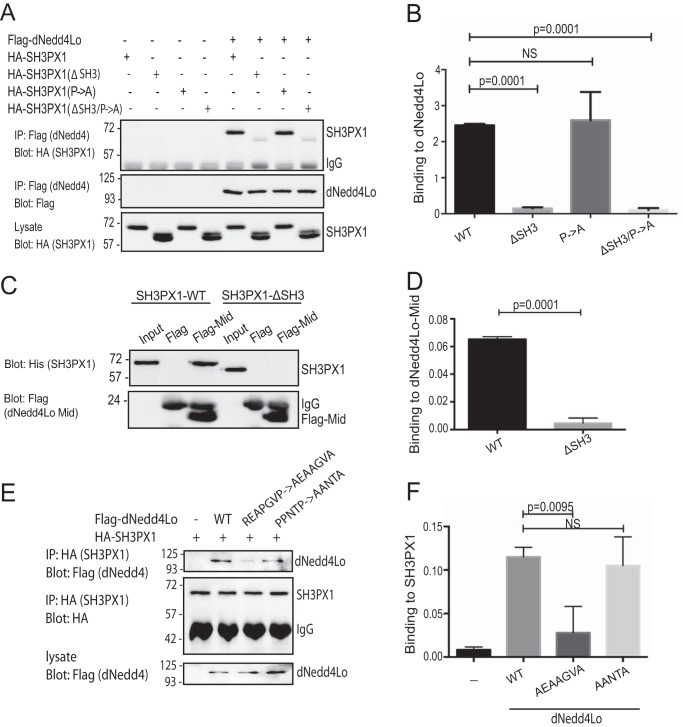
SH3PX1 contains a N-terminal SH3 domain that is known to bind proline-rich motifs (23). The unique Mid region of dNedd4Lo contains 2 proline-rich motifs (ReaPgvP and PPntP) matching or similar to the consensus sequence for potential SH3 domain binding (RXXPXXP or PPXXP) (26). We thus first generated mutant SH3PX1 lacking its SH3 domain (SH3PX1-ΔSH3). This construct was transiently transfected into S2 cells along with FLAG-dNedd4Lo, and subjected to co-IP. Immunoblotting revealed that the SH3 deletion mutant had strongly reduced binding to dNedd4Lo compared with SH3PX1-WT (Fig. 2, A and B), although binding was not completely abolished.
Figure 2.
SH3PX1-SH3 domain binds the REAPGVP motif in dNedd4Lo Middle region. A, Drosophila S2 cells were transfected with FLAG-dNedd4Lo and HA-tagged SH3PX1 WT, Pro → Ala, ΔSH3, or SH3PX1-ΔSH3/Pro → Ala. dNedd4Lo was immunoprecipitated with FLAG beads and the IP immunoblotted with HA to detect co-IP of SH3PX1 or its mutants. Actin was used as a loading control. B, quantification of the co-IP of the experiment in A. The amount of SH3PX1 or its mutants pulled down was normalized to input (lysate) SH3PX1 and to the amount of immunoprecipitated dNedd4Lo (Student's t test: n = 4 separate experiments; NS, not significant). C, purified His-tagged WT and ΔSH3-SH3PX1 were incubated with purified FLAG-tagged dNedd4Lo-Mid, and then subjected to FLAG pulldown followed by SDS-PAGE and immunoblotting for SH3PX1. D, quantification of the pulldown experiment in C. The amount of SH3PX1 or its mutants pulled down was normalized to the input and to the amount of immunoprecipitated dNedd4Lo (Student's t test, n = 4 separate experiments). E, Drosophila S2 cells were transfected with HA-SH3PX1 and FLAG-dNedd4Lo, REAPGVP→AEAAGVA, or PPNTP→AANTA mutants. SH3PX1 was immunoprecipitated with anti-HA antibodies and the IP was immunoblotted with anti-FLAG antibodies to detect co-IP of SH3PX1 with dNedd4Lo proline mutants. F, quantitation of co-IP in E (Student's t test, n = 3 independent experiments).
We also tested whether SH3PX1 binds directly to dNedd4Lo. For this, we purified His-tagged full-length SH3PX1, as well as the FLAG-tagged dNedd4Lo-Mid region, from bacteria. Purified His-SH3PX1-WT and SH3PX1-ΔSH3 were incubated with purified FLAG-Mid-dNedd4Lo in vitro. We found that whereas the full-length SH3PX1-WT was able to bind the dNedd4Lo-Mid region in vitro, this binding was abolished in the SH3PX1-ΔSH3 mutant (Fig. 2, C and D). This indicates that SH3PX1 binds directly to the dNedd4Lo unique Mid region via its SH3 domain.
To test where in the dNedd4Lo-Mid region the SH3PX1-SH3 domain binds, we individually mutated the Pro-rich motifs in the dNedd4Lo-Mid region (REAPGVP→AEAAGVA and PPNTP→AANTA). As shown in Fig. 2, E and F, we found that mutating the REAPGVP (but not the PPNTP) sequence severely impaired binding to SH3PX1.
In addition, dNedd4Lo contains three WW domains that can bind PY motifs (L/PPXY) on substrate proteins (14). Analysis of the SH3PX1 sequence revealed the presence of a PY motif (LPRY). To determine whether dNedd4Lo could bind to this sequence, a HA-tagged single PY motif mutant of SH3PX1 was generated (SH3PX1-Pro → Ala), as well as a double mutant of SH3PX1 that contained a PY motif mutation along with deletion of the SH3 domain (SH3PX1-ΔSH3/Pro → Ala). These constructs were transiently transfected along with FLAG-tagged dNedd4Lo in S2 cells. Our results show that co-IP of FLAG-dNedd4Lo with SH3PX1-Pro → Ala did not affect binding to dNedd4Lo (Fig. 2, A and B). Furthermore, there was no significant reduction in binding of dNedd4Lo to the combined SH3PX1-ΔSH3/Pro → Ala mutant compared with the SH3PX1-ΔSH3 mutant alone. Taken together, these results indicate that the SH3 domain of SH3PX1 (but not its PY motif) binds the REAPGVP motif in the dNedd4Lo-Mid region.
dNedd4Lo reduces SH3PX1 levels at the cell periphery of S2 cells
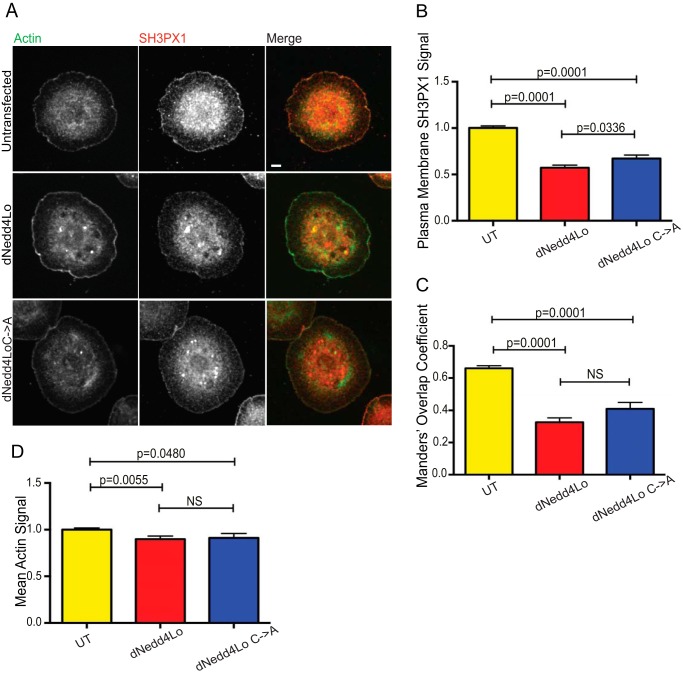
In S2 cells, SH3PX1 was shown to localize to the cell cortex adjacent to lamellipodia, and just distal to the cortical F-actin band at the cell periphery, where it co-localizes with actin nucleating proteins Scar and Wasp (27). Because we observed that SH3PX1 binds dNedd4Lo in S2 cells, we tested the effect of ectopic expression of WT-dNedd4Lo on the levels and localization of SH3PX1 in these cells. Cells were transfected with FLAG-WT dNedd4Lo, or its catalytically inactive mutant dNedd4Lo Cys → Ala, and analyzed by confocal microscopy. In cells overexpressing dNedd4Lo, SH3PX1 levels at the plasma membrane region were significantly reduced compared with control cells (Fig. 3, A and B). Additionally, quantification of the Mander's 1 (M1) correlation coefficient showed a decrease in overlap between SH3PX1 and cortical actin in dNedd4Lo-overexpressing cells (M1 = 0.661 for control cells; M1 = 0.327 for dNedd4Lo-overexpressing cells) (Fig. 3C). Although peripheral actin was also reduced in the dNedd4Lo-overexpressing S2 cells (Fig. 3D), this reduction was much smaller than that of SH3PX1, suggesting that dNedd4Lo does not target SH3PX1 indirectly by reducing actin levels. Surprisingly, we also found a decrease in intensity of SH3PX1 at the plasma membrane in the presence of catalytically inactive dNedd4Lo Cys → Ala (Fig. 3C). Together, these results indicate that dNedd4Lo down-regulates or removes SH3PX1 from the plasma membrane region in S2 cells, an effect that may be independent of (or only partially dependent on) ubiquitination.
Figure 3.
SH3PX1 is removed from the cell periphery upon dNedd4Lo expression in S2 cells. A, S2 cells were transiently transfected with either FLAG-dNedd4Lo or FLAG-dNedd4Lo Cys → Ala and spotted onto concanavalin A-coated coverslips. Cells were stained and imaged to visualize actin (green) and SH3PX1 (red). Scale bar, 4 μm. B, quantification of plasma membrane SH3PX1 intensity in A. Levels of SH3PX1 at the plasma membrane were significantly reduced in cells overexpressing both dNedd4Lo and dNedd4Lo Cys → Ala. C, quantification of Mander's 1 (M1) overlap coefficient for SH3PX1 and phalloidin (actin) in A. Overlap of SH3PX1 signal and actin signal are significantly reduced in cells overexpressing both dNedd4Lo and dNedd4Lo Cys → Ala. D, quantification of peripheral actin staining intensity. (WT, n = 203 cells; dNedd4Lo, n = 75 cells dNedd4Lo Cys → Ala, n = 60 cells. Student's t test; NS, not significant.)
dNedd4Lo reduces levels of SH3PX1 post-synaptically at the SSR, but not throughout the muscle
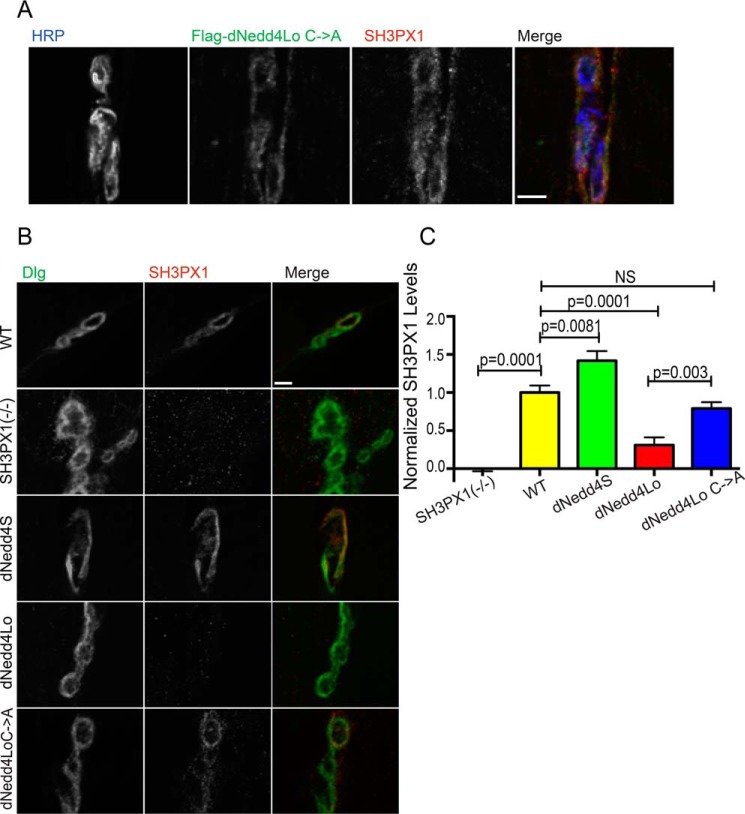
Our previous work showed that dNedd4 is enriched endogenously post-synaptically at the muscle and SSR, where it co-localizes with the post-synaptic marker Discs large (Dlg) (8, 9). As well, overexpression of dNedd4Lo in muscle with the 24B-Gal4 driver results in localization of dNedd4Lo to the SSR as well as throughout the muscle. SH3PX1 is also present at third instar larval NMJs, at both the pre-synaptic terminals and the post-synaptic SSR (25). Since we showed that SH3PX1 binds dNedd4Lo in vitro and in S2 cells (Figs. 1 and 2), we investigated whether the two proteins co-localize in vivo at larval NMJs. As dNedd4Lo is a ubiquitin ligase that can degrade substrates at NMJs (18), we used the catalytically inactive dNedd4Lo Cys → Ala mutant for these experiments to prevent potential degradation of endogenous SH3PX1 that would prevent its detection. To test for co-localization of SH3PX1 and dNedd4Lo at NMJs, we expressed FLAG-dNedd4Lo Cys → Ala in the muscle using the early muscle driver 24B-Gal4 (UAS-Flag-dNedd4Lo C→A/24B-Gal4) and examined NMJs on muscle 6/7 in third instar larvae by immunohistochemistry. We used antibodies against FLAG (dNedd4Lo Cys → Ala), endogenous SH3PX1, and horseradish peroxidase (HRP) to label pre-synaptic regions. Our results show that SH3PX1 co-localizes with FLAG-dNedd4Lo C → A at the post-synaptic SSR in NMJs (Fig. 4A).
Figure 4.
Post-synaptic dNedd4Lo overexpression leads to reduced SH3PX1 levels at the SSR. A, SH3PX1 and dNedd4Lo Cys → Ala co-localize at the post-synapse of third instar larval NMJs at muscle 6/7. Scale bar, 3 μm. B, larvae that were a driver control line (+/24B-Gal4), SH3PX1 mutant, and overexpressing FLAG-tagged dNedd4S (UAS-Flag-dNedd4S/24B-Gal4), dNedd4Lo (UAS-Flag-dNedd4Lo/24B-Gal4), and dNedd4Lo Cys → Aal (UAS-Flag-dNedd4Lo Cys → Ala/24B-Gal4) with 24B-Gal4 were stained with anti-Dlg (green) and anti-SH3PX1 (red) antibodies and analyzed for post-synaptic SH3PX1 levels at the bouton on muscle 6/7. Driver control was included to show WT SH3PX1 levels, and the SH3PX1 mutant was included to demonstrate anti-SH3PX1 antibody specificity. Scale bar, 3 μm. C, quantification of SH3PX1 intensity (levels) at the SSR shown in B, identified as the area marked by Dlg labeling. SH3PX1 intensity is significantly reduced in larvae overexpressing dNedd4Lo with 24B-Gal4. (+/24B-Gal4, n = 56 boutons; SH3PX1 mutant, n = 49 boutons; UAS-Flag-dNedd4S/24B-Gal4, n = 45 boutons; UAS-Flag-dNedd4Lo/24B-Gal4, n = 60 boutons; UAS-Flag-dNedd4Lo Cys → Ala/24B-Gal4, n = 64 boutons. Student's t test; NS, not significant.)
Next, we tested whether dNedd4Lo can regulate SH3PX1 levels in vivo. We expressed FLAG-dNedd4Lo, or the controls FLAG-dNedd4S (which lacks the unique Mid region and thus does not bind SH3PX1) or FLAG-dNedd4Lo Cys → Ala, with the 24B-Gal4 driver. We then performed immunohistochemistry with antibodies against endogenous SH3PX1 and the post-synaptic marker Dlg, and examined NMJs on muscle 6/7 for SH3PX1 intensity in the regions marked by Dlg staining. We used w1118 flies crossed to 24B-Gal4 as WT controls, and homozygous null SH3PX1 mutant flies as negative controls for SH3PX1 signal. Interestingly, expression of dNedd4Lo with 24B-Gal4 caused a significant decrease in SH3PX1 levels at the SSR compared with WT control larvae, dNedd4S, or the catalytically-inactive dNedd4Lo Cys → Ala (Fig. 4, B and C). This suggests that dNedd4Lo degrades SH3PX1 at the SSR, which requires its catalytic activity. Surprisingly, overexpression of dNedd4S with 24B-Gal4 led to an increase in the SH3PX1 signal at the SSR compared with WT controls. Collectively, these data indicate that dNedd4Lo decreases the level of SH3PX1 post-synaptically at the SSR, likely through ubiquitination and degradation.
To determine whether the loss of SH3PX1 observed in larvae that overexpress dNedd4Lo also resulted in ultrastructure changes we used transmission EM to examine NMJs from muscle 6/7 in SH3PX1 mutant and UAS-Flag-dNedd4Lo/24B-Gal4 larvae. Analysis of the post-synapse showed that the apparent ultrastructure of the post-synaptic SSR is not altered in both SH3PX1 mutant and dNedd4Lo-overexpressing larvae compared with WT controls (Fig. S1), indicating that neither SH3PX1 nor dNedd4Lo regulate ultrastructure at the post-synapse.
Since we observed a decrease in SH3PX1 levels at the SSR upon dNedd4Lo overexpression, we tested whether the loss of SH3PX1 was specific to the SSR, or if it occurs throughout the muscle. We examined the steady-state levels of SH3PX1 in body wall muscles in larvae expressing FLAG-dNedd4Lo, -dNedd4S or -dNedd4Lo Cys → Ala with 24B-Gal4. Larvae were dissected to isolate muscle tissue, and muscle fillet preps were lysed and subjected to SDS-PAGE and immunoblotting with antibodies against SH3PX1. No significant changes in SH3PX1 levels were detected in the muscles of any of the three dNedd4 forms (Fig. 5, A and B). This suggests that dNedd4Lo regulates SH3PX1 specifically at the SSR, and not broadly throughout the muscle.
Figure 5.
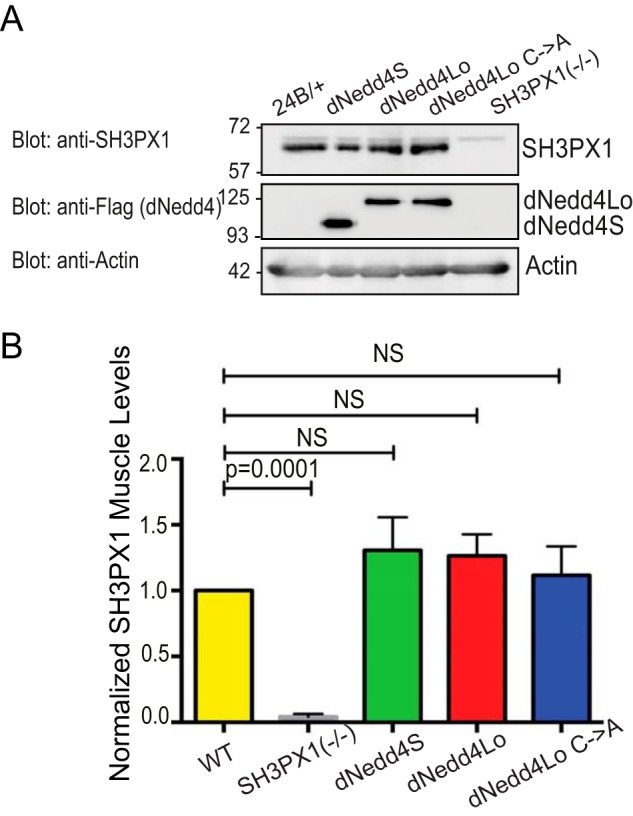
Steady-state levels of SH3PX1 in the muscle are not altered upon dNedd4 overexpression. A, 10 larvae of each WT (driver control line, +/24B-Gal4), SH3PX1 mutant, and overexpressing FLAG-tagged dNedd4S, dNedd4Lo, and dNedd4Lo Cys → Ala with the 24B-Gal4 driver, were collected and dissected to isolate muscle tissue. Muscles were homogenized and subjected to SDS-PAGE and immunoblotting with anti-FLAG (dNedd4; middle panel) and anti-SH3PX1 (top panel) antibodies. Driver control was included to show WT muscle SH3PX1 levels, and SH3PX1 mutant was included to demonstrate anti-SH3PX1 antibody specificity. Actin was used as a loading control. B, quantification of SH3PX1 signal in A. Levels of SH3PX1 in muscle overexpressing all dNedd4 isoforms are not significantly altered from WT controls. SH3PX1 levels were normalized to actin (Student's t test: n = 3 separate experiments, 30 muscle samples total for each genotype; NS: not significant).
dNedd4Lo does not regulate muscle T-tubule architecture or SNb branch innervation via SH3PX1
We showed previously that dNedd4 is expressed throughout Drosophila muscles where it plays a role in T-tubule formation and is required for proper innervation of the SNb branch from muscle 13 to 12 (8, 9, 18). If dNedd4Lo regulates either of these processes via SH3PX1, we would expect SH3PX1 mutant larvae to display similar defects. Thus, we first tested if homozygous null SH3PX1 mutants displayed abnormal innervation patterns at muscle 13/12. WT (w1118) and SH3PX1 mutant third instar larvae were dissected and stained with phalloidin to visualize muscles and HRP to visualize nerves, and examined using immunofluorescence. SNb branch innervation from muscle 13 to 12 was scored as either abnormal or normal. No significant differences in the percentage of abnormal innervation were observed in SH3PX1 mutant larvae compared with WT controls (Fig. S2, A and B). We also examined potential defects in T-tubule architecture in SH3PX1 mutant larvae. WT (w1118) and SH3PX1 mutant third instar larvae were dissected and stained with Dlg to visualize T-tubules in the muscle. No morphological or architectural differences in the muscle and T-tubules were observed between WT and SH3PX1 mutant larvae (Fig. S2C). Thus, it is unlikely that dNedd4Lo regulates SNb branch innervation or T-tubule architecture via SH3PX1.
dNedd4Lo does not regulate the levels of SH3PX1 at pre-synaptic terminals
SH3PX1 is also present at pre-synaptic terminals (25). Since we observed that dNedd4Lo regulates SH3PX1 levels at the post-synapse, we tested if this effect also occurs at pre-synaptic terminals. Thus, FLAG-dNedd4Lo, -dNedd4S, or -dNedd4Lo Cys → Ala were expressed in neurons by crossing the UAS lines to the pre-synaptic neuronal driver elavc155-Gal4. Third instar larvae were stained with antibodies to SH3PX1, Dlg, and HRP, and NMJs on muscle 6/7 were examined. Our results show no significant differences in SH3PX1 levels and localization between the three lines (Fig. 6, A and B). These results suggest that dNedd4Lo regulates SH3PX1 levels at the post-synaptic SSR, but not at pre-synaptic terminals.
Figure 6.
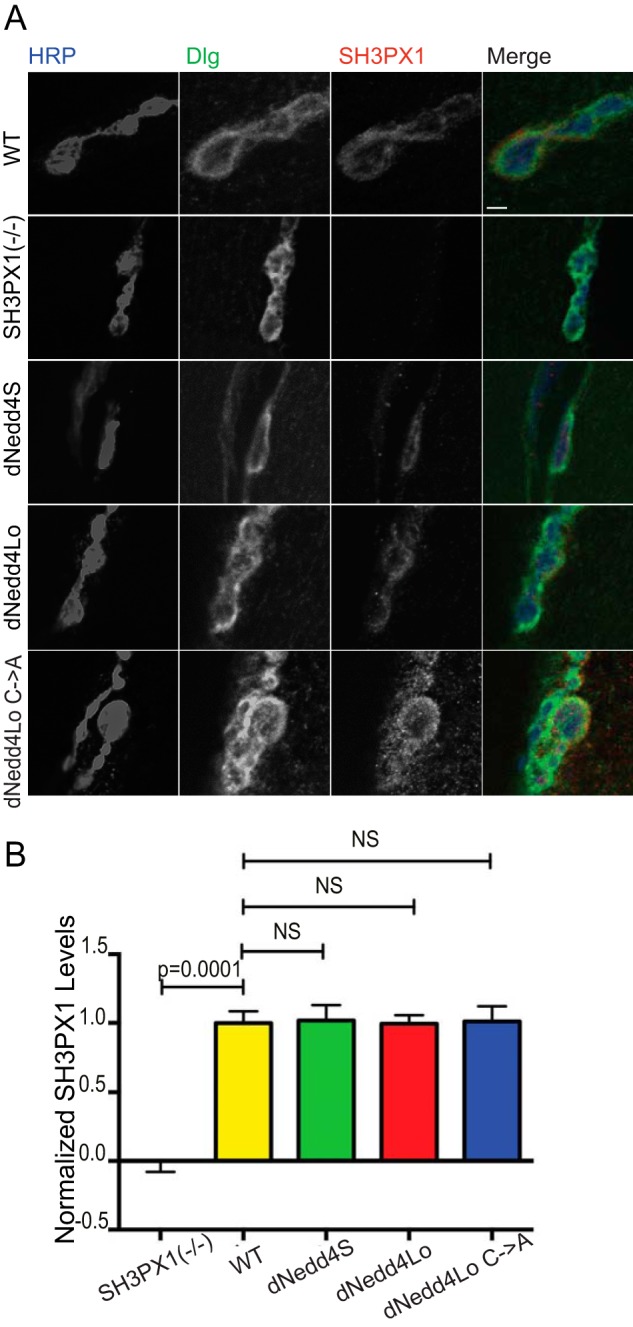
Pre-synaptic dNedd4Lo overexpression does not alter SH3PX1 levels. A, larvae that were a driver control line (+/elavC155-Gal4), SH3PX1 mutant, and overexpressing FLAG-tagged dNedd4S (UAS-Flag-dNedd4S/elavC155-Gal4), dNedd4Lo (UAS-Flag-dNedd4Lo/elavC155-Gal4), and dNedd4Lo Cys → Ala (UAS-Flag-dNedd4Lo Cys → Ala/elavC155-Gal4) at the pre-synapse with the CNS driver elavC155-Gal4 were stained with anti-Dlg (green), anti-HRP (blue), and anti-SH3PX1 (red) antibodies and analyzed for pre-synaptic SH3PX1 levels at boutons on muscle 6/7. Driver control was included to show WT SH3PX1 levels, and the SH3PX1 mutant was included to demonstrate anti-SH3PX1 antibody specificity. Scale bar, 3 μm. B, quantification of SH3PX1 intensity (levels) at the pre-synapse in B, identified as the area marked by HRP labeling. SH3PX1 signal intensity is not reduced in larvae overexpressing dNedd4Lo at the pre-synapse (+/elavC155-Gal4, n = 56 boutons; SH3PX1 mutant, n = 20 boutons; UAS-Flag-dNedd4S/elavC155-Gal4, n = 46 boutons; UAS-Flag-dNedd4Lo/elavC155-Gal4, n = 48 boutons; UAS-Flag-dNedd4Lo C→Ala/elavC155-Gal4, n = 36 boutons. Student's t test; NS, not significant).
Loss of SH3PX1 impairs larval locomotion
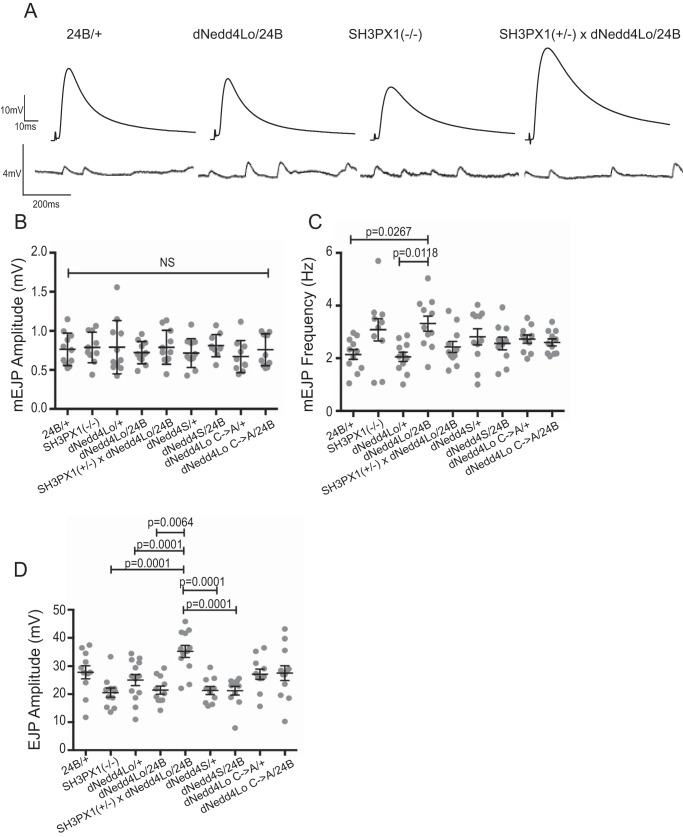
Our previous work showed that third instar larvae overexpressing dNedd4Lo in the muscle and post-synapse with 24B-Gal4 display impaired larval locomotion (9) (Fig. 7). As well, our analysis of homozygous null SH3PX1 mutant third instar larvae revealed a statistically significant (32%) reduction in total path length traveled compared with WT controls, whereas heterozygous SH3PX1(+/−) larvae showed an ∼14.7% reduction (although not statistically significant) (Fig. 7). Since we observed a decrease in post-synaptic SH3PX1 levels upon overexpression of dNedd4Lo, we tested if dNedd4Lo genetically interacts with SH3PX1 post-synaptically to regulate larval locomotion. We crossed UAS-Flag-dNedd4Lo flies to SH3PX1(+/−) flies to generate a line that is heterozygous for SH3PX1 and overexpresses dNedd4Lo (UAS-Flag-dNedd4Lo;SH3PX1(+/−)). Since we observed a decrease in SH3PX1 levels by dNedd4Lo at the SSR, we would expect that a genetic interaction between dNedd4Lo and SH3PX1 would cause degradation of SH3PX1 in UAS-Flag-dNedd4Lo;SH3PX1(+/−) larvae overexpressing dNedd4Lo with 24B-Gal4, causing a further decrease in locomotion relative to larvae overexpressing dNedd4Lo alone (with WT SH3PX1 levels). However, our results show a small increase (not the expected decrease) in locomotion in the UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 larvae, which was not statistically significant relative to SH3PX1 mutants or dNedd4Lo larvae (Fig. 7). Analysis of the post-synaptic levels of SH3PX1 at the SSR by immunofluorescence showed that whereas SH3PX1 levels are significantly decreased in UAS-Flag-dNedd4Lo/24B-Gal4 and UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 flies compared with SH3PX1(+/−) larvae alone, there is no significant difference in SH3PX1 levels between UAS-Flag-dNedd4Lo/24B-Gal4 and UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 larvae, similar to the observed trend in their locomotor activity (Fig. S3). These results suggest that SH3PX1 and dNedd4Lo do not cooperate to regulate larval locomotion.
Figure 7.
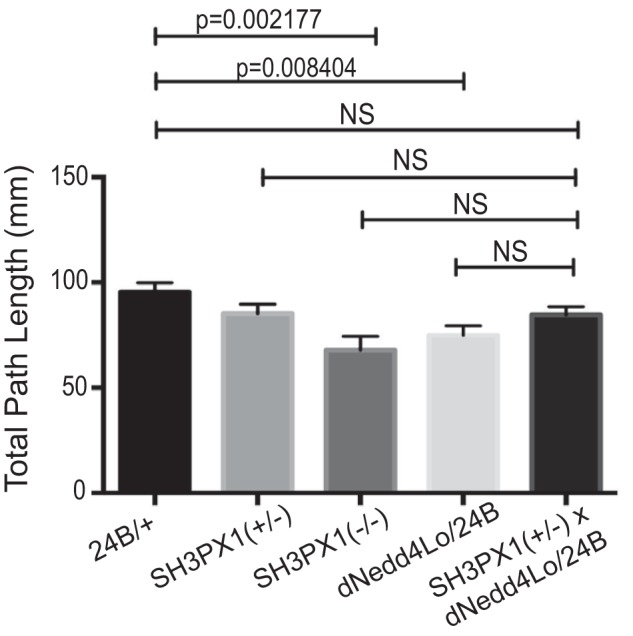
SH3PX1 does not genetically interact with dNedd4Lo to regulate larval locomotion. UAS-dNedd4Lo flies were crossed with SH3PX1 mutant flies to generate larvae overexpressing dNedd4Lo with 24B-Gal4 and heterozygous for SH3PX1 (UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4). Larvae that were driver control line (+/24B-Gal4), SH3PX1(+/−), SH3PX1 mutant, UAS-Flag-dNedd4Lo/24B-Gal4, and UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 were examined for their larval locomotion activity. The graph represents quantification of mean total path length in the indicated larvae (one-way ANOVA followed by Tukey's post hoc multiple comparisons; NS, not significant). n = 25 larvae for SH3PX1(+/−), n = 29 for UAS-Flag-dNedd4Lo/24B-Gal4; n = 30 for (+/24B), n = 24 for (UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4), n = 20 for SH3PX1(−/−).
dNedd4Lo-overexpressing larvae have impaired neurotransmission that is rescued in dNedd4Lo;SH3PX1(+/−) larvae
SH3PX1 mutant larvae were previously shown to regulate neurotransmission in third instar larvae, and have reduced evoked excitatory junction potentials (EJPs) and decreased quantal content. Muscle-specific expression of SH3PX1 can partially rescue these defects, indicating that there is post-synaptic contribution by SH3PX1 to neurotransmission (25). We thus tested if this effect is regulated by dNedd4Lo. We first analyzed larvae overexpressing dNedd4Lo with 24B-Gal4 for potential defects in spontaneous and evoked potentials. Similar to SH3PX1 mutant larvae, no change in mini EJP (mEJP) amplitude was detected in dNedd4Lo-overexpressing larvae (Fig. 8, A and B). Interestingly, however, we detected a significant increase in mEJP frequency in dNedd4Lo overexpressing larvae compared with controls (Fig. 8, A and C). However, SH3PX1 mutant larvae, as well as UAS-Flag-dNedd4Lo;SH3PX1(+/−) larvae did not display significant differences in mEJP frequency (Fig. 8C), suggesting that whereas dNedd4Lo may play a role in spontaneous vesicle fusion, it does not likely do this through SH3PX1.
Figure 8.
Electrophysiological analysis of neurotransmitter release in dNedd4-overexpressing and SH3PX1 mutant larvae. A, representative traces of EJPs and mEJPs in control (+/24B-Gal4), UAS-Flag-dNedd4Lo/24B-Gal4, SH3PX1 mutant, and UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 larvae. B, mEJP amplitude measured in the indicated genotypes. No significant differences in mEJP amplitude were detected across lines. C, mEJP frequency measured in the indicated genotypes. UAS-Flag-dNedd4Lo/24B-Gal4 larvae have a significantly increased mEJP frequency compared with relevant controls (one-way ANOVA with Tukey's post hoc multiple comparisons; NS, not significant). D, evoked EJPs measured in the indicated genotypes. (One-way ANOVA with Tukey's post hoc multiple comparisons; NS, not significant.) (n = 11 larvae for +/24B, n = 11 larvae for SH3PX1(−/−), n = 13 larvae for UAS-Flag-dNedd4Lo/+, n = 11 larvae for UAS-Flag-dNedd4Lo/24B-Gal4, n = 12 larvae for UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4, n = 10 larvae for UAS-Flag-dNedd4S/+, n = 11 for UAS-Flag-dNedd4S/24B-Gal4, n = 11 for UAS-Flag-dNedd4Lo Cys → Ala/+, n = 12 for UAS-Flag-dNedd4Lo Cys → Ala/24B-Gal4).
We next examined evoked EJPs in dNedd4Lo-overexpressing larvae. dNedd4Lo larvae crossed to 24B-Gal4 displayed decreased EJP amplitude compared with WT controls, as did SH3PX1 mutant larvae (9). Surprisingly, UAS-Flag-dNedd4Lo;SH3PX1(+/−) larvae showed the opposite trend, with EJP amplitude greater than WT controls, above that of dNedd4Lo-overexpressing, or SH3PX1 mutant larvae (Fig. 8D). Interestingly, this effect is consistent with the trend we observed in our analysis of larval locomotion (Fig. 7), suggesting that although both dNedd4Lo and SH3PX1 regulate EJP amplitudes on their own, it appears that dNedd4Lo overexpression can rescue the partial loss of SH3PX1.
Discussion
Our results show that dNedd4Lo reduces levels of SH3PX1 at the cell periphery of S2 cells and at the post-synaptic SSR in vivo. Previous studies from our lab showed that dNedd4Lo functions as a negative regulator of NMS and larval locomotion in Drosophila third instar larvae (9). We also showed that regulation of these events required not only the catalytic activity of dNedd4Lo, but also the presence of both the unique N-terminal and Middle regions of dNedd4Lo. Because the catalytic HECT domain of dNedd4Lo is also present in dNedd4S, a positive regulator of NMS (8), we hypothesized that this negative regulation of NMS by dNedd4Lo is the result of the unique regions of dNedd4Lo specifically interacting with other proteins or substrates. In the present study, we identified Drosophila SH3PX1 as an interactor of the unique Middle region of dNedd4Lo. In vivo expression of dNedd4Lo post-synaptically reduced SH3PX1 levels at the SSR specifically, but not throughout the muscle, nor pre-synaptically.
Interestingly, whereas the SH3PX1 mutant and dNedd4Lo-overexpressing larvae both show reduced locomotor activity, overexpression of dNedd4Lo in heterozygous mutant SH3PX1 larvae did not give rise to any further defects in locomotion but instead rescued synaptic transmission defects. These results indicate that dNedd4Lo regulates SH3PX1 levels at the post-synaptic SSR, and both proteins modulate NMJ function.
It is possible that dNedd4Lo and SH3PX1 do not interact genetically to regulate locomotion or neurotransmission. Alternatively, because there is still one copy of SH3PX1 present in the dNedd4Lo;SH3PX1(+/−) larvae, it is possible that genetic compensation, whereby transcription of the remaining copy is increased to maintain SH3PX1 at necessary physiological levels, is occurring. Furthermore, EJP analysis of neurotransmission revealed a reduction in EJP in the homozygous SH3PX1 mutant or UAS-Flag-dNedd4Lo/24B-Gal4 larvae relative to controls. However, this reduction was abolished in the UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 larvae, which have significantly greater EJPs compared with SH3PX1 mutant or UAS-Flag-dNedd4Lo/24B-Gal4, suggesting a compensation effect. One possible explanation for this effect could be genetic compensation of the remaining allele of SH3PX1, as above, or retrograde homeostatic synaptic compensation from the pre-synapse, where SH3PX1 plays an important role in EJP stimulation (25). Indeed, this type of homeostatic compensation is commonly observed at the Drosophila NMJ (28). Because our immunofluorescence analysis of SH3PX1 levels in UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 larvae showed that SH3PX1 levels at the SSR in this line are not significantly different from that of dNedd4Lo-overexpressing larvae alone (Fig. S3), it appears most likely that retrograde homeostatic synaptic compensation is occurring in the UAS-Flag-dNedd4Lo;SH3PX1(+/−)/24B-Gal4 line, in which pre-synaptic SH3PX1 activity is increased in response to post-synaptic loss of SH3PX1 by dNedd4Lo. This effect is not observed in UAS-Flag-dNedd4Lo/24B-Gal4 larvae alone because although SH3PX1 levels at the SSR are extremely low, the pre-synaptic pool of SH3PX1 is present at WT levels and synaptic function is not placed under enough stress to trigger a compensation mechanism. This may implicate dNedd4Lo in homeostatic regulation of synaptic SH3PX1 function. It is also possible that in addition to direct interactions between dNedd4Lo and SH3PX1, dNedd4Lo is a negative regulator and SH3PX1 a positive regulator of two different mechanisms that converge on the regulation of locomotion or neurotransmission.
Because previous studies implicated dNedd4Lo as a negative regulator of NMS and larval locomotion, we sought to determine how it regulates these processes through its interacting partners. dNedd4Lo was shown to genetically interact with dAmph in the body wall muscles, where it regulates T-tubule architecture and larval locomotion at least partially through dAmph degradation (18). In the current study we tested whether the dNedd4Lo·SH3PX1 interaction could help elucidate how dNedd4Lo regulates NMS and muscle innervation, which we previously showed to be impaired in dNedd4Lo (9). However, analysis of SH3PX1 mutant larvae did not reveal any apparent defects in innervation, indicating that dNedd4Lo does not regulate this process via SH3PX1.
Nedd4 family proteins most commonly bind substrates containing PY motifs with their WW domains (17). However, mutations of a PY motif within SH3PX1 did not alter the ability of SH3PX1 to co-IP with dNedd4Lo in S2 cells. It thus appears that the SH3 domain of SH3PX1 provides the sole binding interface to dNedd4Lo, a less common mode of interaction between a Nedd4 family member and its substrates. A similar phenomenon was observed with the dNedd4Lo·dAmph interaction (18). Interestingly, SNX9 was identified as a substrate of the Nedd4 family member Itch in melanoma cells, where the SNX9 SH3 domain binds a proline-rich region in Itch (29). It thus appears that interactions between Nedd4 family members and BAR-SH3 sorting nexins are potentially conserved.
To investigate the role of dNedd4Lo in NMJ development, we expressed dNedd4Lo in muscles using the 24B-Gal4 driver. In the embryonic stage of Drosophila development, when NMS takes place beginning at Stage 11, dNedd4S expression remains high, whereas that of dNedd4Lo is reduced until NMS is complete (9). Our results here, along with our previous work (18), illustrate the importance of ensuring dNedd4Lo expression remains low during NMS, to ensure that levels of Nedd4Lo substrates, such as dAmph or SH3PX1, are high. These substrates are regulated by active dNedd4Lo, suggesting it normally could degrade them through ubiquitination. However, our inability to treat live larvae with proteasome inhibitors, the low amount of proteins that can be harvested from the post-synapse region, as well as the dramatic reduction of SH3PX1 signal upon dNedd4Lo overexpression, pose challenges to our ability to detect in vivo SH3PX1 ubiquitination at the SSR by dNedd4Lo.
It also remains unclear whether the loss in SH3PX1 signal that we observed at the SSR is due to degradation or degradation secondary to trafficking effects. Our S2 cells data suggest that dNedd4Lo can traffic from or degrade SH3PX1 at the cell periphery. Curiously, this effect, although to a lesser extent, is also seen in the catalytically-inactive dNedd4Lo Cys → Ala mutant, suggesting ubiquitination-independent trafficking of SH3PX1 in these cells. In accord, we were not able to detect SH3PX1 ubiquitination by dNedd4Lo in S2 cells. Our attempts to perform pulse-chase analysis in S2 cells to investigate the role of dNedd4Lo-mediated degradation of SH3PX1 on its stability were not successful because dNedd4Lo itself was degraded faster than endogenous SH3PX1 in these cells. Nevertheless, at the post-synapse, the in vivo loss of SH3PX1 clearly requires dNedd4Lo catalytic activity. The mammalian Itch·SNX9 interaction leads to direct ubiquitination of SNX9 by Itch in vitro and in HEK293 cells, resulting in SNX9 degradation (29). In Drosophila, dNedd4 was shown to regulate proteins both through trafficking and degradation. During Notch signaling, dNedd4 ubiquitinates Notch to promote its ligand-independent internalization and inactivation, controlling its cellular localization as well as targeting it for proteasomal degradation within the wing disc (30). dNedd4 also ubiquitinates Deltex (Dx), a positive regulator of Notch signaling, targeting it for degradation (30). In contrast, dNedd4 was shown to promote internalization of Comm from the muscle surface, to enable NMS (8), implicating a role for this E3 ligase in trafficking. Thus, protein trafficking and/or degradation are both potential ways that dNedd4Lo may regulate SH3PX1 at the post-synapse.
Unexpectedly, we observed a significant increase in SH3PX1 levels at the SSR upon overexpression of dNedd4S. It is possible that dNedd4S regulates another as-yet unidentified protein that itself is a negative regulator of SH3PX1 localization or levels in this compartment. Indeed, dNedd4S and dNedd4Lo have been shown to have opposing functions in the regulation of NMS (8, 9), as indicated above. The relevance of the opposite effects on post-synaptic SH3PX1 protein levels to the process of NMS remains unknown.
Although overexpression of dNedd4Lo in muscles resulted in a robust reduction of SH3PX1 levels at the SSR, we did not observe the same effect upon pre-synaptic dNedd4Lo overexpression using the neuronal driver elavC155-Gal4. Immunostaining for endogenous dNedd4 at the Drosophila NMJ and body wall musculature reveal that dNedd4 is highly enriched in the SSR of NMJs as well as in the muscle, with very low expression at the pre-synapse (18). Additionally, overexpression of dNedd4S or dNedd4Lo individually in the muscle with the 24B-Gal4 driver results in localization of both isoforms to the SSR. It is thus not surprising that dNedd4Lo appears to function specifically on the post-synaptic side of the NMJ. Surprisingly, no significant change in muscle SH3PX1 levels were observed upon expression of dNedd4Lo there. Although dNedd4Lo does function in the muscle to regulates T-tubule formation via dAmph (18), that function is independent from its regulation of SH3PX1 levels. This suggests a truly SSR-specific regulation of SH3PX1 by dNedd4Lo.
Mammalian SNX9 is expressed in the pre-synaptic compartment of cultured hippocampal neurons, where it binds to dynamin-1 and N-WASP via its SH3 domain (24). When SNX9 is overexpressed in these cells, it retards clathrin-mediated synaptic vesicle endocytosis primarily due to its interaction with dynamin-1 (24). SH3PX1 also interacts with Drosophila N-WASP and the clathrin coat adaptor protein AP-50 in S2 cells, implicating it in cytoskeletal remodeling and clathrin-mediated endocytosis (23, 31). Whether or not these protein–protein interactions occur at the SSR, and thus contribute in some way to the post-synaptic function of SH3PX1, is unknown.
In summary, our work here identified SH3PX1 as an in vivo substrate for dNedd4Lo at the post-synaptic SSR, and showed that SH3PX1 and dNedd4Lo regulate neurotransmission and larval locomotion in Drosophila.
Experimental procedures
Fly stocks
All fly strains and crosses are depicted in Table S1. The UAS-Gal4 system was used to drive tissue-specific overexpression of dNedd4 isoforms or mutants in all transgenic lines. SH3PX1 mutant larvae, kindly provided by Dr. K. O'Connor-Giles (Wisconsin University), were generated using CRISPR-mediated homology-directed repair to replace a portion of the coding sequence from the SH3PX1 locus with a 3×P3-DsRed marker (25). All flies were maintained at room temperature on standard food and all experiments were performed at 25 °C. The dNedd4 null mutant is lethal (9) and hence could not be used in our current studies.
Plasmid constructs
The cDNA for SH3PX1 was obtained from the Berkeley Drosophila Genome Project (sequence tag LD47602) (Berkeley, CA). To generate HA-tagged SH3PX1 for expression in mammalian cells, PCR-amplified full-length SH3PX1 was subcloned into pcDNA3.1 with an N-terminal HA tag using the Gateway cloning system (Life Technologies, Invitrogen). For expression of WT, 2Trp → Ala, Pro → Ala, and SH3PX1-ΔSH3 in S2 cells, PCR-amplified sequences for full-length SH3PX1 and SH3PX1-ΔSH3 (NCBI accession number AY069705, amino acid residues 62–565) were subcloned into pAHW (Drosophila Genome Resource Center, Indiana University, IN) containing an N-terminal HA tag using the Gateway cloning system. SH3PX1 2Trp → Ala and Pro → Ala and dNedd4Lo REAPGVP→AEAAGVA and PPNTP→AANTA mutants were generated using the QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA).
For bacterial purification of SH3PX1 and SH3PX1-ΔSH3, PCR-amplified sequences for full-length SH3PX1 and SH3PX1-ΔSH3 (NCBI accession number AY069705, residues 62–565) were subcloned into pDEST17 containing an N-terminal His6 tag using the Gateway cloning system. For bacterial purification of FLAG (Flag)-tagged unique Middle region of dNedd4Lo, the PCR-amplified DNA fragment corresponding to the dNedd4Lo Middle region (NCBI accession number NP_001137965, residues 315–474) was subcloned into pDEST14 using the Gateway cloning system. An N-terminal FLAG tag was then inserted using the QuikChange Site-directed Mutagenesis kit, as above. Generation of GST-tagged N-terminal and Middle regions of dNedd4Lo for bacterial expression, and FLAG-tagged dNedd4S and dNedd4Lo for S2 cell expression, were previously described (9, 18).
Preparation of S2 cells for immunofluorescent staining
S2 cells were grown in Schneider's media (Life Technologies) supplemented with 10% fetal bovine serum and 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 25 μg/ml of amphotericin B (Wisent Bioproducts, Quebec, CA). The cells were seeded into 6-well plates (1.0 × 106 cells/well), grown overnight, and then transiently transfected with 0.4 μg of FLAG-dNedd4Lo or FLAG-dNedd4Lo Cys → Ala using Effectene (Qiagen, Germantown, MD). 24 h after transfection, protein expression was induced with 500 μm CuSO4 for 24 h. Cells were then seeded onto coverslips coated in 50 μg/ml of concanavalin A (Sigma) and allowed to adhere for 2 h. Coverslips were then washed in 1× PBS and fixed in 2% paraformaldehyde (PFA) for 15 min at room temperature, followed by washing 3 times in 1× PBS. Cells were then permeablized with 0.1% saponin, blocked in 2% normal goat serum and 2% donkey serum, and stained with primary antibodies for 1.5 h at 24 °C. Primary antibodies used include mouse anti-FLAG and rabbit anti-SH3PX1 to detect dNedd4 and SH3PX1, respectively (Table S2). Coverslips were then washed three times in 1× PBS and stained with secondary antibodies for 1 h at room temperature. Phalloidin staining was performed for the last 20 min of secondary antibody staining (Table S2). Coverslips were washed three times in 1× PBS and mounted using Dako mounting medium (Sigma) for confocal imaging. Images of cells were taken with a Zeiss objective lens set to ×40 (water imaging medium). Images were acquired using a Hamamatsu C9100-13-EM-CCD camera and were analyzed using Volocity 6.3.0 (PerkinElmer Life Sciences). The cell periphery was defined as the outer band of F-actin marked by phalloidin staining in cells. Intensity of SH3PX1 signal in FLAG-dNedd4Lo and FLAG-dNedd4Lo Cys → Ala-overexpressing cells was normalized to SH3PX1 signal in WT cells.
In vitro binding of dNedd4Lo N-term/Mid and SH3PX1
Drosophila S2 cells were transiently transfected with HA-SH3PX1 using the Effectene reagent. 24 h post-transfection, cells were lysed in lysis buffer (150 mm NaCl, 50 mm HEPES, 1% Triton X-100, 10% glycerol, 1.5 mm MgCl2, and 1.0 mm EGTA) with protease inhibitors (1 mm phenylmethanesulfonyl fluoride, 1 μg/ml of each aprotinin, leupeptin, and pepstatin A). 2 μg of GST alone, GST-N-term, or GST-Mid conjugated to anti-GST–agarose beads was added to 1 mg of cleared cell lysate for 1.5 h at 4 °C. Beads were washed four times in HNTG solution (20 mm HEPES, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100), proteins were separated on SDS-PAGE and transferred onto nitrocellulose for immunoblotting. Immunoblotting was performed using anti-HA and anti-GST antibodies to detect SH3PX1 and dNedd4Lo N-term/Mid, respectively (Table S2). All blots were imaged using the Odyssey Imaging system.
Co-IP of dNedd4Lo and SH3PX1 mutants in S2 cells
S2 cells were transiently transfected with 0.4 μg of FLAG-dNedd4S, FLAG-dNedd4Lo, FLAG-dNedd4Lo (AEAAGVA) or FLAG-dNedd4Lo (AANTA) mutants and 0.4 μg of HA-SH3PX1, HA-SH3PX1-ΔSH3, HA-SH3PX1-Pro → Ala or HA-SH3PX1-ΔSH3/Pro → Ala, where indicated, using Effectene and proteins expression was induced as above. Cells were lysed in lysis buffer with protease inhibitors as above. For co-IP of dNedd4Lo to SH3PX1 mutants, 10 μl of M2 anti-FLAG–agarose beads (Sigma) were added to 1 mg of cleared lysate for 2 h at 4 °C to IP FLAG-dNedd4S or FLAG-dNedd4Lo. For co-IP of dNedd4Lo proline mutants to SH3PX1, 10 μl of protein-G beads coupled with anti-HA antibodies were added to 1 mg of cleared lysate for 2 h at 4 °C to IP HA-SH3PX1. Beads were washed three times in HNTG solution, proteins were separated on SDS-PAGE and transferred onto nitrocellulose for immunoblotting. Immunoblotting was performed using anti-HA and anti-FLAG antibodies to detect SH3PX1 and FLAG-dNedd4 (short or long), respectively (Table S2), and imaged as above. All blots were quantified using Image Studio. For all co-IP experiments, binding was normalized to both the amount of SH3PX1 present in the cell lysate and the amount of dNedd4 that was immunoprecipitated.
In vitro binding of bacterially-purified SH3PX1 and dNedd4Lo-Middle
Bacterially-expressed and purified FLAG-dNedd4Lo-Mid (conjugated to M2 anti-FLAG–agarose beads) was incubated with bacterially-expressed and purified His-SH3PX1-WT or His-SH3PX1-ΔSH3 in 1× PBS + 10% Triton X-100 for 1 h at 4 °C. The beads were then spun down and washed four times with HNTG solution followed by three times with 1× PBS, and proteins were separated on SDS-PAGE, transferred onto nitrocellulose, immunoblotted with anti-His and anti-FLAG antibodies to detect SH3PX1 and dNedd4Lo-Mid, respectively (Table S2), and imaged as above.
Third instar larval fillet preparations and immunofluorescent staining of NMJs and muscles
Wandering third instar larvae were dissected in 1× PBS using a standard fillet preparation protocol (32). Larval fillets were fixed in 4% PFA for 20 min, washed three times with 1× PBT (0.1% Tween 20 in 1× PBS), and blocked overnight in 1× PBT containing 2% normal goat serum and 2% donkey serum at 4 °C. They were then incubated with primary antibody at 24 °C for 1.5 h, washed three times for 10 min in 1× PBT, and stained with secondary antibody at 24 °C for 2 h. Phalloidin staining to visualize muscle F-actin was performed for 20 min at the end of the secondary antibody staining. Larvae were washed for another three times for 10 min in 1× PBT and mounted onto coverglass with Dako mounting medium (Sigma) for confocal imaging.
Images of NMJs at muscle 6/7 were taken using the Zeiss AxioVert 200M confocal microscope with a Zeiss objective lens set to ×63. Images of SNb innervation patterns at muscle 12/13 were taken with a Zeiss objective lens set to ×40. Images were acquired using a Hamamatsu C9100–13-EM-CCD camera and were analyzed and deconvolved using Volocity 6.3.0 (PerkinElmer Life Sciences).
For analysis of post-synaptic SH3PX1 levels at the SSR, third instar larvae were stained with mouse anti-Dlg (post-synaptic marker) followed by Alexa Fluor 488-conjugated goat anti-mouse antibody, and rabbit anti-SH3PX1 followed by Cy3-conjugated donkey anti-rabbit antibody (Table S2). For analysis of pre-synaptic SH3PX1 levels, larvae were stained with mouse anti-Dlg followed by Alexa Fluor 488-conjugated goat anti-mouse antibody, rabbit anti-SH3PX1 followed by Cy3-conjugated donkey anti-rabbit antibody, or Alexa Fluor 647-conjucated anti-HRP (pre-synaptic marker). For quantification, the intensity of the SH3PX1 signal in the SH3PX1 mutant line was subtracted from the intensity of SH3PX1 in each measured sample to eliminate background. SH3PX1 intensity for each genotype was then normalized to that of 24B-Gal4 driver control lines. Crosses performed for these experiments are depicted in Table S1. For analysis of SNb innervation at muscle 13/12, larvae were stained with phalloidin (to visualize muscle F-actin) and Cy3-conjugated anti-HRP (Table S2).
Analysis of SH3PX1 protein levels in muscles of dNedd4Lo-overexpressing larvae
Ten 3rd instar larvae of each genotype: WT, SH3PX1 homozygous mutant, and overexpressing FLAG-tagged dNedd4S, dNedd4Lo, or dNedd4Lo Cys → Ala with the 24B-Gal4 driver, were dissected in 1× PBS on ice to isolate muscle tissue using a standard fillet preparation protocol (32). Larval fillets were homogenized on ice in standard lysis buffer with protease inhibitors, as above, 40 μm MG-132 (Selleck Chemicals, Houston, TX) and 0.2 mm chloroquine (Bioshop, Burlington, CA), and incubated on ice for 10 min. 30 μg of cleared lysate was then separated on SDS-PAGE, transferred onto nitrocellulose, and immunoblotted with anti-FLAG and anti-SH3PX1 antibodies to detect FLAG-dNedd4 and SH3PX1, respectively (Table S2).
Larval locomotion
The following genotypes were used to analyze larval locomotion: 1) Drosophila SH3PX1 heterozygous SH3PX1C1/+; 2) Drosophila SH3PX1 homozygous SH3PX1C1/SH3PX1C1 (null mutant); 3) the muscle-specific driver line 24B-Gal4; 4) overexpression of dNedd4Lo at the post-synapse and in muscles, UAS-dNedd4Lo;24B-Gal4; and 5) overexpression of dNedd4Lo in muscles in Drosophila SH3PX1 heterozygous background, UAS-dNedd4Lo/SH3PX1C1;24B-Gal4.
All behavioral experiments were performed at a 4-h time window (between 15.00 and 19.00 h) in an environmentally controlled room (40–45% relative humidity, 25 °C). Each wandering third instar larva was taken out of the wall of the vial, washed briefly with distilled water, and gently transferred using a paintbrush to the center of a 92 × 16-mm Petri dish plate (Sarstedt, Numbrecht, Germany) containing 3% agarose. The plate was placed on a cold-light X-ray film illuminator (model 140001; S&S X-Ray Products) with a homogeneous emission. A color camera (EverFocus EQ610, Polistar II) was fitted with a CCTV lens (Computar, Vari Focal TG4Z2813 FCS-IR) and fixed on a mounting bracket ∼50 cm above the plate. The distance of the camera to the object, zoom, and focus and iris aperture were optimized for video tracking. The path of freely crowding larvae within the arena was tracked during 2 min with Ethovioson XT, version 10 (Noldus Information Technology, Leesburg, VA). Video recording was started right after the first full wave of contraction was observed. All video recordings obtained from video tracking were saved as MPEG movies.
Transmission electron microscopy
Third instar larvae were dissected and fixed in 0.1 m sodium cacodylate buffer containing 2% PFA and 2.5% glutaraldehyde. Samples were washed in 0.1 m sodium cacodylate buffer with 0.2 m sucrose, and post-fixed in 1% osmium tetroxide for 1.5 h at room temperature. Samples were again washed in 0.1 m sodium cacodylate buffer with 0.2 m sucrose, and then subjected to ethanol dehydration. Segments A2 to A4 were isolated from larval samples, and were infiltrated with propylene oxide and embedded in Quetol-Spurr resin at 65 °C. Trimmed blocks were sectioned on a Leica Ultracut RMC MT6000 ultramicrotome. Ultrathin sections were collected on copper mesh grids and stained with lead citrate and uranyl acetate. Images were collected using a Tecnai 20 transmission electron microscope.
Electrophysiology
Wandering third instar larvae were dissected in hemolymph-like solution 3 (HL3) saline containing 1.0 mm Ca2+ (33). Recordings were taken using an AxoClamp 2B amplifier (Axon Instruments, Burlingame, CA). A recording electrode was filled with 3 m KCl and inserted into muscle 6 at abdominal segments A3 or A4. Resting membrane potentials were between −50 and −70 mV and were not different between genotypes. A stimulating electrode filled with saline was used to stimulate the severed segmental nerve. Miniature excitatory junctional potentials (mEJPs; minis) were recorded for 2 min and 16 nerve-evoked potentials (EJPs) were recorded at 1 Hz. Muscle resistance was assessed in each muscle and was not different between genotypes. Analyses were performed using Clampfit 10.0 software (Molecular Devices, Sunnyvale, CA).
Statistical analysis
All immunoblotting and immunofluorescence experiments were analyzed using a Student's t test. Histograms and error bars represent mean ± S.E. Innervation experiments were analyzed with Fisher's exact test in a 2 × 2 contingency table. Electrophysiology and larval locomotion experiments were analyzed using one-way ANOVA followed by Tukey's post hoc multiple comparisons. For all experiments, a p value of <0.05 was considered significant.
Author contributions
S. S. W., A. S.-K., K. H., K. G. I., A. P., Y. Zhong, and Y. Zhang data curation; S. S. W., A. S.-K., K. H., K. G. I., Y. Zhang, and D. R. formal analysis; S. S. W., K. H., K. G. I., A. P., G. L. B., and B. S. investigation; S. S. W. and K. H. methodology; S. S. W. and A. S.-K. writing-original draft; A. S.-K. and D. R. conceptualization; A. S.-K., X. F., G. L. B., B. S., and D. R. supervision; Y. Zhong resources; D. R. funding acquisition; D. R. project administration; D. R. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Drs. K. O'Connor-Giles for SH3PX1 mutant flies, and Jack Dixon for SH3PX1 antibodies.
This work was supported by a Ontario Graduate Scholarship and RESTRACOMP studentship (to S. W.), and NSERC Discovery Grants RGPIN-2017-06475 (to D. R.), RGPIN 250078-12 (to B. S.), and RGPIN05229 and RGPAS462171 (to G. L. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1–S2.
- NMS
- neuromuscular synaptogenesis
- NMJ
- neuromuscular junction
- dNedd4S
- dNedd4-short
- dNedd4Lo
- dNedd4-long
- Comm
- Commissureless
- GST
- glutathione S-transferase
- dAmph
- amphiphysin
- PX
- PHOX homology
- SH3
- Src homology
- SSR
- subsynaptic reticulum
- IP
- immunoprecipitate
- EJP
- excitatory junction potential
- HRP
- horseradish peroxidase
- HA
- hemagglutinin
- PFA
- paraformaldehyde
- mEJP
- miniature EJP
- ANOVA
- analysis of variance
- M1
- Mander's 1 correlation coefficient
- SNb
- segmental nerve b.
References
- 1. Johansen J., Halpern M. E., and Keshishian H. (1989) Axonal guidance and the development of muscle fiber-specific innervation in Drosophila embryos. J. Neurosci. 9, 4318–4332 10.1523/JNEUROSCI.09-12-04318.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Broadie K. S., and Bate M. (1993) Development of the embryonic neuromuscular synapse of Drosophila melanogaster. J. Neurosci. 13, 144–166 10.1523/JNEUROSCI.13-01-00144.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keshishian H., Chiba A., Chang T. N., Halfon M. S., Harkins E. W., Jarecki J., Wang L., Anderson M., Cash S., and Halpern M. E. (1993) Cellular mechanisms governing synaptic development in Drosophila melanogaster. J. Neurobiol. 24, 757–787 10.1002/neu.480240606 [DOI] [PubMed] [Google Scholar]
- 4. Menon K. P., Carrillo R. A., and Zinn K. (2013) Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip, Rev. Dev. Biol. 2, 647–670 10.1002/wdev.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCabe B. D., Hom S., Aberle H., Fetter R. D., Marques G., Haerry T. E., Wan H., O'Connor M. B., Goodman C. S., and Haghighi A. P. (2004) Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron 41, 891–905 10.1016/S0896-6273(04)00073-X [DOI] [PubMed] [Google Scholar]
- 6. Wan H. I., DiAntonio A., Fetter R. D., Bergstrom K., Strauss R., and Goodman C. S. (2000) Highwire regulates synaptic growth in Drosophila. Neuron 26, 313–329 10.1016/S0896-6273(00)81166-6 [DOI] [PubMed] [Google Scholar]
- 7. Glickman M. H., and Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 10.1152/physrev.00027.2001 [DOI] [PubMed] [Google Scholar]
- 8. Ing B., Shteiman-Kotler A., Castelli M., Henry P., Pak Y., Stewart B., Boulianne G. L., and Rotin D. (2007) Regulation of Commissureless by the ubiquitin ligase DNedd4 is required for neuromuscular synaptogenesis in Drosophila melanogaster. Mol. Cell Biol. 27, 481–496 10.1128/MCB.00463-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhong Y., Shtineman-Kotler A., Nguyen L., Iliadi K. G., Boulianne G. L., and Rotin D. (2011) A splice isoform of DNedd4, DNedd4-long, negatively regulates neuromuscular synaptogenesis and viability in Drosophila. PLoS ONE 6, e27007 10.1371/journal.pone.0027007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunn R., Klos D. A., Adler A. S., and Hicke L. (2004) The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 165, 135–144 10.1083/jcb.200309026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plant P. J., Lafont F., Lecat S., Verkade P., Simons K., and Rotin D. (2000) Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J. Cell Biol. 149, 1473–1484 10.1083/jcb.149.7.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Persaud A., Alberts P., Mari S., Tong J., Murchie R., Maspero E., Safi F., Moran M. F., Polo S., and Rotin D. (2014) Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci. Signal. 7, ra95 10.1126/scisignal.2005290 [DOI] [PubMed] [Google Scholar]
- 13. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., and Forman-Kay J. D. (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 10.1016/j.cell.2007.06.050 [DOI] [PubMed] [Google Scholar]
- 14. Kanelis V., Bruce M. C., Skrynnikov N. R., Rotin D., and Forman-Kay J. D. (2006) Structural determinants for high-affinity binding in a Nedd4 WW3* domain-Comm PY motif complex. Structure 14, 543–553 10.1016/j.str.2005.11.018 [DOI] [PubMed] [Google Scholar]
- 15. Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., and Rotin D. (1996) WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15, 2371–2380 10.1002/j.1460-2075.1996.tb00593.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huibregtse J. M., Scheffner M., Beaudenon S., and Howley P. M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563–2567 10.1073/pnas.92.7.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rotin D., and Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- 18. Safi F., Shteiman-Kotler A., Zhong Y., Iliadi K. G., Boulianne G. L., and Rotin D. (2016) Drosophila Nedd4-long reduces Amphiphysin levels in muscles and leads to impaired T-tubule formation. Mol. Biol. Cell 27, 907–918 10.1091/mbc.E15-06-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lundmark R., and Carlsson S. R. (2003) Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J. Biol. Chem. 278, 46772–46781 10.1074/jbc.M307334200 [DOI] [PubMed] [Google Scholar]
- 20. Pylypenko O., Lundmark R., Rasmuson E., Carlsson S. R., and Rak A. (2007) The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 26, 4788–4800 10.1038/sj.emboj.7601889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teasdale R. D., and Collins B. M. (2012) Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem. J. 441, 39–59 10.1042/BJ20111226 [DOI] [PubMed] [Google Scholar]
- 22. Yarar D., Waterman-Storer C. M., and Schmid S. L. (2007) SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev. Cell 13, 43–56 10.1016/j.devcel.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 23. Worby C. A., Simonson-Leff N., Clemens J. C., Kruger R. P., Muda M., and Dixon J. E. (2001) The sorting nexin, DSH3PX1, connects the axonal guidance receptor, Dscam, to the actin cytoskeleton. J. Biol. Chem. 276, 41782–41789 10.1074/jbc.M107080200 [DOI] [PubMed] [Google Scholar]
- 24. Shin N., Lee S., Ahn N., Kim S. A., Ahn S. G., YongPark Z., and Chang S. (2007) Sorting nexin 9 interacts with dynamin 1 and N-WASP and coordinates synaptic vesicle endocytosis. J. Biol. Chem. 282, 28939–28950 10.1074/jbc.M700283200 [DOI] [PubMed] [Google Scholar]
- 25. Ukken F. P., Bruckner J. J., Weir K. L., Hope S. J., Sison S. L., Birschbach R. M., Hicks L., Taylor K. L., Dent E. W., Gonsalvez G. B., and O'Connor-Giles K. M. (2016) BAR-SH3 sorting nexins are conserved interacting proteins of Nervous wreck that organize synapses and promote neurotransmission. J. Cell Sci. 129, 166–177 10.1242/jcs.178699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng S., Chen J. K., Yu H., Simon J. A., and Schreiber S. L. (1994) Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science 266, 1241–1247 10.1126/science.7526465 [DOI] [PubMed] [Google Scholar]
- 27. Hicks L., Liu G., Ukken F. P., Lu S., Bollinger K. E., O'Connor-Giles K., and Gonsalvez G. B. (2015) Depletion or over-expression of Sh3px1 results in dramatic changes in cell morphology. Biol. Open 4, 1448–1461 10.1242/bio.013755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frank C. A. (2014) Homeostatic plasticity at the Drosophila neuromuscular junction. Neuropharmacology 78, 63–74 10.1016/j.neuropharm.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baumann C., Lindholm C. K., Rimoldi D., and Lévy F. (2010) The E3 ubiquitin ligase Itch regulates sorting nexin 9 through an unconventional substrate recognition domain. FEBS J. 277, 2803–2814 10.1111/j.1742-4658.2010.07698.x [DOI] [PubMed] [Google Scholar]
- 30. Sakata T., Sakaguchi H., Tsuda L., Higashitani A., Aigaki T., Matsuno K., and Hayashi S. (2004) Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 14, 2228–2236 10.1016/j.cub.2004.12.028 [DOI] [PubMed] [Google Scholar]
- 31. Worby C. A., Simonson-Leff N., Clemens J. C., Huddler D. Jr, Muda M., and Dixon J. E. (2002) Drosophila Ack targets its substrate, the sorting nexin DSH3PX1, to a protein complex involved in axonal guidance. J. Biol. Chem. 277, 9422–9428 10.1074/jbc.M110172200 [DOI] [PubMed] [Google Scholar]
- 32. Brent J. R., Werner K. M., and McCabe B. D. (2009) Drosophila larval NMJ dissection. J. Vis. Exp. p1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stewart B. A., Atwood H. L., Renger J. J., Wang J., and Wu C. F. (1994) Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A 175, 179–191 10.1007/BF00215114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.