Abstract
This JBC Review on the discoveries of yeast phosphatidate (PA) phosphatase genes is dedicated to Dr. Herbert Tabor, Editor-in-Chief of the Journal of Biological Chemistry (JBC) for 40 years, on the occasion of his 100th birthday. Here, I reflect on the discoveries of the APP1, DPP1, LPP1, and PAH1 genes encoding all the PA phosphatase enzymes in yeast. PA phosphatase catalyzes PA dephosphorylation to generate diacylglycerol; both substrate and product are key intermediates in the synthesis of membrane phospholipids and triacylglycerol. App1 and Pah1 are peripheral membrane proteins catalyzing an Mg2+-dependent reaction governed by the DXDX(T/V) phosphatase motif. Dpp1 and Lpp1 are integral membrane proteins that catalyze an Mg2+-independent reaction governed by the KX6RP–PSGH–SRX5HX3D phosphatase motif. Pah1 is PA-specific and is the only PA phosphatase responsible for lipid synthesis at the nuclear/endoplasmic reticulum membrane. App1, Dpp1, and Lpp1, respectively, are localized to cortical actin patches and the vacuole and Golgi membranes; they utilize several lipid phosphate substrates, including PA, lyso-PA, and diacylglycerol pyrophosphate. App1 is postulated to be involved in endocytosis, whereas Dpp1 and Lpp1 may be involved in lipid signaling. Pah1 is the yeast lipin homolog of mice and humans. A host of cellular defects and lipid-based diseases associated with loss or overexpression of PA phosphatase in yeast, mice, and humans, highlights its importance to cell physiology.
Keywords: phosphatidic acid, diacylglycerol, triacylglycerol, phosphatase, lipid, phosphatidate, Saccharomyces cerevisiae, yeast, lipid metabolism, APP1, DPP1, LPP1, PA phosphatase, PAH1
Herbert Tabor and my association with the Journal of Biological Chemistry (JBC)
This JBC Review is a reflection on my association with Dr. Herbert Tabor and the JBC, and the discoveries of the PA2 phosphatase genes in yeast. I dedicate this JBC Review to Herb on the occasion of his 100th birthday. I also express my esteem and gratitude to Herb for the support he has given me as an author, reviewer, board editor, and associate editor of the JBC.
The JBC is the venue for the seminal work on the biochemistry and molecular biology of membrane lipids. William Dowhan (postdoc advisor) and Eugene P. Kennedy (Dowhan's postdoc advisor) published their best work in the JBC, and emulating them, my best work is also published here. My first JBC paper (postdoctoral studies) was on the interfacial kinetics of the phosphatidylserine synthase of Escherichia coli (1), and as an independent investigator, my first JBC paper was on the purification and characterization of the yeast phosphatidylserine synthase (2). The reviewing editors of these papers were thorough indeed, and I learned that the conclusions of our work had to be rigorously supported by sound experimental approaches. This has become a guiding principle of my research and mentoring.
As my career progressed, I aspired to become associated with the JBC, just like the icons in my field (e.g. Eugene P. Kennedy, William Dowhan, Susan A. Henry, Edward A. Dennis, Christian R. H. Raetz, Dennis E. Vance, and Robert M. Bell) who were either members of the Editorial Board or Associate Editors. It was during the late 1980s when I received an invitation from Herb to review a lipid enzymology paper. Of course, I accepted the invitation, and this began my association with the journal as a reviewer. I had continued to receive invitations to review papers for the journal, primarily from Associate Editor Robert M. Bell. After “paying my dues” as a reviewer, I was appointed to the editorial board for a 5-year term in 1992 and then again in 1998. The associate editors that I worked closely with were Robert M. Bell, Stephen M. Prescott, Claudia Kent, and William L. Smith. I reviewed ∼6 papers per month and submitted my reviews within a few days. I was rewarded for this service by being appointed associate editor in 2006. Then the real work began; Herb was assigning me 30–40 papers a month. Fortunately, the JBC has an amazing editorial board whose members are dedicated to the journal. Making an editorial decision on the good and the bad manuscripts is relatively easy, but it is those papers that fall in the “gray zone” that make decisions difficult. I have tried to follow Herb's guiding principle to give the benefit of doubt to the authors with the opportunity to make their science better and thus worthy of publication in the JBC. A few years ago, Herb stepped down as Editor-in-Chief, but he still assigns manuscripts as an emeritus editor of the journal. I am pleased to work closely with Herb on the “Classics and Reflections” committee.
Why we study PA phosphatase
Research in my laboratory utilizes biochemical and molecular genetics approaches to study the regulation of lipid synthesis in the yeast Saccharomyces cerevisiae (3–6). The purification and characterization of lipid synthesis enzymes and the molecular characterization of lipid synthesis regulation are facilitated in yeast because of their genetic tractability and ease of growth. Importantly, our work with yeast has proven to be relevant to the mechanisms that regulate lipid synthesis in humans (5–7).
Several papers published in the JBC by Eugene P. Kennedy (8–18), William Dowhan (19–24), Christian R. H. Raetz (25–28), Edward A. Dennis (29–33), and Robert M. Bell (34–39) have had a major influence on the way I think about science. Their work has provided me with a framework and high standard for performing well-designed experiments to address basic questions on the enzymology and regulation of lipid metabolism.
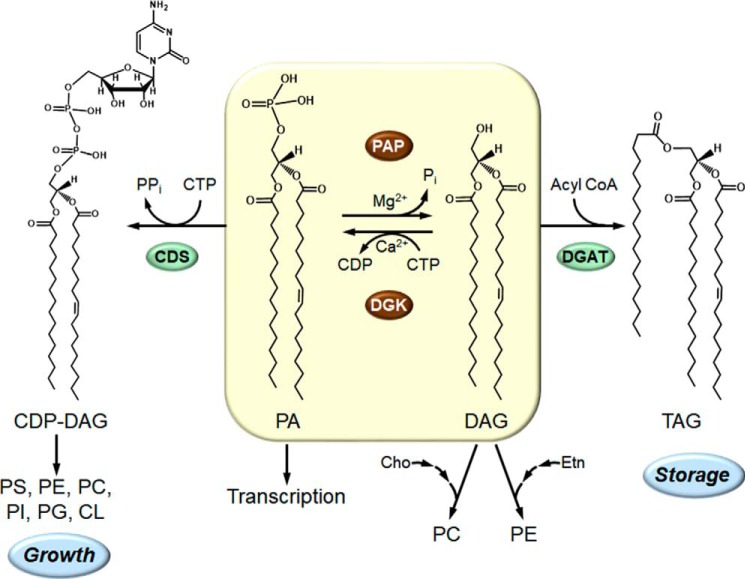
Studies from several laboratories, primarily that of Susan A. Henry, have indicated that a key step in the regulation in lipid synthesis occurs at the point where PA is bifurcated to CDP–DAG and DAG (Fig. 1). CDP–DAG is utilized for the synthesis of all membrane phospholipids, whereas DAG is primarily used for the synthesis of TAG (Fig. 1) (reviewed in Refs. 4, 6, 40, 41). The DAG is also used to synthesize phosphatidylcholine or phosphatidylethanolamine via the Kennedy pathway when cells are supplemented with choline or ethanolamine (Fig. 1). Moreover, the PA molecule itself regulates the expression of phospholipid synthesis genes by controlling the cellular location of the transcriptional repressor protein Opi1 (42), a key component of the Henry regulatory circuit (4, 6). The microsomal CDP–DAG synthase, which utilizes PA as a substrate to form CDP–DAG (43), is not a highly-regulated enzyme, and thus, the important regulator of PA utilization is the other branch point enzyme PA phosphatase (Fig. 1). By the nature of its reaction, PA phosphatase controls the synthesis of TAG and membrane phospholipids and the abundance of the important signaling molecule PA (4, 44). The reaction of the enzyme, namely the dephosphorylation of PA to generate DAG (Fig. 1), was first characterized by Kennedy and co-workers in 1957 (10). In 1987, when we initiated our studies on PA phosphatase, the enzyme had not been purified; the identity of the gene encoding the enzyme was unknown; and no mutants were available. How the PA phosphatase genes in yeast were discovered is discussed in the remainder of this JBC Review.
Figure 1.
Central role of the PA phosphatase in the synthesis of TAG and membrane phospholipids in yeast. The structures of CDP–DAG, PA, DAG, and TAG are shown with fatty acyl groups of 16 and 18 carbons with and without a single double-bond where indicated. The PA phosphatase (PAP) plays a major role in governing whether cells utilize PA for the synthesis of TAG via DAG or whether they utilize PA for the synthesis of membrane phospholipids via CDP–DAG. The PA phosphatase reaction is counterbalanced by the CTP-dependent DAG kinase (DGK) reaction. When the CDP–DAG pathway for phospholipid synthesis is blocked, phosphatidylcholine and phosphatidylethanolamine may be synthesized from the DAG derived from the PA phosphatase reaction when cells are supplemented with choline and ethanolamine, respectively, via the CDP-choline and CDP-ethanolamine branches of the Kennedy pathway. In addition to its role in lipid synthesis, PA signals the transcriptional regulation of phospholipid synthesis genes via the Henry (Opi1/Ino2/Ino4) regulatory circuit. More comprehensive pathways of lipid synthesis, along with details of the Henry regulatory circuit, may be found in Refs. 5, 6. CDS, CDP–DAG synthase; DGAT, DAG acyltransferase; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol; PG, phosphatidylglycerol; CL, cardiolipin; Cho, choline; Etn, ethanolamine.
Identification of the DPP1 and LPP1 genes
In 1989, my graduate student Yi-Ping Lin successfully purified PA phosphatase from a total membrane fraction of yeast (45). The eight-step procedure developed by Yi-Ping yielded a 91-kDa protein that was used to carry out enzymological and kinetic studies of the enzyme (45, 46). Another graduate student, Wen-I Wu, joined the laboratory; her project was to purify a sufficient quantity of the 91-kDa enzyme to obtain amino acid sequence information to be used in a reverse-genetics approach to isolate the PA phosphatase gene. To improve the purification, Wen-I substituted the detergent Triton X-100 for sodium cholate and made changes to the column resins and chromatography conditions. The result was a purified protein with a molecular mass of 34 kDa, not the 91-kDa protein that Yi-Ping had purified. Moreover, PA phosphatase activity in this preparation was not dependent on Mg2+ ions as it was for the PA phosphatase purified by Yi-Ping (45). The pH optima of the two PA phosphatase preparations also differed. I suggested to Wen-I that she abandon her modified scheme to purify PA phosphatase and use the established procedure developed by Yi-Ping. Wen-I complied with my suggestion and used the 91-kDa enzyme to study the regulation of PA phosphatase activity by phospholipids (47), sphingolipids (48), and nucleotides (49). Unfortunately, we did not obtain unambiguous sequence information for the 91-kDa protein to synthesize a well-defined oligonucleotide probe to “fish out” the PA phosphatase gene from a genomic library.
In the meantime, Wissing and Behrbohm (50, 51) had just discovered a novel PA kinase in plants that converts PA to DGPP. This novel phospholipid, which contains a pyrophosphate group attached to DAG, is a metabolite of PA that is synthesized during G-protein activation in response to stress (52). In a collaboration with Wissing, we discovered that a PA phosphatase activity in plants, yeast, bacteria, and mammals could dephosphorylate DGPP (53). This activity has an acidic pH optimum and does not require Mg2+ ions like the PA phosphatase activity associated with the 34-kDa protein that Wen-I Wu had purified from yeast. Wen-I showed that the 34-kDa enzyme utilized DGPP as a substrate with a specificity constant 10-fold greater when compared with that of PA (54). Thus, we called the 34-kDa enzyme DGPP phosphatase (54). Through a kinetic analysis, we found that the enzyme first dephosphorylates DGPP to form PA, and then it dephosphorylates PA to form DAG (54). In fact, in the presence of DGPP, the enzyme does not dephosphorylate PA (54). We also found that the enzyme utilizes a variety of lipid phosphate substrates that include DGPP, PA, lyso-PA, ceramide phosphate, sphingosine phosphate, farnesyl pyrophosphate, geranylgeranyl pyrophosphate, dolichyl pyrophosphate, and dolichyl phosphate (54–56). In contrast, the 91-kDa PA phosphatase is specific for PA (45, 54, 57).
We had obtained unambiguous sequences in the 34-kDa protein by Edman degradation (58). At this time, the yeast genome was supposedly sequenced, and the gene encoding the enzyme should have been in the database. However, this was not the case. It was suggested by the curators of the Saccharomyces database that perhaps we had not purified a yeast protein. So, to be sure we had not purified DGPP phosphatase from an organism other than S. cerevisiae, we purified the enzyme again and obtained the same protein sequences. We had heard through the “grapevine” that several bona fide yeast genes were not in the database. Eventually, however, all the genes were deposited in the database, and the gene (DPP1 for diacylglycerol pyrophosphate phosphatase) was identified (58). Cloning and expression studies, fronted by David Toke (postdoctoral associate), along with a biochemical analysis confirmed the DPP1–DGPP phosphatase relationship (58). Shortly after publishing the identification of the DPP1 gene, it was shown to be one of the most highly expressed genes in response to zinc deprivation (59, 60).
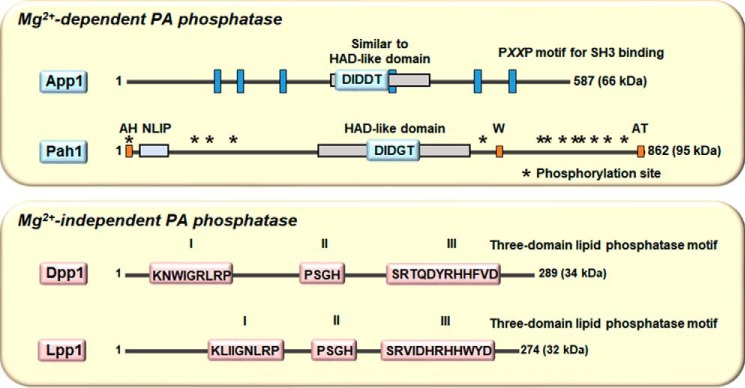
During this time, Joseph Stukey (postdoctoral associate) discovered a novel phosphatase sequence motif through a bioinformatics analysis using the E. coli pgpB-encoded phosphatidylglycerophosphate phosphatase as the query (61). The motif consists of three domains with the consensus sequences KX6RP (domain 1)–PSGH (domain 2)–SRX5HX3D (domain 3) (61). Dpp1 was shown to contain this phosphatase motif (Fig. 2) (58), and subsequently, the pgpB-encoded protein was shown to exhibit DGPP phosphatase activity (62).
Figure 2.
Mg2+-dependent and -independent PA phosphatase enzymes in yeast have distinct catalytic motifs. The diagrams show linear representations of App1 and Pah1 (upper panel) and Dpp1 and Lpp1 (lower panel). The Mg2+-dependent PA phosphatase activities of App1 and Pah1 are governed by the DXDX(T/V) motifs in the HAD-like domain, whereas the Mg2+-independent PA phosphatase activities of Dpp1 and Lpp1 are governed by the three-domain phosphatase motif. For Pah1, the approximate positions are indicated for the amphipathic helix (AH) required for ER membrane interaction (77), the NLIP and HAD-like domains that are required for PA phosphatase activity (116), the acidic tail (AT) required for interaction with Spo7 of the Pah1 phosphatase (70, 77, 80), the tryptophan (W) residue within the C-terminal conserved sequence WRDPLVDID required for Pah1 function (117), and the sites phosphorylated by Pho85–Pho80, Cdc28–cyclin B, protein kinase A, protein kinase C, and casein kinase II that regulate the location, activity, and proteasomal degradation of Pah1 (76, 78, 79, 83, 84, 118).
A search of the protein database using Dpp1 as the query identified a closely related protein with a subunit size of 32 kDa that contains the novel phosphatase sequence motif (Fig. 2) (63). We named the gene encoding this protein LPP1 for lipid phosphate phosphatase because, like Dpp1, Lpp1 utilizes a variety of lipid phosphate substrates (e.g. PA, lyso-PA, and DGPP) (63). Orthologs of Dpp1 and Lpp1 in mammalian cells, named lipid phosphate phosphatases, also exhibit a broad substrate specificity and play roles in lipid signaling (64–66). A series of cloning, expression, and biochemical studies confirmed the LPP1–lipid phosphate phosphatase relationship (63). We constructed a mutant that lacks both the DPP1 and LPP1 genes; the analysis of PA phosphatase activity in the mutant indicated that other PA phosphatase genes exist in yeast, including the gene coding for the 91-kDa enzyme (Fig. 3A) (63, 67).
Figure 3.
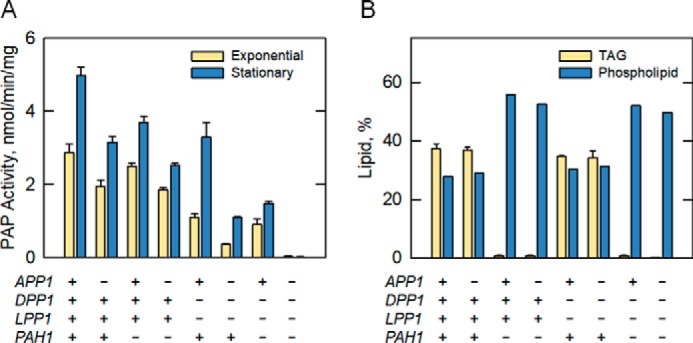
Contributions of the APP1, DPP1, LPP1, and PAH1 genes to the PA phosphatase activity and lipid content in yeast. Cells expressing or lacking the indicated gene (+ and −, respectively) were examined for PA phosphatase (PAP) activity (A) or for lipid (TAG and phospholipids) content (B). The PA phosphatase in yeast cells, which is elevated in the stationary phase, is contributed by the APP1, DPP1, LPP1, and PAH1 genes. The quadruple mutant with deletions in all four genes lacks detectable PA phosphatase activity. Cells with the deletion of PAH1 exhibit a massive reduction in TAG content. The data were taken from Ref. 67.
Identification of the PAH1 gene
In the summer of 2005, we were cleaning the −80 °C freezer in the laboratory. Gil-Soo Han, a postdoctoral associate, found some Mono Q chromatography fractions used for the purification of the 91-kDa PA phosphatase that were placed there in 1993 by Wen-I Wu. According to Wen-I's laboratory notebook, some of the fractions had PA phosphatase activity, whereas others did not have activity. Gil-Soo subjected the fractions to SDS-PAGE and was able to match up the enrichment of the 91-kDa protein with the elution profile of PA phosphatase activity. A gel slice containing the 91-kDa protein was subjected to trypsin digestion followed by amino acid sequence analysis of peptide fragments by MS (a more sensitive method to obtain protein sequence when compared with Edman degradation). Unambiguous sequence information obtained from 23 peptides matched perfectly with the deduced amino acid sequence of the SMP2 gene (57). SMP2 was originally identified as a gene involved in plasmid maintenance and respiration (68). Later, Siniossoglou and co-workers (69) identified SMP2 as a gene whose overexpression complements the abnormal expansion of the nuclear/ER membrane that is caused by mutations (nem1Δ or spo7Δ) in the subunits of a membrane-associated protein phosphatase (70). The deletion of SMP2 was shown to cause the same membrane expansion phenotype as the protein phosphatase mutants, as well as the derepression of several phospholipid synthesis genes (69). Collectively, the data raised the suggestion that Smp2 might be a transcription factor (69). Gil-Soo went on to express and purify His6-tagged Smp2 from E. coli and showed that it possessed PA phosphatase activity; it had the same enzymological properties as the 91-kDa enzyme originally purified by Yi-Ping in 1989 (45, 57). Because the name SMP2 has no meaning in a functional sense, we renamed the gene PAH1 for phosphatidic acid phosphohydrolase. (We could not use the acronym PAP for the PA phosphatase gene because it was already being used for the poly(A) polymerase gene.) A bioinformatics analysis of Pah1 indicated that the protein contains a conserved haloacid dehalogenase (HAD)–like domain with the DXDX(T/V) motif found in a superfamily of Mg2+-dependent phosphatase enzymes with diverse substrate specificity (Fig. 2) (71, 72). To determine whether we had identified all the PA phosphatase genes in yeast, a mutant lacking DPP1, LPP1, and PAH1 was constructed and analyzed for PA phosphatase activity (57, 67). This analysis showed that there was still another PA phosphatase gene to be identified (Fig. 3A) (57, 67).
Identification of the APP1 gene
Attempts to identify the remaining PA phosphatase gene through bioinformatics and by genetic screens were unsuccessful. Accordingly, we set forth to identify the gene through the reverse genetics approach. A new graduate student, Minjung Chae, joined the laboratory and was convinced to take on the project. Minjung was told that all she had to do was purify PA phosphatase from the dpp1Δ lpp1Δ pah1Δ triple mutant, get a protein sequence, and the rest would be easy. Minjung spent about 2 years developing an eight-step procedure that did not result in the complete purification of the enzyme (67). The PA phosphatase activity in the triple mutant was relatively low (e.g. 5-fold lower when compared WT cells), and the purification was hampered by the lability of the PA phosphatase activity (67). Nonetheless, a peak of activity from the last column of the procedure correlated with the enrichment of a minor protein band upon SDS-PAGE (67). The band was excised from the SDS-polyacrylamide gel and digested with trypsin, and the resulting peptides were analyzed by LC–MS. The analysis yielded protein sequences of several proteins of unknown function, one of which we had hoped would be PA phosphatase. Based on the genetic information in the yeast database, we expressed and purified His6-tagged versions of these proteins from E. coli, but none of them exhibited PA phosphatase activity. This suggested that the enzyme in the preparation had to be very low in abundance. Accordingly, we used more sensitive LC–MS/MS to detect low abundance proteins. To our dismay, the analysis showed that the excised protein band contained 112 proteins (or proteolytic fragments thereof)! We were devastated by this result and Minjung returned home for a break.
Upon Minjung's return to the laboratory, we devised a plan to focus on the proteins with unknown function that had a phosphatase motif and proteins predicted to be involved in lipid synthesis. Cell extracts from 20 mutants in the yeast gene-knockout collection were assayed for loss in PA phosphatase activity. Of the mutants, one lacking the APP1 gene showed a small (30%) but reproducible decrease in PA phosphatase activity (67). APP1 stands for actin patch protein because App1 is a component of cortical actin patches and interacts with endocytic proteins. The molecular function of App1 had been unknown. To provide evidence that APP1 encodes a PA phosphatase, His6-tagged and protein A-tagged, respectively, versions of the App1 protein were expressed and purified from E. coli and yeast and assayed for PA phosphatase activity (67, 73). Both purified preparations of App1 exhibited PA phosphatase activity (67, 73). App1 has a subunit size of 66 kDa, and it contains the DXDX(T/V) catalytic motif in a region that is similar to the HAD-like domain in Pah1. App1 lacks the NLIP domain found in Pah1 but contains six copies of the PX2P motif for interaction with proteins with Src homology 3 domains (Fig. 2) (67). App1 is a fungus-specific PA phosphatase as no orthologs exist in mammalian cells. The analysis of the app1Δ dpp1Δ lpp1Δ pah1Δ quadruple mutant showed no PA phosphatase activity, and thus, all PA phosphatase activity in yeast is encoded by APP1, DPP1, LPP1, and PAH1 (Fig. 3A) (67).
Cellular locations and roles of the PA phosphatases
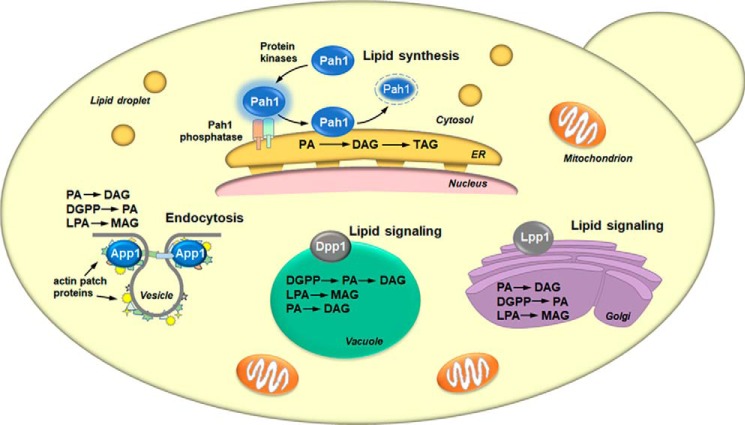
Fig. 4 depicts the cellular locations and roles of the four PA phosphatases in yeast. All PA phosphatase enzymes must associate with a membrane, the location of their phospholipid substrates. App1 and Pah1 associate with the membrane in a peripheral manner. App1 associates with actin patch proteins at sites of endocytic vesicle formation (74), whereas Pah1 associates with the nuclear/ER membrane upon its dephosphorylation by the Nem1–Spo7 protein phosphatase (e.g. Pah1 phosphatase) (69, 70, 75–82). Pah1 in the cytosol is a phosphoprotein whose phosphorylation is carried out by multiple protein kinases that include Pho85–Pho80 (78), Cdc28–cyclin B (76), protein kinase A (79), protein kinase C (83), and casein kinase II (84). Dpp1 and Lpp1, respectively, are integral membrane proteins that reside in the vacuole (85–87) and Golgi apparatus (87). The contribution of each PA phosphatase to lipid metabolism has been assessed by the analysis of mutants lacking the APP1, DPP1, LPP1, and PAH1 genes (Fig. 3B) (67). This analysis clearly shows that Pah1 is the major regulator of TAG content; the enzymes encoded by APP1, DPP1, and LPP1 have little effect on the relative amounts of TAG and membrane phospholipids. The association of App1 with cortical actin patches (74) suggests that it may regulate the local concentrations of PA and DAG through its PA phosphatase activity (67). These lipids are known to facilitate membrane fission/fusion events in model systems, and they are also known to interact with and regulate enzymes (e.g. phosphatidylinositol 4-phosphate kinase, protein kinase C, and protein kinase D) that play important roles in vesicular trafficking (88–92). Dpp1 and Lpp1, respectively, are thought to control the signaling function of PA, DAG, DGPP, and lyso-PA in vacuole and Golgi membranes (54, 58, 63, 86, 87).
Figure 4.
Cellular locations and roles of yeast PA phosphatases. The figure depicts a yeast cell with the cellular locations of the App1, Dpp1, Lpp1, and Pah1 PA phosphatase enzymes and their physiological roles. The lipid phosphate phosphatase reaction catalyzed by each enzyme is indicated in the order of substrate preference. For Pah1, the enzyme in the cytosol is phosphorylated by multiple protein kinases. The phosphorylated Pah1 (indicated by the blue halo) translocates to the ER membrane through its dephosphorylation by the Pah1 phosphatase, which is composed of Nem1 (catalytic subunit, green) and Spo7 (regulatory subunit, peach). Dephosphorylated Pah1 that is associated with the ER membrane catalyzes the conversion of PA to DAG, which is then acylated to form TAG. Dephosphorylated Pah1 is degraded by the proteasome (indicated by the dashed lines). App1 associates with actin patch proteins at cortical actin patches. The App1 enzyme is postulated to be involved in the formation of endocytic vesicles. The vesicle diagram is adapted from Ref. 74. Dpp1 and Lpp1, respectively, are integral membrane proteins in the vacuole and Golgi apparatus. They are postulated to be involved with lipid signaling by controlling the amounts of bioactive lipids.
Epilogue
Of the four PA phosphatases, our laboratory has focused more on Pah1 because of its importance to lipid metabolism and lipid-based disease. The discovery that yeast PAH1 encodes PA phosphatase has led to the identification of genes encoding the enzyme in mice (93, 94) and humans (57, 95). The protein sequence analysis of Pah1 has revealed that it is the yeast homolog of the mouse fat-regulating protein known as lipin 1 (93). In 2001, Reue and co-workers (93) coined the name lipin 1 as the product of the Lpin1 gene whose mutation was responsible for fatty liver dystrophy (fld) mice at birth (96). It has been known that loss of lipin 1 in mice causes lipodystrophy and that overexpression of lipin 1 causes obesity (93, 97). However, the molecular function of lipin 1 had not been established. Knowing that PA phosphatase was the product of yeast PAH1 and that Pah1 shares sequence homology with lipin 1 at the N-terminal NLIP and within the HAD-like (CLIP) domains (Fig. 2) led Gil-Soo Han to the hypothesis that lipin 1 is in fact a PA phosphatase (57). The expression, purification, and biochemical analysis of human lipin 1 confirmed this hypothesis (57, 95). There are three spliced variant forms of Lpin1, as well as Lpin2 and Lpin3, all of which that encode proteins with PA phosphatase activity (57, 93–95).
The importance of PA phosphatase to lipid homeostasis and cell physiology is exemplified in yeast, mice, and humans by a host of cellular defects and lipid-based diseases associated with loss or overexpression of the enzyme activity. In yeast, loss of PAH1 (SMP2, see above) results in a massive expansion of the nuclear/ER membrane (69); this is ascribed to increases in PA content and phospholipid synthesis that occur at the expense of TAG synthesis (57, 98). The increase in phospholipid synthesis is associated with the induction of phospholipid synthesis gene expression (69, 99), whereas the reduction in the synthesis of TAG is associated with a decrease in lipid droplet formation (100–102). The imbalance of lipid homeostasis in cells lacking PAH1 results in a multitude of other phenotypes that include fatty acid-induced toxicity (100), hypersensitivity to oxidative stress (103), loss in cell wall strength (104, 105), reduction in chronological life span (103), the inability to fuse vacuoles (106), the inability to degrade cellular components (e.g. autophagy) (107), and the inability to grow on nonfermentable carbon sources (57, 68) and at elevated temperatures (57, 68, 69). Some of these phenotypes are suppressed by the loss of the DGK1 gene that encodes the CTP-dependent DAG kinase (100, 101, 108). These observations emphasize the importance of maintaining a proper balance of PA and DAG to lipid metabolism and cell physiology. Lipin 1 deficiency in human and mouse causes rhabdomyolysis (109, 110), and deficiency in the mouse is also characterized by hepatic steatosis during the neonatal period causing lipodystrophy, insulin resistance, and peripheral neuropathy (93, 111). Lpin1 overexpression results in increased lipogenesis and obesity (97). Polymorphisms in the human LPIN1 gene are associated with insulin resistance and the metabolic syndrome (112). Human lipin 2 deficiency causes chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anemia (113, 114), whereas genetic variations in the human LPIN2 gene are associated with type 2 diabetes (115).
What has become clear from our work on yeast Pah1 is that too little or too much PA phosphatase activity is detrimental to cell physiology. Thus, the current focus of our work is to more fully understand the mode of action and regulation of activity with the goal of identifying effector molecules that will “fine-tune” the enzyme function.
Acknowledgments
I am grateful to the graduate students and postdoctoral associates who have worked on the PA phosphatase genes and enzymes in my laboratory. Michael Homann, a graduate student who worked on the regulation of CDP–DAG synthase, is acknowledged for the recommendation to study PA phosphatase, and Robert M. Bell is acknowledged for the suggestion to enzymatically synthesize the 32P-labeled PA substrate using the E. coli DAG kinase. I acknowledge Symeon Siniossoglou for making seminal contributions to the understanding of Pah1 and its regulation by phosphorylation and dephosphorylation and for being a great collaborator. I acknowledge Gil-Soo Han for discovering the identities of Pah1 and Dgk1 as PA phosphatase and DAG kinase enzymes, respectively, and for helping me move our research forward. Finally, I thank Herb Tabor for being the quintessential editorial mentor. Happy 100th birthday Herb!
This work was supported, in whole or in part, by National Institutes of Health Grants GM028140 and GM050679 from the USPHS. This JBC Review is part of a collection honoring Herbert Tabor on the occasion of his 100th birthday. The author declares that he has no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PA
- phosphatidate
- DAG
- diacylglycerol
- TAG
- triacylglycerol
- DGPP
- diacylglycerol pyrophosphate
- HAD
- haloacid dehalogenase
- ER
- endoplasmic reticulum.
References
- 1. Carman G. M., and Dowhan W. (1979) Phosphatidylserine synthase from Escherichia coli: the role of Triton X-100 in catalysis. J. Biol. Chem. 254, 8391–8397 [PubMed] [Google Scholar]
- 2. Bae-Lee M., and Carman G. M. (1984) Phosphatidylserine synthesis in Saccharomyces cerevisiae. Purification and characterization of membrane-associated phosphatidylserine synthase. J. Biol. Chem. 259, 10857–10862 [PubMed] [Google Scholar]
- 3. Carman G. M., and Zeimetz G. M. (1996) Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 271, 13293–13296 10.1074/jbc.271.23.13293 [DOI] [PubMed] [Google Scholar]
- 4. Carman G. M., and Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 10.1074/jbc.R700038200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carman G. M., and Han G.-S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang Y.-F., and Carman G. M. (2008) CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res. 47, 333–339 10.1016/j.plipres.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kennedy E. P., and Weiss S. B. (1956) The function of cytidine coenzyme in the biosynthesis of phospholipids. J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 9. Kennedy E. P. (1956) The synthesis of cytidine diphosphate choline, cytidine diphosphate ethanolamine, and related compounds. J. Biol. Chem. 222, 185–191 [PubMed] [Google Scholar]
- 10. Smith S. W., Weiss S. B., and Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 11. Borkenhagen L. F., and Kennedy E. P. (1957) The enzymatic synthesis of cytidine diphosphate choline. J. Biol. Chem. 227, 951–962 [PubMed] [Google Scholar]
- 12. Weiss S. B., Smith S. W., and Kennedy E. P. (1958) The enzymatic formation of lecithin from cytidine diphosphate choline and d-1,2-diglyceride. J. Biol. Chem. 231, 53–64 [PubMed] [Google Scholar]
- 13. Paulus H., and Kennedy E. P. (1960) The enzymatic synthesis of inositol monophosphatide. J. Biol. Chem. 235, 1303–1311 [PubMed] [Google Scholar]
- 14. Weiss S. B., Kennedy E. P., and Kiyasu J. Y. (1960) The enzymatic synthesis of triglycerides. J. Biol. Chem. 235, 40–44 [PubMed] [Google Scholar]
- 15. Kanfer J. N., and Kennedy E. P. (1964) Metabolism and function of bacterial lipids. II. Biosynthesis of phospholipids in Escherichia coli. J. Biol. Chem. 239, 1720–1726 [PubMed] [Google Scholar]
- 16. Colodzin M., and Kennedy E. P. (1965) Biosynthesis of diphosphoinositide in brain. J. Biol. Chem. 240, 3771–3780 [PubMed] [Google Scholar]
- 17. Chang Y. Y., and Kennedy E. P. (1967) Pathway for the synthesis of glycerolphosphatides in Escherichia coli. J. Biol. Chem. 242, 516–519 [PubMed] [Google Scholar]
- 18. Dowhan W., Wickner W. T., and Kennedy E. P. (1974) Purification and properties of phosphatidylserine decarboxylase from Escherichia coli. J. Biol. Chem. 249, 3079–3084 [PubMed] [Google Scholar]
- 19. Aitken J. F., van Heusden G. P., Temkin M., and Dowhan W. (1990) The gene encoding the phosphatidylinositol transfer protein is essential for cell growth. J. Biol. Chem. 265, 4711–4717 [PubMed] [Google Scholar]
- 20. Clancey C. J., Chang S.-C., and Dowhan W. (1993) Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268, 24580–24590 [PubMed] [Google Scholar]
- 21. Bogdanov M., and Dowhan W. (1995) Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J. Biol. Chem. 270, 732–739 10.1074/jbc.270.2.732 [DOI] [PubMed] [Google Scholar]
- 22. Chang S. C., Heacock P. N., Clancey C. J., and Dowhan W. (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9829–9836 10.1074/jbc.273.16.9829 [DOI] [PubMed] [Google Scholar]
- 23. Chang S. C., Heacock P. N., Mileykovskaya E., Voelker D. R., and Dowhan W. (1998) Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273, 14933–14941 10.1074/jbc.273.24.14933 [DOI] [PubMed] [Google Scholar]
- 24. Ostrander D. B., Zhang M., Mileykovskaya E., Rho M., and Dowhan W. (2001) Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J. Biol. Chem. 276, 25262–25272 10.1074/jbc.M103689200 [DOI] [PubMed] [Google Scholar]
- 25. Raetz C. R., and Newman K. F. (1978) Neutral lipid accumulation in the membranes of Escherichia coli mutants lacking diglyceride kinase. J. Biol. Chem. 253, 3882–3887 [PubMed] [Google Scholar]
- 26. Esko J. D., and Raetz C. R. (1980) Mutants of Chinese hamster ovary cells with altered membrane phospholipid composition. Replacement of phosphatidylinositol by phosphatidylglycerol in a myo-inositol auxotroph. J. Biol. Chem. 255, 4474–4480 [PubMed] [Google Scholar]
- 27. Icho T., Sparrow C. P., and Raetz C. R. (1985) Molecular cloning and sequencing of the gene for CDP-diglyceride synthetase of Escherichia coli. J. Biol. Chem. 260, 12078–12083 [PubMed] [Google Scholar]
- 28. Sparrow C. P., and Raetz C. R. (1985) Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. J. Biol. Chem. 260, 12084–12091 [PubMed] [Google Scholar]
- 29. Deems R. A., and Dennis E. A. (1975) Characterization and physical properties of the major form of phospholipase A2 from cobra venom (Naja naja naja) that has a molecular weight of 11,000. J. Biol. Chem. 250, 9008–9012 [PubMed] [Google Scholar]
- 30. Deems R. A., Eaton B. R., and Dennis E. A. (1975) Kinetic analysis of phospholipase A2 activity toward mixed micelles and its implications for the study of lipolytic enzymes. J. Biol. Chem. 250, 9013–9020 [PubMed] [Google Scholar]
- 31. Jarvis A. A., Cain C., and Dennis E. A. (1984) Purification and characterization of a lysophospholipid from human amniotic membranes. J. Biol. Chem. 259, 15188–15195 [PubMed] [Google Scholar]
- 32. Hendrickson H. S., and Dennis E. A. (1984) Kinetic analysis of the dual phospholipid model for phospholipase A2 action. J. Biol. Chem. 259, 5734–5739 [PubMed] [Google Scholar]
- 33. Lombardo D., and Dennis E. A. (1985) Cobra venom phospholipase A2 inhibition by manoalide. A novel type of phospholipase inhibitor. J. Biol. Chem. 260, 7234–7240 [PubMed] [Google Scholar]
- 34. Coleman R., and Bell R. M. (1976) Triacylglycerol synthesis in isolated fat cells. Studies on the microsomal diacylglycerol acyltransferase activity using ethanol-dispersed diacylglycerols. J. Biol. Chem. 251, 4537–4543 [PubMed] [Google Scholar]
- 35. Lightner V. A., Larson T. J., Tailleur P., Kantor G. D., Raetz C. R., Bell R. M., and Modrich P. (1980) Membrane phospholipid synthesis in Escherichia coli. Cloning of a structural gene (plsB) of the sn-glycerol-3-phosphate acyltransferase. J. Biol. Chem. 255, 9413–9420 [PubMed] [Google Scholar]
- 36. Green P. R., and Bell R. M. (1984) Asymmetric reconstitution of homogeneous Escherichia coli sn-glycerol-3-phosphate acyltransferase into phospholipid vesicles. J. Biol. Chem. 259, 14688–14694 [PubMed] [Google Scholar]
- 37. Hannun Y. A., Loomis C. R., and Bell R. M. (1985) Activation of protein kinase C by Triton X-100 mixed micelles containing diacylglycerol and phosphatidylserine. J. Biol. Chem. 260, 10039–10043 [PubMed] [Google Scholar]
- 38. Hannun Y. A., Greenberg C. S., and Bell R. M. (1987) Sphingosine inhibition of agonist-dependent secretion and activation of human platelets implies that protein kinase C is a necessary and common event of the signal transduction pathways. J. Biol. Chem. 262, 13620–13626 [PubMed] [Google Scholar]
- 39. Hjelmstad R. H., and Bell R. M. (1990) The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene. J. Biol. Chem. 265, 1755–1764 [PubMed] [Google Scholar]
- 40. Henry S. A., and Patton-Vogt J. L. (1998) Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acids Res. 61, 133–179 10.1016/S0079-6603(08)60826-0 [DOI] [PubMed] [Google Scholar]
- 41. Carman G. M., and Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38, 361–399 10.1016/S0163-7827(99)00010-7 [DOI] [PubMed] [Google Scholar]
- 42. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., and Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 10.1126/science.1096083 [DOI] [PubMed] [Google Scholar]
- 43. Carter J. R., and Kennedy E. P. (1966) Enzymatic synthesis of cytidine diphosphate diglyceride. J. Lipid Res. 7, 678–683 [PubMed] [Google Scholar]
- 44. Carman G. M., and Han G.-S. (2009) Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284, 2593–2597 10.1074/jbc.R800059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Y.-P., and Carman G. M. (1989) Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264, 8641–8645 [PubMed] [Google Scholar]
- 46. Lin Y.-P., and Carman G. M. (1990) Kinetic analysis of yeast phosphatidate phosphatase toward Triton X-100/phosphatidate mixed micelles. J. Biol. Chem. 265, 166–170 [PubMed] [Google Scholar]
- 47. Wu W.-I., and Carman G. M. (1996) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by phospholipids. Biochemistry 35, 3790–3796 10.1021/bi952808f [DOI] [PubMed] [Google Scholar]
- 48. Wu W.-I., Lin Y.-P., Wang E., Merrill A. H. Jr., and Carman G. M. (1993) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by sphingoid bases. J. Biol. Chem. 268, 13830–13837 [PubMed] [Google Scholar]
- 49. Wu W.-I., and Carman G. M. (1994) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by nucleotides. J. Biol. Chem. 269, 29495–29501 [PubMed] [Google Scholar]
- 50. Wissing J. B., and Behrbohm H. (1993) Phosphatidate kinase, a novel enzyme in phospholipid metabolism. Purification, subcellular localization, and occurrence in the plant kingdom. Plant Physiol. 102, 1243–1249 10.1104/pp.102.4.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wissing J. B., and Behrbohm H. (1993) Diacylglycerol pyrophosphate, a novel phospholipid compound. FEBS Lett. 315, 95–99 10.1016/0014-5793(93)81141-L [DOI] [PubMed] [Google Scholar]
- 52. Munnik T., de Vrije T., Irvine R. F., and Musgrave A. (1996) Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J. Biol. Chem. 271, 15708–15715 10.1074/jbc.271.26.15708 [DOI] [PubMed] [Google Scholar]
- 53. Riedel B., Morr M., Wu W.-I., Carman G. M., and Wissing J. B. (1997) Metabolism of diacylglycerol pyrophosphate by suspension cultured Catharanthus roseus cells. Identification and characterization of diacylglycerol pyrophosphatase phosphatase in plants. Plant Sci. 128, 1–10 10.1016/S0168-9452(97)00120-9 [DOI] [Google Scholar]
- 54. Wu W.-I., Liu Y., Riedel B., Wissing J. B., Fischl A. S., and Carman G. M. (1996) Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 271, 1868–1876 10.1074/jbc.271.4.1868 [DOI] [PubMed] [Google Scholar]
- 55. Dillon D. A., Chen X., Zeimetz G. M., Wu W.-I., Waggoner D. W., Dewald J., Brindley D. N., and Carman G. M. (1997) Mammalian Mg2+-independent phosphatidate phosphatase (PAP2) displays diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 272, 10361–10366 10.1074/jbc.272.16.10361 [DOI] [PubMed] [Google Scholar]
- 56. Faulkner A., Chen X., Rush J., Horazdovsky B., Waechter C. J., Carman G. M., and Sternweis P. C. (1999) The LPP1 and DPP1 gene products account for most of the isoprenoid phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 274, 14831–14837 10.1074/jbc.274.21.14831 [DOI] [PubMed] [Google Scholar]
- 57. Han G.-S., Wu W.-I., and Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Toke D. A., Bennett W. L., Dillon D. A., Wu W.-I., Chen X., Ostrander D. B., Oshiro J., Cremesti A., Voelker D. R., Fischl A. S., and Carman G. M. (1998) Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 273, 3278–3284 10.1074/jbc.273.6.3278 [DOI] [PubMed] [Google Scholar]
- 59. Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., and Eide D. J. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 7957–7962 10.1073/pnas.97.14.7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuan D. S. (2000) Zinc-regulated genes in Saccharomyces cerevisiae revealed by transposon tagging. Genetics 156, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stukey J., and Carman G. M. (1997) Identification of a novel phosphatase sequence motif. Protein Sci. 6, 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dillon D. A., Wu W.-I., Riedel B., Wissing J. B., Dowhan W., and Carman G. M. (1996) The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 271, 30548–30553 10.1074/jbc.271.48.30548 [DOI] [PubMed] [Google Scholar]
- 63. Toke D. A., Bennett W. L., Oshiro J., Wu W.-I., Voelker D. R., and Carman G. M. (1998) Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 273, 14331–14338 [DOI] [PubMed] [Google Scholar]
- 64. Brindley D. N., and Waggoner D. W. (1998) Mammalian lipid phosphate phosphohydrolases. J. Biol. Chem. 273, 24281–24284 10.1074/jbc.273.38.24281 [DOI] [PubMed] [Google Scholar]
- 65. Brindley D. N., English D., Pilquil C., Buri K., and Ling Z. C. (2002) Lipid phosphate phosphatases regulate signal transduction through glycerolipids and sphingolipids. Biochim. Biophys. Acta 1582, 33–44 10.1016/S1388-1981(02)00135-X [DOI] [PubMed] [Google Scholar]
- 66. Brindley D. N. (2004) Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer. J. Cell. Biochem. 92, 900–912 10.1002/jcb.20126 [DOI] [PubMed] [Google Scholar]
- 67. Chae M., Han G.-S., and Carman G. M. (2012) The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme. J. Biol. Chem. 287, 40186–40196 10.1074/jbc.M112.421776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Irie K., Takase M., Araki H., and Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 [DOI] [PubMed] [Google Scholar]
- 69. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Koonin E. V., and Tatusov R. L. (1994) Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 244, 125–132 10.1006/jmbi.1994.1711 [DOI] [PubMed] [Google Scholar]
- 72. Madera M., Vogel C., Kummerfeld S. K., Chothia C., and Gough J. (2004) The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32, D235–D239 10.1093/nar/gkh117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chae M., and Carman G. M. (2013) Characterization of the yeast actin patch protein App1p phosphatidate phosphatase. J. Biol. Chem. 288, 6427–6437 10.1074/jbc.M112.449629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weinberg J., and Drubin D. G. (2012) Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22, 1–13 10.1016/j.tcb.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. O'Hara L., Han G.-S., Peak-Chew S., Grimsey N., Carman G. M., and Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Choi H.-S., Su W.-M., Morgan J. M., Han G.-S., Xu Z., Karanasios E., Siniossoglou S., and Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 10.1074/jbc.M110.155598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Karanasios E., Han G.-S., Xu Z., Carman G. M., and Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 107, 17539–17544 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Choi H.-S., Su W.-M., Han G.-S., Plote D., Xu Z., and Carman G. M. (2012) Pho85p–Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287, 11290–11301 10.1074/jbc.M112.346023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Su W.-M., Han G.-S., Casciano J., and Carman G. M. (2012) Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p–Pho80p and Cdc28p–cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287, 33364–33376 10.1074/jbc.M112.402339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G.-S., Reggiori F., Carman G. M., and Siniossoglou S. (2013) Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 24, 2124–2133 10.1091/mbc.e13-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu Z., Su W.-M., and Carman G. M. (2012) Fluorescence spectroscopy measures yeast PAH1-encoded phosphatidate phosphatase interaction with liposome membranes. J. Lipid Res. 53, 522–528 10.1194/jlr.M022798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Barbosa A. D., Sembongi H., Su W.-M., Abreu S., Reggiori F., Carman G. M., and Siniossoglou S. (2015) Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 26, 3641–3657 10.1091/mbc.e15-03-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Su W.-M., Han G.-S., and Carman G. M. (2014) Cross-talk phosphorylations by protein kinase C and Pho85p–Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289, 18818–18830 10.1074/jbc.M114.581462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2016) Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291, 9974–9990 10.1074/jbc.M116.726588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Han G.-S., Johnston C. N., Chen X., Athenstaedt K., Daum G., and Carman G. M. (2001) Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J. Biol. Chem. 276, 10126–10133 10.1074/jbc.M011421200 [DOI] [PubMed] [Google Scholar]
- 86. Han G.-S., Johnston C. N., and Carman G. M. (2004) Vacuole membrane topography of the DPP1-encoded diacylglycerol pyrophosphate phosphatase catalytic site from Saccharomyces cerevisiae. J. Biol. Chem. 279, 5338–5345 10.1074/jbc.M311779200 [DOI] [PubMed] [Google Scholar]
- 87. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 88. Roth M. G. (2008) Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239 10.1111/j.1600-0854.2008.00742.x [DOI] [PubMed] [Google Scholar]
- 89. Morris A. J. (2007) Regulation of phospholipase D activity, membrane targeting and intracellular trafficking by phosphoinositides. Biochem. Soc. Symp. 247–257 [DOI] [PubMed] [Google Scholar]
- 90. Maissel A., Marom M., Shtutman M., Shahaf G., and Livneh E. (2006) PKCη is localized in the Golgi, ER, and nuclear envelope and translocates to the nuclear envelope upon PMA activation and serum-starvation: C1b domain and the pseudosubstrate containing fragment target PKCη to the Golgi and the nuclear envelope. Cell. Signal. 18, 1127–1139 10.1016/j.cellsig.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 91. Lehel C., Oláh Z., Jakab G., Szállási Z., Petrovics G., Harta G., Blumberg P. M., and Anderson W. B. (1995) Protein kinase Cϵ subcellular localization domains and proteolytic degradation sites. A model for protein kinase C conformational changes. J. Biol. Chem. 270, 19651–19658 10.1074/jbc.270.33.19651 [DOI] [PubMed] [Google Scholar]
- 92. Baron C. L., and Malhotra V. (2002) Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295, 325–328 10.1126/science.1066759 [DOI] [PubMed] [Google Scholar]
- 93. Péterfy M., Phan J., Xu P., and Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- 94. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., and Reue K. (2007) Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282, 3450–3457 10.1074/jbc.M610745200 [DOI] [PubMed] [Google Scholar]
- 95. Han G.-S., and Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 10.1074/jbc.M110.117747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Langner C. A., Birkenmeier E. H., Ben-Zeev O., Schotz M. C., Sweet H. O., Davisson M. T., and Gordon J. I. (1989) The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J. Biol. Chem. 264, 7994–8003 [PubMed] [Google Scholar]
- 97. Phan J., and Reue K. (2005) Lipin, a lipodystrophy and obesity gene. Cell Metab. 1, 73–83 10.1016/j.cmet.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 98. Pascual F., Soto-Cardalda A., and Carman G. M. (2013) PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288, 35781–35792 10.1074/jbc.M113.525766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Han G.-S., and Carman G. M. (2017) Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J. Biol. Chem. 292, 13230–13242 10.1074/jbc.M117.801720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fakas S., Qiu Y., Dixon J. L., Han G.-S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M. (2011) Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 10.1074/jbc.M111.258798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., and Goodman J. M. (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Han S., Bahmanyar S., Zhang P., Grishin N., Oegema K., Crooke R., Graham M., Reue K., Dixon J. E., and Goodman J. M. (2012) Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem. 287, 3123–3137 10.1074/jbc.M111.324350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Park Y., Han G. S., Mileykovskaya E., Garrett T. A., and Carman G. M. (2015) Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290, 25382–25394 10.1074/jbc.M115.680314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., Southard S. B., Horenstein C. I., Chen-Weiner J., Ram A. F., Kapteyn J. C., Roemer T. W., Vo D. H., Bondoc D. C., Hall J., Zhong W. W., et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ruiz C., Cid V. J., Lussier M., Molina M., and Nombela C. (1999) A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast 15, 1001–1008 [DOI] [PubMed] [Google Scholar]
- 106. Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., and Fratti R. A. (2012) The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287, 2221–2236 10.1074/jbc.M111.317420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rahman M. A., Mostofa M. G., and Ushimaru T. (2018) The Nem1/Spo7–Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 285, 1840–1860 10.1111/febs.14448 [DOI] [PubMed] [Google Scholar]
- 108. Han G.-S., O'Hara L., Carman G. M., and Siniossoglou S. (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283, 20433–20442 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y, Eshel G., Vaz F. M., Pines O., and Elpeleg O. (2008) Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83, 489–494 10.1016/j.ajhg.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang P., Verity M. A., and Reue K. (2014) Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20, 267–279 10.1016/j.cmet.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nadra K., de Preux Charles A.-S., Médard J.-J., Hendriks W. T., Han G.-S., Grès S., Carman G. M., Saulnier-Blache J.-S., Verheijen M. H., and Chrast R. (2008) Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22, 1647–1661 10.1101/gad.1638008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wiedmann S., Fischer M., Koehler M., Neureuther K., Riegger G., Doering A., Schunkert H., Hengstenberg C., and Baessler A. (2008) Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes 57, 209–217 10.2337/db07-0083 [DOI] [PubMed] [Google Scholar]
- 113. Ferguson P. J., and El-Shanti H. I. (2007) Autoinflammatory bone disorders. Curr. Opin. Rheumatol. 19, 492–498 10.1097/BOR.0b013e32825f5492 [DOI] [PubMed] [Google Scholar]
- 114. Ferguson P. J., Chen S., Tayeh M. K., Ochoa L., Leal S. M., Pelet A., Munnich A., Lyonnet S., Majeed H. A., and El-Shanti H. (2005) Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 42, 551–557 10.1136/jmg.2005.030759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Aulchenko Y. S., Pullen J., Kloosterman W. P., Yazdanpanah M., Hofman A., Vaessen N., Snijders P. J., Zubakov D., Mackay I., Olavesen M., Sidhu B., Smith V. E., Carey A., Berezikov E., Uittenlinden A. G., et al. (2007) LPIN2 is associated with type 2 diabetes, glucose metabolism and body composition. Diabetes 56, 3020–3026 10.2337/db07-0338 [DOI] [PubMed] [Google Scholar]
- 116. Han G.-S., Siniossoglou S., and Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 10.1074/jbc.M705777200 [DOI] [PubMed] [Google Scholar]
- 117. Park Y., Han G. S., and Carman G. M. (2017) A conserved tryptophan within the WRDPLVDID domain of yeast Pah1 phosphatidate phosphatase is required for its in vivo function in lipid metabolism. J. Biol. Chem. 292, 19580–19589 10.1074/jbc.M117.819375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2015) Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 290, 11467–11478 10.1074/jbc.M115.648659 [DOI] [PMC free article] [PubMed] [Google Scholar]