Abstract
Self-assembled peptide and protein amyloid nanostructures have traditionally been considered only as pathological aggregates implicated in human neurodegenerative diseases. In more recent times, these nanostructures have found interesting applications as advanced materials in biomedicine, tissue engineering, renewable energy, environmental science, nanotechnology and material science, to name only a few fields. In all these applications, the final function depends on: (i) the specific mechanisms of protein aggregation, (ii) the hierarchical structure of the protein and peptide amyloids from the atomistic to mesoscopic length scales and (iii) the physical properties of the amyloids in the context of their surrounding environment (biological or artificial). In this review, we will discuss recent progress made in the field of functional and artificial amyloids and highlight connections between protein/peptide folding, unfolding and aggregation mechanisms, with the resulting amyloid structure and functionality. We also highlight current advances in the design and synthesis of amyloid-based biological and functional materials and identify new potential fields in which amyloid-based structures promise new breakthroughs.
1. Introduction
The self-assembly and aggregation of peptides and proteins play crucial roles in many of the human’s body functions.1 For instance, networks of collagen fibrils provide a biochemical scaffold with many functions governing the morphology and mechanical properties of biological tissue.2,3 Self-assembled actin fibrils are essential elements for many key functions in eukaryotic cells, such as motility, morphology, maintenance of cell polarity and the regulation of transcription.4 In blood coagulation, wound healing proceeds through the aggregation of fibrin into sealing clots, allowing tissue repair. In addition, there are a number of diseases associated with errant protein aggregation. The misfolding of proteins and their subsequent assembly into amyloid fibrils are pathological hallmarks of a number of devastating degenerative diseases, including Parkinson’s, Alzheimer’s, Type II diabetes and others.5
Historically, due to the discovery of their association with disease states, the study of amyloid fibrils has been largely centred on those associated with neurodegenerative disorders. A great deal of research has been performed to elucidate the formation mechanisms and to understand the mechanisms of toxicity arising from various amyloid species ranging from oligomers to mature amyloid nanofibrils.6 Consequently, a large number of biomedical studies have been devoted to uncovering how to inhibit amyloid formation, and a multitude of biomedical, biochemical, biophysical and nanotechnological processes have been investigated in an attempt to design therapies that can slow down the progress of amyloid-related diseases.7–10
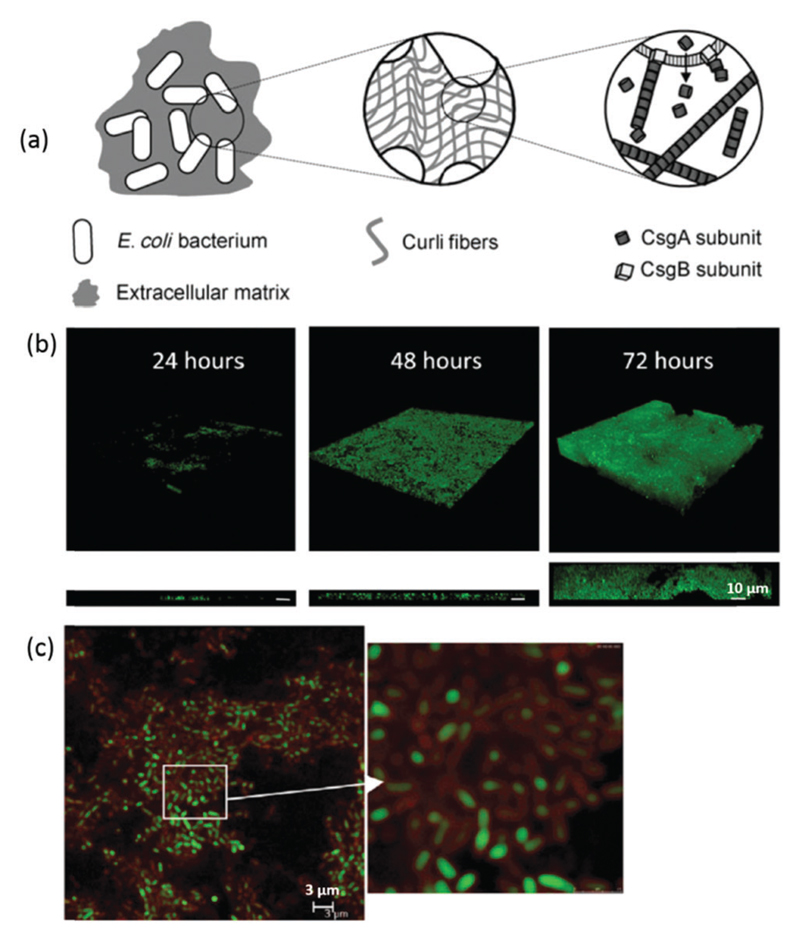
The discovery that functional amyloid fibrils in living organisms also play vital physiological roles within and on the surface of living cells has introduced a new paradigm for the study of amyloid fibrils. Examples of the physiological roles of functional amyloids include, curli fibrils,11 which are associated with the adhesive properties of E. coli biofilms, catalysis of melanin synthesis in mammalian melanosomes12 and human peptide hormone storage.13
In addition to toxic and functional amyloids, in recent years there has been a growing interest in the applications of amyloid fibrils as templates or building blocks in ordered nanomaterials for biomedical, biomaterial and nanotechnological applications.14 Amyloid nanofibrils have been successfully employed as a fundamental component in biomembranes,15 functional nanodevices,16,17 hydrogels for cell culture and drug delivery,18,19 biosensors,20 functional materials with high biocompatibility and unique bio-recognition ability21,22 and as energy conversion materials.23 All the above functions and applications of amyloid fibrils arise due to their unique structural features, enabling them to serve in an extremely vast context of fundamental and applied sciences, spanning from biology to materials science and nanotechnology.
At the atomistic length-scale, the structural features of amyloid fibrils are remarkably similar,24,25 with amino acids arranged into β-strands (separated by ~4 Å) running orthogonal to the fibril axis and closely packed into β-sheets running parallel to the fibril axis (typical intersheet distance ~10–12 Å). In sharp contrast, the mesoscopic structure of amyloid fibrils shows a remarkable diversity, with a multitude of shapes and topologies, depending on the specific aggregation pathways followed.26,27 To date, nanoparticles, nanofibrils, nanotubes, ribbons, nanosheets and 3D scaffolds or multilayers represent just some of the amyloid morphologies observed.28–31 A wide spectrum of available morphologies and free energies, high surface-to-volume ratio, high density of hydrogen bonds and the presence of biocompatible amino acids on their surfaces gives amyloid fibrils a remarkable range of nanomechanical properties and applications across many scientific fields.32,33
In this review, we comprehensively analyse the relationship between the molecular mechanisms of assembly into amyloid fibrils, the resulting amyloid structure and polymorphism, and the ensuing physical properties. We then discuss how the structure and the physical properties of amyloids can be harnessed to provide applications as advanced materials in nanotechnology. In Section 2, we discuss the self-assembly and aggregation mechanisms of amyloids from unfolded proteins (e.g. α-synuclein), folded globular proteins (e.g. β-lactoglobulin) and peptides (e.g. Amyloid β, Aβ). In Section 3, we review the main structural traits of amyloids from the atomic to the mesoscopic length scale. In Sections 4 and 5, we discuss the functionality of natural and artificial amyloid materials and present the current biomedical, material and nanotechnological applications of amyloid-based hybrid materials. We conclude by highlighting the current challenges and future perspectives of amyloid-based materials and discuss the emerging fields in which amyloid fibrils are ideal candidates to contribute to their development. It is expected that this comprehensive review will forge new directions for the design, synthesis and wider applications of protein and peptide amyloid-based biological and functional materials.
2. Self-assembly and aggregation mechanisms of amyloids
The formation of amyloids can be achieved with either native folded proteins, which often have to undergo activation reactions, such as unfolding and hydrolysis before aggregating, or unfolded proteins and peptides, which under a broad range of conditions exhibit 1D growth after a fast nucleation step. In this part, we would like to introduce key information on the self-assembly and aggregation mechanisms of peptides and proteins, which include a series of microscopic events, such as protein unfolding, hydrolysis and aggregation. We describe the application of chemical kinetic studies to identify different microscopic mechanisms and we discuss the connections between aggregation mechanisms and the length distribution of the aggregate population, which are key properties defining the function of the final amyloid product.
2.1. Amyloidogenic proteins and peptides
2.1.1. Protein folding and unfolding
The formation of well-folded protein structures is central to the function of every living cell.34 Based on the information coded by DNA molecules and transcribed into messenger RNA, the ribosome synthesizes polypeptide chains of a specific amino acid sequence, which undergo internal organization to form distinct conformational arrangements. The spontaneous arrangement of the amino acid chain based on the physicochemical properties of its constituents is denoted as “protein folding”.35 The correct organization of the polypeptide chain into well-folded three-dimensional (3D) arrangements allows the proper activity of proteins, including enzymatic activity, storage, transport, sensing, signalling and structural functions. The folding of proteins into the distinct thermodynamically favourable conformations is achieved through folding pathways that hierarchically direct the protein into the lowest energy thermodynamic state. Several canonical secondary structures, such as alpha-helix and beta-sheet, constitute the tertiary folding, which is a much more complex energy-minimized molecular organization. As will be further discussed, this dogma is currently challenged as the amyloid state of proteins and polypeptides may actually represent the true energetic minimum of protein assemblies.
Even though correct protein folding is required for their biological function, under certain conditions, proteins can undergo an unfolding process, losing their tertiary as well as secondary structure.36 The unfolding can be induced physically, especially by temperature changes (mostly heating and in some cases cooling), as well as by hydrostatic pressure or chemically, by the addition of chaotropic agents (such as urea, guanidinium chloride, magnesium chloride, alcohols and detergents) that are able to disrupt the hydrogen bonding network between water molecules.
Protein unfolding can be either reversible or irreversible, depending on potential irregular interactions of the unfolded protein either within the same polypeptide chain (intramolecular interactions) or with neighbouring molecules (intermolecular interactions). The association of exposed hydrophobic surfaces is the main driving force of intermolecular interactions. Instead of a normal folding process in which the hydrophobic parts of a protein are buried in its core, many of these parts are held together by non-covalent interactions. In the case of intermolecular interactions, the molecular assemblies can form ordered assemblies, such as crystals or amyloid fibrils.37
2.1.2. Unfolded proteins
Unlike the more common case of folded proteins, many intrinsically unfolded proteins exist in a natively unfolded state, either for the entire molecule or at specific regions of the peptide chain.38–40 In such a case, the hydrophobic parts of the proteins are exposed without any external unfolding reaction. Solution-based analytical methods, including nuclear magnetic resonance (NMR) and circular dichroism (CD), have allowed the determination of the unfolded state of proteins. The use of temperature-controlled experiments allows the melting temperature to be determined. Indeed, in some cases, proteins were found to be unfolded at physiological temperature (37 °C) but completely folded at a lower temperatures (e.g. 4 °C).41,42
The basis for the existence of proteins in an unfolded state might have a clear physiological significance. For instance, one of the roles of protein unfolding is to control the physiological stability of such proteins. The fact that unfolded proteins expose hydrophobic patches, which are otherwise buried within the core of the protein, results in their identification as damaged proteins, leading to their degradation by the protein quality control machinery.43 The existence of proteins in this state allows the modulation of their half-life. In extreme cases, some protein molecules could be degraded in a few minutes. This property is useful for two-component systems, such as bacterial toxin–antitoxin systems, in which the instability of one component is critical for the physiological control of the system.44
Protein unfolding plays a critical role in at least three biological processes, such as protein translocation, protein degradation and the passive elasticity of striated muscle,45 in which the unfolding is thought to be induced by the cellular machinery pulling the polypeptide chain to better disentangle the native domains. For the amyloid fibrils formed by globular proteins, the unfolding is a necessary step to change the conformation of the protein from one minimum to another in the protein folding energy landscape.46
2.1.3. Misfolded proteins
As mentioned above, proteins can exist in an aggregated amyloid organization which was first associated with human diseases, including Alzheimer’s disease (AD), Parkinson’s disease and Type II diabetes.47 The intermolecular interaction between unfolded or partially unfolded proteins can lead to the formation of supramolecular β-sheet structures consisting of more than one protein molecule. This could be considered an abnormal organization in which a handful of proteins or polypeptide molecules join together to form ordered assemblies.
Amyloids were first identified more than a century ago in association with disease. In 1901, Dr Eugene L. Opie, an American physician, identified the formation of deposits in the pancreas of Type II diabetes patients.48 A few years later, in 1906, the deposition of biomaterial in the brain of a demented patient post mortem was found by Aloysius “Alois” Alzheimer in Germany.49 The deposits were denoted as “amyloids” (starch-like) due to their positive staining by iodine as carbohydrate deposits. Only decades later were these aggregative forms found to be made of proteins. With the advancement of electron microscopy in the 1950s, it was discovered that these protein assemblies have a typical nanoscale order. This regularity at the nanoscale underlies some of the properties that were later utilized for various technological applications, as described in this review.
The structure of amyloids, regardless of their source, is a very typical one comprising elongated supramolecular filaments with a diameter of 7–10 nm. Amyloids have a predominantly β-sheet secondary structure as determined by X-ray fibre diffraction (XRFD) and infrared or CD spectroscopy. Interestingly, both parallel and antiparallel β-sheet structures have been observed within amyloid fibrils of different sources in spite of the very uniform ultrastructure. Amyloid fibrils were also found to bind to specific dyes. Beyond their ability to be stained with iodine, later on, other amyloid-specific dyes were identified, including Thioflavin T (ThT) and Congo red.50,51 The use of Congo red is especially interesting due to the typical birefringence that is observed upon the staining of the amyloid fibrils when placed between cross-polarizers. It was suggested that proteins of unrelated origin could form remarkably similar structures in the disease state. The hypothesis was that the formation of such structures plays a role in the damage caused to various organs and tissues observed in these diseases. Indeed this “amyloid hypothesis” was supported by the observation of notable toxicity of the amyloids or their earlier soluble intermediates.
A very important extension of the “amyloid hypothesis” was provided by Dobson and co-workers who realized that non-disease-related proteins could also form typical amyloid fibrils with all the common structural and biophysical characteristics of disease-associated amyloid assemblies.52 It was thus suggested that the amyloid structure may actually reflect a generic minimal energy organization of polypeptide chains and that the structure of folded proteins is essentially a meta-stable kinetically trapped state, suggesting that most or all proteins would reach the favourable amyloidal organization at infinite time.53 This hypothetical notion was later supported empirically as it was found that most cellular proteins are at the verge of aggregation (the “life at the edge” phenomenon) and that the proteostasis of the biological system requires an advanced cellular machinery that can keep the proteins and polypeptides in a soluble state.54
2.1.4. Peptide-based amyloids
The formation of amyloid fibrils has also been identified in various functional peptides, including the islet amyloid polypeptide (37 amino acids), Amyloid β (40–42 amino acids) and human calcitonin (31 amino acids), all associated with human diseases. The aggregated form of the islet amyloid peptide is found in the pancreas of Type II diabetes patients,55 whilst aggregated forms of Aβ are found in the brain of Alzheimer’s disease patients and similar structures made of calcitonin are found in the thyroid of patients with thyroid carcinoma, all identified using electron microscopy (EM). Furthermore, the peptide amyloids were found to share all other biophysical properties of protein amyloids, including the spectroscopic features, XRFD patterns and the staining with specific dyes.
As noted above, the formation of amyloid fibrils was initially identified in naturally occurring proteins and polypeptides. A reductionist approach has since been applied to identify the minimal peptide fragments that can form amyloid fibrils. Tenidis and co-workers were able to identify hexapeptide fragments of the islet amyloid polypeptide that form amyloid fibrils.56 Later studies identified the ability of a pentapeptide fragment of calcitonin, as well as heptapeptide fragments of Aβ, to form such ordered assemblies.57
2.1.5. Peptide nanostructures and non-protein amyloids
Further reductionist approaches were used in order to identify even shorter amyloid-forming peptide motifs. It was found that the dipeptides diphenylalanine (FF) can form amyloid-related assemblies. The diphenylalanine motif is at the centre of the Aβ polypeptide associated with Alzheimer’s disease. As noted above, it was demonstrated that a heptapeptide fragment of Aβ, specifically KLVFFAE (Lys-Leu-Val-Phe-Phe-Ala-Glu), could form fibrillar assemblies, and two pentapeptide fragments, specifically KLVFF and LVFFA, are inhibitors of amyloid formation by the full-length protein.58 It was found that the nanostructures made by FF shares many functional properties with amyloid assemblies,59,60 including the intrinsic luminescence properties, the binding of amyloid-specific dyes, mechanical rigidity and the production of reactive oxygen species. This suggests that the FF nanostructures indeed represent a highly simplified model that reflects the structural, biophysical and biochemical properties of amyloid structures.28,61
Later studies identified the ability of various short peptides to form ordered assemblies. Frederix and co-workers screened over 8000 naturally occurring tripeptides for the formation of supramolecular entities using molecular dynamics simulations.62 The most aggregation-prone peptide screened was PFF (Pro-Phe-Phe), whilst many other highly aggregating peptides contained the FF motif, and to a lower extent other diaromatic motifs (including FW, WF, FY and WW). This comprehensive non-biased analysis of all peptides is consistent with the observation of the high occurrence of aromatic amino acids in short peptides that can form typical amyloid fibrils.
In order to identify the minimal requirement for amyloid formation, amino acids were also tested for their ability to self-associate. Very surprisingly, it was found that phenylalanine could form amyloid fibrils with all the characteristics of protein fibrils, including nanoscale fibrillar morphology, the binding of ThT and Congo red and notable cytotoxicity.63 X-ray crystallography suggested the formation of β-sheet like structures by this amino acid. Similar to protein amyloids, the phenylalanine amyloids were also found to bind to phospholipid membranes. Later on, it was found that other amino acids (including Trp, Tyr and Cys), as well as nucleotides and other metabolites, could also form typical amyloid-like structures.64,65 The formed assemblies reveal many ultrastructural similarities among themselves and to protein and peptide amyloids. It was therefore suggested that the amyloid hypothesis could be even further extended to include nonproteinaceous building blocks.
The simple synthesis, chemical diversity, small size and low cost make very short motifs, including tripeptides, dipeptides and single amino acids, ideal building blocks for various applications in nanoscience and nanotechnology.66,67 Moreover, the mechanical, optical, electric and piezoelectric properties of some self-assembled structures should allow their use as alternatives to inorganic components in electronic, electrooptic and electromechanical systems. The bottom-up assembly of complex nanostructures from these simple building blocks allows the utilization of fabrication techniques that were previously used in surface modification applications, including physical vapour deposition, printing using inkjet technology and unidirectional axial growth by the controlled evaporation of volatile solvents.
2.2. Factors controlling amyloid growth and kinetics
2.2.1. Solution property-mediated amyloid formation
2.2.1.1. pH-Mediated amyloids
The growth of amyloid fibrils is highly sensitive to solution conditions, including pH and the presence of salts or denaturing agents. Very careful preparation protocols have to be followed in studying the fibrillization of Aβ42, for example starting from a well-defined state of the unaggregated peptide (achieved by initial dissolution in a hydrophobic solvent) and then carefully by controlling the addition of water or buffer to a dried film.68
Typically, amyloid fibril formation by proteins is induced by reducing the pH. The aggregation of short peptides depends on the pI of the peptide or the pKa of its constituent residues and its relationship to the solution pH. It has been suggested that fibril formation is favoured when the net charge of the peptide is not too large (in the range of −1 to +1).69
A study of amylin peptide fibrillization indicates the presence of two ionizable residues: the α-amino group at the N-terminus and His18.70 The pKa values of the former unit in the amylin peptide were found to be similar to the random coil value (pKa = 8); however, the His18 residue had a pKa = 5.0, significantly lower than the random coil value pKa = 6.5. This was ascribed to the local influence of hydrophobic residues. His18 was found to act as an electrostatic switch hindering fibrillization in its charged state. An apparent pKa = 4.0 for an amylin fragment peptide, NAc-SNNFGAILSS-NH2, which contains no titratable groups, was instead ascribed to the pH-induced ionization of the amyloid-sensitive dye, ThT.
In another example, the aggregation of the amyloid β peptide Aβ1–42 was studied as a function of pH.71 This was investigated experimentally and the analysis was facilitated by molecular mechanics modelling of the fragment peptide Aβ17–42, which revealed favourable electrostatic interactions between Asp23 and Lys28 (i.e. salt bridge formation) above the pI of the peptide. The aggregation of the Aβ1–42 peptide itself was analyzed at lower pH. At pH > 9.5, aggregation was not observed because Lys28 was uncharged.71
The self-assembly of another type of amyloid-forming building block, so-called peptide amiphiphiles (PAs), can also be mediated by the solution property. PAs are designed amyloidogenic peptides modified by the attachment of hydrophobic lipid tails,72 which then show combined surfactant-like properties and a self-assembly ability.73 The pH value has a pronounced effect on the self-assembly of PAs. For example, Stupp’s group demonstrated that PAs containing acidic amino acids could be triggered to self-assemble into nanofibrils at acidic pH74 or with the use of divalent cations.75
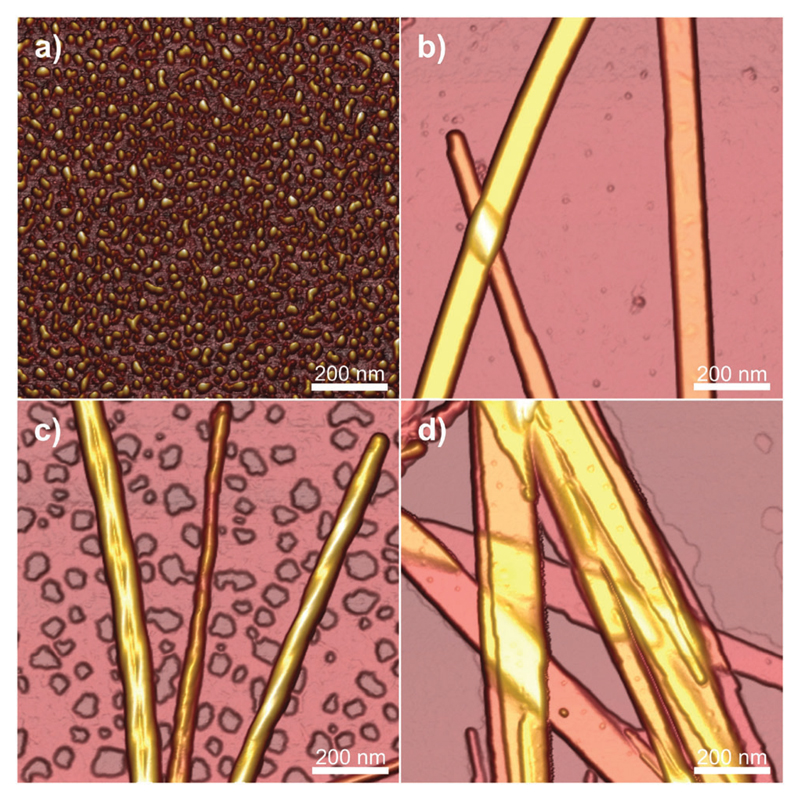
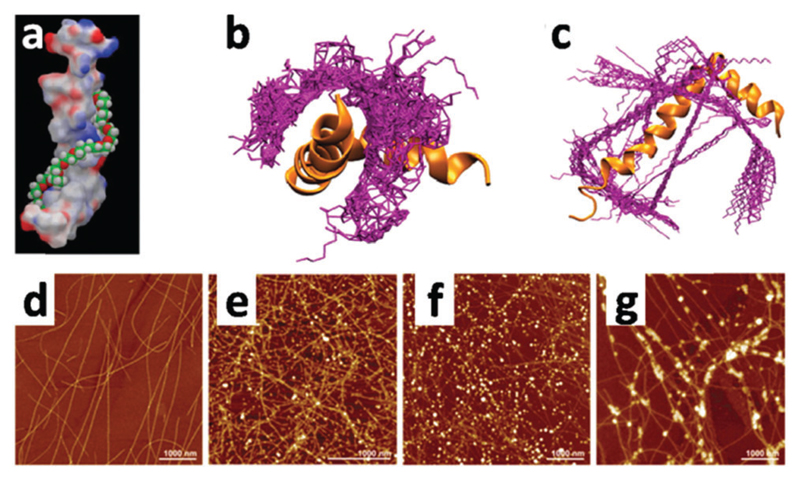
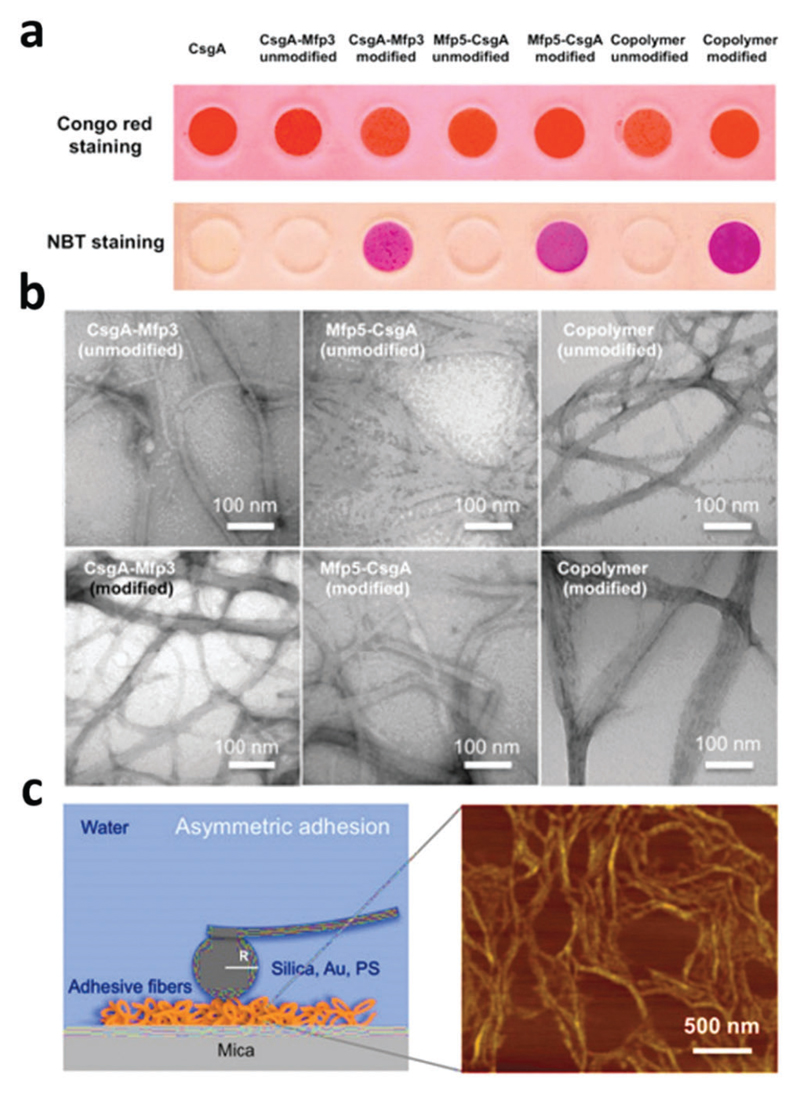
In another case, Dehsorkhi et al. found that PAs containing the pentapeptide KTTKS sequence, i.e. containing two cationic lysine residues, could be assisted to form adjustable nanostructures ranging from spherical micelles to tape-like and twisted structures by simply adjusting the pH of the solution to 2, 3, 4 and 7, respectively.76 Atomic force microscopy (AFM) was used to image the morphology at the selected pH values. It can be seen from Fig. 1 that fibrils, tapes or spherical micelles form depending on the pH (the net charge is approximately +1 at pH 7 and +2 at low pH). Small-angle X-ray scattering (SAXS) confirmed these morphology transitions and CD indicated a transition from a β-sheet conformation in fibrils and tapes to a disordered conformation at pH 2 for the spherical micelles.
Fig. 1.
Tapping mode AFM height images showing the morphology of C16-KTTKS at pH values of (a) pH 2, (b) pH 3, (c) pH 4, (d) pH 7. Reprinted with permission from ref. 76. Copyright 2013, Royal Society of Chemistry.
2.2.1.2. Ionic strength-mediated amyloids
Along with the pH dependence of amyloid formation, we also highlight a few pertinent examples in which the ionic strength of the solution also shows close effects on the formation of amyloids.
Hoyer et al. investigated, together with the effects of pH, the role of salt concentration on the in vitro aggregation of α-synuclein, and observed aggregates of varying morphologies depending on both the pH and salt concentration (NaCl and MgCl2).77 Their results indicated that the morphology of α-synuclein aggregates is highly sensitive to the solution conditions (both pH value and ionic strength). In another case, Raman et al. investigated the effect of salts such as NaCl, NaI, NaClO4 and Na2SO4 on the formation of β2-microglobulin amyloid.78 The presence of salts increased the hydrophobicity of proteins, whilst increasing the anion concentration affected the interplay between the electrostatic and hydrophobic interactions during amyloid formation. In particular SO42− ions were identified as having strong effects on amyloid formation, and this was suggested to be important in terms of the role of glycosaminoglycans and proteoglycans in amyloidogenesis. The critical aggregation concentration of β-lactoglobulin also depends on the ionic strength, and below this concentration mainly “dead-end” species are formed that consist of irreversibly denatured proteins.79 The morphology of the fibrils also changes and shorter and more flexible fibrils are formed at higher ionic strength.
The formation of peptide amyloids is also be affected by varying the ionic strength. For example, Marek et al. investigated the ionic strength effects on the formation of islet amyloid polypeptide (IAPP) fibrils.80 They suggested that the kinetics of IAPP amyloid formation is strongly dependent on the ionic strength in the range of 20–600 mM at pH 8.0. Recently, Abelein and co-workers characterized the explicit effect of ionic strength on the microscopic aggregation rates of Aβ40,81 and found that physiological ionic strength could accelerate the aggregation kinetics of Aβ40 by promoting the surface-catalyzed secondary nucleation reactions. Their results indicated that the salts could decrease the free-energy barrier for Aβ40 folding to a mature stable state, thereby favouring the formation of mature fibrils.
A recent model, based on DLVO-type colloid theory, accounts for the stability of amyloid fibril dispersions and allows for the influence of ionic strength and salt concentration (as well as the presence of organic reagents).82 The theory was used to calculate certain quantities (fibril hydrodynamic radius and Fuchs stability ratio, which describes the energy barrier between two interacting fibrils) that were then compared to experimental data for a model amphiphilic peptide (RADA 16-I).82
2.2.2. Temperature-mediated fibrillation
Heating (with or without pH adjustment to acidic conditions) is another common method of inducing amyloid formation with proteins and peptides.25 Again, there are too many studies involving heat treatment to review them all and space permits only selected examples to be discussed herein. At sufficiently high concentration, the fibrillization of peptides is accompanied by gelation. For example, β-lactoglobulin forms fibrillar gels on heating at low pH values. Particulate gels are formed at higher pH values, close to the isoelectric point where the protein has a low net charge.83,84 Cold denaturation is generally a milder form than hot denaturation and leads only to a partial unfolding of proteins85 and so, rarely, amyloid formation is reported under these conditions. However, cold denaturation can be used to dissociate amyloid fibrils, as discussed below.
PAs can show thermal transitions mediated by lipid chain melting behaviour as well as changes in the hydrogen bonding of amino acid residues, and temperature-dependent changes in solubility. In one example, conjugates of C23 or C25 alkyl chains (both containing one diacetylene unit) and the bio-derived GANPNAAG peptide sequence were shown to have very different disassembly transition temperatures on heating, as well as distinct thermosreversibility properties.86 The longer chain PA reassembled on cooling, the shorter one did not.86 The same PA C16-KTTKS discussed in the previous section also exhibited interesting temperature-mediated fibrillization below 30 °C (depending on concentration), which may be associated with the palmitoyl chain melting temperature.87 At low temperature, this compound (Tradename: Matrixyl) forms extended tape-like fibrils, but at high temperature, small spherical micelles are observed.87 The thermoresponsiveness of other lipopeptides and peptides, for example, elastin-like peptides, which undergo LCST (lower critical solution temperature) behaviour, has been reviewed elsewhere.88
The formation of amyloid fibrils by the egg-white protein ovalbumin occurs at high temperature (90 °C) and low pH, and the fibril morphology has been examined with or without NaCl.89 Two types of aggregate were observed: thin flexible worm-like fibrils or thicker periodically twisted ribbons. Differences in β-sheet content between the two were studied by CD, WAXS and Fourier transform infrared spectroscopy (FTIR) (the latter aggregate lacks amyloid characteristics). The stiffness of the two types of fibril also differs, as quantified by peak force-quantitative nanomechanical AFM.89
In another example, the fibrillization and defibrillization (‘depolymerization’) of β2-microglobulin was followed by detailed ThT fluorescence measurements.90 Incubation at 99 °C for 10 min was found to lead to complete dissociation of the fibrils into monomers. This occurred via both fibril breakage and dissociation of monomers from the fibril ends. Repolymerization experiments revealed that the number of extendable fibril ends increased significantly upon incubation at elevated temperatures. Stabilization of the fibrils using a number of additives (salts or surfactant) was examined and it was found that the anionic surfactant SDS (sodium dodecyl sulfate) could prevent fibril dissociation up to 99 °C.90
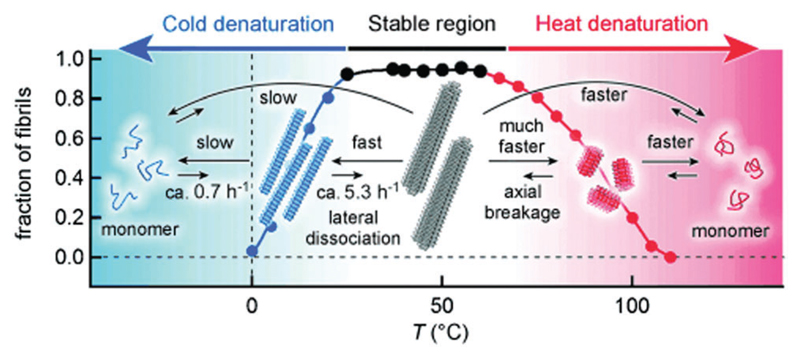
Whilst the amyloid fibrils of many peptides, such as β2-microglobulin and Aβ1–40 or Aβ1–42, undergo heat-induced breakup, cold denaturation (dissociation into monomers) was additionally observed for α-synuclein (Fig. 2).91 CD spectroscopy was used to monitor the loss of the β-sheet structure on cooling (to 0 °C), and the temperature dependence was analyzed, together with additional isothermal titration calorimetric (ITC) measurements, to provide thermodynamic information. The results suggested that cold denaturation results from the burial of charged residues in the core of α-synuclein fibrils, opposite to the case of protein folding.91 The dissociation of amyloid fibrils under cold conditions is exemplified by a study on α-synuclein in supercooled water at –15 °C.86
Fig. 2.
Schematic of the defibrillization of α-synuclein upon either cold or hot denaturing. Reprinted with permission from ref. 91. Copyright 2014, Wiley-VCH Verlag GmbH & Co.
The denaturation of insulin under extreme temperature conditions up to 140 °C was probed via CD and ThT fluorescence experiments.92 The amyloid structure was gradually replaced with a random coil structure above 80 °C until no amyloid structure was detected at 140 °C. Fibrillization was observed when the sample was cooled down to 100 °C and incubated, showing that even exposure to very high temperature, which favours full unfolding, does not lead to a completely irreversible denaturing.92
2.2.3. Organic reagent-induced fibrillation
Protein denaturing agents, such urea, salts, guanidinium hydrochloride, or surfactants (e.g. SDS) can cause amyloid fibril formation. For example, many studies on amyloid fibril formation by the prion protein PrP have involved chemical denaturants that promote non-native conformational states.93 Alcohol co-solvents generally lead to an increase in the β-sheet structure associated with the fibril formation of peptides and proteins. On the other hand, high concentrations of hexafluoroisopropanol (HFIP) or 2,2,2-trifluoroethanol (TFE) inhibit aggregation (and are widely used to disperse peptides and proteins in an unaggregated form). Acetonitrile is also reported to have an effect in inhibiting fibrillization.7
In a further example, the formation of fibrils (so-called protein nanofibres) by the extracellular matrix (ECM) adhesion protein fibronectin was observed after incubation at 37 °C in water/ethanol mixtures.94 The fibrils were used as scaffolds to deposit N-hydroxysulfosuccinimide (NHS)-modified CdSe–ZnS core–shell quantum dots (QDs), which had potential applications as biophotonic nanohybrid materials. Fibrinogen also forms fibrils by incubation at pH 2, and these were used as templates for biomineralization.95
2.2.4. Metal ion-induced fibrillation
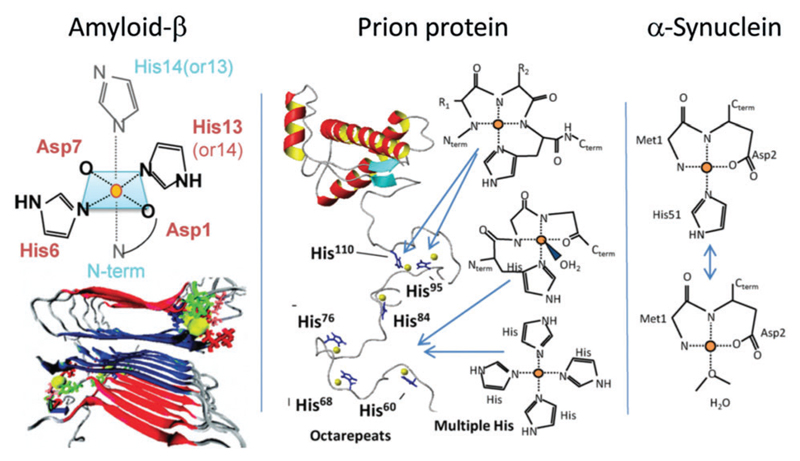
Metal ions are associated with amyloid deposits in several neurodegenerative disorders, including Alzheimer’s, Parkinsons and prion diseases. Metal ion coordination (through residues such as histidine) may cause inter-peptide cross-linking (Fig. 3 shows some possible structures), which in turn influences oligomerization and fibrillization. In Parkinson’s disease, elevated levels of copper and iron ions are found in the cerebrospinal fluid and Lewy bodies (which are intracellular inclusion bodies containing β-sheet-rich aggregates of α-synuclein).96 An early characteristic of prion disease is a metal imbalance. Here, Cu2+ has been found in scrapie isolates and confers the prion strain type.96 Furthermore, copper ions, found in trace quantities in the bloodstream, are known to bind to PrP in vivo and in vitro and to influence PrP levels in the brain. The aggregation of Aβ in Alzheimer’s disease may also be promoted by metal ions.97–101 Metal ions (e.g. Cu2+, Fe3+, Zn2+, Al3+) were found to be co-localized at abnormally high concentrations with senile plaques in AD brains.102–106 Furthermore, Aβ rapidly aggregates in the presence of physiological concentrations of Zn2+ at pH 7.4.107–109 In addition, metal ion-mediated amyloid formation is thought to be associated with inflammation in AD patients. For example, Cu2+-induced aggregation was enhanced at mildly acidic pH values associated with inflammation.109 The apparent interdependence of metal ions and amyloid assembly in AD opens up potential therapeutic targets. For instance, treatment with metal ion chelators can reduce the deposition of Aβ in brains.110–113 The majority of studies to date have focused on metal ions ability to enhance Aβ fibrillation (e.g. Cu2+, Zn2+ Al3+ and Fe3+);114 however, some studies have proposed that under certain conditions copper111,115,116 and zinc111,115 (but not iron111) ions are non-fibrillogenic. It should be noted, however, that amorphous and/or oligomeric aggregates may still be promoted through increased intramolecular bridging. This was exemplified by a study of different fragments of Aβ peptides, some of which promote fibrillization, whilst others reduce fibril formation.116 Apart from Aβ, di- and tri-valent metal ions have been shown to cause significant increases in the rate of fibril formation of α-synuclein and there appears to be a correlation to the ion charge density.117 The binding between protein and metal ions occurs via metal–ligand supramolecular interactions, which has been studied in detail for model metal ions by Bolisetty et al., both by molecular dynamic simulations and binding isotherms.118
Fig. 3.
Proposed modes of metal ion binding involved in the aggregation of the three proteins or peptides indicated. Reprinted with permission from ref. 96. Copyright 2012, Elsevier Ltd.
The subject of peptide self-assembly triggered by metal ions has been reviewed in depth.119 Many artificial ligands for metal ions have been incorporated into peptide-based molecules, and in addition the influence of metal ions on peptides incorporating natural ion-binding residues (histidine, cysteine, tryptophan or glutamic acid) has been examined. Self-assembly into different structures, including α-helix-based structures, β-turns, has been reviewed,119 but this is outside the scope of the current review.
2.2.5. Biopolymer-induced formation
Proteoglycans are an essential component of the ECM and they have important effects on amyloid aggregation in vivo, and have also been investigated in vitro. Glycosaminoglycans (GAGs) or proteoglycans are thought to be associated with AD since sulfated GAGs, such as heparin or chondroitin sulfate, are present in neuritic plaques, neurofibrillary tangles and vascular amyloid deposits.113,120–122 The binding of some sulfated GAGs can prevent the proteolytic degradation of fibrillar Aβ. Sulfated GAGs can interact with histidine residues on peptides, such as Aβ. Interestingly, sulfated GAGs can promote fibril formation due to a charge templating effect.123 It has been reported that the sulfate spacing in heparin and several other GAGs is ideal for β-sheet formation (with an associated 4.8 Å strand spacing), but this is not the case for some other polysaccharides.123 In parallel studies, it has been reported that heparin or heparin sulfate can accelerate the fibrillization of Aβ in vitro,121 probably due to electrostatic binding to a specific domain in the Aβ11–28 region.124 The influence of uncharged polysaccharides on fibril formation has been less studied, although one study suggests that glycogen can promote β-sheet formation of the prion protein.125
In a few cases, the inhibition of amyloid fibrillization by polysaccharides has been reported; for instance, κ-carrageenan forms a complex with positively charged β-lactoglobulin that partly hindered high temperature fibril formation.126 Uncomplexed β-lactoglobulin still formed fibrils, but protein–carrageenan complexes did not. Chitosan and poly(vinylsulfate) have been reported to have an inhibitory effect on Aβ1–42 fibrillization.123 The influence of proteoglycans on amyloid fibrillization is reviewed elsewhere.7,114
Linse’s group investigated the effects of polyamino acids and polyelectrolytes on Aβ fibrillization.127 They investigated the kinetics of Aβ1–42 aggregation using ThT fluorescence measurements and observed a concentration-dependent accelerating effect on the aggregation process from all the positively charged polymers examined (polyglutamic acid and polyacrylic acid). In contrast, no effect was seen for the negative polymers polylysine or poly(ethylenimine) or poly(diallyldimethyl ammonium chloride) or the neutral polymers polythreonine.127
The interaction between nucleic acids and amyloid fibrils has been investigated by several groups. DNA is known to be a powerful promotor of fibrillization due to electrostatic interactions with negatively charged residues on the DNA. The complexation of DNA with two arginine-containing molecules – one bola-amphiphile and one PA – has been examined.128 Both of these peptide-based compounds self-assemble into layered β-sheet structures with incorporated DNA, with the structural integrity of the DNA maintained.
In another example, it was shown that DNA origami nanotubes can sheathe transthyretin fragment amyloid fibrils formed within them.129 A DNA origami construct was used to form 20-helix DNA nanotubes with sufficient space for the fibrils inside.
2.2.6. Nanoparticle-induced/inhibited amyloid fibrillation
Nanoparticles can significantly influence amyloid formation as they may catalyze fibril formation due to increased local protein concentration or may inhibit aggregation when there is strong binding or a large particle/protein interaction surface area.130 In the context of a high local amyloid concentration, nanoparticle/amyloid hybrids have been proposed as model systems to understand amyloid formation under crowded conditions relevant to those observed in vivo.131 The effects of nanoparticles on amyloid formation may also be related to aspects of protein adsorption on nanoparticles in the blood stream, with relevance to nanoparticle toxicity.132 The ability of a nanoparticle to influence amyloid aggregation is dependent on the stability of the protein and its intrinsic aggregation rate.130 Amyloid fibrillization in the presence of nanoparticles with varying hydrophobicity and other surface chemistries has been examined. Polymeric nanoparticles can either increase or decrease the fibrillization of amyloid proteins, depending on the nanoparticle hydrophobicity and the unfolding behaviour of the protein and the hydrogen bonding capacity of the subunits within it.133
Polymeric nanoparticles (uncharged acrylamide-based copolymers) inhibit the fibrillization of Aβ1–40, an observation ascribed to the binding of Aβ (in monomeric or oligomeric form) to the nanoparticles.134 The binding mainly affects nucleation, and the lag time was found to be strongly influenced by the copolymer composition. The binding is due to a combination of hydrophobicity (controlled via copolymer composition) and hydrogen bonding between polar groups on the polymer and in Aβ.134 In the case of cationically (amide)-functionalized polystyrene nanoparticles, the inhibition of fibrillization is observed for high particle surface areas, whereas fibrillization is accelerated for low particle surface areas due to reduction of the lag phase.135
Inorganic nanoparticles can function as Aβ fibrillization inhibitors, although this was demonstrated with cytotoxic CdTe nanoparticles.136 On the other hand, it seems that TiO2 nanoparticles can promote Aβ fibril formation by reducing the nucleation period;137 however, the precise mechanism of this is unclear. Polyoxometalates that comprise inorganic early transition metal clusters also inhibit the aggregation of Aβ.138 Surprisingly, inorganic nanoparticles based on porous silica have been shown to penetrate the brains of fruit flies (D. melanogaster), without exhibiting neurotoxic effects, thus potentially enabling delivery across the blood–brain barrier (BBB).139 BBB permeability can be modelled using the parallel artificial membrane permeability assay, which measures the passive diffusion of small molecule through an artificial lipid membrane.140,141
There is growing evidence that metal nanoparticles may act as seeds for amyloid nucleation and growth,142 an observation which could be relevant for amyloid-related neurodegenerative diseases in the light of the fact that nanoparticles may be able to pass through the BBB. For instance, the nucleation of amyloid oligomers has been reported on gold nanorods143 for a model synthetic bacterial protein that was functionalized with a hexa-histidine tag for binding to the gold surface. Conformational changes in the bound protein were probed using surface-enhanced Raman spectroscopy (SERS), and the nucleation was ascribed to the formation of immobilized pre-amyloidogenic monomers.143 Gold and silver nanoparticles have been shown to accelerate fibril growth of the NNFGAIL peptide from human islet amyloid polypeptide and the prion protein Sup35 peptide GNNQQNY in physiological aqueous solutions.144 Large-scale molecular dynamics simulations highlighted the role of the structural reorganization of the peptide corona around the gold nanoparticles as being the rate-limiting step in the aggregation process.145
Both metal ions and metal nanoparticles could promote the formation of amyloid fibrils, but the nature of their binding with proteins is different. Metal nanoparticles adhere with amyloids predominantly through electrostatic interactions and surface tension reduction, whereas metal ions bind to amyloids via supramolecular metal–ligand interactions.
2.2.7. Interface- and mechanical-force-mediated amyloid formation
It has been suggested that fibrillization kinetics can be influenced during mixing by mass transfer effects. In addition, mixing leads to shear forces, which can then influence the growth of fibrils by perturbing the equilibrium between soluble protein molecules and proteins incorporated into fibrils, since fibrils can fragment and create new nuclei.146 The possibility to fragment fibrils by mechanical forces has been commonly exploited to produce monodispersed short amyloid fibrils by the sonication of long filaments. For instance, Chatani et al. studied this process for β2-microglobulin.147 However, (ultra)sonication is not always required to produce low dispersity (in width) amyloid fibrils, as exemplified by the protocol used by the Mezzenga group to prepare well-defined β-lactoglobulin fibrils, which does not involve sonication.148
The influence of shear on the structure and mechanical properties of amyloid fibrils of this protein has been investigated using both controlled (steady shear in a Couette cell) or uncontrolled (stirring) shear flows. It has been observed that distinct morphologies (with different mechanical behaviour) can be obtained depending on the shear conditions.149 Couette shear induces amyloid fibril formation in β-lactoglobulin starting from a spheroidal seed-like species.150 In contrast, bovine serum albumin undergoes irreversible unfolding (without amyloid formation) in Couette flow.151 The influence of mechanical stress (linear shaking) on the fibrillization kinetics and on the morphology of glucagon has been examined.152 Studies of this type highlight the need for great care in the interpretation and comparison of amyloid formation kinetic data.
Many amyloid-forming proteins and peptides have surfactant-like properties and are active at the air–water interface. This leads to the possibility of using amyloid fibrils as emulsifying agents.153,154 For example, a designed β-sheet-forming peptide containing alternating phenylalanine and charged residues was able to act as a water–oil emulsifier.153 The fibrillization of α-synuclein is enhanced at the air–water interface compared to that at a solid–liquid interface because fibrils are selectively adsorbed at the air–water interface.155 Fibril nucleation is observed even without the presence of seeds, although fibril elongation is faster in bulk when seeds are added at a sufficient concentration.155 A designed coiled-coil peptide forms α-helices at the air–water interface, which can then transform into β-sheets, either with intrinsic slow kinetics or stimulated by the addition of metal ions, such as Zn2+.156 At the interface, increasing peptide conformation or parallel alignment (by compression) of the α-helical intermediates (which tends to pre-align the β-strands) has the greatest effect on β-sheet aggregation. The metal ions actually hinder aggregation of this peptide in bulk but not at the interface.156
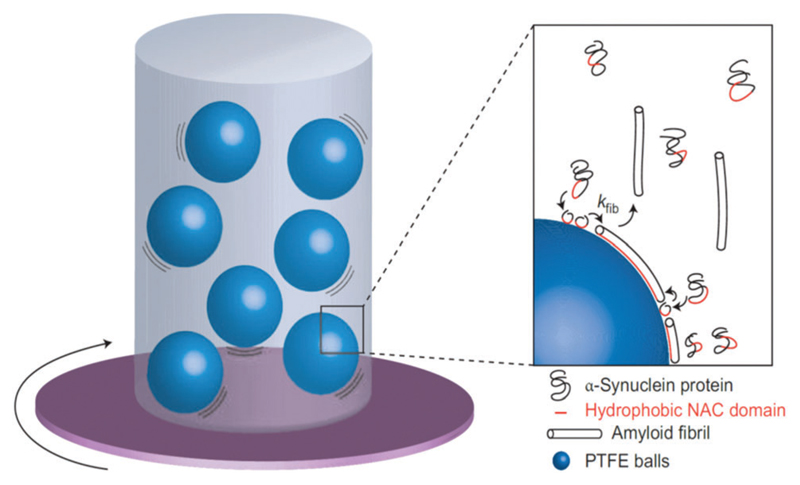
The presence of hydrophobic interfaces can influence the fibrillization of amyloid-forming proteins. Pronchik et al. studied the fibrillization of α-synuclein using a standard fluorescence dye technique used to assay amyloid formation.157 The kinetics of fibrillization in dilute aqueous solutions of the protein were monitored as a function of incubation time, the samples being subjected to agitation in the presence of different types of particles 1–2 mm in diameter (Fig. 4).158 The particles were made of borosilicate glass, which is largely inert, polymethyl-methacrylate (PMMA), which is slightly hydrophilic, or polytetrafluoroethylene (PTFE), which is hydrophobic. Some samples were also agitated in the presence of controlled volumes of air (which is hydrophobic). The fibrillization kinetics were found to depend strongly on the number of PTFE particles, i.e. to the hydrophobic surface area. The inverse lag time also increased in a non-linear fashion with the number of PTFE particles. Further nucleation and growth of fibrils was induced by the addition of PTFE particles to a sample containing fibrils that had already developed upon agitation in the presence of PTFE particles. An increase in dye fluorescence was observed in the presence of air, although fibrils were not observed using AFM. In the case of glass particles, no fibril formation was observed. Fibrillization was observed using PMMA particles, but to a much lower extent than with PTFE particles. As a further control, quiescent samples were examined and these showed no increase in ThT fluorescence in the absence of agitation. The fibrillization kinetics were proportional to the PTFE surface area, but not to the surface area of glass or PMMA. The contact angle of PTFE decreases dramatically in the presence of protein, showing that the protein coats the PTFE surface, progressively reducing the amount of available catalytically active interface. Moreover, the fact that the addition of more PTFE particles leads to the re-initiation of growth indicates that saturation of adsorption had not occurred since fibril-capable protein was still present in solution. Accelerated fibrillization was also observed in the presence of air, although the morphology of fibrils was different (globular aggregates were observed). These results clearly show the importance of hydrophobic interfaces in accelerating the fibrillization of the amyloid-forming protein α-synuclein. These findings provide an important insight to understanding the issues of sample-to-sample reproducibility that plague in vitro studies of amyloid fibrillization. The variability in morphology resulting from mixing in the presence of hydrophobic interfaces may also be important since fibril polymorphism, resulting, for instance, from sonication, has a profound effect on toxicity.159
Fig. 4.
Formation of amyloid fibrils by α-synuclein. A solution was agitated in the presence of particles of hydrophobic PTFE, slightly hydrophilic PMMA or chemically inert glass; experiments with air injected at the top of the cuvette were also performed. Inset: Proposed mechanism of the fibrillization. The aggregation of proteins into fibrils (at the rate constant kfib), caused by association of the protein hydrophobic NAC domains, is enhanced in the presence of the hydrophobic PTFE interface. Reprinted with permission from ref. 158. Copyright 2010, Nature Publishing Group.
The effect of lipid membranes on amyloid aggregation has been examined for several peptides including Aβ and α-synuclein.160,161 The importance of lipid interactions with Aβ is highlighted by the fact that apolipoprotein E (ApoE), (especially the ε4 allele) a key genetic risk factor for AD, is involved in lipid metabolism.162,163 Lipid membranes have a number of important roles in modulating amyloid fibrillization. These include: (partially) unfolding the peptide, increasing the local concentration of peptide bound to the membrane, orienting the bound protein in an aggregation-prone manner and by variation of the penetration depth into the membrane thus affecting the nucleation propensity.164 Lipid rafts are implicated in Aβ dimer and oligomer formation,165–167 and may provide platforms for the selective deposition of different Aβ aggregates (this also depends on the ordering of the lipids within the membranes, which may be different in the rafts168).169 Further information on the interaction of Aβ with membranes is available elsewhere.114
Lipid membranes are influenced by amyloid peptides and vice versa. Advanced fluorescence imaging techniques enabled membrane disruption caused by native and mutant forms of α-synuclein to be examined.170 It was shown that171 α-synuclein partially inserts into the outer leaflet of the lipid bilayer,172 and it was thought that this was due to the interaction with anionic lipid membranes.171,173 However, it was demonstrated that the protein was able to remodel lipid membranes from vesicles to tubules, even when the lipid membrane had no net charge.171
The cross-interaction of IAPP and Aβ peptides at lipid membranes has also been investigated.174 Mixed fibrils are formed at the anionic lipid raft membranes.174
2.3. Aggregation mechanisms and kinetics of proteins and peptides
2.3.1. Aggregation mechanisms of unfolded and folded proteins
In the previous sections we discussed the effect of several physicochemical parameters on the conversion of soluble monomeric peptides and proteins into insoluble amyloid fibrils. In most of these systems, the formation of amyloids is the consequence of an aggregation process under kinetic control. In this perspective, amyloid formation differs from other types of protein aggregation phenomena that are under thermodynamic control, such as protein oligomerization, precipitation and liquid–liquid phase separation. In light of this observation, the kinetics and the mechanisms of amyloid formation play a key role in determining the properties and functions of the final fibrillar products. Therefore, in order to design amyloid products with tailored functions, an understanding of the microscopic mechanisms underlying the aggregation process represents a crucial component.154
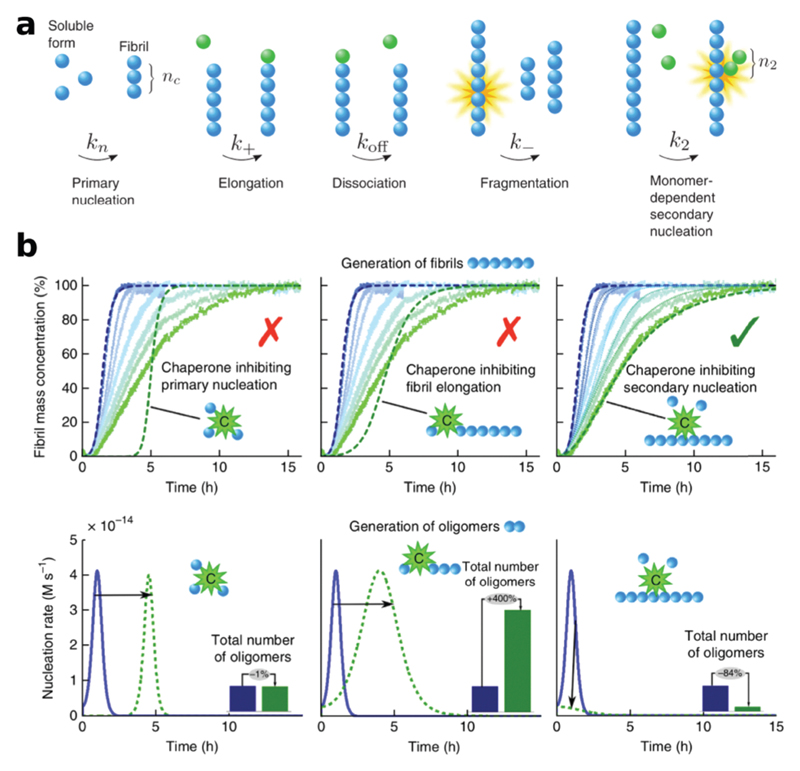
This task exhibits several challenges, since the formation of amyloids is the consequence of a complex aggregation network represented by several elementary reactions of nucleation and growth (Fig. 5a).175,176 The formation of amyloids is triggered initially by primary nucleation processes, which generate the first nuclei from soluble monomers. These nuclei can be represented by a variety of different small soluble species, which can be defined with different terminologies depending on their size, structure and reactivity. In the context of this section, we define generically this broad class of small assemblies as oligomers. Some of these species are non-reactive and off-pathway with respect to the transition into amyloids and accumulate in the system, whilst other oligomers are on-pathway and can further grow into protofibrils and eventually into mature filaments.177–179
Fig. 5.
(a) Individual microscopic events underlying the aggregation mechanisms of amyloids. (b) The identification of the aggregation mechanisms and the specific intervention on targeted microscopic reactions are fundamental for the rational design of tailored functions. This concept is illustrated here with the example of the peptide Aβ1–42, for which the generation of particularly active species can be modulated by inhibiting different microscopic steps. Reprinted with permission from ref. 225. Copyright 2015 Nature Publishing Group.
In the vast majority of amyloid systems, fibril growth occurs via elongation reactions, where a monomeric unit is incorporated at the end of an existing fibril via a diffusion motion over an energetic barrier.180 This process can in principle be reversible. However, the dissociation of monomers from fibrils is typically negligible, given the high thermodynamic stability of the fibrillar structure. In a very few cases, fibril growth can occur via fibril–fibril aggregation, as observed for amphiphilic peptides exhibiting complementary defects at the fibril ends.181
In addition to primary nucleation reactions, secondary nucleation processes have been increasingly identified in the aggregation of several amyloidogenic peptides. Such secondary nucleation processes involve typically the fragmentation of fibrils induced by either thermal energy or mechanical forces.146,182 These breakage events multiply the number of fibrils and increase the concentration of reactive fibril ends that can recruit monomers and elongate. Another common secondary nucleation process, originally identified in seminal studies on sickle haemoglobin,183 involves the generation of new oligomers catalyzed by the presence of the surfaces of existing fibrils.184–187 Such a surface-induced secondary process has been demonstrated to account for most of the production of toxic species during the aggregation of the peptide Aβ185 and, under certain conditions, also of α-synuclein.188 Importantly, these mechanisms of aggregation, commonly identified in vitro, are recently starting to find correlations also in in vivo studies performed using worms189 and mice models.190
The generic aggregation mechanism described above applies to both unfolded and globular proteins forming amyloids. A key difference between these two classes of proteins, however, is related to the monomeric form responsible for initiating and propagating aggregation. Indeed, short peptides and largely unstructured proteins are typically prone to form amyloid fibrils without the requirement of major conformational changes.54,191 In contrast, the formation of amyloids from proteins that are largely folded follows typically a pre-aggregation event that triggers the conversion of the native form into an aberrant conformation that is more aggregation-prone.
One of the most common events is protein misfolding, i.e. the conformational change of the initially folded state into an unfolded or partially folded intermediate.192–194 This is the case, for instance, for insulin,195–198 lysozyme,199,200 β2-microglobulin,201,202 enzyme superoxide dismutase (SOD1)203 and light chain immunoglobulin,204 which have been observed to form fibrils under conditions that promote the formation of partially folded species.
Other reactions that can trigger the formation of amyloids from globular proteins involve truncation of the protein205 or hydrolysis of the original polypeptide sequence into smaller fragments. For instance, lysozyme206,207 and β-lactoglobulin206,208,209 have been shown to form amyloid fibrils under acidic conditions after hydrolysis of the full-length protein into smaller peptides. In particular, two classes of peptides can be identified based on their reactivity: a series of peptides that convert over time into amyloids and a second sub-class that does not aggregate and remains soluble.208,209 An analogous system is represented by the amyloidogenic peptides Aβ1–40 and Aβ1–42, which are generated from the enzymatic cleavage of the amyloid precursor protein (APP).114 The higher aggregation propensity of short peptides with respect to the precursor globular protein is not surprising, since steric constraints disfavour the thermodynamic stability of amyloid fibrils with respect to the soluble state for polypeptide sequences longer than 100 residues.54 Indeed, for large globular proteins, such as immunoglobulins, the formation of amorphous fractal-like aggregates rather than fibrils is typically more favoured.210–212
The formation of individual amyloid filaments can be followed by additional supramolecular events, leading to the generation of 2D and 3D amyloids. These additional processes include lateral fibril–fibril association206 as well as the formation of nematic phases213,214 and gels.154,215 This rich phase behaviour opens a route to finely tuning the morphology and the mechanical properties of supramolecular fibrillar hydrogels and other soft materials by carefully controlling the individual events underlying the aggregation process. This observation highlights once more the importance of identifying the aggregation mechanisms to allow rational design in structure–function studies of amyloid materials.
An attractive strategy to modulate the aggregation mechanisms in a tailored way consists of introducing into the system suitable reactive species. For instance, aggregation reactions can be seeded by adding preformed fibrils or other non-native species, which can trigger the aggregation of physiological monomers following prion-like mechanisms. As discussed previously, other important heterogeneous nucleation events involve the presence of air–water interfaces,155 hydrophobic surfaces157,216 and vesicles,160 which are particularly prone to trigger the formation of amyloid fibrils, although the exact mechanisms underlying these effects are only starting to be elucidated.
We conclude this paragraph by highlighting two emerging directions in amyloid mechanistic studies: a first activity is aimed at increasing our understanding of the microscopic steps underlying the generation of the oligomers.217 This topic is clearly relevant to understand the toxicity associated with the aggregation process in biological systems, and is also crucial to clarify the safety issues associated with the use of amyloid biomaterials for healthcare applications. A second important direction is the description of the behaviour of proteins at high concentrations, which underlies several biotechnological and biological applications. Indeed, under these conditions, the quaternary state of proteins is governed by complex physics, since an increase in protein concentration can both change the phase diagrams and accelerate the rate of nucleation and growth reactions by increasing the activities of the reagents. There is therefore a need to correlate the thermodynamic phase behaviour with the kinetic aspects of the aggregation processes.
2.3.2. Identifying aggregation mechanisms from kinetic studies
In order to identify the microscopic aggregation mechanisms in a particular system, it would be convenient to fully characterize the large class of intermediate species populated during the aggregation process. However, several of these intermediate species are transient and present at low concentrations, and therefore are challenging to characterize experimentally. Indeed, biophysical assays for structural studies are typically well suited to characterize only the initially soluble monomeric state and the final insoluble fibrillar aggregates.
To address this limitation, in analogy with other branches of chemistry and protein biophysics, chemical kinetics is emerging as a powerful tool to investigate amyloid aggregation mechanisms at the molecular level from the measurements of macroscopic rate laws.146,193,218 By recording the global aggregation profiles at different protein concentrations, the reaction orders can be extracted, and compared with integrated laws based on mathematical models describing different microscopic mechanisms.219 One of the greatest advantages of this method is the possibility to extract information on multiple microscopic events of nucleation and growth from a limited number of experimental macroscopic read-outs, which typically include the monomer conversion or the total amount of aggregates formed over time.
For instance, high-throughput assays have been well established to monitor the formation over time of the total fibril content. A conventional method is based on a ThT fluorescent assay,220,221 which relies on an increase in the fluorescence yield of the dye upon binding to the characteristic β-sheet structure of the fibrils. The time evolution of the total fibril content typically exhibits a sigmoidal profile, where a lag phase is followed by an exponential growth regime and eventually by a plateau related to consumption of the soluble monomer. It is important to note that the microscopic reactions described in the previous section are present during all the stages of the aggregation process, even at the early beginning of the reaction. Indeed, although it may be tempting to consider the lag phase as a waiting time, this period represents the time required by the fibrils to reach a critical concentration that is detectable by the experimental assay.222,223
The application of chemical kinetics in amyloids has been hampered for a long time by the complexity of the non-linear aggregation scheme described in the previous paragraph, which has challenged the derivation of analytical rate laws. Moreover, the high sensitivity of amyloids to several physicochemical factors complicates the establishment of robust kinetic assays, which often suffer from irreproducibility issues. Advances in theoretical analysis,146 and the development of optimized experimental protocols,224 however, have recently opened the possibility to apply the kinetic platform to several amyloidogenic systems, leading to the identification of the aggregation mechanisms under a broad range of conditions.
An attractive advantage of kinetic studies is the high sensitivity in detecting, also with high resolution, changes in the aggregation mechanisms derived from modulation of the reagent composition or of intrinsic and extrinsic factors.160,185,188,189,225–227
Of particular interest is the analysis of the changes in the aggregation mechanisms in the presence of inhibitors of amyloid formation. This information is particularly important in the biomedical context of the search for drugs to fight against amyloid-related disorders, where a kinetic inhibition of the aggregation process can represent an effective strategy to avoid the onset and development of the associated disorders over a characteristic lifespan.114 It is becoming apparent, however, that this approach cannot be achieved simply by a generic inhibition of the aggregation process but requires a specific intervention aimed at targeting specific microscopic events that are most responsible for the formation of toxic species, in particular oligomers.228 In this context, the application of chemical kinetics is fundamental to identifying the specific processes that are affected by the presence of different modulators.228,229
A recent example of the importance of this activity has been demonstrated with the peptide Aβ1–42, whereby the application of chemical kinetics and the understanding of the microscopic mechanisms underlying the aggregation process has opened the possibility to tune in a controlled way the generation of specific intermediates characterized by a particularly high level of toxicity, as shown in Fig. 5b.225 This platform allowed the identification of a biological molecule that can selectively suppress the secondary nucleation reaction and therefore the generation of the oligomers. By contrast, the specific targeting of primary nucleation and the elongation rate, although equally efficient in delaying the formation of the fibril amount (top panels), cannot deplete the oligomer formation (bottom panels). This outcome would have not been achievable by means of experimental characterization alone, and thus highlights the importance of theoretical mechanistic frameworks in the structure–activity studies of amyloids.
It is envisioned that in the near future improvements in experimental assays to detect oligomers217,230,231 will enable researchers to perform kinetic studies specific to these species, thereby improving the understanding of the microscopic steps responsible for the formation of these important intermediates.
2.3.3. Aggregation mechanisms and fibril length distribution
In the previous paragraphs, we discussed the importance of identifying aggregation mechanisms in amyloids and we described the use of chemical kinetics as one of the major tools to perform this operation. A particularly important aspect of these activities that deserves special attention in material sciences is the characterization of the time evolution of the fibril length distribution. Indeed, fibrils with different lengths are associated with drastically different mechanical properties and activities, including different toxicity in biological systems.232
From an experimental point of view, different techniques have been successfully applied to characterize the fibril length of amyloids. Single-molecule imaging techniques, including AFM,209,233–237 EM225 and super resolution fluorescence microscopy,238,239 provide a high level of resolution by analysis of a large number of individual filaments. Alternative bulk methods have also been recently developed based on the indirect evaluation of the fibril length from measurements of the physicochemical properties, such as the rotational240 or translational diffusion coefficient241,242 or the sedimentation coefficient.243–247
In addition to the improvements in the experimental characterization, recent progress in the analytical treatment of kinetic models has allowed the derivation of compact expressions describing the dependence of the fibril length distribution on key kinetic parameters.248,249 Experimental information on the full-length distribution provides a large number of constraints for comparisons to be made between model simulations and experimental data. Thereby, the robustness and the refinement of the derived aggregation mechanisms are significantly increased with respect to kinetic analysis that relies only on the comparison with a limited number of average quantities of the fibril population. These more refined models lead to a better understanding of the relationship between aggregation mechanisms, the fibril length distribution and product functions.
3. Atomic to mesoscopic structure of protein and peptide amyloids
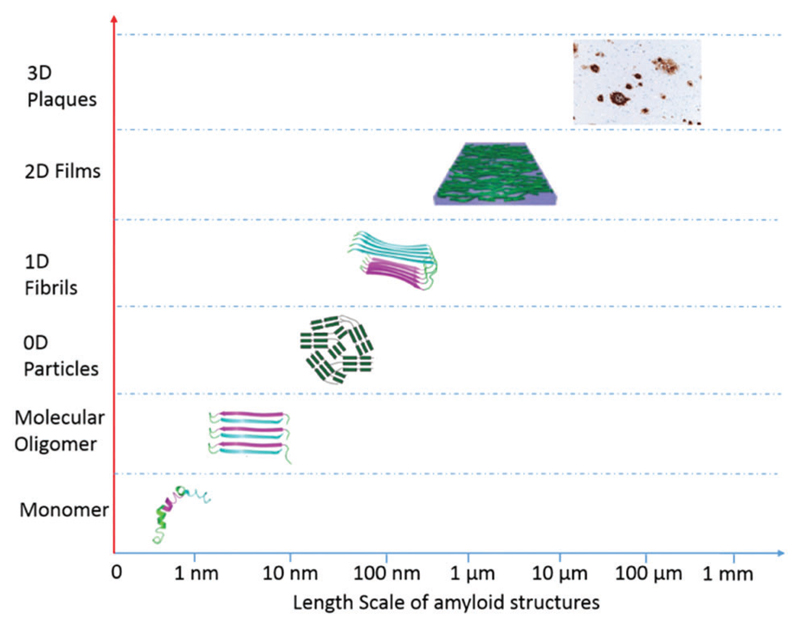
In the above part, we demonstrated and discussed the various self-assembly and aggregation mechanisms of amyloid-forming proteins and peptides. It is clear that small differences in molecular aggregation and self-assembly are responsible for the formation of a wide variety of amyloid nanostructures. Recently, Luo and co-workers reviewed recent advances in the protein assembly for the fabrication of various nanostructures by biotechnological and chemical strategies.250 Although that review did not focus specifically on amyloid fibrils, it reviewed the state of the art on the use of protein assemblies as versatile platforms for designing attractive functional nanostructures. In this section, we focus on the atomistic to mesoscopic structures of a number of amyloid assemblies, including molecular oligomers, 0D aggregates (nanoclusters, nanoparticles, nanotriangles, squares and loops), 1D aggregates (protofibrils, nanofibrils, nanoribbons and nanotubes), 2D aggregates (sheets, films and membranes) and 3D amyloid plaques and scaffolds.
3.1. Amyloid oligomers
Due to their ubiquitous presence in the brains of patients suffering from many neurodegenerative diseases and their apparent cytotoxicity in vitro, insoluble peptide and protein amyloid aggregates were assumed to be the cytotoxic culprit in these diseases.251 However, new evidence suggests that prefibrillar soluble amyloid oligomers with low molecular weight could be the primary toxic species responsible for neuron death in both Alzheimer’s and Parkinson’s disease.114,252,253 Although there is no universal consensus on what constitutes an amyloid oligomer, they can typically be considered to possess some or all of the following biochemical and biophysical characteristics:254 they are molecular aggregates with β-sheet-rich structures composed of between 2 and 30 assembled monomers; they possess various sizes and morphologies, are soluble in aqueous solutions; their morphology is polymorphic and time-dependent and they can aggregate into long, stable mature amyloid fibrils. Previously, a number of excellent review articles on the structure, formation mechanism and toxicity of natural and artificial amyloid oligomers have been reported.114,255,256
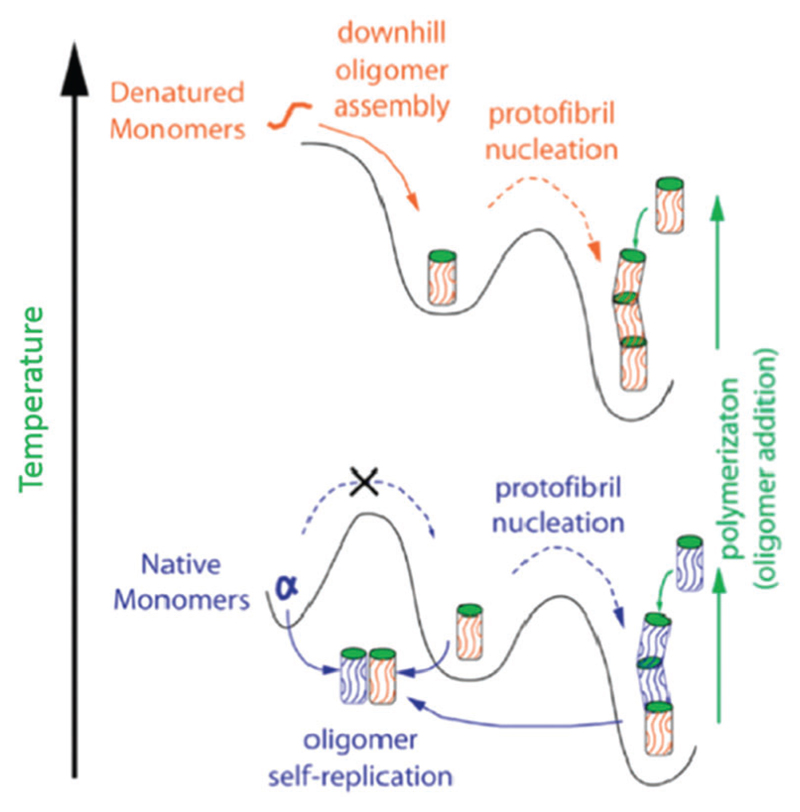
A range of techniques, including AFM,257 EM,258 attenuated total reflection (ATR)-FTIR,259 NMR260 and single particle confocal microscopy,261 have been used to investigate oligomeric structures and the conformational transition between oligomers and mature fibrils. Mulaj et al. combined ThT fluorescence assays, static and dynamic light scattering (S/DLS), AFM and ATR-FTIR to assess the stability, kinetics and structure of hen egg-white lysozyme during its transition from oligomeric species to protofibrils and nanofibrils.257 They found that the amyloid oligomers and protofibrils but not latter stage filaments were responsible for the amyloid growth at both physiological and denaturing temperatures, as shown in Fig. 6. Their results led them to suggest that at physiological temperatures amyloid seeds cannot form spontaneously from native lysozyme monomers. Oligomer self-replication from native monomers at physiological temperature is required to promote protofibril nucleation and the further assembly into mature nanofibrils (bottom part of Fig. 6), which can also be created by using the denatured monomers and thus elevated temperatures (upper part of Fig. 6). This study outlined the self-replication ability of amyloid oligomers and protofibrils as distinct assembly pathways, and is important for understanding the molecular mechanisms and aggregation behaviour of both pathological and functional amyloid materials.
Fig. 6.
Schematic assembly pathways of lysozyme oligomers at both denaturing and native temperatures. Reprinted with permission from ref. 257. Copyright 2014, American Chemical Society.
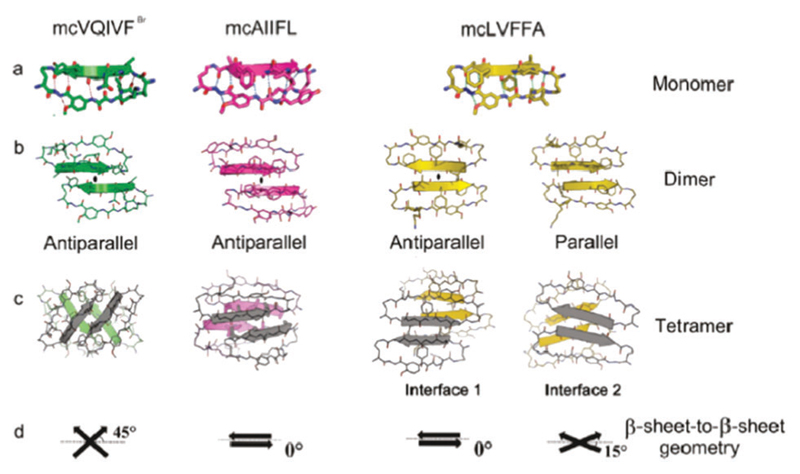
X-ray crystallography and computer simulations have been widely utilized to investigate the atomic structure of amyloid oligomers that can assemble into crystalline morphologies.254,262,263 Liu and co-workers designed a series of macrocyclic peptides based on Aβ and Tau proteins (associated with AD) that formed amyloid oligomers with a crystalline morphology.254 Fig. 7 shows the atomistic structures of three macrocyclic peptides: mcVQIVFBr, mcAIIFL and mcLVFFA, as monomers (Fig. 7a), dimers (Fig. 7b) and tetramers (Fig. 7c) as determined by X-ray crystallography and atomistic simulations. In the monomers, the interactions between the neighbouring strands are limited to the backbone hydrogen bonding (a typical β-sheet structure). For the dimers, the formed intermolecular β-sheets could be aligned in either parallel or antiparallel orientations via hydrogen bonding interactions. In addition, tetramers could be formed by the complementary side-chain interactions of dimers with different molecular packing geometries, as shown in Fig. 7c and d. These findings are helpful for understanding the assembly of amyloidogenic oligomers at the atomic level and offer clues for the design of structure-based therapeutics against amyloid diseases. More recently, the same authors solved the crystal structure of a toxic amyloid oligomer of an 11-residue segment (KVKVLGDVIEV) from the Aβ protein.262 Separately, Domanska et al. demonstrated the utility of nanobodies to trap and characterize the crystalline intermediates of β2-microglobulin amyloids by X-ray crystallography.263
Fig. 7.
(a–c) Crystal structure of macrocyclic peptides with (a) monomer, (b) dimer, and (c) tetramer. (d) Several interaction modes of dimers to form a tetramer. Reprinted with permission from ref. 254. Copyright 2011 American Chemical Society.
Amyloid oligomers with distinct molecular structure and morphology can be created by altering the assembly environment (e.g. temperature, pH) and changing the pathways of monomer aggregation. Alternatively, aggregation can be modified by promoting interactions between protein/peptide monomers with additional biomolecules (e.g. non-amyloid proteins or macromolecular sugars).264 Elucidating the structure of these oligomeric species at the atomic level will promote an understanding of the formation mechanisms and toxicity of amyloid structures.
3.2. 0D amyloid aggregates
By 0D objects we refer to aggregates/clusters, in which there is not a dominant dimensional feature, as in 1D or 2D objects, but for which self-limiting size is observed, differently from 3D aggregates. Numerous 0D amyloid aggregates have been generated in vitro. Observed morphologies include nanoparticles,265–268 nanospheres,269,270 loops, triangles, squares and rings.271–273 In this section, we review the main preparation strategies for these nanostructures and their corresponding formation mechanisms.
3.2.1. Nanoparticles and nanospheres
Prefibrillar amyloid structures, such as spheroidal aggregates (nanoparticles and nanospheres), similar to the molecular oligomers introduced previously, have been frequently proposed to be a highly cytotoxic species in many neurodegenerative diseases.265,274 Silveira and co-workers degraded large prion protein (PrP) aggregates into smaller PrP nanoparticles with sizes ranging from 17 to 27 nm,265 and then characterized the PrP nanoparticles with DLS, non-denaturing gel electrophoresis and transmission electron microscopy (TEM). Their findings suggested that the PrP nanoparticles with masses equivalent to 14–28 PrP molecules are the most infectious initiators for prion diseases. In another study, EI Moustaine et al. formed amyloid nanofibrils and nanoparticles from recombinant PrP at high pressure.266 This study provided insights into the initial molecular processes that lead to misfolding and eventually self-assembly into higher order structures.
Other strategies besides elevated pressure and the degradation of larger aggregates have been investigated for amyloid–nanoparticle fabrication. Fändrich et al. reported the creation of Aβ1–40 amyloid peptide nanoparticles by a simple self-aggregation method (Fig. 8a).267,268 Typically, Aβ1–40 with a concentration of 2.5 mg mL−1 was first dissolved in 100% HFIP, and the solution was diluted 10-fold with ultrapure water after 10 min incubation. Amyloid nanoparticles with sizes ranging from 15 to 30 nm were formed after a further 15 min incubation, as shown in the TEM image in Fig. 8a. In another study, Guo and co-workers reported that it is theoretically possible for triphenylalanine (FFF)-based peptides to self-assemble into nanospheres using large-scale coarse-grained molecular dynamics simulations.269 The simulations showed that the FFF nanospheres were formed and stabilized by peptide–peptide electrostatic interactions, vdW interactions and strong peptide–solvent interactions.
Fig. 8.
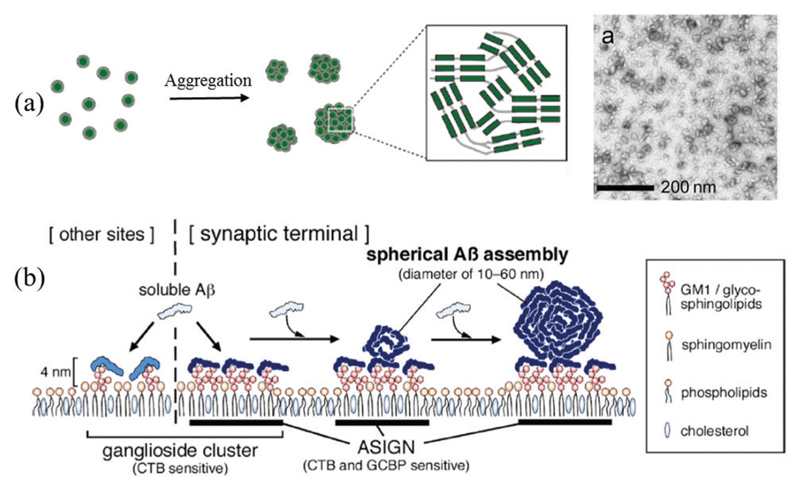
Typical strategies for creating 0D amyloid (a) nanoparticles and (b) nanoclusters: (a) Aβ oligomer self-aggregation, (b) lipid bilayer membrane-induced Aβ assembly. CTB is cholera toxin B subunit. ASIGN is Aβ-sensitive ganglioside nanocluster. Images (a and b) are reproduced with permission from (a) ref. 268, Copyright 2014, American Chemical Society, and (b) ref. 270, Copyright 2013, American Chemical Society.
Matsubara and co-workers demonstrated that Aβ1–40 peptides can be induced to self-assemble into nanospheres on synapsemimicking lipid membranes (Fig. 8b).270 They showed that Aβ binding and assembly was promoted on GM1 lipid domains. This was found to be due to the presence of an Aβ-sensitive ganglioside nanocluster (ASIGN) within the glycosphingolipids (GM1) domain. The corresponding AFM images indicated that a thin Aβ layer and Aβ nanospheres were formed simultaneously. This study outlines a possible lipid-mediated assembly mechanism of Aβ proteins that may occur in the AD brain.
3.2.2. Annular oligomers: loops, triangles, squares and rings
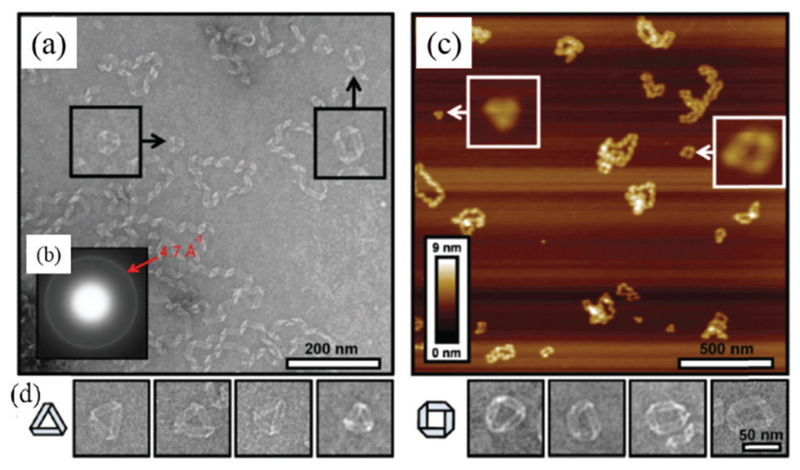
Other more complex 0D amyloid nanostructures have also been observed for a variety of amyloidogenic sequences, including loops, triangles, squares and rings.
Conway and co-workers reported the creation of annular oligomers of α-synuclein when comparing the aggregation behaviour of wild-type (WT) and a homo-mutant form (A53T) of α-synuclein connected to early-onset Parkinson’s disease.271 The nanoscale annular oligomers were formed from equimolar mixtures of WT and A53T protein. It was found that the acceleration of oligomerization but not fibrillization is a shared property of α-synuclein mutations, which suggests that the nonfibrillar intermediates, including annular oligomers, may be critical in pathogenesis. Elsewhere, Hatters et al. reported the preparation of annular oligomers from the aggregation of human apolipoprotein C-II (ApoC-II).272,273 CD indicated a time-dependent increase in the amount of β-sheet structure. After incubation for 48 h, the amyloid aggregates were measured by TEM and AFM, and ordered closed loops with a diameter of 50–75 nm were observed. The above studies demonstrated an alternative folding pathway of human apolipoproteins. Two loop formation models, specifically the worm-like chain and randomwalk approaches confirmed that the formation of annular oligomers is critically dependent on the fibril flexibility, which allows fibrils with appropriate lengths to bend back and anneal end-to-end to form a loop.273 In addition, Wong et al. found that another apolipoprotein, ApoA-I, can also form loop-like structures with a periodicity in the range of 25–60 nm.275
Recently, de Messieres et al. reported that ApoC-III protein can aggregate into 0D amyloid triangles, small squares and loops.276 The formed amyloid loops (Fig. 9a and c) show ribbon-like structures consistent with a helical twist, which is similar to the loop structures formed by α-synuclein,271 ApoC-II272,273 and ApoA-I.275 The corresponding electron diffraction pattern (Fig. 9b) shows a typical interstrand spacing for the β-sheet conformation, suggesting the amyloid loops are formed by the self-assembly of β-sheets. Other amyloid structures, including triangles and squares, were also observed, as shown in Fig. 9d.
Fig. 9.
Amyloid triangles, squares and loops of ApoC-III. (a and b) TEM image of loops and the electron diffraction pattern, (c) AFM image of loops and (d) TEM images of triangles and polyhedra. Reproduced with permission from ref. 276. Copyright 2014, American Chemical Society.
The formation of loop-like structures in amyloid oligomers is related to the structures and properties of the constituent proteins. For example, the apolipoproteins (i.e. ApoA-I, ApoC-II and ApoC-III) have similar helical conformations in an annular morphology as they have when they are bound to lipid membranes,277 and all three proteins are found on high-density lipoprotein and can mediate lipids to form plaques with a similar size and shape to lipoprotein particles.
Studies have suggested that the formation of an annular amyloid structure may be related to the “channel hypothesis”.278 This hypothesis proposes that cell death in neurodegenerative diseases occurs via a disruption of cellular homeostasis due to unregulated calcium (and other ion) transport across the cell membrane. This is thought to occur due to the presence of annular oligomers of Aβ peptides or proteins (e.g. α-synuclein) that insert themselves into the cell membrane and create aberrant ion channels. Ding et al. found that annular α-synuclein protofibrils could be produced when incubating spherical amyloid oligomers in solution or by allowing them to adsorb onto ex vivo brain-derived lipid membranes.279 In their study, two distinct oligomeric morphologies were formed, namely spherical and annular protofibrils. The annular protofibrils were formed by incubating spherical oligomers for prolonged periods. In addition, membrane-associated annular protofibrils were observed by binding the spherical protofibrils to brain-derived lipid membranes. Further studies indicated that annular oligomers of α-synuclein resulted in a more rapid formation of pores or ion-permeable channels than the soluble monomeric α-synuclein, providing strong evidence for the neurotoxicity of small annular α-synuclein oligomers.280–282 Kayed et al. studied the formation of annular Aβ and α-synuclein protofibrils in solution and on lipid membranes and proposed a possible formation mechanism of the annular protofibrils at the surface of lipid membranes.283 Their mechanism states that spherical oligomers first interact with the membrane, and then the additional oligomers are recruited to the lipid bilayer, conjugate with the bound oligomers to form a β-barrel pore.
Whilst the “channel hypothesis” does provide a compelling explanation for neurotoxicity via uncontrolled calcium influx into cells, little direct evidence has been provided either in vitro or in vivo. Additionally a number of alternative mechanisms by which amyloid oligomers can induce similar cell membrane disruption have been proposed. These include membrane thinning,284 excessive membrane tubulation285,286 or membrane extraction through amyloid–lipid co-aggregation.170,287 Further research into the mechanisms driving the assembly of annular amyloid oligomers, both in solution and on model cell membranes, will help to elucidate the importance and relevance of the ”channel hypothesis“ in relation to the alternative proposed mechanisms of cell membrane deregulation by amyloid oligomers.
3.3. 1D amyloid superstructures
In this section, the self-assembly and formation of 1D peptide and protein nanofibrils, twisted and untwisted nanoribbons, helical ribbons and nanotubes are introduced and discussed in detail.
3.3.1. Amyloid protofilaments, protofibrils and nanofibrils
Here, we focus on the formation and superstructures of amyloid protofibrils and nanofibrils,288 and place particular emphasis on fibrils possessing twisted, helical and chiral morphologies.289
In general, all 1D amyloid superstructures are found to have a highly hierarchical morphology in which constitutive 1D fibril precursors combine to form mature amyloid structures. The terminology used for the 1D fibrillar precursors is not consistent throughout the literature, with the terms protofilaments or protofibrils loosely interchangable in most reports. Here, we adopt the view that protofilaments are the simplest mature 1D building block of amyloids, whilst protofibrils are a form of amyloid that has not yet reached the mature fully-formed stage. According to this terminology, protofilaments form individually first, then assemble into loosely packed protofibrils, which further assemble into more ordered mature amyloid fibrils. Fig. 10 shows an example of such a process through individual snapshots resolved by AFM.240 The white arrows highlight the points at which different protofilaments start to attach and overlap to form protofibrils.
Fig. 10.
Individual protofilaments of β-lactoglobulin aligning and starting to attach at specific points to form first protofibrils and finally mature fibrils. Reproduced with permission from ref. 240. Copyright 2011 Royal Society of Chemistry.
Mature (1D) amyloid fibrils are then generally formed from a number of intertwined protofilaments, each being 2–5 nm in diameter and up to a few mm in length.235 Variations in the packing of these protofilaments result in a wide variety of morphologies, all with potentially different functions. Insight into the packing mechanisms can be achieved with various nanoscale analytical techniques.
Stroud et al. studied the structure and properties of oligomers of Aβ1–42 with XRD, TEM, CD, FTIR and chromatography.290 They found that the peptide molecules could stack into short protofilaments consisting of pairs of helical β-sheets, which wrapped around each other to form a superhelical structure. In another example, Dearborn and co-workers utilized cryo-TEM and scanning transmission electron microscopy (STEM) to study the in vitro self-assembly of α-synuclein.291 They found that protofibrils have a typical size with a length of 7.5 nm and width of 2.5 nm.
In the following part of this section, we discuss the various 1D morphologies observed and how they can affect the biological, biomedical or nanotechnological functions.
3.3.2. Twisted ribbons and helical ribbons
Many multifilamentous morphologies have been observed, including twisted ribbons, helical ribbons and rippled structures. Twisted ribbons are characterized by a saddle-like (Gaussian) curvature; helical ribbons on the other hand are characterized by a mean curvature but zero Gaussian curvature: in other words, they can be wrapped around a cylinder. The transition from a twisted to helical ribbon in amyloids is now well understood and has already been reviewed extensively.154,292 In general, a twisted to helical transition is observed upon an increase in the number of protofilaments (width to thickness ratio), as a consequence of a different way to store bending and torsional energy by twisted vs. helical ribbons. Helical ribbons can finally close into nanotubes, eliminating the extra energy associated with edge line tension. This mechanism and the associated entire series of transitions have been observed both in peptide- and protein-based amyloids.29,206,293 More common, however, is the presence of one individual polymorphic form observed at distinct time-points.
α-synuclein fibrils have been shown to co-exist as left- and right-handed helical ribbons with differing pitches.77 Adamcik et al. examined the different stages of aggregation throughout heat-denatured β-lactoglobulin amyloid fibril assembly with single-molecule AFM and theoretical analysis.233 They found that the mature fibrils have a multistranded left-handed twisted morphology. Fig. 11a presents the typical AFM images and corresponding coarse-grain molecular dynamics reconstructions of the left-handed helical β-lactoglobulin fibrils. The β-lactoglobulin nanofibrils were shown to have persistence lengths of between 1 and 4 μm and maximum heights of 2, 4, 6, 8 and 10 nm for 1, 2, 3, 4 and 5 twisted filaments, respectively. In addition, the helical pitch of nanofibrils showed a clear proportional increase from 35 to 135 nm with increasing filament number. This work provided a general model for understanding amyloid fibril assembly into a twisted ribbon morphology. In another study, Fitzpatrick and co-workers investigated the atomic structure and assembly of fibrils formed from the TTR105–115 peptide. As with α-synuclein and β-lactoglobulin, TTR105–115 was found to assemble into mature fibrils via the hierarchical assembly of β strands of peptide molecules into protofilaments, which further intertwined to form mature fibrils with a twisted ribbon structure (Fig. 11b).294
Fig. 11.
Twisted and helical amyloid ribbons: (a) left-handed β-lactoglobulin nanofibrils with multistranded twisted filaments. Reprinted with permission from ref. 233. Copyright 2010, Nature Publisher. (b) Cross-β amyloid TTR105–115 fibril with triplet atomic-resolution structure. Reprinted with permission from ref. 294. Copyright 2013, National Academy of Sciences. (c) Twisted right-handed helical ILQINS hexapeptide ribbon. Reproduced with permission from ref. 297. Copyright 2014, American Chemical Society. (d) Twisted double-helical peptide ribbon, Reprinted with permission from ref. 298. Copyright 2009, Wiley-VCH Verlag GmbH & Co. (e) Amyloid-inspired rippled β-sheet ribbons by the co-assembly of enantiomeric amphipathic peptides. Reprinted with permission from ref. 299. Copyright 2012, American Chemical Society.
In general, there is a well-defined linear relationship L ∝ n between the periodicity L of twisted ribbon amyloids and the number of constitutive protofilaments n.233 Using coarse-grained simulations, Assenza et al. showed that the relationship between the twisted ribbon periodicity and the number of protofilaments has a pseudo-linear behaviour at small n, i.e. L ~ (3n2 − 7)1/2. When the number of protofilaments increases and the fibrils approaches maturity, the relationship with periodicity approaches a truly linear behaviour.295 This has led to the conclusion that this behaviour is a universal mesoscopic signature of amyloid fibril polymorphism.
Twisted and helical ribbons with different structures have also been created by selecting specific peptide sequences and controlling their molecular self-assembly. For example, Uesaka et al. investigated the self-assembly of the histidine (his)-containing helical peptides of the form A3-Hisn-B, where A is a hydrophilic polysarcosine chain and B is a hydrophobic helical dodecapeptide.296 Dependent on the pH of the peptide solutions, the molecular assemblies formed different morphologies, including twisted ribbons, helical ribbons and nanotubes. The A3-His2-B peptide formed twisted ribbons, helical ribbons and nanotubes at pH 3.0, 5.0 and 7.4, respectively, whilst A3-His-B only formed helical ribbons at pH 3.0. The pH-dependent morphological changes were explained by variations in the electrostatic repulsion, which in turn affected the molecular packing of the peptides. This study showed a very effective pH-responsive strategy for creating adjustable 1D amyloid structures by controlling the intermolecular interactions.
Lara and co-workers reported that a hexapeptide (ILQINS) identified as being an amyloidogenic sequence in left-handed helical ribbons formed from hen egg-white lysozyme self-assembles into right-handed helical ribbons and crystals.297 At short incubation times, multistranded right-handed helical ribbon structures were observed (Fig. 11c). At longer incubation times, the right-handed ribbons were almost entirely replaced by aggregates with a crystalline structure. Hamley and co-workers demonstrated the formation of left- and right-handed twisted helical amyloid ribbons by assembling the peptide fragment (KLVFF) modified with two β2-alanine residues (β2Aβ2A-KLVFF), as shown in Fig. 11d.298 In some cases, the left- and right-handed twisted helices were intertwined into a double-helix amyloid ribbon, as shown in the inset image. The formation of helical ribbons is ascribed to both functional motifs of the designed peptide, in which the KLVFF motif is responsible for the self-assembly to cylindrical fibrils and the β-amino acids are crucial for the formation of helical nanoribbons. A sequence of twisted fibrils, helical ribbons and nanotubes were observed as kinetic states during the aggregation of the capped version of this peptide, where globular structures were also observed as the initial state.293 The closure into nanotubes was very slow, being observed only after several weeks.
Swanekamp and co-workers reported the formation of rippled β-sheet l/d-cofibrils from the co-assembly of two enantiomeric amphipathic peptides: l-(FKFE)2 and d-(FKFE)2, as shown in the inset of Fig. 11e.299 The l-(FKEF)2 peptide self-assembled in water into left-handed helical fibrils, whilst the d-(FKFE)2 peptide self-assembled into enantiomeric right-handed fibrils (TEM image of Fig. 11e). The equimolar mixing of l-(FKFE)2 and d-(FKFE)2 created a new fibril type, which contained alternating l- and d-peptides in a rippled β-sheet orientation.
Lashuel and co-workers demonstrated the creation of polymorphic β-sheet quaternary structures, including fibrils and ribbons, by using a small number of peptidomimetics.300 They found that the distribution of quaternary amyloid structures could be adjusted by manipulating the pH, buffer conditions and ionic strength. Their study indicated that it is possible to control both the self-assembly of designed peptide structures and their lateral interactions to create untwisted amyloid ribbons.
Adamcik and co-workers created large multistranded amyloid ribbons from the microtubule-binding fragment (VQIVYKPVD LSKVTSKCGSLGNIHHK, known as R3) of Tau protein.301 Tau does not aggregate spontaneously in vitro, but it undergoes aggregation in the presence of polyanions, such as heparin.302 The peptide motif, VQIVYK, plays critical roles in the self-assembly and formation of β-sheet amyloid aggregates of Tau. Therefore, the self-assembly of R3 in both the presence and absence of heparin was investigated. In the presence of heparin, R3 fibrils with a normal twisted fibrillar morphology were rapidly formed, as seen by AFM, TEM and strong ThT binding. In the absence of heparin, aggregation was much reduced, as evidenced by decreased ThT binding; however, amyloid ribbons consisting of large numbers of laterally associated protofilaments were observed (Fig. 12a). Increasing incubation times up to 1 week demonstrated the continuous growth of the multifilamentous ribbons, eventually forming giant multistranded amyloid ribbons with 2D laminated structures (composed of over 45 laterally associated protofilaments of 350 nm total width).
Fig. 12.
Multistranded amyloid ribbons: (a) Tau protein R3 ribbon, reprinted with permission from ref. 301. Copyright 2016 Wiley-VCH Verlag GmbH & Co. (b) Multistranded hIAPP20–29 ribbon, reprinted with permission from ref. 303. Copyright 2013, National Academy of Sciences. (c and d) Lysozyme (c) and β-lactoglobulin (d) ribbons, reprinted with permission from ref. 206. Copyright 2011, American Chemical Society. (e) Amelogenin ribbon, reprinted with permission from ref. 305. Copyright 2016, Nature Publishing Group.
Recently, Zhang et al. studied the self-assembled structures of hIAPP20–29, the amyloidogenic core fragment relevant to Type II diabetes, by quantitative nanomechanical AFM.303 They provided strong evidence of the coexistence of fibrils with flat ribbon and helical ribbon morphologies, as shown in Fig. 12b. As in the work of Adamcik et al.,301 the ribbons show clear striations along their long axis, indicating the presence of multiple parallel protofilaments (inset of Fig. 12b). AFM-based force–volume and nanoindentation measurements indicated that the flat ribbon structure had higher stiffness than helical ribbons, suggesting that the core of the helical ribbons were hollow.
Some proteins, such as lysozyme,206 β-lactoglobulin206 and amelogenins,304–306 can also form multistranded amyloid ribbons over time. Lara et al. introduced a general self-assembly mechanism for converting hydrolyzed globular lysozyme and β-lactoglobulin into multistranded amyloid ribbons (Fig. 12c and d).206 After long periods of time (up to 100 hours), multistranded ribbons with widths up to 173 nm were observed due to a modular lateral assembly of around 17 protofilaments. This study provided novel insight into the fibrillation mechanisms of globular proteins and amyloid polymorphism. Recently, Carneiro and co-workers showed that recombinant human full-length amelogenin protein (rH174) can self-assemble into amyloid-like ribbons, both in vitro and in vivo.305 In the presence of calcium and phosphate, rH174 assembled into highly aligned ribbons on a glass substrate (Fig. 12e) with widths of 23 nm and a regular peak-to-peak distance of approximately 69 nm.
The above studies show that multistranded amyloid ribbons can be fabricated from a range of proteins or amyloidogenic peptides (i.e. R3, KLVFF and VQIVYK). Examples such as these increase our understanding of how peptide sequences and the reaction conditions affect the final morphology of the assembled structures. A detailed knowledge of the factors underpinning assembly into mature fibrils should allow the routine rational design of amyloid-forming peptides containing specified amino acid sequences additional to the amyloidogenic core. This will enable us to tailor the structure and function of the fabricated assemblies for a range of applications. To date, only a few examples of such functionalized peptide sequences have been reported,307,308 largely due to the difficulty in predicting the amyloidogenicity of the modified peptides.
3.3.3. Amyloid nanotubes
Amyloid protein and peptide nanotubes are a particularly interesting class of amyloid nanostructure, due to their uniform dimensions, hollow architecture and potential for modification, which have led to many potential applications for materials science, nanotechnology and biomedicine. A comprehensive review of the synthesis and bionanotechnological applications of various protein and peptide nanotubes is available,309 in which the structure, design and corresponding nanotube assembly mechanisms of proteins (lysozyme, Hcp1, TRAP and others) and surfactant-like peptides have been demonstrated and discussed. In this section, we focus on the studies of several novel amyloid protein and peptide nanotubes first described after the publication of the above review.
Amyloid nanotubes can be created by either the closure of helical ribbons precursors, as discussed above (Section 3.3.2), or by the molecular design of single proteins or peptide building blocks.310–312 For example, Zhao et al. reported that a symmetric amphiphilic peptide with the sequence of KI4K could assemble into nanotubes in aqueous solution.311 The created peptide nanotubes have typical diameters in the range of 80–160 nm and lengths in the order of μm. In addition, they found that the peptide assemblies could be converted from nanotubes to nanofibrils by increasing the acetonitrile concentration in the assembly system. In another study, Brodin and co-workers reported the design and synthesis of protein nanotubes with adjustable diameters by using a single tetrameric Zn8R4 building block that was created by mixing disulphide-linked protein dimer (R2) with 4 equivalents of Zn2+.312 The formation and morphology of the nanotubes was mediated by altering the concentration of Zn2+, causing the rapid formation of nanotubes with widths of 48 ± 3 and 20 ± 2 nm, respectively. The initial formation of the Zn8R4 building blocks was crucial for the formation of nanotubes as the direct mixing of 10-fold Zn2+ with R2 in the first step resulted in only amorphous aggregates. This work showed that it is possible to kinetically dictate the self-assembly of protein building blocks to desired nanostructures by tuning the intermolecular interactions with metal coordination.
Amyloid nanotubes can also be prepared by the supramolecular co-assembly of two peptide-based building blocks.313,314 The integration of two types of building blocks allows the fabrication of nanomaterials with a complex structure and extended biophysical properties. Adler-Abramovich and co-workers formed peptide nanotubes with controllable physical dimensions313 from the co-assembly of two diphenylalanine (FF)-based building blocks. Aqueous solutions of FF and Boc-FF (N-(tert-butoxy-carbonyl)-l-Phe-l-Phe-COOH) were mixed at various molar ratios (Fig. 13a). Increasing the Boc-FF concentration in the co-assembly system resulted in a systematic decrease in the length of the assembled nanotubes, as shown in Fig. 13b and c. This work revealed a simple and effective strategy for creating peptide nanotubes with a controllable length distribution through the co-assembly of two peptide building blocks, providing a template for molecular engineering at the nanoscale.
Fig. 13.
(a–c) Supramolecular co-assembly for the formation of amyloid peptide nanotubes with controllable dimensions: (a) co-assembly mechanism of FF and Boc-FF, (b) SEM image of peptide nanotubes with a FF/Boc-FFF ratio of 5 : 1 and (c) length distribution. Reproduced with permission from ref. 313. Copyright 2016, American Chemical Society. (d and e) Co-assembly of amyloid amphiphilic peptide-based molecules to form multiwalled nanotubes: (d) co-assembly mechanism, and (e) TEM image of peptide nanotubes. Reproduced with permission from ref. 314. Copyright 2014, American Chemical Society.
Lin and co-workers produced multiwalled amyloid nanotubes by mixing two oppositely charged drug–peptide amphiphilic molecules.314 The peptide moiety GNNQQNY, a key β-sheet forming sequence derived from the yeast prion Sup35, was modified by an anticancer drug camptothecin (CPT) to design the drug–peptide building blocks. Two lysine (K) and two glutamic acid (E) residues were added to the C-terminal of the drug–peptide chain to adjust the overall amphiphilicity and pKa, as shown in Fig. 13d. They found that qCPT-Sup35-K2 (here “q” means four CPTs are bound to the peptide moiety) and qCPT-Sup35-E2 could co-assemble into nanotubes, but dCPT-Sup35-K2 (here “d” means two CPTs are bound onto the peptide moiety) and dCPT-Sup35-E2 could only co-assemble into nanofibrils (Fig. 13d). When qCPT-Sup35-K2 and qCPT-Sup35-E2 were mixed, nanotubes with uniform morphology were clearly observed, as shown in Fig. 13e. The created CPT-peptide nanotubes had an outer diameter of about 123 nm and wall thickness of approximately 25 nm, which suggested the presence of multiple bilayers in the assembled structure. Theoretically, the created drug–peptide nanotubes possess a 36% fixed CPT loading, and therefore could have promising application in drug delivery and cancer therapeutics.
3.4. 2D amyloids (sheets, films and membranes)
3.4.1. 2D protein amyloids
Amyloid fibrils generally possess very high mechanical strength and adhere well to various substrates. These features make it possible to create 2D amyloid assemblies using simple post-assembly treatment techniques.
A number of different post-assembly processes have been investigated to fabricate 2D amyloid films. These include amyloid stacking,315 filtration316 and self-assembly at the air–water interface.317,318 Lysozyme nanofibril films were created by stacking preformed nanofibrils onto a polytetrafluoroethylene film (Fig. 14a),315 and β-lactoglobulin nanofibril films were fabricated via the vacuum filtration of solutions of β-lactoglobulin nanofibrils (Fig. 14b).316
Fig. 14.
Typical methods for the fabrication of 2D protein amyloids: (a) stacking, (b) filtration and (c) self-assembly at an air–water interface. Image (a–c) are reproduced by permission from (a) ref. 315, Copyright 2010, Nature publishing Group, (b) ref. 316, Copyright 2015, American Chemical Society, and (c) ref. 317, Copyright 2016, Royal Society of Chemistry.
Jordens and co-workers reported the formation of β-lactoglobulin amyloid fibril films at an air–water interface.318 The assembly mechanism was found to follow a complex non-equilibrium process leading to a crowded interface and a viscoelastic 2D film.319,320 The structure of the interface could be further defined by combining long protein nanofibrils with short protein linear aggregates, as shown in Fig. 14c.317 Experimental and simulation results indicated that the short protein aggregates orient perpendicular to the long nanofibrils at very short distances and parallel to the axis of nanofibrils at intermediate distances, as shown in the AFM image in Fig. 14c. Liquid crystalline 2D bimodal systems such as this may have interesting technological applications but are complex and hard to control due to a large number of long-range non-covalent interactions affecting the self-assembly process. Thus studies such as this are very helpful to better understand and guide liquid crystal structures at an interface, and to unveil the process of formation of amyloid biofilms, as discussed later in this review.
Recently, Wang et al. exploited the conformation change of lysozyme after breaking down its disulfide bond by tris(2-carboxyethyl) phosphine (TCEP) to induce heterogeneous nucleation and assembly (Fig. 15a).321 The assembled lysozyme fibrils spontaneously concentrated at both the solid–water and water–air interfaces (Fig. 15a and b), and the films formed at the water–air interface could easily be converted to free-floating films (Fig. 15c), transferred to the surface of a hydrogel and then contact printed onto a water-sensitive substrate (Fig. 15d).
Fig. 15.
(a) Schematic illustration of the proposed mechanism for amyloid nanofilm formation. (b–d) Schematic strategies for lysozyme fibril nanofilm formation: (b) on the surface of immersed materials (solid–liquid interface) and (c) at the aqueous solution surface (vapour–liquid interface). (d) The contact-printing technique to deposit the free-floating amyloid nanofilm onto water-sensitive substrates. Reproduced with permission from ref. 321. Copyright 2015, Wiley-VCH Verlag GmbH & Co.
3.4.2. 2D peptide amyloids
Amyloid nanofibrils fabricated from short peptide sequences have also been utilized to fabricate 2D structures, including membranes,322 nanosheets323,324 and films.325,326 This is typically achieved by adjusting the assembly conditions (ionic strength, pH or physical stimulations) to promote the formation of 2D substrates.
Zhang’s group reported the spontaneous self-assembly of a self-complementary oligopeptide (EAK16) to form a stable macroscopic membrane.322 They found that peptides with alternating hydrophilic and hydrophobic amino acid residues readily form β-sheet structures and then aggregate into stable membranes dependent on the ionic strength of the solution. The assembled membranes have very high stability to heat and extreme pH (acid and alkaline) due to the formation of complementary ionic bonds between glutamic acid (E) and lysine (K) residues. This work paved the way for the fabrication of nanofibrous peptide structures with controllable self-assembly dependent on their sequence.
Dai and co-workers demonstrated the formation of amyloid nanosheets from a mutated form of Aβ16–22 (KLVFFAK) (Fig. 16a).323 The nanosheets were typically a few microns long, several hundred nm wide and around 2.2 nm-thick (Fig. 16b). Increasing the ionic strength of the peptide solution promoted the formation of more uniform nanosheets. Based on their experimental observations and molecular dynamics simulations, the authors hypothesized that the peptide molecules within the nanosheet stand upright to form a monolayer (peptide length = 2.2 nm). The nanosheet then grows in two dimensions along both the fibril axis (a′) via the main-chain hydrogen bonds and the zippering axis (b′) via the side-chain steric hydrophobic interactions (Fig. 16c). This work shows that the functionality of amyloid nanosheets can be tuned by replacing the amino acid side chains at the periphery but retaining the core nanosheet-forming sequence (LVFFA). In another study, Hamley and co-workers reported that the amphiphilic peptide (Ala)6Arg could self-assemble into 3 nm-thick peptide sheets at low concentration in aqueous solutions.324 The self-assembly of the peptide and the formation of sheets are driven by the unique conformation generated by the designed amphiphilic sequence and the electrostatic properties of the arginine headgroup.
Fig. 16.
(a) Amino acid sequence of Aβ1–42 and the molecular structure of Aβ16–22; (b) AFM image and height analysis of self-assembled amyloid nanosheets; (c) structural model of the KLVFFAK nanosheet. Reproduced with permission from ref. 323. Copyright 2015, National Academy of Sciences.
Physical stimulations can also mediate the self-assembly of peptide molecules and promote the subsequent formation of 2D amyloid films. For instance, Pan and co-workers developed a simple, effective and environmentally friendly method of fabricating nanofibrous films from Aβ16–22, via argon glow discharge.326
3.5. 3D amyloid plaques and scaffolds
3D insoluble deposits in the brain (i.e. Lewy bodies in PD, and Aβ plaques or Tau tangles in AD) are major pathological hall-marks in many neurodegenerative diseases.327 These deposits typically have length scales spanning from several to a few hundred microns and are composed predominantly of amyloid fibrils, with other assorted biomolecules (lipids, sugars, etc.). Currently, visualization of these deposits (often post mortem) represents one of the only definitive methods to diagnose a number of diseases, including Alzheimer’s and Parkinson’s. Therefore, a considerable amount of research has focused on developing in vivo and in vitro brain-imaging protocols to detect and observe these deposits. A number of different imaging techniques, including magnetic resonance imaging (MRI),328 positron emission tomography (PET),329 near infrared (NIR) imaging330 and FTIR microscopy,331 have been utilized to investigate the formation and growth of amyloid plaques. Benseny-Cases and co-workers used micro-FTIR spectroscopy to observe the in situ co-localization of amyloid senile plaques in tissue samples of human brains affected by Alzheimer’s disease.331 They found that the oxidization of lipids in tissues is associated with the aggregation of peptides and the formation of amyloid plaques. The tissue samples from non-Alzheimer’s disease samples showed a lower level of lipid oxidation, which indicated that the oxidative capacity of amyloid peptides or proteins may play a crucial role in the formation of amyloid plaques.
In addition to natural amyloid plaques, artificial amyloid 3D structures (multilayers332 and microgels333,334) have been reported. Qin et al. utilized an Aβ peptide derivative with a diphenylalanine moiety (Ne-RGDFF-OH) to create peptide nanofibrils that assembled into supramolecular hydrogels at both pH 8 and 6.5.332 The self-assembly of the designed peptide was defined by three distinct motifs, namely the naphthyl group (Ne), which provides the hydrophobic force to enhance the self-assembly ability in aqueous solutions; the FF motif, which serves as the core amyloidogenic sequence; and the RGD sequence (a sequence found in the ECM protein fibronectin), which acts as both an acceptor and a donor of hydrogen bonds. In addition, the RGD sequences can promote biocompatibility and cell attachment. Their results indicated that the morphology of the peptide nanofibrils could be further adjusted into catenulate microfibres (forming a row or chain), multilayered amyloid plaques and hydrogels by controlling the drying conditions of the amyloid nanofibril solution.
Amyloid nanofibrils can also be fabricated to form 3D microgels.333 Recently, Knowles’ group established a class of microgels based on amyloid protein fibrils by combining their inherent self-assembly process with microscale structuring techniques.334 Both water-in-oil and oil-in-water microgels were prepared by forming microdroplets of a concentrated aqueous solution of lysozyme protein, as shown in Fig. 17a and b. For the formation of water-in-oil microgels, a microfluidic device was fabricated to create microdroplets of lysozyme solution encapsulated in an immiscible oil phase (Fig. 17a). After incubation, protein monomers assembled into protein nanofibril gels, which were recovered from the oil phase by extensive washing. Confocal fluorescence microscopy (Fig. 17c) and cryo-SEM (Fig. 17e) images indicated that networks of protein nanofibrils were formed in the interior of the microgels. When the phases were reversed (i.e. water-in-oil becomes oil-in-water) hollow microgels were formed (Fig. 17b). The corresponding confocal fluorescence microscopy (Fig. 17d) and cryo-SEM (Fig. 17f) images showed that in this case the lysozyme monomers were located at the oil–water interface, and thus the fibrillar network structure was formed on the outer shell of the oil-in-water microgels. Protein microgels based on amyloid nanofibrils may have interesting applications for enhanced drug delivery due to their high biocompatibility and biodegradability.
Fig. 17.
Synthesis of amyloid 3D lysozyme microgels from amyloid fibril networks: (a) water-in-oil microgels, and (b) oil-in-water microgels. (c–f) Typical (c and d) 3D reconstructions of the confocal images and (e and f) cryo-SEM images of: (a) water-in-oil and (b) oil-in-water microgels, respectively. Reproduced with permission from ref. 334. Copyright 2015, American Chemical Society.
3.6. Summary of the characteristic length scale of various amyloid structures
In parts 3.1–3.5, we comprehensively reviewed the formation of various amyloid structures with morphologies including prefibrillar oligomers, nanoparticles, fibrils, films and plaques. In summary, the various amyloid systems discussed and their dominant morphologies, length scale and assembly conditions are presented in Table 1.
Table 1. Summary of the types, species, length scales and formation conditions of amyloid nanostructures.
| Amyloid type | Species | Length scale | Assembly condition | Ref. |
|---|---|---|---|---|
| Oligomers | Lysozyme | Monomer to fibril | pH 3, 70 °C | 257 |
| Cyclic peptide | Monomer, dimer, tetramer | 20 mM PBS, pH 7 | 254 | |
| KCKCLGDVIEV | 6-mers/2.2 nm | — | 262 | |
| β2-Microglobulin | — | pH 7.5, 20 mM Tris | 263 | |
| 0D | ||||
| NPs | PrP | 17–27 nm | Self-assembly | 265 |
| NPs | Recom-PrP | 20 nm | Self-assembly | 266 |
| NPs | Aβ1–40 | 15–30 nm | Self-aggregation | 268 |
| Sphere | FFF-peptide | 10–13 nm | Self-assembly | 269 |
| Sphere | Aβ1–40 | 10–60 nm | Membrane-assembly | 270 |
| Loop | α-synu-A53T | H = 2–4 nm, D = 23–55 nm | Self-assembly | 271 |
| Loop | ApoC-II | H = 2.1 nm, W = 12 nm, D = 50–75 nm | Self-aggregation in PBS buffer | 272 |
| Triangle | ApoC-III | D = 35–77 nm | NaPi buffer | 276 |
| 1D | ||||
| Protofibril | β-Lactoglobulin | H = 3 nm, L = 500 nm | pH 2, 90 °C, D2O | 240 |
| Protofibril | Aβ1–42 | 13–28 nm | PBS, 37 °C | 290 |
| Protofibril | α-synuclein | L = 7.5, W = 2.5 nm | HEPES, 37 °C | 291 |
| Helical fibril | β-Lactoglobulin | L = 0.5–15 μm, H = 2–6 nm | pH 2, self-assembly | 233 |
| Fibril | TTR105–115 | H = 7–16 nm, L = 1–3 μm | Acetone/water, pH 2 | 294 |
| Ribbon | ILQINS | W = 63–87 nm, L = 5.57 μm | pH 2, 90 °C | 297 |
| Ribbon | β2Aβ2AKLVFF | W = 17.5 nm | Self-assembly | 293 and 298 |
| Cofibril | (FKFE)2 | D = 8.2 ± 1.0 nm, L = a few 100 nm | Self-assembly in water | 299 |
| Ribbon | R3 | W = 147 nm, L = a few μm | Self-assembly in buffer | 301 |
| Ribbon | hIAPP20–29 | H = 7.4 nm, L = several 100 nm | Self-assembly | 303 |
| Nanotube | KI4K | D = 80–160 nm, L = a few μm | Self-assembly in water | 311 |
| Nanotube | Zn8R4 | D = 20–48 nm, L = a few μm | Zn2+-Induced assembly | 312 |
| Nanotube | FF, Boc-FF | L = 10 μm | 90 °C, H2O, assembly | 313 |
| Nanotube | CPT-Sup35 | D = 123 ± 28 nm, L = a few μm | 1 : 1 MeCN/H2O | 314 |
| 2D | ||||
| Film | Lysozyme | A few mm | Stacking | 315 |
| Film | β-Lactoglobulin | Adjustable | Filtration | 316 |
| Film | β-Lactoglobulin | Adjustable | Self-assembly at air–water interface | 317 |
| Nanosheet | Aβ16–22 | H = 2.3 nm, W = 500 nm, L = a few μm | Self-assembly | 323 |
| Sheet | A6R | Thick = 3 nm, L/W = a few 100 nm | Self-assembly in solution | 324 |
| 3D | ||||
| Plaques | Aβ1–42 | Several-100 μm | Aggregation | 331 |
| Multilayer | Ne-RGDFF-OH | Hydrogel | Assembly at pH 8/6.5 | 332 |
| Microgels | Lysozyme | 2–60 μm | Microdroplet | 334 |
In addition, referring to previous reports by Knowles and Mezzenga,32,33 an illustrative chart of amyloid materials across all relevant length scales is shown in Fig. 18.
Fig. 18.
Length scale of various amyloid structures from monomer to oligomers, 0D, 1D, 2D and 3D.
4. Manifold functionality of biological and artificial protein/peptide amyloid materials
In this section, we present and discuss the fabrication of biological (biofilms) and functional hybrid amyloid materials.
4.1. Fabrication and functions of biological amyloids
Natural protein and peptide amyloid materials show higher stability, mechanical strength, and increased resistance to protease biodegradation compared to their corresponding protein and peptide monomers. These properties have been utilized in nature to create “functional” amyloids with beneficial physiological functions. Numerous examples of functional amyloid systems have been found in bacteria, plants and mammals.335–338 Additionally, the above physical characteristics make synthetic amyloids attractive structural components for the formation of functional biomaterials with wide-ranging applications.
4.1.1. Amyloid-based biofilms
Biofilms are complex bacterial communities embedded in a predominantly proteinaceous ECM that protects bacteria from the surrounding environments.339 Amyloid protein fibrils have been identified as a major structural component of many biofilms.340 To better understand the fundamental processes underpinning biofilm formation, amyloid formations in bacterial biofilms (curli fibrils) have been investigated for growing films of E. coli.14,341 In the formation process of the biofilm, CsgA subunits are secreted by E. coli and self-assemble into amyloid curli fibrils, whilst CsgB subunits serve as nucleator proteins to anchor the curli fibrils to the bacterial membrane (Fig. 19a).14
Fig. 19.
(a) Formation mechanism of curli fibrils in E. coli biofilm. Reprinted with permission from ref. 14. Copyright 2008, Wiley-VCH. (b) 3D projection (upper) and side view (lower) CLSM images of S. typhimurium biofilms with a growth period of 24, 48 and 72 h. (c) CLSM image of a 72 h biofilm with DNA staining. Reproduced with permission from ref. 345. Copyright 2015, Elsevier Inc.
Biofilm formation can also be affected by a range of other protein subunits other than CsgA and CsgB.342–344 For example, Ostrowski et al. found that a small protein named YuaB serves both an exopolysaccharide and an accelerator for mediating the formation of TasA amyloid fibrils during biofilm formation.342 Herbst et al. investigated the formation of biofilms with the Gram-negative bacterium, P. aeruginosa, and found that the proteome of this bacterium is tightly associated with amyloid fibril formation, the distribution of fibrils, biofilm formation and proteolytic activity.344 Studies have also shown that DNA may play an important role in the formation of biofilms, Gallo and co-workers reported the formation and roles of curli–DNA composites in S. typhimurium pellicle biofilms formed at the air–liquid interface (Fig. 19b).345 Microscopy images of the 72 h S. typhimurium biofilms showed high concentrations of stained DNA (Fig. 19c), indicating that extracellular DNA (eDNA) either released by dying bacteria or actively released into the ECM serves as an important component for the formation of these biofilms. Furthermore, ThT fluorescent assays showed that eDNA accelerates the polymerization of curli fibrils, inhibiting their degradation by DNAase and creating potent immunogenic complexes, which were seen to activate a number of different immune cell types. This work highlighted a role of curli–DNA composites in stimulating the innate and the adaptive immune system.
The formation of amyloid-based biofilms can be mediated by adjusting the physiochemical environment of the growing bacteria. For instance, Wu and co-workers investigated the formation of biofilms of E. coli at the air–liquid interface,346 and found that the curli fibrils present in the biofilms result in a significant increase in their strength, viscoelasticity and electrical resistance. The same authors studied the effects of chemicals such as dimethyl sulfoxide and ethanol on amyloid biogenesis and biofilm formation. They showed that the presence of these small molecules increased the formation of the E. coli biofilm.347 Other environmental signals (e.g. ionic strength and pH) can also affect the formation of biofilms. Taglialegna et al. reported that the Bap protein of S. aureus assembles into functional amyloid nanofibrils and can be induced to form a biofilm matrix at low pH and in the presence of low concentrations of Ca2+ ions.348 Increased Ca2+ concentration favours a more stable Bap conformation, leading to the inhibition of amyloid formation. This relationship between Ca2+ concentration and amyloid formation could have implications in the design of antibacterial materials and therapeutics.
4.1.2. Amyloid hydrogels and aerogels
Hydrogels are water swollen polymeric networks formed by long-range non-covalent interactions of self-assembled nanofibrils. Amyloidogenic proteins and peptides have been shown to make useful molecular building blocks for the design and synthesis of hydrogels.349 Due to their relative ease of synthesis and biocompatibility, these hydrogels show a wide variety of applications in diverse fields, including cellular therapies, drug delivery and tissue repair.67,350 In order to successfully fabricate amyloid hydrogels, two conditions must be met: first, the protein or peptide monomers must have the ability to form amyloid fibrils in aqueous solutions and second, the fibrils must be able to be synthesized at sufficient concentration to promote gelation.19
Amyloid hydrogels have been formed from a variety of amyloid proteins, including elastin,351 α-synuclein,352 lysozyme353,354 and β-lactoglobulin.355–357 Bhak et al. reported the synthesis of amyloid hydrogels derived from α-synuclein fibrils with a curled morphology (the so-called CAF, or curly amyloid fibrils). They showed that CAF hydrogels have potential applications as a matrix for enzyme entrapment.352 In their study, they found that under normal self-assembly conditions (200 rpm, 37 °C and 100 h), α-synuclein preferred to form straight amyloid fibrils (SAF) that did not readily form hydrogels (Fig. 20a). CAFs that did undergo gelation were created by isolating α-synuclein granules formed in the middle of the lag phase of the aggregation pathway. The granules were subjected to centrifugal membrane filtration to form CAFs (Fig. 20b). These hydrogels were characterized via ThT binding assays and confocal microscopy (Fig. 20c). The authors proposed that these α-synuclein hydrogels could have potential applications in fields as diverse as tissue engineering, drug delivery, nanofiltration and biosensing. Knowles and co-workers fabricated lysozyme hydrogels loaded with drugs.315 The addition of beta-adrenoceptor antagonists into the lysozyme hydrogel altered the nanostructure of lysozyme amyloids and affected the drug-release profiles. This study suggests that a hydrogel-based drug carrier architecture can be adjusted to obtain the desirable release performance by careful selection of the structural promoters and disruptors of amyloids.
Fig. 20.
(a–c) Amyloid protein nanofibre hydrogel: (a) formation mechanism, (b) SEM and (c) fluorescence images. Reproduced with permission from ref. 352. Copyright 2010 Elsevier Ltd. (d–f) Amyloid peptide nanofibre hydrogel: (d) formation mechanism, (e) optical image of hydrogel and (f) AFM image. Reproduced with permission from ref. 19. Copyright 2015 Elsevier Ltd.
Bolisetty et al. discussed the gelation of β-lactoglobulin amyloid fibrils and proposed that β-lactoglobulin fibrils could undergo both an isotropic–nematic and a sol–gel phase transition with increasing fibril concentration or ionic strength.357 This work shed light on the dynamic behaviour of biological colloidal system and opens new directions in the fabrication of amyloid gels.
Hydrogels have also been prepared from Aβ amyloid fibrils. Jacob et al. studied the formation of self-healing amyloid hydrogels from a series of peptides based on the high aggregationprone C-terminus of Aβ1–40 and Aβ1–42.19 They found that a number of sequences only formed hydrogels when the amino acid sequence was conjugated to an aromatic Fmoc-protecting group. Additional long-range aromatic stabilization (π–π stacking) offered by the Fmoc groups (Fig. 20d) was found to be a major driving force promoting gelation of these peptides. The addition of a fluorescent dye (Nile Red) into the peptide amyloid hydrogel inhibited gelation, suggesting that Nile Red blocks exposed hydrophobic sites on the formed amyloid fibrils and reduces their non-covalent interactions. The corresponding optical (Fig. 20e) and AFM (Fig. 20f) images identified the formation of amyloid hydrogels with a 3D structure composed of nanoscale fibrous networks.
The additional stabilization offered by the conjugated Fmoc group allows very short peptide fragments from Aβ to form amyloid-like hydrogels. For instance Fmoc-dipeptide hydrogels (FF and KK) have been formed based on β-sheet self-assembly and intermolecular π–π association.358 Hydrogels have also been formed through the self-assembly of non-Fmocs containing short peptides.359–361 Tena-Solsona et al. demonstrated that the self-assembly and co-aggregation of tetrapeptides containing alternate aromatic and polar amino acid residues (Z-FDFD, Z-DFDF, and others, where Z denotes a benzyloxycarbonyl group) can lead to hydrogel formation, which can be used to screen positively charged Lys residues involved in amyloid misfolding.359 In a further study, pH-responsive hydrogels were formed from Z-FDFD, Z-FKFK and Z-KFKF.360
An interesting extension of the hydrogel is its “dry” homologue, the so-called aerogel. In a recent study, Nystrom et al. demonstrated that β-lactoglobulin amyloids can form ultralight aerogels composed solely of β-lactoglobulin amyloid fibrils and air.362 This new material is among the lightest materials ever created and fully exploits the rigidity and versatility of β-lactoglobulin amyloid fibrils. To create the dry form, the water content of β-lactoglobulin amyloid hydrogels was solvent-exchanged with ethanol to produce an alchogel. Then, exploiting the fact that ethanol is fully miscible with supercritical CO2, they removed the liquid phase by supercritical CO2 processing, leading to the final amyloid aerogels. Excitingly, these aerogels can be used to template the formation of hybrid materials with unique properties, leading, for example, to the lightest form of gold ever produced to date. These gold aerogels were formed by combining β-lactoglobulin amyloid fibrils and gold single crystals and fabricating the aerogels as outlined above. The resultant aerogels contained no more than 2% solid mass. Beside being the lightest gold material ever produced, the gold aerogels have additional properties unseen in solid gold, such as photonic, fluorescent and catalytic properties. This example and the others preceding it illustrate the endless possibilities available when designing materials using amyloids as building blocks.
4.2. Fabrication and functions of artificial amyloid-based hybrid materials
Amyloid fibrils generally display multiple, identical and periodically spaced binding sites for small molecules along their surface. This makes them an exciting prospect for the design of amyloid materials functionalized with specific chemistries post-assembly. In this section, we introduce the preparation and fabrication of 1D, 2D and 3D functionalized or hybrid nanomaterials.
4.2.1. 1D amyloid-based hybrid materials
Lysozyme, Aβ, curli proteins and a number of other systems have been used to fabricate 1D amyloid hybrid systems. Whilst amyloids make ideal substrates for hybridization with additional functional materials, care must be taken that additional materials do not modify the amyloid aggregation and alter the balance of hydrophobic and electrostatic interactions within the forming fibrils.
4.2.1.1. Amyloid–nanoparticle
One of the most studied amyloid hybrid systems is the amyloid–nanoparticle hybrid. Such hybrids allow the fabrication of nanoparticle-decorated amyloids with specified material properties. Alternatively, nanoparticles have been used as therapeutic targets for disease-related amyloids with the aim of inhibiting amyloid aggregation. Typically, the formation of amyloid–nanoparticle hybrids is based on metal or metal compound nanoparticles.363–365 Bolisetty et al. reported the synthesis of hybrids formed by dispersing negatively charged iron oxide (Fe3O4) magnetic nanoparticles in positively charged β-lactoglobulin solutions at acidic pH. Depending on the pH, different hybrid aggregates are formed: at pH 3 amyloid fibrils with their surface decorated with nanoparticles were observed, whilst at pH 4.5 only spherical nanoclusters were observed.366 Liao et al. studied the influence of gold nanoparticles (AuNPs) on Aβ fibrillation and found that they could alter the observed fibrillization pathways.367 AuNPs added to preformed Aβ fibrils, caused fragmentation of the fibrils. AuNPs possessing negative surface potential inhibited Aβ fibrillation and redirected assembly into off-pathway intermediates. Such anionic AuNPs could potentially have applications as AD therapeutics, preventing assembly into toxic species.367
More complex nanoparticles have also been investigated. For instance, Yoo et al. reported that thioglycolic acid-stabilized CdTe nanoparticles are capable of the efficient inhibition of amyloid formation.136 Inhibition occurs as CdTe nanoparticles preferably bind to oligomers but not monomers, halting the aggregation process at this stage. This paper highlights a potential issue with such amyloid inhibitors; indeed, preventing fibrillization may be relatively simple, but if it is at the expense of creating more amyloid oligomers, extreme care must be taken to ensure that the oligomers formed are not themselves highly cytotoxic. Other nanoparticles have also been used to reduce amyloid aggregation kinetics. Anand et al. designed capsaicin-capped silver nanoparticles (AgNPs) to suppress albumin aggregation.364 Interestingly, assembly inhibition was not observed in the presence of isolated capsaicin molecules or unmodified nanoparticles, suggesting a cooperative effect between the silver nanoparticles and capsaicin coating.
Polymeric nanoparticles have also been shown to affect the fibrillation kinetics of amyloid fibrils. Brambilla et al. showed that PEGylated nanoparticles with long in vivo retention times bind strongly to the Aβ1–42 peptide.368 Structural analysis revealed that binding occurred through a non-specific binding of the PEG polymer on the surface of the NP (Fig. 21a). Simulations revealed that the PEG chains wind around the helical peptide between residues 1–25 and more loosely interact with the helix at residues 26–42 and at the β-turn (Fig. 21b and c). To evaluate the role of oxygen atoms in the PEG, it was replaced with a fully alkylated polymer chain. This fully saturated polymer chain did not wind around the helix (1–25) but bound only with the termini of both helices, clearly indicating the importance of the oxygen atoms in such polymers.
Fig. 21.
(a) Atomistic model of a PEG chain docked to Aβ1–42. The chain interacts with both hydrophobic as well as hydrophilic residues of the peptide and forms a spiral structure. (b) Best 50 conformations of the PEG chain (purple) docked to Aβ1–42 (orange), and (c) alkyl PE chain (purple) docking on Aβ1–42 (orange). Images (a–c) are reproduced with permission from ref. 368. Copyright 2012, American Chemical Society. (d–g) AFM (bottom row) images of the β-lactoglobulin amyloid fibrils (d) and fibrils decorated by gold (e), silver (f) and palladium (g) nanoparticles after the respective metal salt reduction by NaBH4. Images (d)–(g) are reproduced with permission from ref. 370. Copyright 2014, American Chemical Society.
Amyloid fibrils have also been used for metallization and mineralization experiments, and for the fabrication of amyloid– nanoparticle nanohybrids. Previously, Wei and co-workers showed it was possible to functionalize the surface of amyloid-like fibrinogen nanofibrils with AuNPs.369 Bolisetty et al. were able to decorate β-lactoglobulin amyloid fibrils (Fig. 21d) with Au (Fig. 21e), Ag (Fig. 21f) and Pd (Fig. 21g) NPs via metal salt reduction by NaBH4.370 Hamley et al. examined the labelling of amyloid nanofibrils created from the self-assembly of a surfactant-like peptide Ala10His6 (hexa-histidine connected with an oligo-alanine sequence) with NTA-functionalized AuNPs in the presence of Ni.371 Pazos and co-workers reported a one-pot synthesis for the nucleation of uniformly sized and spatially ordered AgNPs using supramolecular nanofibrils formed by PAs.372 Aldehyde moieties at the N-terminus could reduce two silver ions to form Ag2 clusters without the need for an external reducing agent or additives to control the nucleation process.
4.2.1.2. Amyloid-quantum dots
Similar to nanoparticles, QDs have been shown to inhibit amyloid fibrillation. Xiao et al. reported that N-acetyl-l-cysteine-capped QDs (NAC-QDs) are capable of disrupting the fibrillation of insulin amyloid assemblies due to hydrogen bonding between the NAC-QDs and the fibrils.373 Ng et al. suggested that NAC-QDs may act as neuro-protective agents through fibrillation inhibition and by a reduction in the formation of reactive oxygen species.374
Liu et al. demonstrated that graphene QDs (GQDs) can efficiently inhibit the aggregation of Aβ peptides.375 This is thought to be either due to binding of the negatively charged GQDs to the central hydrophobic motif and/or positively charged histidine residues present on Aβ1–42 (Fig. 22a and b). Further studies revealed that the inhibition efficiency of the GQDs decreased with increasing surface negative charge, indicating that the hydrophobic interactions are likely to be dominant. In some cases, QDs can accelerate the aggregation of amyloid proteins. Roberti et al. demonstrated that the aggregation of α-synuclein at high concentrations was enhanced by adding multivalent QDs (Fig. 22c).376 This is likely due to aggregation seeding initiated by the high local concentrations displayed at the surface of the QDs. Fig. 22c(i–iii) shows that the QDs functionalized with biotinylated α-synuclein (bAS) are incorporated into α-synuclein fibrils, resulting in increased aggregation.
Fig. 22.
(a) Schematic representation of the GQDs used for inhibiting the aggregation of Aβ1–42 peptides. (b) The kinetics of Aβ1–42 aggregation as monitored by the thioflavin T fluorescence in the absence or presence of GQD. Reproduced with permission from ref. 375. Copyright 2015 Royal Society of Chemistry. (c) AFM images of the fibrils obtained from mixtures of (i–iii) QD–bAS (QD/bAS = 1/40), and (iv) α-synuclein alone. Scale bars: 10 μm. Reproduced with permission from ref. 376. Copyright 2009, American Chemical Society.
The inherent fluorescence of QDs opens many new potential applications for amyloid fibrils–QD hybrids, for example they could be used as fluorescent biosensors, or for bioimaging or diagnostics. NHS modification of CdSe–ZnS core–shell QDs has been used to detect the formation of amyloid protein fibrils in solution by fluorescent measurements after binding to the fibrils.94 Quan et al. reported a red-emitting fluorescent probe constructed by PEGylated QDs further functionalized with benzotriazole (BTA) that facilitated targeting to the β2 position of Aβ fibrils with high affinity, enabling the sensitive detection of amyloid fibrils.377 Due to a combination of the greater fluorescent quenching of QDs and the higher affinity of the BTA, QD–PEG–BTA probes were able to achieve more sensitive detection than conventional thioflavin derivatives. Su et al. used peptide nanofibrils decorated with QDs as an intracellular fluorescent imaging agent.378 Cellular internalization of the nanofibril–QD hybrids was promoted by the increased interactions between the cationic fibrils and the slightly negatively charged outer leaflet of the cell membrane. In a further study, they created novel functional hybrid materials based on the conjugation of GO, peptide nanofibrils and GQDs.379
4.2.1.3. Amyloid–hydroxyapatite
Due to their broad functionalization possibilities, long contour lengths, exceptional mechanical properties, accurately controlled growth and ease of synthesis, amyloid fibrils are ideal candidates to act as biomimetic materials, performing similar biological functions to fibrous networks such as fibronectin or collagen. In vivo bone regrowth occurs in the presence of collagen–hydroxyapatite (HA) composites; thus in the fields of tissue engineering and regenerative medicine there is a strong desire for biomaterials that can mimic the properties of these composites.380 Stupp’s group demonstrated that one-dimensional (1D) cylindrical and fibrillar nanostructures were able to direct the growth of oriented HA crystals, whilst flatter nanostructures failed to reproduce the orientation found in biological systems.74,380 By adjusting the pH, concentration and ionic strength, Wei et al. prepared amyloid-like fibrinogen fibrils in the absence of Ca2+ and thrombin.95 These fibrils were mineralized to form amyloid–HA composites, proving that self-assembled fibrinogen nanofibrils can mimic collagen and serve as a building block for HA-based biomaterials for bone tissue regeneration applications. Wang et al. fabricated a HA scaffold via a layer-by-layer assembly of graphene oxide (GO) nanosheets and preformed fibrinogen nanofibrils.381 After 7 days of mineralization in a simulated body fluid, the GO–nanofibril surface was uniformly covered with branch-like apatite minerals that did not appear on a bare GO substrate. Therefore, it appeared that the amyloid nano-fibrils provided a structural foundation and surface functional groups for the growth of HA crystals. A second generation of amyloid peptide nanofibrils was designed with specific motifs present on the surface of the fibril to provide the nucleation sites for HA mineralization.382 Using these fibrils and GO nano-sheets, they fabricated a nanohybrid scaffold designed to promote mineralization. The second generation nanohybrids enhanced nanoscale apatite crystal growth, leading to the formation of HA spheres with a diameter of 4 μm.
In order to be useful biomimetic scaffolds for bone regrowth, amyloid–HA hybrids should not only promote mineralization but they must also display the high levels of mechanical stiffness required to provide the load-bearing ability of newly formed bone. Adopting a simple filtration procedure, Li et al. fabricated a highly laminated structure composed of β-lactoglobulin and/or lysozyme amyloid fibrils and HA platelets (Fig. 23a).383 The formed fibril network possessed a homogenous and compact surface, and the resultant biomimetic bone grown on the networks had a similar Young’s modulus and density to human bone (Fig. 23b–e). More importantly, these scaffolds promoted the growth of human osteoblast cells, providing a compelling example of amyloid-based biomimetic bones.
Fig. 23.
(a) Schematic representation of the fabrication of bone-mimetic composites based on amyloid fibril and HA platelets. (b) TEM image of β-lactoglobulin amyloid fibrils. (c–e) SEM image of HA platelets (c), surface (d) and fracture sections (e) of the amyloid-based composite with 60 wt% brushite platelets. Reproduced with permission from ref. 383. Copyright 2014, Wiley-VCH Verlag GmbH & Co.
4.2.1.4. Amyloid–carbon materials
Various carbon nanomaterials, including fullerenes (C60), carbon nanotubes (CNTs), graphene and GO, have been investigated for their ability to inhibit amyloid assembly.384,385 Due to the presence of multiple benzene rings in the majority of carbon nanomaterial structures, hydrophobic interactions are thought to play a key role in the inhibitory ability of carbon nanomaterials. In a comprehensive review, Li and Mezzenga discussed how C60, CNT and GO affect amyloid fibrillation and the formation of amyloid–carbon material hybrids.384 It was expected that the conformation and chemical behaviour of the Aβ peptide would change in the presence of single-walled carbon nanotubes (SWNTs) due to the adsorption of the peptide onto the hydrophobic surface of the SWNTs. Importantly, the cytotoxicity of cells cultured in the presence of SWNTs coated with Aβ peptides was drastically reduced compared to cells cultured with the Aβ peptide alone.386
To enhance the binding of amyloids to carbon nanomaterials, attempts have been made to modify the nanomaterials with reactive polymers or small molecules. For example, Li and Mezzenga introduced sulfonic functional groups on the surfaces of multiwalled carbon nanotubes (MWNTs), promoting interactions with β-lactoglobulin fibrils (Fig. 24a).355 The negatively charged functionalized MWNT surfaces promote complex formation with the positively charged β-lactoglobulin fibrils. Beside complementary chemistry, other factors, such as the persistence length (of both MWNTs and fibrils) and the ionic strength, are important for the fabrication of hybrid amyloid–carbon materials. If the persistence lengths of both MWNTs and amyloid fibrils are not similar, the mismatch leads to a misalignment of the fibrils along the MWNTs contour SO3− (Fig. 24b and c). However, an appropriate content of SO3− would make the individual MWNTs match several amyloid fibrils at different positions, as shown in Fig. 24d.
Fig. 24.
Formation of amyloid nanofibril–MWNT hybrid: (a) schematic illustrations of the functionalization of CNTs with amyloid fibrils. (b–d) TEM images of hybrids consisting of functionalized MWNTs (black arrows) and amyloid fibrils (white arrows) at 0.1 wt% with: (b and c) covalent and (d) non-covalent functionalization. Reproduced with permission from ref. 355. Copyright 2012, American Chemical Society.
The surface modification of carbon nanomaterials plays an important role in the design of multifunctional amyloid–carbon material composites.381,382,387 For instance, positively charged lysozyme fibrils can bind to GO sheets, providing binding sites for negative charged AuNPs.388 The charges from GO and lysozyme then become partially nullified, making the hybrids ideal platforms for immobilizing additional molecules, such as enzymes, to afford additional functions.
Mahmoudi et al. found that the large available surface area of GO sheets is able to delay the process of Aβ fibrillation via the adsorption of amyloid monomers.389 In addition, the protein coating layer can create a protective shell on the surface of the GO sheets, resulting in an increase in the amyloid assembly lag time. Yang et al. reported that graphene nanosheets could penetrate and extract a large number of peptides from preformed amyloid fibrils.390 This leads to the intriguing question of whether graphene or GO sheets could have therapeutic applications and could potentially reverse the amyloid formation process in neurodegenerative diseases. However, it is likely that such therapies would either need to be injected directly into the brain or be able to cross the BBB, factors, which complicates the development of such therapeutics. Very recently, Castelletto and co-workers reported the fabrication of a hybrid biomaterial based on the cooperative self-assembly of polysaccharide sodium alginate with PA (C16-KKFF) and by subsequently adding GO.391 The created hybrid biomaterial showed excellent bioactivity and high mechanical strength.
4.2.1.5. Amyloid-biomacromolecules
Functionalizing amyloid fibrils with specific biomolecules, such as proteins and sugars, may have significant biomedical applications. For instance, networks of amyloid-based materials can be produced that mimic various biological tissues, such as muscles or the cornea.392,393
Zhong et al. reported strong underwater adhesives constructed by co-polymers of tyrosine containing mussel foot proteins (Mfps) of Mytilus galloprovincialis and CsgA protein (which forms curli fibrils in E. coli).394 The co-polymers all formed fibrous structures, but the nitro blue tetrazolium (NBT) assay turned purple (to confirm the presence of the adhesive moiety DOPA) only when the Mfps proteins were modified with tyrosinase (which converts tyrosine residues to DOPA) (Fig. 25a and b). To assess the underwater adhesion of the adhesive fibres, AFM tips modified with silica, gold or polystyrene surfaces were used. The strongest adhesion was found with silica tips and tyrosinase-modified fibres, suggesting that the amyloid domains of the adhesive fibres not only provided a high surface area for contact but could also modulate how the Mfp domains interact with the substrates and achieve different adhesion levels (Fig. 25c). In another case, Dubey et al. identified that amyloid nanofibrils can promote the co-aggregation of other proteins, which also explained the mechanism of coexistence of two amyloid-linked diseases in individual patients.395 However, both aggressive co-aggregation and cross-seeding reactions between different proteins occurred only at 70 °C.
Fig. 25.
(a) CR and NBT assays confirm the amyloidogenic features and the formation of an amyloid–DOPA hybrid. (b) TEM images of unmodified and modified amyloid nanofibrils with biomolecules. (c) Schematic presentation of the measurement of the adhesion force between amyloid nanofibrils and biomolecules with AFM force spectroscopy. Representative AFM image showing modified Mfp5-CsgA fibres 1 h after deposition on a mica surface. Reproduced with permission from ref. 394. Copyright 2014, Nature Publishing Group.
There are many studies based on the physiochemical properties and functions of amyloid-fibril-based materials.396 Nikiforov et al. studied the electromechanical coupling of amyloid fibrils in both prokaryotic and eukaryotic cells.397 They found that the mechanical properties of the fibril and the electrical double layer at the fibril–water interface are responsible for its electromechanical response. Ling and co-workers combined regenerated silk fibrils from Bombyx mori (B. mori) fibroin with amyloid fibrils produced in vitro from β-lactoglobulin.398 By either adding inorganic nanoparticles or by selectively removing one of the fibrous constituents via enzymatic reactions, they further explored how the Young’s moduli, porosity and stimuli-responsive features of the resulting hybrid materials could be controlled. The authors also showed that these membranes could be used for molecular-weight-dependent separation, pointing at possible future applications of these porous scaffolds as protein-based separation membranes.
4.2.2. 2D amyloid-based hybrid materials
The Young’s modulus of many amyloidogenic 2D films are similar to those of many rigid proteinaceous materials found in nature, (e.g. keratin and collagen).315 Thus, when functionalized with additional biologically relevant molecules, 2D amyloid hybrid materials can be fabricated that mimic both chemical and structural facets of biology.
Based on amyloid fibrils and silk fibroin fibrils, Ling et al. designed a multifunctional membrane decorated with magnetic nanoparticles.398 The formed membrane was transparent and homogenous under cross-polarized light, with β-strands of silk fibroin running parallel to the film plane and the amyloid fibrils in a perpendicular orientation (Fig. 26a and b) as detected by wide-angle X-ray scattering (Fig. 26c–e). The addition of positively charged magnetic nanoparticles resulted in the fabrication of a homogenously distributed film combining dual magnetic and moisture responsiveness (Fig. 26f and g). Hydration of the membrane resulted in a reduction in rigidity and allowed for cyclic shape printing and recovery on repeated wetting and drying cycles. Upon applying a magnetic field to the hydrated film, the membrane underwent a bending motion and maintained its shape after removal of the field and drying. Layer-by-layer deposition of silk proteins and amyloid fibrils has also been employed to create an immunosensing platform capable of detecting Aβ1–40.399 Multilayers containing 1, 3 and 5 bilayers of silk fibroin and Aβ1–40 fibrils were fabricated that showed linear responses to increasing Aβ1–40 antibody concentrations via cyclic voltammetry.
Fig. 26.
(a) Schematic orientation of β-sheets and β-strands in silk fibroin fibrils and amyloid fibrils. (b) Schematic illustration and cross-polarized light observation image of silk fibroin fibrils and amyloid fibrils composite film. (c–e) 2D-WAXS patterns of the films containing: (c) 100% silk fibroin fibrils, (d) 100% amyloid fibrils and (e) 5 : 5 silk fibroin fibrils : amyloid fibrils. (f) Magnetic functionalization and tensile properties of the film (silk fibroin fibrils : amyloid fibrils : magnetic nanoparticles weight ratio of 70 : 10 : 20), as prepared by vacuum filtration. (g) Shape-memory properties of the magnetic composite film when exposed to the combined presence of an external magnetic field and water. Reproduced with permission from ref. 398. Copyright 2014 Wiley-VCH Verlag GmbH & Co.
Wu et al. used the previously discussed amyloid–GO immobilization platform to immobilize Au nanocatalysts and enzymes.388 This integrated system maintained the catalytic activity of the immobilized AuNPs, and was used as an electrochemical sensor for colorimetric glucose sensing. Yan et al. also reported a new approach combining electron-induced molecular self-assembly with simultaneous metal nanoparticle formation.400 The peptide motif KLVFF (Aβ16–20) was combined with metal ions and assembled into membranes in the presence of an argon plasma. The argon plasma resulted in the reduction of the metal ions, forming homogenously dispersed metal NPs that decorated the underlying film of amyloid fibrils.326 Such hybrid amyloid–nanoparticle films may have important applications in heterogeneous metal catalysis. This was recently demonstrated with amyloid–nanoparticle porous membranes that were capable of securing a continuous flow of a feeding solution, which then undergoes instantaneous heterogeneous catalysis within the hybrid membranes.316
2D amyloid hybrids have also been used to elucidate a better understanding of the mechanisms of bacterial biofilm formation. Nguyen et al. fused functional peptide domains onto the CsgA protein, resulting in a self-assembled network of curli fibres resembling the wild-type system.401 This molecular engineering strategy should provide diverse artificial functions to synthetic hybrid amyloid biofilms. Additional peptide sequences could be introduced to the biofilm to provide various functionalities, including biomineralization, substrate adhesion and protein immobilization. Using similar strategies, robust 2D materials with programmed functions could be fabricated with applications as large-scale engineered biomaterials.
4.2.3. 3D amyloid-based hybrid materials
Utilizing amyloid-based molecules as building blocks to develop 3D functional materials with well-defined architectures and chemistry is a growing area of interest.402,403
3D hybrid amyloid materials are typically designed based on one or several kinds of lower dimensional materials. For example, Lara and co-workers fabricated 3D structures from single crystal nanoplatelets, protein nanotubes and ribbons.29 By changing the procedure by which water is removed from the system, the same building blocks could be organized into organic–inorganic hybrid films of unique physical properties, composed of well-organized layered 2D single crystal gold nanoplatelets and 1D amyloid fibrils (Fig. 27a).404 Adhikari and Banerjee incorporated graphene and reduced graphene oxide (RGO) into amyloid networks, resulting in stable transparent hydrogels.405 In this system, two kinds of Fmoc-protected dipeptides, namely Fmoc-YD-OH and Fmoc-FD-OH, formed composite hydrogels without the presence of any external stabilizing agent. The hydrogels were stabilized via inherent π–π interactions between the Fmoc group, the aromatic side chains of the Tyr and Phe residues and the RGO sheets. A number of stimulus-responsive amyloid hydrogels have also been investigated. For instance, by combining MWNTs decorated with sulfonic groups and β-lactoglobulin nanofibrils, pH-responsive hydrogels were fabricated that reversibly form gels at acid pH.355
Fig. 27.
(a) Layered organization of amyloid fibrils and gold platelet hybrid nanocomposites. Reproduced with permission from ref. 404. Copyright 2013, Wiley-VCH Verlag GmbH & Co. (b) An illustrative description of the development of a photoluminescent peptide–QD hydrogel through the self-assembly of Fmoc-FF building blocks and their PL quenching associated with the enzymatic detection of analytes. Reprinted with permission from ref. 406. Copyright 2014, Wiley-VCH Verlag GmbH & Co.
Metallized and small molecule-modified nanofibrils also offer the possibility to create hydrogels for applications in nanotechnology. In a recent study conducted by Stupp’s group,372 bacterial growth was inhibited by hydrogels formed from peptide nanofibrils decorated with AgNPs. Kim et al. formed meso-tetra(4-pyridyl) porphine (TPyP) and Fmoc-diphenylalanine (Fmoc-FF) composite nanofibrils.406 The four pyridyl groups at the meso-functional position of TPyP and the carboxylic and hydroxyl groups of Fmoc-FF promoted hydrogel formation (Fig. 27b).
5. Various applications of amyloid-based hybrid nanomaterials
Thanks to their unique physico-chemical properties, amyloid-based nanomaterials, have many potential applications in a number of fields, including biomedical engineering, tissue engineering, energy storage and catalysis. In this section, we review some of the most promising and exciting examples of where amyloid materials have been incorporated into functional devices.
5.1. Biomedical engineering
Historically, research on amyloids has largely focused on obtaining an improved understanding of the mechanisms causing toxicity in neurodegenerative diseases. However, in more recent times, there has been a growing interest in using non-toxic amyloid assemblies for biomedical engineering applications.33,407 Some of the key studies in this area are outlined in this section.
Due to the regular distribution of labile groups on their surface, many amyloid fibrils are amenable to functionalization with a broad range of moieties and molecules. For instance, Bolisetty et al. showed that amyloid fibrils can be used to enhance nanoparticle transfection into living organisms by incubating gold, silver and palladium nanoparticle-decorated amyloid fibrils in the presence of living dendritic or MCF7 cells and confirmed their ability to cross the cell membrane and be taken up into living cells (Fig. 28a).370 Mains et al. explored lysozyme amyloid hydrogels loaded with a series of small molecule beta-adrenoceptor antagonists, such as atenolol, propranolol and timolol.354 Atenolol was shown to disrupt β-sheet content within the hydrogel, whilst propranolol had the opposite effect and timolol had little effect. This research highlighted the need for the careful selection of structure promoters and disruptors for drug-delivery applications. The conjugation of amyloid fibrils with drug compounds may also be a useful strategy to create long-acting drug depots and to provide controlled release systems, preventing rapid clearance from the body.17
Fig. 28.
(a) Schematic representation of the internalization and transport of metal nanoparticle-decorated amyloid fibrils into living cancer cells. Reprinted with permission from ref. 370. Copyright 2014, American Chemical Society. (b) Proposed coating mechanism of SEVI fibrils with amyloid-binding oligomers. These coatings prevent the direct interaction of HIV-1 with SEVI fibrils and prevent SEVI-mediated enhancement of viral infection in cells. Reprinted with permission from ref. 413. Copyright 2012, American Chemical Society.
Cheetham et al. designed a drug–peptide conjugate from Tau fibrils and the anticancer drug CPT (mCPT-mal-Tau), which formed discrete, stable well-defined nanofibrils and nanotubes capable of quantitative drug loading.408 The drug loading in the peptide nanostructures increased from 23% in non-fibrous controls to 38%. The corresponding drug-release experiments indicated that the self-assembled drug-loaded peptide nanostructures released the bioactive form of the drug and enhanced the in vitro efficacy against a number of cancer cell lines.
Further examples of amyloid-based drug delivery systems include a camptothecin (KCK-CPT) prodrug that self-assembles initially into nanotubes, and upon complexation with hyaluronic acid into arrays of nanofibrils, which then form micelles upon dilution.409 The hyaluronic acid-coated micelles were then seen to undergo efficient endocytosis into cancer cells in vitro, where they were degraded back to the nanofibrous morphology in the endosome. Finally, the KCK-CPT prodrug was converted to the active form by glutathione (GSH) in the cytoplasm, inducing apoptosis in the cancer cell. Waku and Tanaka showed that self-assembling peptide nanofibrils can be used as potential delivery agents for vaccines.410,411 Highly antigen-loaded nano-assemblies were formed by conjugating antigens to β-sheet-forming peptides. The antigen-loaded peptide nanofibrils were taken up more efficiently by murine RAW264 cells compared to monomeric cell penetrating peptide-modified antigenic peptides, possibly because their size is more suitable for cellular uptake. Improved intracellular antigen delivery using a system such as this may promote a more efficient induction of the immune response required for effective vaccination to occur.
Due to their small size and nanoscale regularity, amyloid oligomers have been explored as potential nanomaterials for improved targeting and medical imaging. By conjugating either fluorescent molecules or superparamagnetic iron oxide (FeO) particles with oligomeric Aβ, Kumar et al. observed that oligomeric hybrids specifically targeted macrophages in an in vitro co-culture of peripheral blood mononuclear cells and macrophages.268 Of particular interest might be both the visualization of the disease-associated accumulation of macrophages in vivo by MRI and the imaging of atherosclerotic plaques to assess the extent of cardiovascular disease. Very recently, Lock and co-workers synthesized an anticancer drug, specifically Pemetrexed (Pem), conjugated onto the peptide sequence (FE).412 The PemFE conjugate spontaneously self-assembles into nanofibrils and hydrogels under physiological conditions. The location, distribution, recovery and drug release of injected PemFE hydrogels was successfully monitored by chemical exchange saturation transfer (CEST) MRI. This work proposed a potential strategy to monitor the in vivo distribution and release of drugs by using supramolecular design and self-assembly.
Capule et al. reported that oligovalent amyloid-binding molecules can reduce the enhancement of human immunodeficiency virus-1 (HIV-1) infection. The designed amyloid-binding molecules competitively bind to semen-derived enhancer of virus infection (SEVI) fibrils preventing SEVI-mediated enhancement of the viral infection in cells (Fig. 28b).413 Yolamanova et al. demonstrated that amyloid fibrils fabricated from a 12-residue peptide (enhancing factor C) boosted the virus infection of HIV-1 by a factor of four compared to naturally occurring SEVI fibrils.414 This reduced virus infection is due to the formation of an electrostatic ‘nanobridge’ between the virion and the cell. Such amyloid-based nanomaterials may significantly improve our ability to direct retroviral gene transfer in basic research and clinical applications.
5.2. Tissue engineering
Materials that can accurately replicate the structure of the micro- and nanoscale fibrous network that surrounds many cell types, i.e. the ECM, have many applications in tissue engineering and cellular therapies. For instance, ECM-mimicking scaffolds may aid the culture of large volumes of clinically relevant therapeutic cells in vitro, or be used as implantable scaffolds for cell growth and tissue regeneration in vivo. Networks of amyloid fibrils assembled from either natural proteins or synthetic peptides are promising candidates for such materials as they possess similar morphological and mechanical properties to the fibrillar proteins that make up the ECM (e.g. fibronectin, collagen and laminin). Additionally, they can be inexpensive to produce in the large volumes required for clinical applications and are well suited to chemical functionalization with peptide sequences (typically from ECM proteins) that can promote cell adhesion, growth or differentiation (in stem cells).
In 2004, Stupp’s group integrated the IKVAV sequence (a cell binding sequence from laminin) with a PA system to fabricate nanofibrous hydrogels. The density of the IKVAV presentation on the surface of the nanofibrils was three orders of magnitude higher than for laminin in the ECM. Furthermore, neural progenitor cells cultured in these hydrogels were seen to efficiently differentiate into neurons.415 Various other amyloid-based systems decorated with cell adhesion sequences have also been reported. For instance, Gras et al. modified a 10-residue peptide fragment (TTR105–115) of the amyloidogenic protein transthyretin (TTR) with the integrin binding sequence RGD from fibronectin. In addition to the RGD-modified sequence, two similar sequences were synthesized with additional tripeptides that have no reported biological activity (RAD and RGE). All three peptides retained their ability to form amyloid fibrils and supported cell growth when fibroblasts were cultured on adsorbed networks of the fibrils. However, only the RGD-presenting fibrils was able to disrupt fibroblast cell adhesion to a fibronectin-coated substrate. This suggested that the RGD-modified fibrils could competitively bind to integrin assemblies present on the cell membrane.308 The biocompatibility of these modified TTR105–115-RGD fibrils has been further explored using a number of membrane integrity and apoptotic markers.416 Despite good cellular attachment (via specific RGD–integrin bonds), the materials were found to show fibril morphology and concentration-dependent cytotoxicity after longer periods of time. Studies such as this highlight the importance of fully understanding how the physical, morphological and chemical properties of amyloid-based materials affect cellular interactions before they can be applied as biomaterials. In addition to TTR105–115-RGD, an analogous amyloidogenic peptide was synthesized with a more physiological cyclic RGDfK sequence attached (TTR105–115-cRGDfK). Compared to the TTR105–115-RGD fibrils, the TTR105–115-cRGDfK fibrils were seen to significantly promote cell adhesion and cell spreading through the development of integrin-mediated focal adhesions between the cRGDfK ligands and the cell membrane.307,417 However, masking of the cRGDfK chemistry with a thin plasma polymer layer revealed that the addition of the bulky cRGDfK sequence results in nanofibrils with a topography less favourable to cell growth.417 Thus, once again, this highlights the importance of considering both the chemical and topographical features when designing amyloid-based biomaterials. In another interesting recent example, RGD-functionalized PAs were used to create enzyme (matrix metalloprotease)-triggerered releasable free-standing collagen-rich films, produced by human stromal corneal fibroblasts.418,419 The PA coating was aligned by use of a lithographic process and this led to aligned collagen deposition, mimicking the alignment of collagen fibres in the cornea.
3D scaffolds based on amyloid nanofibril systems functionalized with cell adhesion moieties have also been used as biomaterials that promote cell growth. Zhou et al. created hydrogels from a mixture of Fmoc-FF and Fmoc-RGD.420 The rapid gelling system promoted the adhesion and 3D cell culture of encapsulated dermal fibroblasts through specific interactions with integrins on the cell membrane. Furthermore, by adjusting the ratios of Fmoc-FF and Fmoc-RGD in the system, the concentration and ligand spacing of the RGD moieties in the hydrogel could be accurately controlled. Based on a similar gelation mechanism, a series of peptides based on Aβ were combined with Fmoc-protecting groups.19,421 By adjusting the peptide sequence, concentration and the ionic strength, the mechanical properties of the formed gels could be controlled, which in turn was used to drive stem cell differentiation. In a similar study, Das and co-workers synthesized a new class of implantable α-synuclein-inspired peptide hydrogels with the ability to direct stem cell differentiation in vivo (Fig. 29a).422 Mesenchymal stem cells (MSCs) were seeded into an α-synuclein-based peptide hydrogel and transplanted into the brains of mice (Fig. 29b). After 7 days in vivo, the brains were harvested and sectioned, and the cells implanted within the hydrogel showed increased evidence of neuronal differentiation compared to cells transplanted in the absence of the hydrogel (Fig. 29c). Furthermore, the presence of the hydrogel resulted in an increased cell number, both within the caudate putamen (Fig. 29d) and in the substantia nigra of the implanted mice (Fig. 29e).
Fig. 29.
Amyloid hydrogels for cell culture: (a) schematic of amyloid hydrogels for 2D and 3D cell cultures. (b) Schematic of the morphology of cells at each stage during the implantation. Stage 1: cultured cells for 24 h; stage 2: cells were primed with a differentiation medium for 5 days; stage 3: cells were then transplanted with hydrogel A5 into the mice brains, and stage 4: the brains were harvested. Scale bars: 200 μm for stage 1–3 and 100 μm for stage 4. (c) Implanted GFP-hMSCs with α-synuclein hydrogel (left) and without a hydrogel (right) at the caudate putamen after 7 days in vivo. (d) Cell viability when implanted with and without a hydrogel. (e) Box plot of the area with survived cells when transplanted with and without a hydrogel. Reproduced with permission from ref. 422. Copyright 2016, Nature Publishing Group.
The section above highlights the considerable amount of research that has gone into designing bespoke amyloidogenic peptide sequences that self-assemble and display specific cell adhesion moieties on their surface. However, the design and synthesis of such sequences is non-trivial and is relatively expensive. Therefore, a different approach has also been investigated whereby inexpensive and abundant raw materials that are easily applicable to scale up to clinically relevant volumes required in the biomedical industry are used for the formation of amyloid-based scaffolds. Healy et al. extracted crystalline proteins from fish eye lenses and studied the influence of temperature on their self-assembly into amyloid fibrils.423 Furthermore, these fibrils were shown to be stable over a wide range of pH and over long periods of time and showed no significant cytotoxicity to Hec-1a endometrial cells.424 This preliminary work suggests that crystalline nanofibrils should be further investigated as 2D or 3D biomaterials. Reynolds et al. fabricated amyloid fibril networks from lysozyme on solid supports with well-defined nanoscale surface features reminiscent of the topography of the ECM.425 By masking the surface chemistry of the fibril networks with a thin layer of inert oligo (ethylene glycol) plasma polymer, they proved that the ECM-mimicking nanotopography alone was enough to promote the attachment and spreading of a fibroblast cell line. In a following study, they showed that the attachment, spreading and cytoskeletal tension of cells could be controlled by adjusting the surface coverage of the amyloid fibril networks, both in the presence and absence of serum proteins.426
Li et al. fabricated poly(N-isopropylacrylamide) (PNIPAM)-decorated amyloid fibrils from β-lactoglobulin. These decorated fibrils underwent a sol–gel transition below body temperature (32 °C).427 This hybrid could be used as an injectable material at room temperature, whilst turning into a gel at body temperature. This finding may contribute to designing thermally controlled “smart” scaffolds for tissue regeneration applications.
5.3. Energy materials
The unique physiochemical properties of amyloid fibrils make them attractive materials, not just in biomedical and regenerative medical fields. For instance, amyloid fibrils have been investigated as potential materials for next-generation energy-harvesting devices,428,429 lithium ion batteries,23,430 organic solar cells,431 photovoltaic432 and catalyst materials.316,433
Hanczyc et al. showed that amyloid fibrils of insulin, lysozyme and α-synuclein specifically enhance multiphoton absorption.428 Channon et al. demonstrated a detailed mechanism of the light-harvesting function resulting from co-assembly of two independent luminescent moieties into amyloid-like fibrils.429
Similarly, light-harvesting peptide nanotubes were synthesized by the co-assembly of FF and porphyrin (Fig. 30a).434 The dipeptide assemblies and porphyrin molecules mimicked the electron separator and mediator in natural photosynthetic systems, respectively. By incorporating PtNPs onto the surface of the FF/porphyrin nanotubes, the self-metallization of PtNPs occurred, enabling an efficient separation and transfer of the exited electrons from the porphyrin to an electron mediator.
Fig. 30.
(a) Biomimetic photosynthesis by light-harvesting peptide nanotubes. Reproduced with permission from ref. 434. Copyright 2012, Wiley-VCH Verlag GmbH & Co. (b) Schematic diagram of the hybrid photovoltaic device prepared using an active layer composed of TiO2-hybrid nanowires blended with polythiophene and AFM image of TiO2 decorating the surface of the amyloid fibrils. Reproduced with permission from ref. 432. Copyright 2012, Wiley-VCH Verlag GmbH & Co.
Park’s group performed studies on the biomimetic mineralization of FePO4 and Co3O4 nanoparticles on self-assembled FF nanofibrils23 and nanotubes.430 They found that biomimetic inorganic–organic hybrid nanomaterials could act as promising cathode materials for rechargeable lithium ion batteries with a high reversible capacity and good capacity retention during cycling.
Inganäs et al. demonstrated the integration of amyloid insulin nanofibrils into organic solar cells to enhance the transport properties of photovoltaic devices.431 They found that the amyloid nanofibrils had a significant effect on the donor–acceptor material organization. At a specific ratio of nanofibrils, donors and acceptors, the hybrid organic solar cells showed improved charge transport compared to the materials without added nanofibrils.
Bolisetty et al. utilized β-lactoglobulin amyloid fibrils as templates to successfully direct the synthesis of closely packed TiO2 hybrid nanowires.432 Due to the well-organized combination of electrostatic and hydrogen bond interactions, TiO2 nanoparticles decorated the surfaces of the protein fibrils uniformly. Subsequently, the TiO2-coated amyloid hybrid nano-wires could be prepared into a photovoltaic active layer by spincoating a blended mixture of polythiophene-coated fibrils and amyloid–TiO2 hybrid nanowires (Fig. 30b). Acar et al. demonstrated the amyloid peptide nanofibril templated synthesis of TiO2 nanostructures via a bottom-up approach.435 By staining the calcinated TiO2 layer with an N719 photosensitizer dye, they fabricated dye-sensitized solar cells with high loading capabilities and improved open circuit voltages, exploiting the high surface area offered by the amyloid fibrils.
Amyloid fibrils also have important potential applications in heterogeneous catalysis. Bolisetty et al. prepared AuNPs and PdNPs, respectively, on β-lactoglobulin amyloid fibrils for the catalytic reduction of 4-nitrophenol.316 In another study, Chaves et al. reported a new biocatalyst based on the photo-immobilization of lipase onto amyloid fibrils via a photo-induced cross-linking reaction.433 The resulting insoluble nanoscale biocatalyst showed higher enzyme stability than the soluble enzyme under several extreme conditions. Scott and co-workers demonstrated the preparation of amyloid (Fmoc-FF) peptide gel microparticles that were emulsified and stabilized with SiO2 nanoparticles.436 The amyloid matrix was used to immobilize the enzyme lipase B (CalB) and the catalytic performance was assessed by monitoring the esterification of octanol in heptane. In the best performing system, the authors observed almost a 4-fold increase in catalytic activity compared to native CalB. Bio-catalytic amyloid microparticles such as these could be very useful for fabricating biosensors and bioinspired solar fuel devices.
5.4. Environmental science and technology
Amyloid fibrils have also found important applications in environmental sciences. For instance, Bolisetty and Mezzenga reported that inexpensive and environmentally friendly β-lactoglobulin amyloid fibrils could be blended with activated carbon and filtered to form membranes capable of purifying a variety of contaminants from wastewater samples.118 The initial study showed that these simple membranes provided remarkable levels of purification for water samples containing heavy metal, organic and bacterial contamination. As shown in Fig. 31a, heavy metal ions purification occurred due to the presence of metal ion-binding sites on the surface of the amyloid fibrils. After filtering, the content of potassium dicyanoaurate(l), mercury chloride and several other tested pollutants was reduced by nearly three orders of magnitude (Fig. 31b and c). Leung et al. modified lysozyme amyloid fibrils with ethylenediamine to reduce their carboxyl content, enabling the adsorption of toxic chromium(vi) ions in water.437 These studies open up new possibilities in fabricating functionalized amyloid-based materials that can efficiently scavenge pollutants from water supplies.
Fig. 31.
Amyloid nanofibril-based materials for water purification. (a) Structure of the β-lactoglobulin protein with the heavy metal-binding motif highlighted, 121-cys, with a lead ion attached and the 121-cys-containing fragment (LACQCL) from β-lactoglobulin with docked Pb2+. (b and c) Concentrations of heavy metal and radioactive pollutants before and after filtration through the amyloid fibril-activated carbon hybrid adsorber membrane: (b) potassium dicyanoaurate(i); (c) mercury chloride. Reproduced with permission from ref. 118. Copyright 2016, Nature Publishing Group.
In a similar manner to the scavenging of waterborne contaminants outlined above, amyloid-based materials could have applications as materials for capturing carbon dioxide from the atmosphere, thus helping to address the global threat of climate change. Li and co-workers reported that amyloid fibrils formed from the peptide sequence VQIVYK were capable of sequestering CO2.438 They found that the ε-amino group of lysine was uncharged at a high pH, which made it thus capable of forming carbamate with atmospheric CO2. The materials were able to capture CO2 in the presence of water, which was later released by heating. In a further study, the same authors used VQIVYK capped with N-terminal acetylation and C-terminal amidation to increase fibril formation and promote the diffusion of small gaseous molecules.439 The binding of CO2 was thought to occur via carbamate formation with amine functional groups present in the lysine residues. In another design, they mutated the glutamine residue in position 2 to lysine, generating the hexapeptide VKIVYK.438,439 However, although there was twice the number of amines in the designed fibrils, a two-fold increase in the binding capacity of CO2 was not observed, indicating that in these fibrils the amine groups are only partially accessible.
5.5. Electronic nanodevices
Inspired by the high efficiency of charge transport in biological systems, the possibility of introducing charged proteins and peptides in electronic devices has been suggested.440 Their unique hybrid conductivity behaviour makes self-assembled peptide nanostructures powerful building blocks for the construction of electronic nanodevices.441 Carny et al. fabricated coaxial gold and silver nanocables by binding metallic NPs onto the self-assembled peptide nanotubes via molecular recognition.442 Dinca et al. successfully fabricated 3D structures from amyloid fibrils with potential application in molecular electronics.443,444 Amit et al. studied the conductance of the thiophene-containing peptide (2-Thi) (2-Thi) VLKAA under a range of humidity conditions.445 They found that the conductivity of the fibrils was increased at higher relative humidity, indicating proton transport rather than electron transport dominates the conductive behaviour. Compared with amyloids from the naturally occurring peptide AAKLVFF, the conductance of (2-Thi) (2-Thi) VLKAA was found to be much higher, and this was attributed to subtle changes in the folding structure.
Self-assembled amyloid nanofibrils have found applications as building blocks for electrochemical transistors.440 For instance, Hamedi et al. designed amyloid insulin fibrils coated with the highly conducting polymer alkoxy sulfonate poly(3,4-ethylene-dioxythiophene) (PEDOT-S).446 The assembled network was dispersed in an acetonitrile electrolyte and probed with a Pt gate electrode (Fig. 32a and b). The fabricated amyloid–PEDOT-S transistor showed high source–drain current and also displayed repeatable switching characteristics with an on/off ratio of >40 when sweeping the gate between 0 and 0.5 V, clearly showing that the PEDOT-S had not lost any electrical or electrochemical properties after self-assembly with amyloid fibrils. Furthermore, the nanowire networks showed Ohmic connection to the metal, and no detachment during the electrochemical reactions. Meier et al. prepared conducting polyaniline (PAni) nanowires with a core–shell structure using amyloid nanofibrils as a template.447 Adsorption of the conducting polymer to the fibrils was facilitated through binding between the hydrophobic aromatic functional groups on the PAni and the exposed hydrophobic pockets on the surface of the growing fibrils (Fig. 32c). The resultant materials were deposited on an array of gold electrodes, as shown in Fig. 32d. The conductivity of the coated hybrid fibrils was found to be far higher than that of uncoated fibrils. In another study, Tu and co-workers reported the fabrication of an electrochromic transistor based on PEDOT-decorated amyloid fibrils.448 These studies show that amyloid fibrils provide an excellent structural template for the directed deposition of conducting polymers, which has major potential applications in the fabrication of next-generation microelectronic devices.
Fig. 32.
Fabrication of amyloid-based transistors. (a and b) PEDOT-S amyloid nanofibrils-based transistor: (a) molecular structure and (b) schematic picture of an electrolyte gated transistor. Reproduced with permission from ref. 446. 2008, American Chemical Society. (c and d) Amyloid–PAni hybrid nanofibril-based transistor: (c) synthesis and AFM image of PNF–PAni hybrid fibrils and (d) deposited hybrid fibrils on gold electrode array for conductivity measurements (PNF: peptide nanofibrils). Reproduced with permission from ref. 447. Copyright 2015, American Chemical Society.
Amyloid fibrils also exhibit promising electronic applications as organic light-emitting diodes (OLEDs).449–453 For instance, amyloid insulin fibrils were conjugated with luminescent conjugated polymers, and used as the active layer in an OLED device.449,450 The introduction of amyloid fibrils into the fabricated OLED device resulted in a 10-fold increase in the external quantum efficiency.450 In other studies, the same amyloid fibrils were functionalized with phosphorescent organometallic (Ir) complexes to fabricate red and yellow OLEDs.451,452 The conjugation of phosphorescent organometallic complexes with amyloid structures was seen to strongly improve the triplet exciton confinement and to make it possible to fabricate white-emitting devices at a low loading of phosphorescent complexes. More information on the modification of amyloid fibrils as well as the fabrication and applications of OLEDs with fibrils have been discussed in a recent review article.453
5.6. Biosensor architectures
Amyloids have been shown to have promising applications in the design of novel biosensors. This is in part due to their high mechanical strength and chemical resistance to the surrounding milieu, their ability to detect nanoscale protein–ligand interactions and their selectivity for low concentrations of biomolecule analytes.454 For instance, self-assembled amyloid peptide nanotubes have been utilized to create novel electrochemical biosensing platforms for detecting hydrogen peroxide and enzymes.455,456
Amyloids functionalized with ligands such as fluorophores, antibodies or enzymes can also be used for biosensing applications.457 Glucose oxidase (GOx)-functionalized whey protein nanofibrils (WPNFs) were seen to promote enzyme immobilization on screen-printed gold electrodes due to the large surface-to-volume ratio of the WPNF nanoscaffold.458 Compared to simple physical enzyme adsorption, the produced cyclic voltammogram exhibited a distinct increase in the anodic peak current response when using WPNF nanoscaffolds. The anodic peak currents were even larger when using the thiol-functionalized WPNFs, due to increased binding of WPNF on the gold surface.
Amyloid protein nanofibrils have shown potential for the fabrication of enzyme-linked immunosorbent assay (ELISA) immunosensors.459,460 For instance, Men and co-workers demonstrated the design of auto-biotinylated bifunctional protein nano-wires (bFPNw) based on the self-assembly of recombinant biotin-modified amyloid protein Sup35 (Fig. 33a).459 The high concentration and regular arrangement of biotin molecules on the surface of the bFPNws allows any of the hundreds of commercially available diagnostic enzymes to be transferred to the surface of the immunosensor via the biotin–avadin reaction (Fig. 33b). These biosensors were able to detect Yersinia pestis F1 antigen with a 2000- to 4000-fold increase in sensitivity compared to traditional ELISAs (Fig. 33c). In addition, the bFPNw-based system was seen to amplify the detection signal, reduce the non-specific binding and improve the stability. In a similar example, gene fusion was used to express recombinant Sup35-E2-GFP-MPH, where E2-GFP-MSH is a fluorescent biosensor based on a green fluorescent protein linked to the enzyme methyl parathion hydrolase. The self-assembled nanofibrils were able to detect the pesticide methyl parathion with a sensitivity around 10 000 times greater than free E2-GFP-MSH.460
Fig. 33.
Amyloid protein nanofibril-based immunosensors. (a) Schematic synthesis of Sup35-BAP nanofibrils. (b) Fabrication of an immunosensor architecture via biotin–streptavidin interaction. (c) Improved sensing performance compared to non-fibril sensors. Reproduced with permission from ref. 459. Copyright 2010, Elsevier Ltd.
Whilst gene fusion approaches such as those discussed above certainly show great promise for the design of amyloids that can act as enzymatic biosensors, their complexity and the specialized expertise required for their development may hinder their development. As an alternative, amyloid β-lactoglobulin nanofibrils were combined with GO to fabricate biodegradable nanocomposites with shape-memory and enzymatic sensing properties.20 Free-standing nanofibril–RGO films were fabricated in a series of steps: after assembly of the fibrils and binding of the fibrils to the GO sheets, the GO sheets were reduced at high temperatures to RGO. RGO-fibril films were formed by vacuum filtration, as shown in Fig. 34a and b. The free-standing hybrid nanocomposite films possessed a film thickness of 40–60 μm (Fig. 34c) and displayed a remarkable and fully reversible shape-memory effect. In addition, the nanocomposites could be totally degraded by simple enzymatic reactions, or, under controlled enzymatic reactions, could be used as a new class of biosensors to measure enzymatic activity (Fig. 34d).
Fig. 34.
Amyloid nanofibril–GO shape-memory materials. (a) Fabrication mechanism, (b) AFM of nanofibril–GO hybrid, (c) SEM image of nanofibril–GO hybrid film. (d) Biosensors for enzymatic activity measurements. Reproduced with permission from ref. 20. Copyright 2012 Nature Publishing Group.
5.7. Other functional nanomaterials
Besides the above-mentioned biomedical and nanotechnological applications, amyloids fibrils have been used for the fabrication of other novel functional nanomaterials, including liquid crystal materials,298,461–465 photoluminescence and optical waveguiding materials.466–468
The formation of a liquid crystal phase by amyloid fibrils is important for the fabrication of nanomaterials with hard structures, which could be reinforced by the rigid and anisotropic amyloid fibrils. Suspensions of amyloid fibrils frequently form liquid crystal phases due to long-range non-covalent interactions between assemblies.160,321–325 For instance, Corrigan et al. demonstrated that the lysozyme nanofibrils readily form liquid crystal phases.462 Zhao et al. created amyloid fibrils suspensions with isotropic–nematic coexistence by freeze–thaw cycling.463 Han and co-workers found that FF rapidly self-assembles into nanowires with a high aspect ratio in volatile organic solvents, which showed a colloidal nematic liquid crystalline phase over a broad concentration range.464 Hamley and co-workers showed that AAKLVFF nanotubes465 and helical peptide βAβAKLVFF ribbons298 and other amyloidogenic peptides can form nematic liquid phases in organic solvents and water, respectively.
Self-assembled amyloids have been used for the synthesis of bioinspired functional materials for optical waveguiding. Ryu et al. reported the in situ conjugation of self-assembled FF nanotubes with photosensitizers (4-acetylbiphenyl) and lanthanide (Tb3+ and Eu3+) ions for fabricating novel photoluminescent nanotube materials.466 Their finding indicated that the FF nanotubes acted not only as a host matrix for lanthanide complexes, but also served as a photosensitizer. By careful adjustment of the composition of lanthanide complexes, various nanotubes with switchable colours (red, green, blue, cyan and purple) were fabricated. In another study, Yan and co-workers demonstrated the optical waveguiding of self-assembled hexagonal FF microtubes and FF fibrous networks.467,468 The fabricated peptide nanotubes exhibited remarkable thermal stability and optical waveguiding, making them novel candidates to design and develop optical and electrical nanodevices.
6. Conclusions and outlook
In this review, we summarized the recent progress in both the fundamental study and applications of amyloid systems. We focused on the elucidation of the self-assembly mechanisms, hierarchical structure and physical properties of a series of amyloid nanostructures, from molecular oligomers to 0D, 1D, 2D and 3D nanomaterials. We elaborated on how the understanding at a very fundamental level of these salient features illuminates their applications in the most diverse areas of artificial and biological materials. A wide range of strategies for fabricating natural and artificial amyloid-based materials were discussed. In addition, the various applications of amyloid-based hybrid nanomaterials for biomedical engineering, tissue engineering, energy materials, environment science, nanodevices, biosensors and others were introduced and discussed in detail.
Looking to the future, there are some areas that would benefit further research. For instance, more work should be focused on using computer simulation techniques, such as molecular dynamics, in order to understand the molecular mechanisms underpinning the formation of amyloid oligomers and other intermediates. Second, work should be performed to elucidate the selection rules and mechanisms responsible for the creation of 2D and 3D amyloid-based materials. Multidimensional amyloid-based nanomaterials show many promising applications for materials science and nanoelectronics. Currently, the controllable growth and formation of 2D and 3D amyloid structures is challenging and very difficult to predict due to a lack of fundamental knowledge. Third, the exploitation of functional motifs from amyloid proteins to create biofunctional-specific amyloid nanostructures should be further investigated. In addition, it is possible to endow other functions, such as material recognition, biomineralization and cell adhesion, to the created amyloid nanostructures by inserting additional peptide motifs. Last but not least, it is to be hoped that the biocompatibility, biodegradation and cellular toxicity of amyloid materials will be understood and tailored to a greater extent as this could potentially unlock an extraordinary large numbers of applications in the field of biotechnologies and biomaterials.
In this respect, the contrast between the disease-relevant instances of amyloids and the various applications of amyloid-derived materials in a biological context deserves some discussion as this raises the question of the potential safety challenges of such materials. In general, the main discriminant in the selection of the amyloid building blocks should be the chemical nature of the peptides or proteins forming the amyloids. When the primary structure of the peptide/protein building block contains motifs from pathologically relevant amyloid-forming peptides, potential dangers need to be carefully considered, although there are also examples where amyloids fibrils composed from fragments derived from pathological amyloidogenic proteins have been successfully used as cell scaffolds.308 A very recent study, however, clearly points to a safe use of amyloid fibrils in vivo – even in the context of food applications – when the fibrils are composed of hydrolyzed edible proteins.469 Shen et al. indeed used hybrids of β-lactoglobulin amyloid fibrils and iron nanoparticles to design a new efficient colloidal form of highly bioavailable nano-sized iron to be used in anaemia treatment.469 The authors exploited the combined acidic and enzymatic environment present in the stomach to allow a fast dissolution of both the amyloid fibrils, digested by pepsin, and the iron nanoparticles, dissolved by the acidic pH, leading to a rapid dissolution of these hybrids into hydrolyzed milk proteins and highly bioavailable iron ions. This work convincingly demonstrates that if amyloid fibrils are made of biodegradable building blocks and are administered through the gastrointestinal tract to enable correct digestion, they could revolutionize fields such as pharmaceutics, food technology and nutrition, thereby tremendously widening the scope of applications of these fascinating materials.
Acknowledgements
GW thanks the financial supports from the Deutsche Forschungsgemeinschaft (DFG) under grant WE 5837/1-1. ZS acknowledges the financial supports from the National Natural Science Foundation of China (Grant No. 51573013). NPR thanks the Australian Research Council (ARC) Training Centre for Biodevices at Swinburne University of Technology (IC140100023) for financial support. IWH would like to acknowledge EPSRC (UK) for Platform grant EP/L020599/1 and the Royal Society (UK) for a Wolfson Research Merit Award. EG thanks the support by a grant from the European Research Council BISON project.
Main abbreviations
- NMR
Nuclear magnetic resonance
- CD
Circular dichroism
- XRFD
X-ray fibre diffraction
- ThT
Thioflavin T
- EM
Electron microscopy
- AFM
Atomic force microscopy
- SAXS
Small-angle X-ray scattering
- LCST
Lower critical solution temperature
- FTIR
Fourier transform infrared spectroscopy
- ITC
Isothermal titration calorimetric
- HFIP
Hexafluoroisopropanol
- TFE
2,2,2-Trifluoroethanol
- ECM
Extracellular matrix
- QDs
Quantum dots
- NHS
N-Hydroxysulfosuccinimide
- GAGs
Sulfated glycosaminoglycans
- BBB
Blood–brain barrier
- PMMA
Polymethylmethacrylate
- PTFE
Polytetrafluoroethylene
- IAPP
Islet amyloid polypeptide
- APP
Amyloid precursor protein
- Aβ
Amyloid β
- PrP
Prion protein
- ATR
Attenuated total reflection
- S/DLS
Static and dynamic light scattering
- PrP
Prion protein
- WT
Wild-type
- TEM
Transmission electron microscope
- MRI
Magnetic resonance imaging
- CPT
Camptothecin
- TCEP
Tris(2-carboxyethyl) phosphine
- CAF
Curly amyloid fibrils
- GQDs
Graphene QDs
- HA
Hydroxyapatite
- GO
Graphene oxide
- SWNTs
Single-walled carbon nanotubes
- NBT
Nitro blue tetrazolium
- CEST
Chemical exchange saturation transfer
- PA
Peptide amphiphiles
- PNIPAM
Poly(N-isopropylacrylamide)
- PEDOT-S
Poly(3,4-ethylenedioxythiophene)
- OLEDs
Organic light-emitting diodes
- GOx
Glucose oxidase
Biographies
 Gang Wei Gang Wei received his PhD
from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, China,
in 2007. Following his doctoral studies, he worked as the Alexander von Humboldt
(2007) and Carl-Zeiss (2009) Postdoctoral Fellow at the
Friedrich-Schiller-University of Jena, Germany. Since 2012, he has worked as a
senior researcher and group leader in the Hybrid Materials Interfaces Group at
University of Bremen, Germany. His research interests include carbon-based
nanomaterials, supramolecular self-assembly, sensors and biosensors, as well as
single-molecule force spectroscopy.
Gang Wei Gang Wei received his PhD
from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, China,
in 2007. Following his doctoral studies, he worked as the Alexander von Humboldt
(2007) and Carl-Zeiss (2009) Postdoctoral Fellow at the
Friedrich-Schiller-University of Jena, Germany. Since 2012, he has worked as a
senior researcher and group leader in the Hybrid Materials Interfaces Group at
University of Bremen, Germany. His research interests include carbon-based
nanomaterials, supramolecular self-assembly, sensors and biosensors, as well as
single-molecule force spectroscopy.
 Zhiqiang Su Zhiqiang Su was born in
1975 and completed his PhD studies in 2005 at the Institute of Chemistry, Chinese
Academy of Sciences. After a postdoctoral stay at the Tsinghua University, he joined
the Beijing University of Chemical Technology in 2007 and was appointed as Full
Professor in 2012. In 2011, he studied at Friedrich-Schiller-University Jena,
Germany as an experienced research fellow of the Alexander von Humboldt Foundation.
His research interests include nanohybrids, biomedical materials, biosensors and
bioelectronics. So far, he has published more than 80 papers in international
peer-reviewed journals.
Zhiqiang Su Zhiqiang Su was born in
1975 and completed his PhD studies in 2005 at the Institute of Chemistry, Chinese
Academy of Sciences. After a postdoctoral stay at the Tsinghua University, he joined
the Beijing University of Chemical Technology in 2007 and was appointed as Full
Professor in 2012. In 2011, he studied at Friedrich-Schiller-University Jena,
Germany as an experienced research fellow of the Alexander von Humboldt Foundation.
His research interests include nanohybrids, biomedical materials, biosensors and
bioelectronics. So far, he has published more than 80 papers in international
peer-reviewed journals.
 Nicholas P. Reynolds Nicholas
Reynolds received his PhD from the Chemistry Department at The University of
Sheffield in 2007; he then went on to complete a postdoctoral position at The
University of Zurich. Following his time in Zurich, he undertook a Swiss National
Science Foundation Fellowship at CSIRO in Melbourne. Nicholas currently works as a
Research Scientist at the Swinburne University of Technology in Melbourne. His
interests cover a wide range of biointerface and biomaterials research. Examples
include investigations into the structure and self-assembly mechanisms of amyloid
fibrils, their applications as biomaterials and the development of lipid- and
polymer-based therapeutic nanoparticles.
Nicholas P. Reynolds Nicholas
Reynolds received his PhD from the Chemistry Department at The University of
Sheffield in 2007; he then went on to complete a postdoctoral position at The
University of Zurich. Following his time in Zurich, he undertook a Swiss National
Science Foundation Fellowship at CSIRO in Melbourne. Nicholas currently works as a
Research Scientist at the Swinburne University of Technology in Melbourne. His
interests cover a wide range of biointerface and biomaterials research. Examples
include investigations into the structure and self-assembly mechanisms of amyloid
fibrils, their applications as biomaterials and the development of lipid- and
polymer-based therapeutic nanoparticles.
 Paolo Arosio Paolo Arosio obtained
his Master’s degree in Chemical Engineering from the Politecnico di Milano in
2007, and his PhD from the Swiss Federal Institute of Technology ETH Zurich in 2011.
After a period as Postdoctoral Fellow at the Department of Chemistry at the
University of Cambridge, UK, he returned to ETH as Assistant Professor in
Biochemical Engineering in 2016. His research interests focus on understanding and
controlling protein self-assembly and protein interaction processes that underlie
problems of key fundamental and practical importance in biology and
biotechnology.
Paolo Arosio Paolo Arosio obtained
his Master’s degree in Chemical Engineering from the Politecnico di Milano in
2007, and his PhD from the Swiss Federal Institute of Technology ETH Zurich in 2011.
After a period as Postdoctoral Fellow at the Department of Chemistry at the
University of Cambridge, UK, he returned to ETH as Assistant Professor in
Biochemical Engineering in 2016. His research interests focus on understanding and
controlling protein self-assembly and protein interaction processes that underlie
problems of key fundamental and practical importance in biology and
biotechnology.
 Ian W. Hamley Professor Ian W. Hamley
is Diamond Professor of Physical Chemistry at the University of Reading. He has more
than 20 years’ experience of research on different types of soft materials,
including peptides, polymers, liquid crystals and surfactants. He received a Royal
Society-Wolfson Research Merit Award in 2011, the RSC Peter Day Award for Materials
Chemistry (2016) and the MacroGroup UK Medal (2017). His research programme focuses
on the self-assembly of peptide materials and its relationship to bioactivity. He
has supervised more than forty postdoctoral and postgraduate researchers and has
published over 350 papers and several edited and authored books.
Ian W. Hamley Professor Ian W. Hamley
is Diamond Professor of Physical Chemistry at the University of Reading. He has more
than 20 years’ experience of research on different types of soft materials,
including peptides, polymers, liquid crystals and surfactants. He received a Royal
Society-Wolfson Research Merit Award in 2011, the RSC Peter Day Award for Materials
Chemistry (2016) and the MacroGroup UK Medal (2017). His research programme focuses
on the self-assembly of peptide materials and its relationship to bioactivity. He
has supervised more than forty postdoctoral and postgraduate researchers and has
published over 350 papers and several edited and authored books.
 Ehud Gazit Ehud Gazit is a Professor
and Endowed Chair at Tel Aviv University. He is also the academic director of the
BLAVATNIK CENTER for Drug Development. Gazit received his BSc (summa cum laude) from
Tel Aviv University and his PhD (with highest distinction) from the Weizmann
Institute of Science in 1997. He has been faculty member at Tel Aviv University
since 2000, following his postdoctoral studies at Massachusetts Institute of
Technology. He is fellow of the Royal Society of Chemistry and member of the
European Molecular Biology Organization. In 2015, he was knighted by the Italian
Republic for his service to science and society.
Ehud Gazit Ehud Gazit is a Professor
and Endowed Chair at Tel Aviv University. He is also the academic director of the
BLAVATNIK CENTER for Drug Development. Gazit received his BSc (summa cum laude) from
Tel Aviv University and his PhD (with highest distinction) from the Weizmann
Institute of Science in 1997. He has been faculty member at Tel Aviv University
since 2000, following his postdoctoral studies at Massachusetts Institute of
Technology. He is fellow of the Royal Society of Chemistry and member of the
European Molecular Biology Organization. In 2015, he was knighted by the Italian
Republic for his service to science and society.
 Raffaele Mezzenga Raffaele Mezzenga
finished his PhD at EPFL Lausanne (2001) and a postdoc at UCSB Santa Barbara, before
joining in 2003 the Nestlé Research Center in Lausanne. In 2005, he was hired
as Associate Professor in Physics at the University of Fribourg, and he then joined
ETH Zurich on 2009 as a Full Professor. His research focuses on the fundamental
understanding of self-assembly processes in soft condensed matter. His work has been
internationally recognized by several prestigious distinctions, such as the
Biomacromolecules/Macromolecules Young Investigator Award (2013, American Chemical
Society), the Dillon Medal (2011, American Physical Society) and the Young Scientist
Research Award (2011, American Oil Chemist Society).
Raffaele Mezzenga Raffaele Mezzenga
finished his PhD at EPFL Lausanne (2001) and a postdoc at UCSB Santa Barbara, before
joining in 2003 the Nestlé Research Center in Lausanne. In 2005, he was hired
as Associate Professor in Physics at the University of Fribourg, and he then joined
ETH Zurich on 2009 as a Full Professor. His research focuses on the fundamental
understanding of self-assembly processes in soft condensed matter. His work has been
internationally recognized by several prestigious distinctions, such as the
Biomacromolecules/Macromolecules Young Investigator Award (2013, American Chemical
Society), the Dillon Medal (2011, American Physical Society) and the Young Scientist
Research Award (2011, American Oil Chemist Society).
Footnotes
Gang Wei: 0000-0002-3838-8659
Nicholas P. Reynolds: 0000-0003-4360-1335
Ian W. Hamley: 0000-0002-4549-0926
Raffaele Mezzenga: 0000-0002-5739-2610
References
- 1.Aguzzi A, O’Connor T. Nat Rev Drug Discovery. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 2.Weber KT, Sun Y, Tyagi SC, Cleutjens JPM. J Mol Cell Cardiol. 1994;26:279–292. doi: 10.1006/jmcc.1994.1036. [DOI] [PubMed] [Google Scholar]
- 3.Cen L, Liu W, Cui L, Zhang WJ, Cao YL. Pediatr Res. 2008;63:492–496. doi: 10.1203/PDR.0b013e31816c5bc3. [DOI] [PubMed] [Google Scholar]
- 4.Reymann AC, Boujemaa-Paterski R, Martiel JL, Guerin C, Cao WX, Chin HF, De La Cruz EM, Thery M, Blanchoin L. Science. 2012;336:1310–1314. doi: 10.1126/science.1221708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles TPJ, Vendruscolo M, Dobson CM. Nat Rev Mol Cell Biol. 2014;15:496. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 6.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, et al. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones OG, Mezzenga R. Soft Matter. 2012;8:876–895. [Google Scholar]
- 8.Hard T. J Phys Chem Lett. 2014;5:607–614. doi: 10.1021/jz4027612. [DOI] [PubMed] [Google Scholar]
- 9.Andreetto E, Malideli E, Yan LM, Kracklauer M, Farbiarz K, Tatarek-Nossol M, Rammes G, Prade E, Neumuller T, Caporale A, Spanopoulou A, et al. Angew Chem, Int Ed. 2015;54:13095–13100. doi: 10.1002/anie.201504973. [DOI] [PubMed] [Google Scholar]
- 10.Doig AJ, Derreumaux P. Curr Opin Struct Biol. 2015;30:50–56. doi: 10.1016/j.sbi.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maji SK, Schubert D, Rivier C, Lee S, Rivier JE, Riek R. PLoS Biol. 2008;6:240–252. doi: 10.1371/journal.pbio.0060017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherny I, Gazit E. Angew Chem, Int Ed. 2008;47:4062–4069. doi: 10.1002/anie.200703133. [DOI] [PubMed] [Google Scholar]
- 15.Romero D, Aguilar C, Losick R, Kolter R. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, Buzhansky L, Rosenman G, Gazit E. Nat Nanotechnol. 2009;4:849–854. doi: 10.1038/nnano.2009.298. [DOI] [PubMed] [Google Scholar]
- 17.Hauser CAE, Maurer-Stroh S, Martins IC. Chem Soc Rev. 2014;43:5326–5345. doi: 10.1039/c4cs00082j. [DOI] [PubMed] [Google Scholar]
- 18.Yan CQ, Pochan DJ. Chem Soc Rev. 2010;39:3528–3540. doi: 10.1039/b919449p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob RS, Ghosh D, Singh PK, Basu SK, Jha NN, Das S, Sukul PK, Patil S, Sathaye S, Kumar A, Chowdhury A, et al. Biomaterials. 2015;54:97–105. doi: 10.1016/j.biomaterials.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Li CX, Adamcik J, Mezzenga R. Nat Nanotechnol. 2012;7:421–427. doi: 10.1038/nnano.2012.62. [DOI] [PubMed] [Google Scholar]
- 21.Buchanan LE, Carr JK, Fluitt AM, Hoganson AJ, Moran SD, de Pablo JJ, Skinner JL, Zanni MT. Proc Natl Acad Sci U S A. 2014;111:5796–5801. doi: 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob RS, George E, Singh PK, Salot S, Anoop A, Jha NN, Sen S, Maji SK. J Biol Chem. 2016;291:5278–5298. doi: 10.1074/jbc.M115.678177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu J, Kim SW, Kang K, Park CB. Adv Mater. 2010;22:5537–5541. doi: 10.1002/adma.201000669. [DOI] [PubMed] [Google Scholar]
- 24.Tycko R, Wickner RB. Acc Chem Res. 2013;46:1487–1496. doi: 10.1021/ar300282r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamley IW. Angew Chem, Int Ed. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 26.Kelly JW. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 27.van der Linden E, Venema P. Curr Opin Colloid Interface Sci. 2007;12:158–165. [Google Scholar]
- 28.Yan XH, Zhu PL, Li JB. Chem Soc Rev. 2010;39:1877–1890. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- 29.Lara C, Handschin S, Mezzenga R. Nanoscale. 2013;5:7197–7201. doi: 10.1039/c3nr02194g. [DOI] [PubMed] [Google Scholar]
- 30.Ulijn RV, Smith AM. Chem Soc Rev. 2008;37:664–675. doi: 10.1039/b609047h. [DOI] [PubMed] [Google Scholar]
- 31.Dehsorkhi A, Castelletto V, Hamley IW. J Pept Sci. 2014;20:453–467. doi: 10.1002/psc.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knowles TPJ, Buehler MJ. Nat Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 33.Knowles TPJ, Mezzenga R. Adv Mater. 2016;28:6546–6561. doi: 10.1002/adma.201505961. [DOI] [PubMed] [Google Scholar]
- 34.Ellis RJ, Hartl FU. Curr Opin Struct Biol. 1999;9:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 35.Anfinsen C, Scheraga H. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- 36.Murphy KP, Privalov PL, Gill SJ. Science. 1990;247:559. doi: 10.1126/science.2300815. [DOI] [PubMed] [Google Scholar]
- 37.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 38.Uversky VN, Gillespie JR, Fink AL. Proteins: Struct, Funct, Bioinf. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Uversky VN. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 41.Greenfield NJ. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croke RL, Sallum CO, Watson E, Watt ED, Alexandrescu AT. Protein Sci. 2008;17:1434–1445. doi: 10.1110/ps.033803.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hebert DN, Molinari M. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 44.Gazit E, Sauer RT. J Biol Chem. 1999;274:2652–2657. doi: 10.1074/jbc.274.5.2652. [DOI] [PubMed] [Google Scholar]
- 45.Matouschek A. Curr Opin Struct Biol. 2003;13:98–109. doi: 10.1016/s0959-440x(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 46.Hartl FU, Hayer-Hartl M. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 47.Stefani M, Dobson CM. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 48.Opie EL. J Exp Med. 1901;5:527. doi: 10.1084/jem.5.5.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lage JMM. J Alzheimer’s Dis. 2006;9:15–26. doi: 10.3233/jad-2006-9s303. [DOI] [PubMed] [Google Scholar]
- 50.LeVine H. Amyloid, Prions, and Other Protein Aggregates. 1999;309:274–284. [Google Scholar]
- 51.Reichhardt C, Jacobson AN, Maher MC, Uang J, McCrate OA, Eckart M, Cegelski L. PLoS One. 2015;10:e0140388. doi: 10.1371/journal.pone.0140388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Proc Natl Acad Sci U S A. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gazit E. Angew Chem, Int Ed. 2002;41:257–259. doi: 10.1002/1521-3773(20020118)41:2<257::aid-anie257>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 54.Baldwin AJ, Knowles TPJ, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, Christodoulou J, et al. J Am Chem Soc. 2011;133:14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 55.Khemtemourian L, Gazit E, Miranker A. J Diabetes Res. 2016;2016 doi: 10.1155/2016/2535878. 2535878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tenidis K, Waldner M, Bernhagen J, Fischle W, Bergmann M, Weber M, Merkle M-L, Voelter W, Brunner H, Kapurniotu A. J Mol Biol. 2000;295:1055–1071. doi: 10.1006/jmbi.1999.3422. [DOI] [PubMed] [Google Scholar]
- 57.Reches M, Porat Y, Gazit E. J Biol Chem. 2002;277:35475–35480. doi: 10.1074/jbc.M206039200. [DOI] [PubMed] [Google Scholar]
- 58.Castelletto V, Ryumin P, Cramer R, Hamley IW, Taylor M, Allsop D, Reza M, Ruokolainen J, Arnold T, Hermida-Merino D, Garcia CI, et al. Sci Rep. 2017;7 doi: 10.1038/srep43637. 43637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reches M, Gazit E. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 60.Reches M, Gazit E. Isr J Chem. 2005;45:363–371. [Google Scholar]
- 61.Nguyen V, Zhu R, Jenkins K, Yang R. Nat Commun. 2016;7 doi: 10.1038/ncomms13566. 13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frederix PW, Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T. Nat Chem. 2015;7:30–37. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- 63.Adler-Abramovich L, Vaks L, Carny O, Trudler D, Magno A, Caflisch A, Frenkel D, Gazit E. Nat Chem Biol. 2012;8:701–706. doi: 10.1038/nchembio.1002. [DOI] [PubMed] [Google Scholar]
- 64.Shaham-Niv S, Adler-Abramovich L, Schnaider L, Gazit E. Sci Adv. 2015;1:e1500137. doi: 10.1126/sciadv.1500137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh P, Brar SK, Bajaj M, Narang N, Mithu VS, Katare OP, Wangoo N, Sharma RK. Mater Sci Eng, C. 2017;72:590–600. doi: 10.1016/j.msec.2016.11.117. [DOI] [PubMed] [Google Scholar]
- 66.Fichman G, Adler-Abramovich L, Manohar S, Mironi-Harpaz I, Guterman T, Seliktar D, Messersmith PB, Gazit E. ACS Nano. 2014;8:7220–7228. doi: 10.1021/nn502240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fichman G, Gazit E. Acta Biomater. 2014;10:1671–1682. doi: 10.1016/j.actbio.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 68.Chromy BA, Nowak RJ, Lambert MP, Viola KL, Chang L, Velasco PT, Jones BW, Fernandez SJ, Lacor PN, Horowitz P, Finch CE, et al. Biochemistry. 2003;42:12749–12760. doi: 10.1021/bi030029q. [DOI] [PubMed] [Google Scholar]
- 69.de la Paz ML, Goldie K, Zurdo J, Lacroix E, Dobson CM, Hoenger A, Serrano L. Proc Natl Acad Sci U S A. 2002;99:16052–16057. doi: 10.1073/pnas.252340199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jha S, Snell JM, Sheftic SR, Patil SM, Daniels SB, Kolling FW, Alexandrescu AT. Biochemistry. 2014;53:300–310. doi: 10.1021/bi401164k. [DOI] [PubMed] [Google Scholar]
- 71.Kobayashi S, Tanaka Y, Kiyono M, Chino M, Chikuma T, Hoshi K, Ikeshima H. J Mol Struct. 2015;1094:109–117. [Google Scholar]
- 72.Hamley IW. Soft Matter. 2011;7:4122–4138. [Google Scholar]
- 73.Niece KL, Hartgerink JD, Donners JJJM, Stupp SI. J Am Chem Soc. 2003;125:7146–7147. doi: 10.1021/ja028215r. [DOI] [PubMed] [Google Scholar]
- 74.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 75.Hartgerink JD, Beniash E, Stupp SI. Proc Natl Acad Sci U S A. 2002;99:5133–5138. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dehsorkhi A, Castelletto V, Hamley IW, Adamcik J, Mezzenga R. Soft Matter. 2013;9:6033–6036. [Google Scholar]
- 77.Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. J Mol Biol. 2002;322:383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 78.Raman B, Chatani E, Kihara M, Ban T, Sakai M, Hasegawa K, Naiki H, Rao CM, Goto Y. Biochemistry. 2005;44:1288–1299. doi: 10.1021/bi048029t. [DOI] [PubMed] [Google Scholar]
- 79.Arnaudov LN, de Vries R. Biomacromolecules. 2006;7:3490–3498. doi: 10.1021/bm060584i. [DOI] [PubMed] [Google Scholar]
- 80.Marek PJ, Patsalo V, Green DF, Raleigh DP. Biochemistry. 2012;51:8478–8490. doi: 10.1021/bi300574r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abelein A, Jarvet J, Barth A, Graslund A, Danielsson J. J Am Chem Soc. 2016;138:6893–6902. doi: 10.1021/jacs.6b04511. [DOI] [PubMed] [Google Scholar]
- 82.Owczarz M, Casalini T, Motta AC, Morbidelli M, Arosio P. Biomacromolecules. 2015;16:3792–3801. doi: 10.1021/acs.biomac.5b01092. [DOI] [PubMed] [Google Scholar]
- 83.Parker R, Noel TE, Brownsey GJ, Laos K, Ring SG. Biophys J. 2005;89:1227–1236. doi: 10.1529/biophysj.105.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bromley EHC, Krebs MRH, Donald AM. Eur Phys J E: Soft Matter Biol Phys. 2006;21:1–8. [Google Scholar]
- 85.Mishra R, Winter R. Angew Chem, Int Ed. 2008;47:6518–6521. doi: 10.1002/anie.200802027. [DOI] [PubMed] [Google Scholar]
- 86.van den Heuvel M, Lowik D, van Hest JCM. Biomacromolecules. 2008;9:2727–2734. doi: 10.1021/bm800424x. [DOI] [PubMed] [Google Scholar]
- 87.Miravet JF, Escuder B, Segarra-Maset MD, Tena-Solsona M, Hamley IW, Dehsorkhi A, Castelletto V. Soft Matter. 2013;9:3558–3564. [Google Scholar]
- 88.Löwik DWPM, Leunissen EHP, van den Heuvel M, Hansen MB, van Hest JCM. Chem Soc Rev. 2010;39:3394–3412. doi: 10.1039/b914342b. [DOI] [PubMed] [Google Scholar]
- 89.Lara C, Gourdin-Bertin S, Adamcik J, Bolisetty S, Mezzenga R. Biomacromolecules. 2012;13:4213–4221. doi: 10.1021/bm301481v. [DOI] [PubMed] [Google Scholar]
- 90.Kardos J, Micsonai A, Pal-Gabor H, Petrik E, Graf L, Kovacs J, Lee YH, Naiki H, Goto Y. Biochemistry. 2011;50:3211–3220. doi: 10.1021/bi2000017. [DOI] [PubMed] [Google Scholar]
- 91.Ikenoue T, Lee YH, Kardos J, Saiki M, Yagi H, Kawata Y, Goto Y. Angew Chem, Int Ed. 2014;53:7799–7804. doi: 10.1002/anie.201403815. [DOI] [PubMed] [Google Scholar]
- 92.Arora A, Ha C, Park CB. Protein Sci. 2004;13:2429–2436. doi: 10.1110/ps.04823504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cobb NJ, Apetri AC, Surewicz WK. J Biol Chem. 2008;283:34704–34711. doi: 10.1074/jbc.M806701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei G, Keller TF, Zhang JT, Jandt KD. Soft Matter. 2011;7:2011–2018. [Google Scholar]
- 95.Wei G, Reichert J, Bossert J, Jandt KD. Biomacromolecules. 2008;9:3258–3267. doi: 10.1021/bm800824r. [DOI] [PubMed] [Google Scholar]
- 96.Viles JH. Coord Chem Rev. 2012;256:2271–2284. [Google Scholar]
- 97.Bush AI. Curr Opin Chem Biol. 2000;4:184–191. doi: 10.1016/s1367-5931(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 98.Zatta P, editor. Metal ions and neurodegenerative disorders. World Scientific; Singapore: 2003. [Google Scholar]
- 99.Adlard PA, Bush AI. J Alzheimer’s Dis. 2006;10:145–163. doi: 10.3233/jad-2006-102-303. [DOI] [PubMed] [Google Scholar]
- 100.Miu AC, Benga O. J Alzheimer’s Dis. 2006;10:179–201. doi: 10.3233/jad-2006-102-306. [DOI] [PubMed] [Google Scholar]
- 101.Brown DR. Dalton Trans. 2009:4069–4076. doi: 10.1039/b822135a. [DOI] [PubMed] [Google Scholar]
- 102.Smith MA, Harris PLR, Sayre LM, Perry G. Proc Natl Acad Sci U S A. 1997;94:9866–9868. doi: 10.1073/pnas.94.18.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sayre LM, Perry G, Smith MA. Curr Opin Chem Biol. 1999;3:220–225. doi: 10.1016/S1367-5931(99)80035-0. [DOI] [PubMed] [Google Scholar]
- 104.Sayre LM, Perry G, Harris PLR, Liu YH, Schubert KA, Smith MA. J Neurochem. 2000;74:270–279. doi: 10.1046/j.1471-4159.2000.0740270.x. [DOI] [PubMed] [Google Scholar]
- 105.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. J Neurol Sci. 1998;158:47–52. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 106.Suh SW, Jensen KB, Jensen MS, Silva DS, Kesslak PJ, Danscher G, Frederickson CJ. Brain Res. 2000;852:274–278. doi: 10.1016/s0006-8993(99)02096-x. [DOI] [PubMed] [Google Scholar]
- 107.Bush AI, Pettingell WH, Multhaup G, Paradis MD, Vonsattel JP, Gusella JF, Beyreuther K, Masters CL, Tanzi RE. Science. 1994;265:1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 108.Bush AI, Pettingell WH, Paradis MD, Tanzi RE. J Biol Chem. 1994;269:12152–12158. [PubMed] [Google Scholar]
- 109.Atwood CS, Moir RD, Huang XD, Scarpa RC, Bacarra NME, Romano DM, Hartshorn MK, Tanzi RE, Bush AI. J Biol Chem. 1998;273:12817–12826. doi: 10.1074/jbc.273.21.12817. [DOI] [PubMed] [Google Scholar]
- 110.Cherny RA, Legg JT, McLean CA, Fairlie DP, Huang XD, Atwood CS, Beyreuther K, Tanzi RE, Masters CL, Bush AI. J Biol Chem. 1999;274:23223–23228. doi: 10.1074/jbc.274.33.23223. [DOI] [PubMed] [Google Scholar]
- 111.Raman B, Ban T, Yamaguchi K, Sakai M, Kawai T, Naiki H, Goto Y. J Biol Chem. 2005;280:16157–16162. doi: 10.1074/jbc.M500309200. [DOI] [PubMed] [Google Scholar]
- 112.Hamaguchi T, Ono K, Yamada M. Cell Mol Life Sci. 2006;63:1538–1552. doi: 10.1007/s00018-005-5599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hawkes CA, Ng V, McLaurin J. Drug Dev Res. 2009;70:111–124. [Google Scholar]
- 114.Hamley IW. Chem Rev. 2012;112:5147–5192. doi: 10.1021/cr3000994. [DOI] [PubMed] [Google Scholar]
- 115.Innocenti M, Salvietti E, Guidotti M, Casini A, Bellandi S, Foresti ML, Gabbiani C, Pozzi A, Zatta P, Messori L. J Alzheimer’s Dis. 2010;19:1323–1329. doi: 10.3233/JAD-2010-1338. [DOI] [PubMed] [Google Scholar]
- 116.Pradines V, Stroia AJ, Faller P. New J Chem. 2008;32:1189–1194. [Google Scholar]
- 117.Fink AL. Acc Chem Res. 2006;39:628–634. doi: 10.1021/ar050073t. [DOI] [PubMed] [Google Scholar]
- 118.Bolisetty S, Mezzenga R. Nat Nanotechnol. 2016;11:365–371. doi: 10.1038/nnano.2015.310. [DOI] [PubMed] [Google Scholar]
- 119.Zou R, Wang Q, Wu J, Wu J, Schmuck C, Tian H. Chem Soc Rev. 2015;44:5200–5219. doi: 10.1039/c5cs00234f. [DOI] [PubMed] [Google Scholar]
- 120.Snow AD, Mar H, Nochlin D, Kimata K, Kato M, Suzuki S, Hassell J, Wight TN. Am J Pathol. 1988;133:456–463. [PMC free article] [PubMed] [Google Scholar]
- 121.McLaurin J, Franklin T, Zhang XQ, Deng JP, Fraser PE. Eur J Biochem. 1999;266:1101–1110. doi: 10.1046/j.1432-1327.1999.00957.x. [DOI] [PubMed] [Google Scholar]
- 122.McLaurin J, Yang DS, Yip CM, Fraser PE. J Struct Biol. 2000;130:259–270. doi: 10.1006/jsbi.2000.4289. [DOI] [PubMed] [Google Scholar]
- 123.Valle-Delgado JJ, Alfonso-Prieto M, de Groot NS, Ventura S, Samitier J, Rovira C, Fernandez-Busquets X. FASEB J. 2010;24:4250–4261. doi: 10.1096/fj.09-153551. [DOI] [PubMed] [Google Scholar]
- 124.Fraser PE, Nguyen JT, Chin DT, Kirschner DA. J Neurochem. 1992;59:1531–1540. doi: 10.1111/j.1471-4159.1992.tb08470.x. [DOI] [PubMed] [Google Scholar]
- 125.Panza G, Stohr J, Birkmann E, Riesner D, Willbold D, Baba O, Terashima T, Dumpitak C. Rejuvenation Res. 2008;11:365–369. doi: 10.1089/rej.2008.0698. [DOI] [PubMed] [Google Scholar]
- 126.Jones OG, Adamcik J, Handschin S, Bolisetty S, Mezzenga R. Langmuir. 2010;26:17449–17458. doi: 10.1021/la1026619. [DOI] [PubMed] [Google Scholar]
- 127.Assarsson A, Linse S, Cabaleiro-Lago C. Langmuir. 2014;30:8812–8818. doi: 10.1021/la501414j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.da Silva ER, Cooney G, Hamley IW, Alves WA, Lee S, O’Connor BF, Reza M, Ruokolainen J, Walls D. Soft Matter. 2016;12:9158–9169. doi: 10.1039/c6sm01618a. [DOI] [PubMed] [Google Scholar]
- 129.Udomprasert A, Bongiovanni MN, Sha RJ, Sherman WB, Wang T, Arora PS, Canary JW, Gras SL, Seeman NC. Nat Nanotechnol. 2014;9:537–541. doi: 10.1038/nnano.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cabaleiro-Lago C, Szczepankiewicz O, Linse S. Langmuir. 2012;28:1852–1857. doi: 10.1021/la203078w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.O’Brien EP, Straub JE, Brooks BR, Thirumalai D. J Phys Chem Lett. 2011;2:1171–1177. doi: 10.1021/jz200330k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cedervall T, Lynch I, Foy M, Berggad T, Donnelly SC, Cagney G, Linse S, Dawson KA. Angew Chem, Int Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 133.Cabaleiro-Lago C, Lynch I, Dawson KA, Linse S. Langmuir. 2009;26:3453–3461. doi: 10.1021/la902980d. [DOI] [PubMed] [Google Scholar]
- 134.Cabaleiro-Lago C, Quinlan-Pluck F, Lynch I, Lindman S, Minogue AM, Thulin E, Walsh DM, Dawson KA, Linse S. J Am Chem Soc. 2008;130:15437–15443. doi: 10.1021/ja8041806. [DOI] [PubMed] [Google Scholar]
- 135.Cabaleiro-Lago C, Quinlan-Pluck F, Lynch I, Dawson KA, Linse S. ACS Chem Neurosci. 2010;1:279–287. doi: 10.1021/cn900027u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yoo SI, Yang M, Brender JR, Subramanian V, Sun K, Joo NE, Jeong SH, Ramamoorthy A, Kotov NA. Angew Chem, Int Ed. 2011;50:5110–5115. doi: 10.1002/anie.201007824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu WH, Sun X, Yu YP, Hu J, Zhao L, Liu Q, Zhao YF, Li YM. Biochem Biophys Res Commun. 2008;373:315–318. doi: 10.1016/j.bbrc.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 138.Geng J, Li M, Ren JS, Wang EB, Qu XG. Angew Chem, Int Ed. 2011;50:4184–4188. doi: 10.1002/anie.201007067. [DOI] [PubMed] [Google Scholar]
- 139.Barandeh F, Nguyen P-L, Kumar R, Iacobucci GJ, Kuznicki ML, Kosterman A, Bergey EJ, Prasad PN, Gunawardena S. PLoS One. 2012;7:e29424. doi: 10.1371/journal.pone.0029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Di L, Kerns EH, Fan K, McConnell OJ, Carter GT. Eur J Med Chem. 2003;38:223–232. doi: 10.1016/s0223-5234(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 141.Choi JS, Braymer JJ, Nanga RPR, Ramamoorthy A, Lim MH. Proc Natl Acad Sci U S A. 2010;107:21990–21995. doi: 10.1073/pnas.1006091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Linse S, Cabaleiro-Lago C, Xue WF, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Proc Natl Acad Sci U S A. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fernández C, González-Rubio G, Langer J, Tardajos G, Liz-Marzán LM, Giraldo R, Guerrero-Martínez A. Angew Chem, Int Ed. 2016;55:11237–11241. doi: 10.1002/anie.201604970. [DOI] [PubMed] [Google Scholar]
- 144.Gladytz A, Wagner M, Haupl T, Elsner C, Abel B. Part Part Syst Charact. 2015;32:573–582. [Google Scholar]
- 145.Gladytz A, Abel B, Risselada HJ. Angew Chem, Int Ed. 2016;55:11242–11246. doi: 10.1002/anie.201605151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Knowles TPJ, Waudby CA, Devlin GL, Cohen SIA, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 147.Chatani E, Lee YH, Yagi H, Yoshimura Y, Naiki H, Goto Y. Natl Proc Acad Sci U S A. 2009;106:11119–11124. doi: 10.1073/pnas.0901422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jung JM, Savin G, Pouzot M, Schmitt C, Mezzenga R. Biomacromolecules. 2008;9:2477–2486. doi: 10.1021/bm800502j. [DOI] [PubMed] [Google Scholar]
- 149.Dunstan DE, Hamilton-Brown P, Asimakis P, Ducker W, Bertolini J. Soft Matter. 2009;5:5020–5028. [Google Scholar]
- 150.Hill EK, Krebs B, Goodall DG, Howlett GJ, Dunstan DE. Biomacromolecules. 2006;7:10–13. doi: 10.1021/bm0505078. [DOI] [PubMed] [Google Scholar]
- 151.Bekard IB, Asimakis P, Teoh CL, Ryan T, Howlett GJ, Bertolini J, Dunstan DE. Soft Matter. 2012;8:385–389. [Google Scholar]
- 152.Macchi F, Hoffmann SV, Carlsen M, Vad B, Imparato A, Rischel C, Otzen DE. Langmuir. 2011;27:12539–12549. doi: 10.1021/la202125c. [DOI] [PubMed] [Google Scholar]
- 153.Dexter AF. Langmuir. 2010;26:17997–18007. doi: 10.1021/la103471j. [DOI] [PubMed] [Google Scholar]
- 154.Mezzenga R, Fischer P. Rep Prog Phys. 2013;76 doi: 10.1088/0034-4885/76/4/046601. 046601. [DOI] [PubMed] [Google Scholar]
- 155.Campioni S, Carret G, Jordens S, Nicoud L, Mezzenga R, Riek R. J Am Chem Soc. 2014;136:2866–2875. doi: 10.1021/ja412105t. [DOI] [PubMed] [Google Scholar]
- 156.Hoernke M, Falenski JA, Schwieger C, Koksch B, Brezesinski G. Langmuir. 2011;27:14218–14231. doi: 10.1021/la203016z. [DOI] [PubMed] [Google Scholar]
- 157.Pronchik J, He X, Giurleo JT, Talaga DS. J Am Chem Soc. 2010;132:9797–9803. doi: 10.1021/ja102896h. [DOI] [PubMed] [Google Scholar]
- 158.Hamley IW. Nat Chem. 2010;2:707–708. doi: 10.1038/nchem.816. [DOI] [PubMed] [Google Scholar]
- 159.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 160.Galvagnion C, Buell AK, Meisl G, Michaels TCT, Vendruscolo M, Knowles TPJ, Dobson CM. Nat Chem Biol. 2015;11:229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fusco G, De Simone A, Gopinath T, Vostrikov V, Vendruscolo M, Dobson CM, Veglia G. Nat Commun. 2014;5 doi: 10.1038/ncomms4827. 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Michikawa M. J Neurosci Res. 2003;72:141–146. doi: 10.1002/jnr.10585. [DOI] [PubMed] [Google Scholar]
- 163.Michikawa M. Mol Neurobiol. 2003;27:1–12. doi: 10.1385/MN:27:1:1. [DOI] [PubMed] [Google Scholar]
- 164.Gorbenko GP, Kinnunen PKJ. Chem Phys Lipids. 2006;141:72–82. doi: 10.1016/j.chemphyslip.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 165.Kawarabayashi T, Shoji M, Younkin LH, Lin WL, Dickson DW, Murakami T, Matsubara E, Abe K, Ashe KH, Younkin SG. J Neurosci. 2004;24:3801–3809. doi: 10.1523/JNEUROSCI.5543-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kim SI, Yi JS, Ko YG. J Cell Biochem. 2006;99:878–889. doi: 10.1002/jcb.20978. [DOI] [PubMed] [Google Scholar]
- 167.Okada T, Ikeda K, Wakabayashi M, Ogawa M, Matsuzaki K. J Mol Biol. 2008;382:1066–1074. doi: 10.1016/j.jmb.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 168.Mandal PK, Pettegrew JW. Neurochem Res. 2004;29:2267–2272. doi: 10.1007/s11064-004-7035-1. [DOI] [PubMed] [Google Scholar]
- 169.Morita M, Hamada T, Tendo Y, Hata T, Vestergaard MC, Takagi M. Soft Matter. 2012;8:2816–2819. [Google Scholar]
- 170.Reynolds NP, Soragni A, Rabe M, Verdes D, Liverani E, Handschin S, Riek R, Seeger S. J Am Chem Soc. 2011;133:19366–19375. doi: 10.1021/ja2029848. [DOI] [PubMed] [Google Scholar]
- 171.Jiang Z, de Messieres M, Lee JC. J Am Chem Soc. 2013;135:15970–15973. doi: 10.1021/ja405993r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pfefferkorn CM, Heinrich F, Sodt AJ, Maltsev AS, Pastor RW, Lee JC. Biophys J. 2012;102:613–621. doi: 10.1016/j.bpj.2011.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Comellas G, Lemkau LR, Zhou DHH, George JM, Rienstra CM. J Am Chem Soc. 2012;134:5090–5099. doi: 10.1021/ja209019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Seeliger J, Evers F, Jeworrek C, Kapoor S, Weise K, Andreetto E, Tolan M, Kapurniotu A, Winter R. Angew Chem, Int Ed. 2012;51:679–683. doi: 10.1002/anie.201105877. [DOI] [PubMed] [Google Scholar]
- 175.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. J Mol Biol. 2012;421:160–171. doi: 10.1016/j.jmb.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 176.Cohen SIA, Vendruscolo M, Welland ME, Dobson CM, Terentjev EM, Knowles TPJ. J Chem Phys. 2011;135 doi: 10.1063/1.3608916. 065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen SW, Drakulic S, Deas E, Ouberai M, Aprile FA, Arranz R, Ness S, Roodveldt C, Guilliams T, De-Genst EJ, Klenerman D, et al. Proc Natl Acad Sci U S A. 2015;112:E1994–E2003. doi: 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cremades N, Cohen SIA, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, Bertoncini CW, et al. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lorenzen N, Nielsen SB, Buell AK, Kaspersen JD, Arosio P, Vad BS, Paslawski W, Christiansen G, Valnickova-Hansen Z, Andreasen M, Enghild JJ, et al. J Am Chem Soc. 2014;136:3859–3868. doi: 10.1021/ja411577t. [DOI] [PubMed] [Google Scholar]
- 180.Buell AK, Dhulesia A, White DA, Knowles TPJ, Dobson CM, Welland ME. Angew Chem, Int Ed. 2012;51:5247–5251. doi: 10.1002/anie.201108040. [DOI] [PubMed] [Google Scholar]
- 181.Owczarz M, Motta AC, Morbidelli M, Arosio P. Langmuir. 2015;31:7590–7600. doi: 10.1021/acs.langmuir.5b01110. [DOI] [PubMed] [Google Scholar]
- 182.Tanaka M, Collins SR, Toyama BH, Weissman JS. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 183.Ferrone FA, Hofrichter J, Eaton WA. J Mol Biol. 1985;183:611–631. doi: 10.1016/0022-2836(85)90175-5. [DOI] [PubMed] [Google Scholar]
- 184.Saric A, Chebaro YC, Knowles TPJ, Frenkel D. Proc Natl Acad Sci U S A. 2014;111:17869–17874. doi: 10.1073/pnas.1410159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cohen SIA, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, Knowles TPJ. Proc Natl Acad Sci U S A. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fodera V, Librizzi F, Groenning M, van de Weert M, Leone M. J Phys Chem B. 2008;112:3853–3858. doi: 10.1021/jp710131u. [DOI] [PubMed] [Google Scholar]
- 187.Ruschak AM, Miranker AD. Proc Natl Acad Sci U S A. 2007;104:12341–12346. doi: 10.1073/pnas.0703306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Buell AK, Galvagnion C, Gaspar R, Sparr E, Vendruscolo M, Knowles TPJ, Linse S, Dobson CM. Proc Natl Acad Sci U S A. 2014;111:7671–7676. doi: 10.1073/pnas.1315346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Habchi J, Arosio P, Perni M, Costa AR, Yagi-Utsumi M, Joshi P, Chia S, Cohen SIA, Muller MBD, Linse S, Nollen EAA, et al. Sci Adv. 2016;2:e1501244. doi: 10.1126/sciadv.1501244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lang L, Zetterstrom P, Brannstrom T, Marklund SL, Danielsson J, Oliveberg M. Proc Natl Acad Sci U S A. 2015;112:9878–9883. doi: 10.1073/pnas.1503328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pappu RV, Wang X, Vitalis A, Crick SL. Arch Biochem Biophys. 2008;469:132–141. doi: 10.1016/j.abb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Andrews JM, Roberts CJ. J Phys Chem B. 2007;111:7897–7913. doi: 10.1021/jp070212j. [DOI] [PubMed] [Google Scholar]
- 193.Roberts CJ. Biotechnol Bioeng. 2007;98:927–938. doi: 10.1002/bit.21627. [DOI] [PubMed] [Google Scholar]
- 194.Nicoud L, Arosio P, Sozo M, Yates A, Norrant E, Morbidelli M. J Phys Chem B. 2014;118:10595–10606. doi: 10.1021/jp505295j. [DOI] [PubMed] [Google Scholar]
- 195.Ahmad A, Millett IS, Doniach S, Uversky VN, Fink AL. Biochemistry. 2003;42:11404–11416. doi: 10.1021/bi034868o. [DOI] [PubMed] [Google Scholar]
- 196.Nielsen L, Khurana R, Coats A, Frokjaer S, Brange J, Vyas S, Uversky VN, Fink AL. Biochemistry. 2001;40:6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 197.Lee CC, Nayak A, Sethuraman A, Belfort G, McRae GJ. Biophys J. 2007;92:3448–3458. doi: 10.1529/biophysj.106.098608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Manno M, Craparo EF, Martorana V, Bulone D, San Biagio PL. Biophys J. 2006;90:4585–4591. doi: 10.1529/biophysj.105.077636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Buell AK, Dhulesia A, Mossuto MF, Cremades N, Kumita JR, Dumoulin M, Welland ME, Knowles TPJ, Salvatella X, Dobson CM. J Am Chem Soc. 2011;133:7737–7743. doi: 10.1021/ja109620d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Raccosta S, Martorana V, Manno M. J Phys Chem B. 2012;116:12078–12087. doi: 10.1021/jp303430g. [DOI] [PubMed] [Google Scholar]
- 201.Chiti F, De Lorenzi E, Grossi S, Mangione P, Giorgetti S, Caccialanza G, Dobson CM, Merlini G, Ramponi G, Bellotti V. J Biol Chem. 2001;276:46714–46721. doi: 10.1074/jbc.M107040200. [DOI] [PubMed] [Google Scholar]
- 202.McParland VJ, Kad NM, Kalverda AP, Brown A, Kirwin-Jones P, Hunter MG, Sunde M, Radford SE. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 203.Nordlund A, Leinartaite L, Saraboji K, Aisenbrey C, Grobner G, Zetterstrom P, Danielsson J, Logan DT, Oliveberg M. Proc Natl Acad Sci U S A. 2009;106:9667–9672. doi: 10.1073/pnas.0812046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Biochemistry. 2001;40:3525–3535. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 205.Esposito G, Michelutti R, Verdone G, Viglino P, Hernandez H, Robinson CV, Amoresano A, Dal Piaz F, Monti M, Pucci P, Mangione P, et al. Protein Sci. 2000;9:831–845. doi: 10.1110/ps.9.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lara C, Adamcik J, Jordens S, Mezzenga R. Biomacromolecules. 2011;12:1868–1875. doi: 10.1021/bm200216u. [DOI] [PubMed] [Google Scholar]
- 207.Mishra R, Sorgjerd K, Nystrom S, Nordigarden A, Yu YC, Hammarstrom P. J Mol Biol. 2007;366:1029–1044. doi: 10.1016/j.jmb.2006.11.084. [DOI] [PubMed] [Google Scholar]
- 208.Akkermans C, Venema P, van der Goot AJ, Gruppen H, Bakx EJ, Boom RM, van der Linden E. Biomacromolecules. 2008;9:1474–1479. doi: 10.1021/bm7014224. [DOI] [PubMed] [Google Scholar]
- 209.Arosio P, Beeg M, Nicoud L, Morbidelli M. Chem Eng Sci. 2012;78:21–32. [Google Scholar]
- 210.Arosio P, Rima S, Lattuada M, Morbidelli M. J Phys Chem B. 2012;116:7066–7075. doi: 10.1021/jp301091n. [DOI] [PubMed] [Google Scholar]
- 211.Sahin E, Grillo AO, Perkins MD, Roberts CJ. J Pharm Sci. 2010;99:4830–4848. doi: 10.1002/jps.22198. [DOI] [PubMed] [Google Scholar]
- 212.Andersen CB, Manno M, Rischel C, Thorolfsson M, Martorana V. Protein Sci. 2010;19:279–290. doi: 10.1002/pro.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jung JM, Mezzenga R. Langmuir. 2010;26:504–514. doi: 10.1021/la9021432. [DOI] [PubMed] [Google Scholar]
- 214.Owczarz M, Bolisetty S, Mezzenga R, Arosio P. J Colloid Interface Sci. 2015;437:244–251. doi: 10.1016/j.jcis.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 215.Manno M, Giacomazza D, Newman J, Martorana V, San Biagio PL. Langmuir. 2010;26:1424–1426. doi: 10.1021/la903340v. [DOI] [PubMed] [Google Scholar]
- 216.Nayak A, Dutta AK, Belfort G. Biochem Biophys Res Commun. 2008;369:303–307. doi: 10.1016/j.bbrc.2008.01.159. [DOI] [PubMed] [Google Scholar]
- 217.Shammas SL, Garcia GA, Kumar S, Kjaergaard M, Horrocks MH, Shivji N, Mandelkow E, Knowles TPJ, Mandelkow E, Klenerman D. Nat Commun. 2015;6 doi: 10.1038/ncomms8025. 7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ferrone F. Amyloid, Prions, and Other Protein Aggregates. 1999;309:256–274. [Google Scholar]
- 219.Meisl G, Kirkegaard JB, Arosio P, Michaels TCT, Vendruscolo M, Dobson CM, Linse S, Knowles TPJ. Nat Protoc. 2016;11:252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 220.LeVine H. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Biancalana M, Koide S. Biochim Biophys Acta, Proteins Proteomics. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Arosio P, Cukalevski R, Frohm B, Knowles TPJ, Linse S. J Am Chem Soc. 2014;136:219–225. doi: 10.1021/ja408765u. [DOI] [PubMed] [Google Scholar]
- 223.Arosio P, Knowles TPJ, Linse S. Phys Chem Chem Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hellstrand E, Boland B, Walsh DM, Linse S. ACS Chem Neurosci. 2010;1:13–18. doi: 10.1021/cn900015v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cohen SIA, Arosio P, Presto J, Kurudenkandy FR, Biverstal H, Dolfe L, Dunning C, Yang XT, Frohm B, Vendruscolo M, Johansson J, et al. Nat Struct Mol Biol. 2015;22:207–213. doi: 10.1038/nsmb.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Meisl G, Yang XT, Frohm B, Knowles TPJ, Linse S. Sci Rep. 2016;6 doi: 10.1038/srep18728. 18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Meisl G, Yang XT, Hellstrand E, Frohm B, Kirkegaard JB, Cohen SIA, Dobson CM, Linse S, Knowles TPJ. Proc Natl Acad Sci U S A. 2014;111:9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Arosio P, Vendruscolo M, Dobson CM, Knowles TPJ. Trends Pharmacol Sci. 2014;35:127–135. doi: 10.1016/j.tips.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 229.Arosio P, Michaels TCT, Linse S, Mansson C, Emanuelsson C, Presto J, Johansson J, Vendruscolo M, Dobson CM, Knowles TPJ. Nat Commun. 2016;7 doi: 10.1038/ncomms10948. 10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Horrocks MH, Tosatto L, Dear AJ, Garcia GA, Iljina M, Cremades N, Serra MD, Knowles TPJ, Dobson CM, Klenerman D. Anal Chem. 2015;87:8818–8826. doi: 10.1021/acs.analchem.5b01811. [DOI] [PubMed] [Google Scholar]
- 231.Iljina M, Garcia GA, Horrocks MH, Tosatto L, Choi ML, Ganzinger KA, Abramov AY, Gandhi S, Wood NW, Cremades N, Dobson CM, et al. Proc Natl Acad Sci U S A. 2016;113:E1206–E1215. doi: 10.1073/pnas.1524128113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xue WF, Hellewell AL, Gosal WS, Homans SW, Hewitt EW, Radford SE. J Biol Chem. 2009;284:34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Adamcik J, Jung JM, Flakowski J, De Los Rios P, Dietler G, Mezzenga R. Nat Nanotechnol. 2010;5:423–428. doi: 10.1038/nnano.2010.59. [DOI] [PubMed] [Google Scholar]
- 234.Adamcik J, Mezzenga R. Soft Matter. 2011;7:5437–5443. [Google Scholar]
- 235.Adamcik J, Mezzenga R. Curr Opin Colloid Interface Sci. 2012;17:369–376. [Google Scholar]
- 236.Xue WF, Homans SW, Radford SE. Protein Eng, Des Sel. 2009;22:489–496. doi: 10.1093/protein/gzp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xue WF, Radford SE. Biophys J. 2013;105:2811–2819. doi: 10.1016/j.bpj.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Pinotsi D, Buell AK, Dobson CM, Schierle GSK, Kaminski CF. ChemBioChem. 2013;14:846–850. doi: 10.1002/cbic.201300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Pinotsi D, Buell AK, Galvagnion C, Dobson CM, Schierle GSK, Kaminski CF. Nano Lett. 2014;14:339–345. doi: 10.1021/nl4041093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bolisetty S, Adamcik J, Mezzenga R. Soft Matter. 2011;7:493–499. [Google Scholar]
- 241.Rogers SS, Venema P, Van der Ploeg JPM, Van der Linden E, Sagis LMC, Donald AM. Biopolymers. 2006;82:241–252. doi: 10.1002/bip.20483. [DOI] [PubMed] [Google Scholar]
- 242.Baldwin AJ, Anthony-Cahill SJ, Knowles TPJ, Lippens G, Christodoulou J, Barker PD, Dobson CM. Angew Chem, Int Ed. 2008;47:3385–3387. doi: 10.1002/anie.200703915. [DOI] [PubMed] [Google Scholar]
- 243.Schuck P. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Schuck P. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 245.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Howlett GJ, Minton AP, Rivas G. Curr Opin Chem Biol. 2006;10:430–436. doi: 10.1016/j.cbpa.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 247.Arosio P, Cedervall T, Knowles TPJ, Linse S. Anal Biochem. 2016;504:7–13. doi: 10.1016/j.ab.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 248.Michaels TCT, Lazell HW, Arosio P, Knowles TPJ. J Chem Phys. 2015;143 doi: 10.1063/1.4927655. 054901. [DOI] [PubMed] [Google Scholar]
- 249.Michaels TCT, Yde P, Willis JCW, Jensen MH, Otzen D, Dobson CM, Buell AK, Knowles TPJ. J Chem Phys. 2015;143 doi: 10.1063/1.4933230. 164901. [DOI] [PubMed] [Google Scholar]
- 250.Luo Q, Hou CX, Bai YS, Wang RB, Liu JQ. Chem Rev. 2016;116:13571–13632. doi: 10.1021/acs.chemrev.6b00228. [DOI] [PubMed] [Google Scholar]
- 251.Haass C, Selkoe DJ. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 252.Sakono M, Zako T, Ueda H, Yohda M, Maeda M. FEBS J. 2008;275:5982–5993. doi: 10.1111/j.1742-4658.2008.06727.x. [DOI] [PubMed] [Google Scholar]
- 253.Conway KA, Harper JD, Lansbury PT. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 254.Liu C, Sawaya MR, Cheng PN, Zheng J, Nowick JS, Eisenberg D. J Am Chem Soc. 2011;133:6736–6744. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Glabe CG. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fandrich M. J Mol Biol. 2012;421:427–440. doi: 10.1016/j.jmb.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 257.Mulaj M, Foley J, Muschol M. J Am Chem Soc. 2014;136:8947–8956. doi: 10.1021/ja502529m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Harte NP, Klyubin I, McCarthy EK, Min S, Garrahy SA, Xie YJ, Davey GP, Boland JJ, Rowan MJ, Mok KH. J Biol Chem. 2015;290:28343–28352. doi: 10.1074/jbc.M115.676072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrene YF, Narayanaswami V, Goormaghtigh E, Ruysschaert JM, Raussens V. Biochem J. 2009;421:415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 260.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Nat Struct Mol Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Renner M, Lacor PN, Velasco PT, Xu JA, Contractor A, Klein WL, Triller A. Neuron. 2010;66:739–754. doi: 10.1016/j.neuron.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao ML, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, et al. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Domanska K, Vanderhaegen S, Srinivasan V, Pardon E, Dupeux F, Marquez JA, Giorgetti S, Stoppini M, Wyns L, Bellotti V, Steyaert J. Proc Natl Acad Sci U S A. 2011;108:1314–1319. doi: 10.1073/pnas.1008560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Maresca M, Derghal A, Carravagna C, Dudin S, Fantini J. Phys Chem Chem Phys. 2008;10:2792–2800. doi: 10.1039/b802594k. [DOI] [PubMed] [Google Scholar]
- 265.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.El Moustaine D, Perrier V, Smeller L, Lange R, Torrent J. FEBS J. 2008;275:2021–2031. doi: 10.1111/j.1742-4658.2008.06356.x. [DOI] [PubMed] [Google Scholar]
- 267.Habicht G, Haupt C, Friedrich RP, Hortschansky P, Sachse C, Meinhardt J, Wieligmann K, Gellermann GP, Brodhun M, Gotz J, Halbhuber KJ, et al. Proc Natl Acad Sci U S A. 2007;104:19232–19237. doi: 10.1073/pnas.0703793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kumar ST, Meinhardt J, Fuchs AK, Aumuller T, Leppert J, Buchele B, Knupfer U, Ramachandran R, Yadav JK, Prell E, Morgado I, et al. ACS Nano. 2014;8:11042–11052. doi: 10.1021/nn503960h. [DOI] [PubMed] [Google Scholar]
- 269.Guo C, Luo Y, Zhou RH, Wei GH. Nanoscale. 2014;6:2800–2811. doi: 10.1039/c3nr02505e. [DOI] [PubMed] [Google Scholar]
- 270.Matsubara T, Iijima K, Yamamoto N, Yanagisawa K, Sato T. Langmuir. 2013;29:2258–2264. doi: 10.1021/la3038999. [DOI] [PubMed] [Google Scholar]
- 271.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT. Proc Natl Acad Sci U S A. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hatters DM, MacPhee CE, Lawrence LJ, Sawyer WH, Howlett GJ. Biochemistry. 2000;39:8276–8283. doi: 10.1021/bi000002w. [DOI] [PubMed] [Google Scholar]
- 273.Hatters DM, MacRaild CA, Daniels R, Gosal WS, Thomson NH, Jones JA, Davis JJ, MacPhee CE, Dobson CM, Howlett GJ. Biophys J. 2003;85:3979–3990. doi: 10.1016/S0006-3495(03)74812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Dong JJ, Apkarian RP, Lynn DG. Bioorg Med Chem. 2005;13:5213–5217. doi: 10.1016/j.bmc.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 275.Wong YQ, Binger KJ, Howlett GJ, Griffin MDW. Proc Natl Acad Sci U S A. 2010;107:1977–1982. doi: 10.1073/pnas.0910136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.de Messieres M, Huang RK, He Y, Lee JC. Biochemistry. 2014;53:3261–3263. doi: 10.1021/bi500502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Teoh CL, Griffin MDW, Howlett GJ. Protein Cell. 2011;2:116–127. doi: 10.1007/s13238-011-1013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kagan BL, Hirakura Y, Azimov R, Azimova R, Lin MC. Peptides. 2002;23:1311–1315. doi: 10.1016/s0196-9781(02)00067-0. [DOI] [PubMed] [Google Scholar]
- 279.Ding TT, Lee SJ, Rochet JC, Lansbury PT. Biochemistry. 2002;41:10209–10217. doi: 10.1021/bi020139h. [DOI] [PubMed] [Google Scholar]
- 280.Zhu M, Li J, Fink AL. J Biol Chem. 2003;278:40186–40197. doi: 10.1074/jbc.M305326200. [DOI] [PubMed] [Google Scholar]
- 281.Srinivasan R, Marchant RE, Zagorski MG. Amyloid. 2004;11:10–13. doi: 10.1080/13506120410001667872. [DOI] [PubMed] [Google Scholar]
- 282.Lasagna-Reeves CA, Glabe CG, Kayed R. J Biol Chem. 2011;286:22122–22130. doi: 10.1074/jbc.M111.236257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kayed R, Pensalfini A, Margol L, Sokolov Y, Sarsoza F, Head E, Hall J, Glabe C. J Biol Chem. 2009;284:4230–4237. doi: 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dante S, Hauss T, Brandt A, Dencher NA. J Mol Biol. 2008;376:393–404. doi: 10.1016/j.jmb.2007.11.076. [DOI] [PubMed] [Google Scholar]
- 285.Pandey AP, Haque F, Rochet JC, Hovis JS. J Phys Chem B. 2011;115:5886–5893. doi: 10.1021/jp1121917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Varkey J, Isas JM, Mizuno N, Jensen MB, Bhatia VK, Jao CC, Petrlova J, Voss JC, Stamou DG, Steven AC, Langen R. J Biol Chem. 2010;285:32486–32493. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hellstrand E, Nowacka A, Topgaard D, Linse S, Sparr E. PLoS One. 2013;8:e77235. doi: 10.1371/journal.pone.0077235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Gilead S, Gazit E. Supramol Chem. 2005;17:87–92. [Google Scholar]
- 289.Usov I, Adamcik J, Mezzenga R. ACS Nano. 2013;7:10465–10474. doi: 10.1021/nn404886k. [DOI] [PubMed] [Google Scholar]
- 290.Stroud JC, Liu C, Teng PK, Eisenberg D. Proc Natl Acad Sci U S A. 2012;109:7717–7722. doi: 10.1073/pnas.1203193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Dearborn AD, Wall JS, Cheng NQ, Heymann JB, Kajava AV, Varkey J, Langen R, Steven AC. J Biol Chem. 2016;291:2310–2318. doi: 10.1074/jbc.M115.698787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Adamcik J, Mezzenga R. Macromolecules. 2012;45:1137–1150. [Google Scholar]
- 293.Adamcik J, Castelletto V, Bolisetty S, Hamley IW, Mezzenga R. Angew Chem, Int Ed. 2011;50:5495–5498. doi: 10.1002/anie.201100807. [DOI] [PubMed] [Google Scholar]
- 294.Fitzpatrick AWP, Debelouchina GT, Bayro MJ, Clare DK, Caporini MA, Bajaj VS, Jaroniec CP, Wang LC, Ladizhansky V, Muller SA, MacPhee CE, et al. Proc Natl Acad Sci U S A. 2013;110:5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Assenza S, Adamcik J, Mezzenga R, De Los Rios P. Phys Rev Lett. 2014;113 doi: 10.1103/PhysRevLett.113.268103. 268103. [DOI] [PubMed] [Google Scholar]
- 296.Uesaka A, Ueda M, Makino A, Imai T, Sugiyama J, Kimura S. Langmuir. 2014;30:1022–1028. doi: 10.1021/la404784e. [DOI] [PubMed] [Google Scholar]
- 297.Lara C, Reynolds NP, Berryman JT, Xu AQ, Zhang AF, Mezzenga R. J Am Chem Soc. 2014;136:4732–4739. doi: 10.1021/ja500445z. [DOI] [PubMed] [Google Scholar]
- 298.Castelletto V, Hamley IW, Hule RA, Pochan D. Angew Chem, Int Ed. 2009;48:2317–2320. doi: 10.1002/anie.200805500. [DOI] [PubMed] [Google Scholar]
- 299.Swanekamp RJ, DiMaio JTM, Bowerman CJ, Nilsson BL. J Am Chem Soc. 2012;134:5556–5559. doi: 10.1021/ja301642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Lashuel HA, LaBrenz SR, Woo L, Serpell LC, Kelly JW. J Am Chem Soc. 2000;122:5262–5277. doi: 10.1021/ja9937831. [DOI] [PubMed] [Google Scholar]
- 301.Adamcik J, Sanchez-Ferrer A, Ait-Bouziad N, Reynolds NP, Lashuel HA, Mezzenga R. Angew Chem, Int Ed. 2016;55:618–622. doi: 10.1002/anie.201508968. [DOI] [PubMed] [Google Scholar]
- 302.Perez M, Valpuesta JM, Medina M, deGarcini EM, Avila J. J Neurochem. 1996;67:1183–1190. doi: 10.1046/j.1471-4159.1996.67031183.x. [DOI] [PubMed] [Google Scholar]
- 303.Zhang S, Andreasen M, Nielsen JT, Liu L, Nielsen EH, Song J, Ji G, Sun F, Skrydstrup T, Besenbacher F, Nielsen NC, et al. Proc Natl Acad Sci U S A. 2013;110:2798–2803. doi: 10.1073/pnas.1209955110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Martinez-Avila OM, Wu SP, Cheng YF, Lee R, Khan F, Habelitz S. Eur J Oral Sci. 2011;119:75–82. doi: 10.1111/j.1600-0722.2011.00907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Carneiro KMM, Zhai HL, Zhu L, Horst JA, Sitlin M, Nguyen M, Wagner M, Simpliciano C, Milder M, Chen CL, Ashby P, et al. Sci Rep. 2016;6 doi: 10.1038/srep23105. 23105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.He XD, Wu SP, Martinez-Avila O, Cheng YF, Habelitz S. J Struct Biol. 2011;174:203–212. doi: 10.1016/j.jsb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Bongiovanni MN, Scanlon DB, Gras SL. Biomaterials. 2011;32:6099–6110. doi: 10.1016/j.biomaterials.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 308.Gras SL, Tickler AK, Squires AM, Devlin GL, Horton MA, Dobson CM, MacPhee CE. Biomaterials. 2008;29:1553–1562. doi: 10.1016/j.biomaterials.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 309.Hamley IW. Angew Chem, Int Ed. 2014;53:6866–6881. doi: 10.1002/anie.201310006. [DOI] [PubMed] [Google Scholar]
- 310.Elgersma RC, Kroon-Batenburg LMJ, Posthuma G, Meeldijk JD, Rijkers DTS, Liskamp RMJ. Eur J Med Chem. 2014;88:55–65. doi: 10.1016/j.ejmech.2014.07.089. [DOI] [PubMed] [Google Scholar]
- 311.Zhao YR, Deng L, Wang JQ, Xu H, Lu JR. Langmuir. 2015;31:12975–12983. doi: 10.1021/acs.langmuir.5b02303. [DOI] [PubMed] [Google Scholar]
- 312.Brodin JD, Smith SJ, Carr JR, Tezcan FA. J Am Chem Soc. 2015;137:10468–10471. doi: 10.1021/jacs.5b05755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Adler-Abramovich L, Marco P, Amon ZA, Creasey RCG, Michaels TCT, Levin A, Scurr DJ, Roberts CJ, Knowles TPJ, Tendler SJB, Gazit E. ACS Nano. 2016;10:7436–7442. doi: 10.1021/acsnano.6b01587. [DOI] [PubMed] [Google Scholar]
- 314.Lin YA, Cheetham AG, Zhang PC, Ou YC, Li YG, Liu GS, Hermida-Merino D, Hamley IW, Cui HG. ACS Nano. 2014;8:12690–12700. doi: 10.1021/nn505688b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Knowles TPJ, Oppenheim TW, Buell AK, Chirgadze DY, Welland ME. Nat Nanotechnol. 2010;5:204–207. doi: 10.1038/nnano.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Bolisetty S, Arcari M, Adamcik J, Mezzenga R. Langmuir. 2015;31:13867–13873. doi: 10.1021/acs.langmuir.5b03205. [DOI] [PubMed] [Google Scholar]
- 317.Jordens S, Schwenke K, Usov I, Del Gado E, Mezzenga R. Soft Matter. 2016;12:1830–1835. doi: 10.1039/c5sm02545a. [DOI] [PubMed] [Google Scholar]
- 318.Jordens S, Isa L, Usov I, Mezzenga R. Nat Commun. 2013;4:1917. doi: 10.1038/ncomms2911. [DOI] [PubMed] [Google Scholar]
- 319.Jung JM, Gunes DZ, Mezzenga R. Langmuir. 2010;26:15366–15375. doi: 10.1021/la102721m. [DOI] [PubMed] [Google Scholar]
- 320.Jordens S, Ruhs PA, Sieber C, Isa L, Fischer P, Mezzenga R. Langmuir. 2014;30:10090–10097. doi: 10.1021/la5020658. [DOI] [PubMed] [Google Scholar]
- 321.Wang DH, Ha Y, Gu J, Li Q, Zhang LL, Yang P. Adv Mater. 2016;28:7414–7423. doi: 10.1002/adma.201506476. [DOI] [PubMed] [Google Scholar]
- 322.Zhang SG, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci U S A. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Dai B, Li D, Xi W, Luo F, Zhang X, Zou M, Cao M, Hu J, Wang WY, Wei GH, Zhang Y, et al. Proc Natl Acad Sci U S A. 2015;112:2996–3001. doi: 10.1073/pnas.1416690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hamley IW, Dehsorkhi A, Castelletto V. Chem Commun. 2013;49:1850–1852. doi: 10.1039/c3cc39057h. [DOI] [PubMed] [Google Scholar]
- 325.Pan YX, Liu CJ, Zhang S, Yu Y, Dong MD. Chem - Eur J. 2012;18:14614–14617. doi: 10.1002/chem.201200745. [DOI] [PubMed] [Google Scholar]
- 326.Pan YX, Cong HP, Men YL, Xin S, Sun ZQ, Liu CJ, Yu SH. ACS Nano. 2015;9:11258–11265. doi: 10.1021/acsnano.5b04884. [DOI] [PubMed] [Google Scholar]
- 327.Staderini M, Martin MA, Bolognesi ML, Menendez JC. Chem Soc Rev. 2015;44:1807–1819. doi: 10.1039/c4cs00337c. [DOI] [PubMed] [Google Scholar]
- 328.Cheng KK, Chan PS, Fan SJ, Kwan SM, Yeung KL, Wang YXJ, Chow AHL, Wu EX, Baum L. Biomaterials. 2015;44:155–172. doi: 10.1016/j.biomaterials.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 329.Watanabe H, Ono M, Iikuni S, Yoshimura M, Matsumura K, Kimura H, Saji H. Bioorg Med Chem Lett. 2014;24:4834–4837. doi: 10.1016/j.bmcl.2014.08.058. [DOI] [PubMed] [Google Scholar]
- 330.Hintersteiner M, Enz A, Frey P, Jaton AL, Kinzy W, Kneuer R, Neumann U, Rudin M, Staufenbiel M, Stoeckli M, Wiederhold KH, et al. Nat Biotechnol. 2005;23:577–583. doi: 10.1038/nbt1085. [DOI] [PubMed] [Google Scholar]
- 331.Benseny-Cases N, Klementieva O, Cotte M, Ferrer I, Cladera J. Anal Chem. 2014;86:12047–12054. doi: 10.1021/ac502667b. [DOI] [PubMed] [Google Scholar]
- 332.Qin SY, Pei Y, Liu XJ, Zhuo RX, Zhang XZ. J Mater Chem B. 2013;1:668–675. doi: 10.1039/c2tb00105e. [DOI] [PubMed] [Google Scholar]
- 333.Peer D, Karp JM, Hong S, FaroKHzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 334.Shimanovich U, Efimov I, Mason TO, Flagmeier P, Buell K, Gedanken A, Linse S, Akerfeldt KS, Dobson CM, Weitz DA, Knowles TPJ. ACS Nano. 2015;9:43–51. doi: 10.1021/nn504869d. [DOI] [PubMed] [Google Scholar]
- 335.Selkoe DJ, Bell DS, Podlisny MB, Price DL, Cork LC. Science. 1987;235:873–877. doi: 10.1126/science.3544219. [DOI] [PubMed] [Google Scholar]
- 336.Fowler DM, Koulov AV, Balch WE, Kelly JW. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 337.Jordal PB, Dueholm MS, Larsen P, Petersen SV, Enghild JJ, Christiansen G, Hojrup P, Nielsen PH, Otzen DE. Appl Environ Microbiol. 2009;75:4101–4110. doi: 10.1128/AEM.02107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Villar-Pique A, Sabate R, Lopera O, Gibert J, Torne JM, Santos M, Ventura S. PLoS One. 2010;5:e13625. doi: 10.1371/journal.pone.0013625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Taglialegna A, Lasa I, Valle J. J Bacteriol. 2016;198:2579–2588. doi: 10.1128/JB.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 342.Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. J Bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrene YF, Lipke PN. PLoS One. 2011;6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Herbst FA, Sondergaard MT, Kjeldal H, Stensballe A, Nielsen PH, Dueholm MS. J Proteome Res. 2015;14:72–81. doi: 10.1021/pr500938x. [DOI] [PubMed] [Google Scholar]
- 345.Gallo PM, Rapsinski GJ, Wilson RP, Oppong GO, Sriram U, Goulian M, Buttaro B, Caricchio R, Gallucci S, Tukel C. Immunity. 2015;42:1171–1184. doi: 10.1016/j.immuni.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wu C, Lim JY, Fuller GG, Cegelski L. Biophys J. 2012;103:464–471. doi: 10.1016/j.bpj.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Lim JY, May JM, Cegelski L. Appl Environ Microbiol. 2012;78:3369–3378. doi: 10.1128/AEM.07743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Taglialegna A, Navarro S, Ventura S, Garnett JA, Matthews S, Penades JR, Lasa I, Valle J. PLoS Pathog. 2016;12:e1005711. doi: 10.1371/journal.ppat.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Jonker AM, Lowik DWPM, van Hest JCM. Chem Mater. 2012;24:759–773. [Google Scholar]
- 350.Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Nat Mater. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Flamia R, Salvi AM, D’Alessio L, Castle JE, Tamburro M. Biomacromolecules. 2007;8:128–138. doi: 10.1021/bm060764s. [DOI] [PubMed] [Google Scholar]
- 352.Bhak G, Lee S, Park JW, Cho S, Paik SR. Biomaterials. 2010;31:5986–5995. doi: 10.1016/j.biomaterials.2010.03.080. [DOI] [PubMed] [Google Scholar]
- 353.Yan H, Saiani A, Gough JE, Miller AF. Biomacromolecules. 2006;7:2776–2782. doi: 10.1021/bm0605560. [DOI] [PubMed] [Google Scholar]
- 354.Mains J, Lamprou DA, McIntosh L, Oswald IDH, Urquhart AJ. Chem Commun. 2013;49:5082–5084. doi: 10.1039/c3cc41583j. [DOI] [PubMed] [Google Scholar]
- 355.Li CX, Mezzenga R. Langmuir. 2012;28:10142–10146. doi: 10.1021/la301541d. [DOI] [PubMed] [Google Scholar]
- 356.Gosal WS, Clark AH, Ross-Murphy SB. Biomacromolecules. 2004;5:2420–2429. doi: 10.1021/bm049660c. [DOI] [PubMed] [Google Scholar]
- 357.Bolisetty S, Harnau L, Jung JM, Mezzenga R. Biomacromolecules. 2012;13:3241–3252. doi: 10.1021/bm301005w. [DOI] [PubMed] [Google Scholar]
- 358.Shao H, Parquette JR. Chem Commun. 2010;46:4285–4287. doi: 10.1039/c0cc00701c. [DOI] [PubMed] [Google Scholar]
- 359.Tena-Solsona M, Miravet JF, Escuder B. Chem - Eur J. 2014;20:1023–1031. doi: 10.1002/chem.201302651. [DOI] [PubMed] [Google Scholar]
- 360.Tena-Solsona M, Alonso-deCastro S, Miravet JF, Escuder B. J Mater Chem B. 2014;2:6192–6197. doi: 10.1039/c4tb00795f. [DOI] [PubMed] [Google Scholar]
- 361.Krysmann MJ, Castelletto V, Kelarakis A, Hamley IW, Hule RA, Pochan DJ. Biochemistry. 2008;47:4597–4605. doi: 10.1021/bi8000616. [DOI] [PubMed] [Google Scholar]
- 362.Nystrom G, Fernandez-Ronco MP, Bolisetty S, Mazzotti M, Mezzenga R. Adv Mater. 2016;28:472–478. doi: 10.1002/adma.201503465. [DOI] [PubMed] [Google Scholar]
- 363.Majzik A, Fulop L, Csapo E, Bogar F, Martinek T, Penke B, Biro G, Dekany I. Colloids Surf, B. 2010;81:235–241. doi: 10.1016/j.colsurfb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 364.Anand BG, Dubey K, Shekhawat DS, Kar K. Biochemistry. 2016;55:3345–3348. doi: 10.1021/acs.biochem.6b00418. [DOI] [PubMed] [Google Scholar]
- 365.Hayne DJ, Lim S, Donnelly PS. Chem Soc Rev. 2014;43:6701–6715. doi: 10.1039/c4cs00026a. [DOI] [PubMed] [Google Scholar]
- 366.Bolisetty S, Vallooran JJ, Adamcik J, Mezzenga R. ACS Nano. 2013;7:6146–6155. doi: 10.1021/nn401988m. [DOI] [PubMed] [Google Scholar]
- 367.Liao YH, Chang YJ, Yoshiike Y, Chang YC, Chen YR. Small. 2012;8:3631–3639. doi: 10.1002/smll.201201068. [DOI] [PubMed] [Google Scholar]
- 368.Brambilla D, Verpillot R, Le Droumaguet B, Nicolas J, Taverna M, Kona J, Lettiero B, Hashemi SH, De Kimpe L, Canovi M, Gobbi M, et al. ACS Nano. 2012;6:5897–5908. doi: 10.1021/nn300489k. [DOI] [PubMed] [Google Scholar]
- 369.Wei G, Reichert J, Jandt KD. Chem Commun. 2008:3903–3905. doi: 10.1039/b806316h. [DOI] [PubMed] [Google Scholar]
- 370.Bolisetty S, Boddupalli CS, Handschin S, Chaitanya K, Adamcik J, Saito Y, Manz MG, Mezzenga R. Biomacromolecules. 2014;15:2793–2799. doi: 10.1021/bm500647n. [DOI] [PubMed] [Google Scholar]
- 371.Hamley IW, Kirkham S, Dehsorkhi A, Castelletto V, Adamcik J, Mezzenga R, Ruokolainen J, Mazzuca C, Gatto E, Venanzi M, Placidi E, et al. Biomacromolecules. 2014;15:3412–3420. doi: 10.1021/bm500950c. [DOI] [PubMed] [Google Scholar]
- 372.Pazos E, Sleep E, Perez CMR, Lee SS, Tantakitti F, Stupp SI. J Am Chem Soc. 2016;138:5507–5510. doi: 10.1021/jacs.6b01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Xiao LH, Zhao D, Chan WH, Choi MMF, Li HW. Biomaterials. 2010;31:91–98. doi: 10.1016/j.biomaterials.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 374.Ng OTW, Wong Y, Chan HM, Cheng J, Qi X, Chan WH, Yung KKL, Li HW. Biomater Sci. 2013;1:577–580. doi: 10.1039/c3bm60029g. [DOI] [PubMed] [Google Scholar]
- 375.Liu YB, Xu LP, Dai WH, Dong HF, Wen YQ, Zhang XJ. Nanoscale. 2015;7:19060–19065. doi: 10.1039/c5nr06282a. [DOI] [PubMed] [Google Scholar]
- 376.Roberti MJ, Morgan M, Menendez G, Pietrasanta LI, Jovin TM, Jares-Erijman EA. J Am Chem Soc. 2009;131:8102–8107. doi: 10.1021/ja900225w. [DOI] [PubMed] [Google Scholar]
- 377.Quan L, Wu JX, Lane LA, Wang JQ, Lu Q, Gu Z, Wang YQ. Bioconjugate Chem. 2016;27:809–814. doi: 10.1021/acs.bioconjchem.6b00019. [DOI] [PubMed] [Google Scholar]
- 378.Su ZQ, Shen HY, Wang HX, Wang JH, Li JF, Nienhaus GU, Shang L, Wei G. Adv Funct Mater. 2015;25:5472–5478. [Google Scholar]
- 379.Li Y, Zhang W, Zhang L, Li J, Su Z, Wei G. Adv Mater Interfaces. 2017;4 1600895. [Google Scholar]
- 380.Newcomb CJ, Bitton R, Velichko YS, Snead ML, Stupp SI. Small. 2012;8:2195–2202. doi: 10.1002/smll.201102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Wang JH, Wang HX, Wang YZ, Li JF, Su ZQ, Wei G. J Mater Chem B. 2014;2:7360–7368. doi: 10.1039/c4tb01324g. [DOI] [PubMed] [Google Scholar]
- 382.Wang JH, Ouyang ZF, Ren ZW, Li JF, Zhang PP, Wei G, Su ZQ. Carbon. 2015;89:20–30. [Google Scholar]
- 383.Li CX, Born AK, Schweizer T, Zenobi-Wong M, Cerruti M, Mezzenga R. Adv Mater. 2014;26:3207–3212. doi: 10.1002/adma.201306198. [DOI] [PubMed] [Google Scholar]
- 384.Li CX, Mezzenga R. Nanoscale. 2013;5:6207–6218. doi: 10.1039/c3nr01644g. [DOI] [PubMed] [Google Scholar]
- 385.Khatayevich D, Page T, Gresswell C, Hayamizu Y, Grady W, Sarikaya M. Small. 2014;10:1505–1513. doi: 10.1002/smll.201302188. [DOI] [PubMed] [Google Scholar]
- 386.Luo JH, Warmlander SKTS, Yu CH, Muhammad K, Graslund A, Abrahams JP. Nanoscale. 2014;6:6720–6726. doi: 10.1039/c4nr00291a. [DOI] [PubMed] [Google Scholar]
- 387.Wang J, Cao YP, Li Q, Liu L, Dong MD. Chem – Eur J. 2015;21:9632–9637. doi: 10.1002/chem.201500577. [DOI] [PubMed] [Google Scholar]
- 388.Wu X, Li M, Li Z, Lv L, Zhang Y, Li C. J Colloid Interface Sci. 2017;490:336–342. doi: 10.1016/j.jcis.2016.11.058. [DOI] [PubMed] [Google Scholar]
- 389.Mahmoudi M, Akhavan O, Ghavami M, Rezaee F, Ghiasi SMA. Nanoscale. 2012;4:7322–7325. doi: 10.1039/c2nr31657a. [DOI] [PubMed] [Google Scholar]
- 390.Yang ZX, Ge CC, Liu JJ, Chong Y, Gu ZL, Jimenez-Cruz CA, Chai ZF, Zhou RH. Nanoscale. 2015;7:18725–18737. doi: 10.1039/c5nr01172h. [DOI] [PubMed] [Google Scholar]
- 391.Castelletto V, Kaur A, Hamley IW, Barnes RH, Karatzas KA, Hermida-Merino D, Swioklo S, Connon CJ, Stasiak J, Reza M, Ruokolainen J. RSC Adv. 2017;7:8366–8375. [Google Scholar]
- 392.Yoshida R. Curr Org Chem. 2005;9:1617–1641. [Google Scholar]
- 393.Dai ZH, Wang YL, Liu LQ, Liu XL, Tan PH, Xu ZP, Kuang J, Liu Q, Lou J, Zhang Z. Adv Funct Mater. 2016;26:7003–7010. [Google Scholar]
- 394.Zhong C, Gurry T, Cheng AA, Downey J, Deng ZT, Stultz CM, Lu TK. Nat Nanotechnol. 2014;9:858–866. doi: 10.1038/nnano.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Dubey K, Anand BG, Temgire MK, Kar K. Biochemistry. 2014;53:8001–8004. doi: 10.1021/bi501333q. [DOI] [PubMed] [Google Scholar]
- 396.Zhang MZ, Zhao J, Zheng J. Soft Matter. 2014;10:7425–7451. doi: 10.1039/c4sm00907j. [DOI] [PubMed] [Google Scholar]
- 397.Nikiforov MP, Thompson GL, Reukov VV, Jesse S, Guo S, Rodriguez BJ, Seal K, Vertegel AA, Kalinin SV. ACS Nano. 2010;4:689–698. doi: 10.1021/nn901127k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Ling SJ, Li CX, Adamcik J, Shao ZZ, Chen X, Mezzenga R. Adv Mater. 2014;26:4569–4574. doi: 10.1002/adma.201400730. [DOI] [PubMed] [Google Scholar]
- 399.Goncalves JM, Lima LR, Moraes ML, Ribeiro SJL. Mater Sci Eng, C. 2016;68:338–342. doi: 10.1016/j.msec.2016.05.101. [DOI] [PubMed] [Google Scholar]
- 400.Yan JM, Pan YX, Cheetham AG, Lin YA, Wang W, Cui HG, Liu CJ. Langmuir. 2013;29:16051–16057. doi: 10.1021/la4036908. [DOI] [PubMed] [Google Scholar]
- 401.Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. Nat Commun. 2014;5 doi: 10.1038/ncomms5945. 4945. [DOI] [PubMed] [Google Scholar]
- 402.Perez CMR, Stephanopoulos N, Sur S, Lee SS, Newcomb C, Stupp SI. Ann Biomed Eng. 2015;43:501–514. doi: 10.1007/s10439-014-1166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Guillame-Gentil O, Semenov O, Roca AS, Groth T, Zahn R, Voros J, Zenobi-Wong M. Adv Mater. 2010;22:5443–5462. doi: 10.1002/adma.201001747. [DOI] [PubMed] [Google Scholar]
- 404.Li CX, Bolisetty S, Mezzenga R. Adv Mater. 2013;25:3694–3700. doi: 10.1002/adma.201300904. [DOI] [PubMed] [Google Scholar]
- 405.Adhikari B, Banerjee A. Soft Matter. 2011;7:9259–9266. [Google Scholar]
- 406.Kim JH, Nam DH, Lee YW, Nam YS, Park CB. Small. 2014;10:1272–1277. [Google Scholar]
- 407.Riek R, Eisenberg DS. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 408.Cheetham AG, Zhang PC, Lin YA, Lock LL, Cui HG. J Am Chem Soc. 2013;135:2907–2910. doi: 10.1021/ja3115983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Choi H, Jeena MT, Palanikumar L, Jeong Y, Park S, Lee E, Ryu JH. Chem Commun. 2016;52:5637–5640. doi: 10.1039/c6cc00200e. [DOI] [PubMed] [Google Scholar]
- 410.Waku T, Kitagawa Y, Kawabata K, Nishigaki S, Kunugi S, Tanaka N. Chem Lett. 2013;42:1441–1443. [Google Scholar]
- 411.Waku T, Tanaka N. Polym Int. 2017;66:277–288. [Google Scholar]
- 412.Lock LL, Li YG, Mao XP, Chen HW, Staedtke V, Bai R, Ma W, Lin Y, Liu GS, Cui HG. ACS Nano. 2017;11:797–805. doi: 10.1021/acsnano.6b07196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Capule CC, Brown C, Olsen JS, Dewhurst S, Yang J. J Am Chem Soc. 2012;134:905–908. doi: 10.1021/ja210931b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Yolamanova M, Meier C, Shaytan AK, Vas V, Bertoncini CW, Arnold F, Zirafi O, Usmani SM, Muller JA, Sauter D, Goffinet C, et al. Nat Nanotechnol. 2013;8:130–136. doi: 10.1038/nnano.2012.248. [DOI] [PubMed] [Google Scholar]
- 415.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 416.Bongiovanni MN, Gras SL. Biomaterials. 2015;46:105–116. doi: 10.1016/j.biomaterials.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 417.Reynolds NP, Charnley M, Bongiovanni MN, Hartley PG, Gras SL. Biomacromolecules. 2015;16:1556–1565. doi: 10.1021/acs.biomac.5b00114. [DOI] [PubMed] [Google Scholar]
- 418.Gouveia RM, Castelletto V, Alcock SG, Hamley IW, Connon CJ. J Mater Chem B. 2013;1:6157–6169. doi: 10.1039/c3tb21031f. [DOI] [PubMed] [Google Scholar]
- 419.Gouveia RM, Castelletto V, Connon CJ, Hamley IW. Tissue Eng, Part A. 2015;21:1772–1784. doi: 10.1089/ten.tea.2014.0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 421.Cheng G, Castelletto V, Moulton CM, Newby GE, Hamley IW. Langmuir. 2010;26:4990–4998. doi: 10.1021/la903678e. [DOI] [PubMed] [Google Scholar]
- 422.Das S, Zhou K, Ghosh D, Jha NN, Singh PK, Jacob S, Bernard CC, Finkelstein DI, Forsythe JS, Maji SK. NPG Asia Mater. 2016;8:e304. [Google Scholar]
- 423.Healy J, Wong K, Sawyer EB, Roux C, Domigan L, Gras L, Sunde M, Larsen NG, Gerrard J, Vasudevamurthy M. Biopolymers. 2012;97:595–606. doi: 10.1002/bip.22045. [DOI] [PubMed] [Google Scholar]
- 424.Kaur M, Healy J, Vasudevamurthy M, Lasse M, Puskar L, Tobin MJ, Valery C, Gerrard JA, Sasso L. Nanoscale. 2014;6:13169–13178. doi: 10.1039/c4nr04624b. [DOI] [PubMed] [Google Scholar]
- 425.Reynolds NP, Styan KE, Easton CD, Li YL, Waddington L, Lara C, Forsythe JS, Mezzenga R, Hartley PG, Muir BW. Biomacromolecules. 2013;14:2305–2316. doi: 10.1021/bm400430t. [DOI] [PubMed] [Google Scholar]
- 426.Reynolds NP, Charnley M, Mezzenga R, Hartley PG. Biomacromolecules. 2014;15:599–608. doi: 10.1021/bm401646x. [DOI] [PubMed] [Google Scholar]
- 427.Li CX, Alam MM, Bolisetty S, Adamcik J, Mezzenga R. Chem Commun. 2011;47:2913–2915. doi: 10.1039/c0cc05126h. [DOI] [PubMed] [Google Scholar]
- 428.Hanczyc P, Samoc M, Norden B. Nat Photonics. 2013;7:969–972. [Google Scholar]
- 429.Channon KJ, Devlin GL, MacPhee CE. J Am Chem Soc. 2009;131:12520–12521. doi: 10.1021/ja902825j. [DOI] [PubMed] [Google Scholar]
- 430.Ryu J, Kim SW, Kang K, Park CB. ACS Nano. 2010;4:159–164. doi: 10.1021/nn901156w. [DOI] [PubMed] [Google Scholar]
- 431.Barrau S, Zhang F, Herland A, Mammo W, Andersson MR, Inganas O. Appl Phys Lett. 2008;93 023307. [Google Scholar]
- 432.Bolisetty S, Adamcik J, Heier J, Mezzenga R. Adv Funct Mater. 2012;22:3424–3428. [Google Scholar]
- 433.Chaves S, Pera LM, Avila CL, Romero CM, Baigori M, Vieyra FEM, Borsarelli CD, Chehin RN. RSC Adv. 2016;6:8528–8538. [Google Scholar]
- 434.Kim JH, Lee M, Lee JS, Park CB. Angew Chem, Int Ed. 2012;51:517–520. doi: 10.1002/anie.201103244. [DOI] [PubMed] [Google Scholar]
- 435.Acar H, Garifullin R, Aygun LE, Okyay AK, Guler MO. J Mater Chem A. 2013;1:10979–10984. [Google Scholar]
- 436.Scott G, Roy S, Abul-Haija YM, Fleming S, Bai S, Ulijn RV. Langmuir. 2013;29:14321–14327. doi: 10.1021/la403448s. [DOI] [PubMed] [Google Scholar]
- 437.Leung WH, Lo WH, Chan PH. RSC Adv. 2016;6:58363–58364. [Google Scholar]
- 438.Li D, Furukawa H, Deng HX, Liu C, Yaghi OM, Eisenberg DS. Proc Natl Acad Sci U S A. 2014;111:191–196. doi: 10.1073/pnas.1321797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Li D, Jones EM, Sawaya MR, Furukawa H, Luo F, Ivanova M, Sievers SA, Wang WY, Yaghi OM, Liu C, Eisenberg DS. J Am Chem Soc. 2014;136:18044–18051. doi: 10.1021/ja509648u. [DOI] [PubMed] [Google Scholar]
- 440.Scheibel T, Parthasarathy R, Sawicki G, Lin XM, Jaeger H, Lindquist SL. Proc Natl Acad Sci U S A. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Zou QL, Liu K, Abbas M, Yan XH. Adv Mater. 2016;28:1031–1043. doi: 10.1002/adma.201502454. [DOI] [PubMed] [Google Scholar]
- 442.Carny O, Shalev DE, Gazit E. Nano Lett. 2006;6:1594–1597. doi: 10.1021/nl060468l. [DOI] [PubMed] [Google Scholar]
- 443.Dinca V, Kasotakis E, Mourka A, Ranella A, Farsari M, Mitraki A, Fotakis C. Phys Status Solidi C. 2008;5:3576–3579. [Google Scholar]
- 444.Dinca V, Kasotakis E, Catherine J, Mourka A, Ranella A, Ovsianikov A, Chichkov BN, Farsari M, Mitraki A, Fotakis C. Nano Lett. 2008;8:538–543. doi: 10.1021/nl072798r. [DOI] [PubMed] [Google Scholar]
- 445.Amit M, Appel S, Cohen R, Cheng G, Hamley IW, Ashkenasy N. Adv Funct Mater. 2014;24:5873–5880. [Google Scholar]
- 446.Hamedi M, Herland A, Karlsson RH, Inganas O. Nano Lett. 2008;8:1736–1740. doi: 10.1021/nl0808233. [DOI] [PubMed] [Google Scholar]
- 447.Meier C, Lifincev I, Welland ME. Biomacromolecules. 2015;16:558–563. doi: 10.1021/bm501618c. [DOI] [PubMed] [Google Scholar]
- 448.Tu DY, Nilsson D, Forchheimer R. J Disp Technol. 2013;9:755–759. [Google Scholar]
- 449.Herland A, Bjork P, Nilsson KPR, Olsson JDM, Asberg P, Konradsson P, Hammarstrom P, Inganas O. Adv Mater. 2005;17:1466–1471. [Google Scholar]
- 450.Tanaka H, Herland A, Lindgren LJ, Tsutsui T, Andersson MR, Inganas O. Nano Lett. 2008;8:2858–2861. doi: 10.1021/nl801510z. [DOI] [PubMed] [Google Scholar]
- 451.Rizzo A, Inganas O, Solin N. Chem – Eur J. 2010;16:4190–4195. doi: 10.1002/chem.201000146. [DOI] [PubMed] [Google Scholar]
- 452.Rizzo A, Solin N, Lindgren LJ, Andersson MR, Inganas O. Nano Lett. 2010;10:2225–2230. doi: 10.1021/nl1012008. [DOI] [PubMed] [Google Scholar]
- 453.Solin N, Inganas O. Isr J Chem. 2012;52:529–539. [Google Scholar]
- 454.Buell AK, Esbjorner EK, Riss PJ, White DA, Aigbirhio FI, Toth G, Welland ME, Dobson CM, Knowles TPJ. Phys Chem Chem Phys. 2011;13:20044–20052. doi: 10.1039/c1cp22283j. [DOI] [PubMed] [Google Scholar]
- 455.Yemini M, Reches M, Rishpon J, Gazit E. Nano Lett. 2005;5:183–186. doi: 10.1021/nl0484189. [DOI] [PubMed] [Google Scholar]
- 456.Yemini M, Reches M, Gazit E, Rishpon J. Anal Chem. 2005;77:5155–5159. doi: 10.1021/ac050414g. [DOI] [PubMed] [Google Scholar]
- 457.Kim S, Kim JH, Lee JS, Park CB. Small. 2015;11:3623–3640. doi: 10.1002/smll.201500169. [DOI] [PubMed] [Google Scholar]
- 458.Sasso L, Suei S, Domigan L, Healy J, Nock V, Williams MAK, Gerrard JA. Nanoscale. 2014;6:1629–1634. doi: 10.1039/c3nr05752f. [DOI] [PubMed] [Google Scholar]
- 459.Men D, Zhang ZP, Guo YC, Zhu DH, Bi LJ, Deng JY, Cui ZQ, Wei HP, Zhang XE. Biosens Bioelectron. 2010;26:1137–1141. doi: 10.1016/j.bios.2010.07.103. [DOI] [PubMed] [Google Scholar]
- 460.Leng Y, Wei HP, Zhang ZP, Zhou YF, Deng JY, Cui ZQ, Men D, You XY, Yu ZN, Luo M, Zhang XE. Angew Chem, Int Ed. 2010;49:7243–7246. doi: 10.1002/anie.201002452. [DOI] [PubMed] [Google Scholar]
- 461.Hamley IW. Soft Matter. 2010;6:1863–1871. [Google Scholar]
- 462.Corrigan AM, Muller C, Krebs MRH. J Am Chem Soc. 2006;128:14740–14741. doi: 10.1021/ja064455l. [DOI] [PubMed] [Google Scholar]
- 463.Zhao JG, Bolisetty S, Adamcik J, Han J, Fernandez-Ronco MP, Mezzenga R. Langmuir. 2016;32:2492–2499. doi: 10.1021/acs.langmuir.6b00276. [DOI] [PubMed] [Google Scholar]
- 464.Han TH, Kim J, Park JS, Park CB, Ihee H, Kim SO. Adv Mater. 2007;19:3924–3927. [Google Scholar]
- 465.Krysmann MJ, Castelletto V, McKendrick JE, Clifton LA, Hamley IW, Harris PJF, King SA. Langmuir. 2008;24:8158–8162. doi: 10.1021/la800942n. [DOI] [PubMed] [Google Scholar]
- 466.Ryu J, Lim SY, Park CB. Adv Mater. 2009;21:1577–1581. [Google Scholar]
- 467.Yan XH, Su Y, Li JB, Fruh J, Mohwald H. Angew Chem, Int Ed. 2011;50:11186–11191. doi: 10.1002/anie.201103941. [DOI] [PubMed] [Google Scholar]
- 468.Yan XH, Li JB, Mowald H. Adv Mater. 2011;23:2796–2801. doi: 10.1002/adma.201100353. [DOI] [PubMed] [Google Scholar]
- 469.Shen Y, Posavec L, Bolisetty S, Hilty FM, Nystrom G, Kohlbrecher J, Hilbe M, Rossi A, Baumgartner J, Zimmermann MB, Mezzenga R. Nat Nanotechnol. 2017 doi: 10.1038/nnano.2017.1058. [DOI] [PubMed] [Google Scholar]