Abstract
Biosurveillance is a proactive approach that may help to limit the spread of invasive fungal pathogens of trees, such as rust fungi which have caused some of the world’s most damaging diseases of pines and poplars. Most of these fungi have a complex life cycle, with up to five spore stages, which is completed on two different hosts. They have a biotrophic lifestyle and may be propagated by asymptomatic plant material, complicating their detection and identification. A bioinformatics approach, based on whole genome comparison, was used to identify genome regions that are unique to the white pine blister rust fungus, Cronartium ribicola, the poplar leaf rust fungi Melampsora medusae and Melampsora larici-populina or to members of either the Cronartium and Melampsora genera. Species- and genus-specific real-time PCR assays, targeting these unique regions, were designed with the aim of detecting each of these five taxonomic groups. In total, twelve assays were developed and tested over a wide range of samples, including different spore types, different infected plant parts on the pycnio-aecial or uredinio-telial host, and captured insect vectors. One hundred percent detection accuracy was achieved for the three targeted species and two genera with either a single assay or a combination of two assays. This proof of concept experiment on pine and poplar leaf rust fungi demonstrates that the genome-enhanced detection and identification approach can be translated into effective real-time PCR assays to monitor tree fungal pathogens.
Introduction
Invasive fungal pathogens of trees represent a global threat to natural forests and tree plantations, which may result in large economic losses and, in some cases, changes to ecosystem functions and a reduction in biodiversity [1–3]. The worldwide spread of invasive species is directly related to anthropogenic activities and the number of new invasive pathogens within a country can be correlated to its economic activity, as measured by its gross domestic product [1, 4]. Prevention, which is supported by biosurveillance, is the most efficient approach in slowing introduction and spread of invasive alien pathogens [5]. However, biosurveillance of plant pathogens is challenging. Some oomycete and fungal pathogens can live on plant material that is asymptomatic and can go undetected during visual inspections. Others can be misidentified given the paucity of discriminant morphological characters in some groups of fungi and the great taxonomic diversity encountered during surveys and inspections. These factors increase the probability of failure to detect alien pathogens which may lead to their establishment and eventual outbreaks.
These challenges are particularly pressing in rust fungi which can cause some of the most devastating diseases of pines, poplars and eucalyptus [6–10]. Heteroecious rust fungi alternate on both susceptible pycnio-aecial and uredinio-telial hosts, further complicating their detection and identification, particularly when a plant is host to more than one rust species occurring in sympatry. For example, Larix spp. may be infected by both Melampsora and Melampsoridium species, which can cause poplar, willow and birch leaf rusts on their uredinio-telial hosts [11, 12].
The white pine blister rust, caused by the fungus Cronartium ribicola J. C. Fisch. (Basidiomycota, Pucciniales), is one of the major factors that contributed to the decline of the North American populations of five needle pines. This exotic pathogen was accidentally introduced to Canada and USA on infected white pine seedlings imported from Europe and intended for reforestation following extensive logging in the 19th century. It has since established and spread in most areas where its pycnio-aecial (Pinus subsection Strobus) and uredinio-telial (Ribes in the Grossulariaceae or less commonly Pedicularis and Castilleja in the Orobanchaceae) hosts cohabit [6]. Melampsora medusae Thüm. f. sp. deltoidae Shain and Melampsora larici-populina Kleb. (Basidiomycota, Pucciniales) cause poplar leaf rust, leading to severe damage in commercial poplar plantations. Melampsora medusae f. sp. deltoidae is native to North America, where it alternates between its pycnio-aecial (Larix spp., Pseudotsuga spp. and young Pinus spp.) and uredinio-telial (Populus spp. and hybrids in sections Aigeiros and Tacamahaca) hosts. It has spread throughout most of the world where poplars are grown and it has been classified as a quarantine pest in Europe [13, 14]. Melampsora larici-populina is an invasive Eurasian fungus that was first reported in Western and Eastern North America in 1991 and 2002, respectively, where it has been able to overwinter and infect some hybrid poplars which had never been affected by poplar leaf rust before [15, 16].
During the last decade, efforts have been made to improve the methods of detection of these important pathogens, using molecular tools. For example, PCR assays, based on single nucleotide polymorphisms located within the nuclear ribosomal internal transcribed spacer (ITS) region, were developed for the detection of Melampsora species (M. medusae f. sp. deltoidae, M. larici-populina and M. allii-populina) that attack cultivated poplars belonging to sections Aigeiros and Tacamahaca [17]. Real-time PCR assays were also developed for the simultaneous detection of both formae speciales of M. medusae, based on polymorphisms found within the ITS region, and also for the detection of the forma specialis deltoidae only, based on the large ribosomal RNA subunit [18, 19]. Regions other than ribosomal DNA were also targeted. For example, two distinct single-copy genes, MS208 and MS277, were used in a duplex real-time assay, to discriminate M. larici-populina and M. allii-populina, respectively [20]. However, due to the low number of polymorphic DNA regions currently identified and suitable for the design of discriminant assays, other means needed to be explored.
An approach that uses whole genome comparisons to identify genes that are unique to targeted species as well as shared within a group of related species is showing promise to address this issue [21, 22]. This approach has the advantage of reducing the risk of false positive or false negative assignment and producing robust assays that are not dependent upon single nucleotide polymorphisms. We present here the use of the GEDI (genome-enhanced detection and identification) approach [21] to design specific real-time assays targeting C. ribicola, M. medusae and M. larici-populina and taxa belonging to Cronartium and Melampsora genera and evaluate their performance for the detection of pine and poplar leaf rust pathogens from environmental samples.
Material and methods
Isolate collection, monosporal culture and DNA isolation
A worldwide collection of samples from target and non-target species among the Pucciniales was established, with the assistance of collaborators or international herbaria (S1–S4 Tables). No collection permits were required for samples collected from Canadian Crown land. In the cases where samples were collected from private plantations, permission was obtained from the owners. For international samples, collaborators obtained permission. No sampling of endangered species was conducted. Most of the samples were DNA barcoded to confirm their species identification or the presence of pathogens on environmental material. Mono-uredinial cultures of Cronartium ribicola and Melampsora spp. were also used, which were obtained as follows: an initial inoculation of a detached blackcurrant or poplar leaf was made with urediospores sampled from a single uredium observed on environmental material. After 8–10 days post-inoculation, urediospores sampled from one of the uredia developed on the detached leaf were inoculated on a fresh detached leaf. This step was repeated one more time before propagation of the inoculum on multiple detached leaves was initiated.
DNA was isolated using a phenol-chloroform extraction method, the DNeasy Plant Mini procedure (Qiagen GmbH, Hilden, Germany) or protocols implemented in the laboratories of collaborators who provided us with DNA samples.
To estimate the sensitivity of the real-time PCR assays, spore suspensions of Cronartium ribicola, Melampsora medusae f. sp. deltoidae and Melampsora larici-populina were prepared by adding absolute ethanol to bulks of dessicated spores and filtering using cell strainers (100 μm Nylon; BD Falcon, USA). Uniform spore suspension was maintained by vortex mixing and serial dilutions (1:10) were prepared with absolute ethanol plus the surfactant SilWet L-77 (1μL/10mL of ethanol; Momentive Performance Materials Inc., USA). Spore concentration of these serially diluted suspensions was estimated with a hemacytometer, under a light microscope at 250X magnification, to enable the transfer of 1.5 X 106, 2.5 X 104, 5 X 103, 1.25 X 103, 5 X 102, 50 and 25 spores to 2 mL Safe-Lock tubes (Eppendorf AG, Hamburg, Germany), in three biological replicates. Ethanol was evaporated using a CentriVap concentrator (Labconco Corp., Kansas City, MO) and spores were stored at -20°C until DNA isolation. The QIAamp DNA Micro kit (Qiagen GmbH, Hilden, Germany) was used for DNA isolation, as described in the Isolation of Genomic DNA from Tissue Samples section of the manufacturer’s handbook (04/2003 version). Briefly, spores resuspended in 180 μL of Buffer ATL and 1 μL of reagent DX (Qiagen GmbH) were mechanically disrupted twice with an acid-washed tungsten carbide bead (3 mm, Qiagen GmbH), using a Mixer Mill 300 (Qiagen GmbH, Haan, Germany) run at 30 beats per sec for 1.5 min. The lysis was carried out overnight in a heating block at 56°C, in the presence of Proteinase K (20 μL of >600 mAU/mL solution). For the first two hours of the lysis, the samples were vortexed for 10 sec every 10 min. To enhance binding of DNA to the QIAamp membrane, carrier RNA was added to Buffer AL. Following the binding and washing steps, elution was carried out with 50 μL of Buffer EB (Qiagen), except for DNA isolated from 25 and 50 spores, for which 25 μL of Buffer EB was applied onto the column. DNA LoBind tubes (Eppendorf AG) were used as collection tubes at the elution step. The QIAamp DNA Micro kit was also used for DNA isolation from single telia of C. ribicola, following the same described method.
Identification of unique genes based on comparative genomics
Identification of gene and genome regions suitable for developing highly specific real-time PCR assays for M. larici-populina, M. medusae, C. ribicola and the Melampsora and Cronartium genera was carried out using the GEDI method described in a previous study [21]. Briefly, seventeen rust genomes were considered for analyses, including three Cronartium/Endocronartium and six Melampsora species obtained previously by pair-end Illumina sequencing (Canada’s Michael Smith Genome Sciences Centre, Vancouver, Canada) [21]. The 122,475 protein models predicted from these de novo assemblies were combined with those obtained from the genomes of M. larici-populina [23], Cronartium quercuum f. sp. fusiforme, Puccinia graminis f. sp. tritici [23], P. triticina, P. striiformis, Mixia osmundae [24], Sporobolomyces roseus and Rhodotorula graminis [25]. Protein sets were submitted to a clustering process with the orthoMCL algorithm (BLASTp e-value cutoff of 1e-05, minimum similarity cutoff of 50%, inflation parameter of 1.5) [26]. Among the 90,135 clusters found, “singlets” i.e. clusters containing putative unique protein models to single species M. larici-populina, M. medusae f. sp. deltoidae or C. ribicola were identified (S1A Fig) and submitted to a false positive filtering by running a TBLASTn (1e-20) against the other rust genomes included in the study and a BLASTp (1e-20) on NCBI-nr. Putative genus-specific protein models to Melampsora and Cronartium were identified by finding clusters of orthologs/paralogs shared among species within the genus but not with any other rust species (S1B Fig). Filtering of the genus-specific clusters for false positives was carried out by running TBLASTn (1e-20) against rust genomes from alternative genera. For all five targeted taxonomic groups, clusters with positive hits were discarded and the remaining gene sequences were retrieved for each of the protein models. They were submitted to Primer3 V4.0.0 [27] for PCR primer and probe design with the following parameters: amplicon size of 100 to 150 bp; primer optimal size of 20 bases (18–27 bases) with an optimal Tm of 60°C (58–62°C); probe optimal size of 20 bases (18–27 bases) with an optimal Tm of 65°C (64–68°C).
Real-time PCR primer and probe design
Among the hundreds of sequence cluster files (Cluster_n.fas) generated by our bioinformatics method for each targeted taxon, up to twenty clusters were selected per taxon, based on the homology spanned over the entire length of sequences (n ≥ 2) forming ortholog/paralog clusters. Oligo Analyzer V1.5 (Gene Link, http://www.genelink.com/tools/gl-oe.asp) was used to improve the primer/probe combinations initially generated by our bioinformatics method. For an optimal amplification at 60°C, the following rules were applied whenever possible: 1) Tm values for primers between 58–60°C and 8–10 °C higher for probes; 2) minimum primer length of 16 bases; 3) probe length between 21–27 bases; 4) no G at the extreme 5’ end of the probe; 5) no probes with four or more identical nucleotides, especially Gs; and 6) secondary structure and 3’ self-complementarity primer-dimer formation avoided or limited. The parameters used for Tm calculation were: salt concentration of 50 mM and DNA concentration of 250 pM. Furthermore, primers (MEL40, MEL100 and MEL176 assays) and probe (MEL176 assay) with degenerate bases were designed due to interspecific variation that was observed within the alignment of targeted region sequences representative of five to six Melampsora spp.
Experimental conditions of PCR
Following design optimization, primer pairs were synthesized (Integrated DNA Technologies Inc., Coralville, IA, USA) and tested against a reduced panel of DNA samples from target and non-target species among the Pucciniales to select those that specifically amplified the targeted taxonomic groups (refer to footnotes a and b in S1 Table and footnotes a, b and c in S2 Table). One μL of purified DNA solution was used as template for the PCR amplification using 1.0 U of Platinum Taq DNA polymerase (Invitrogen, Life Technologies, Carlsbad, CA, USA), in a 25 μL reaction mix comprising 1X PCR buffer minus Mg, 1.6 mM of MgCl2, 200 μM of dNTPs and 1 μM of each primer. Thermocycling conditions included an initial heat activation step of 3 min at 94°C followed by 35 cycles of denaturation at 92°C for 30 sec, primer annealing at 60°C for 30 sec and extension at 72°C for 1 min. A final extension step at 72°C for 10 min completed the program. DNA fragment analysis was performed with capillary electrophoresis using the QIAxcel Advanced system (Qiagen GmbH, Hilden, Germany). Products of specific amplification were purified on glass microfiber UniFilter (GE Healthcare Whatman) and sequenced (Sanger’s method) at the Genome sequencing and genotyping platform of the CHUL medical research centre (Québec, QC, Canada). Sequences were edited using Sequencher V4.8 (Gene Codes Corporation, Ann Arbor, MI, USA) and deposited in GenBank: MH171727-MH171903.
Primer pairs that resulted in the specific amplification of targeted taxonomic groups were evaluated in combination with TaqMan probes (Tables 1 and 2) on a larger panel of DNA samples (all samples displayed in S1 and S2 Tables). Real-time PCR was performed in a 10 μL reaction mix comprising 1X QuantiTect Multiplex PCR NoRox Master Mix (Qiagen, Valencia, CA), 0.6 μM of each primer, 0.1 μM of double-quenched TaqMan probe (5’FAM/ZEN/3’IB-FQ; Integrated DNA Technologies Inc.) and 1 μL of template DNA, using the 7500 Fast Real-Time PCR system (Applied Biosystems, Life Technologies, Foster City, CA). Thermocycling conditions included an initial heat activation step of 15 min at 95°C followed by 50 cycles of denaturation at 95°C for 15 sec and primer annealing/extension at 60°C for 1 min. Fluorescence was read at the end of each extension step.
Table 1. Genus- and species-specific assays for hierarchical detection of Cronartium spp. and Cronartium ribicola.
| Assay | Targeted taxon | Primer or TaqMan probea | Sequence (5’ → 3’) | Tm (NN)b | Amplicon length | GenBank accession no. |
|---|---|---|---|---|---|---|
| CRO30 | Cronartium spp. | Cronartium30F | ATCGTTCTCATCGTCAGCG | 57.1 °C | 104 bp | MH171752-MH171772 |
| Cronartium30R | TTAAAGCTTTGAAGGCGACG | 56.0 °C | ||||
| Cronartium30T (47–70) | TCTTCACTGCCGTCTACTTTCTGC | 64.2 °C | ||||
| CRO46 | Cronartium spp. | Cronartium46F | CCGGTTATGGCGATCTCAC | 60.4 °C | 117 bp | MH171773-MH171780 |
| Cronartium46R | AAATCGGGCACTCTGTTCTG | 58.9 °C | ||||
| Cronartium46T (72–98) | ATCGCTTTCGATGGACGTGATCATGGC | 67.4 °C | ||||
| CRIB65 | Cronartium ribicola | Crib65Fm | CTTTTGCTACTTGCGCATTTGA | 57.0 °C | 102 bp | MH171727-MH171734 |
| Crib65Rm | CTCTGGTGAGGAGCTATCG | 60.1 °C | ||||
| Crib65T (61–83) | CTACAGGGTTCACCTGCTTCGCC | 68.7 °C | ||||
| CRIB146 | Cronartium ribicola | Crib146F | TCAGGTTTCGGATTTTGAGG | 55.8 °C | 115 bp | MH171735-MH171742 |
| Crib146R | TGCTTCTGAGGCTTTTTGGT | 58.5 °C | ||||
| Crib146T (37–63) | CATACCTGATACGAGTGGACTGAATGA | 63.6 °C | ||||
| CRIB190 | Cronartium ribicola | Crib190Fm | CTCCAGCTACAGTGGGTA | 59.9 °C | 121 bp | MH171743-MH171751 |
| Crib190Rm | CCTTGTCTGTTGGTGAGGT | 59.2 °C | ||||
| Crib190T (50–77) | ACATGGGAACGACAAGGACAATTTGGAC | 66.0 °C |
a In parentheses: probe position within amplicon.
b Values estimated by Oligo Analyzer V1.5.
Table 2. Genus- and species-specific assays for hierarchical detection of Melampsora spp., Melampsora medusae and Melampsora larici-populina.
| Assay | Targeted taxon | Primer or TaqMan probea | Sequence (5’ → 3’) | Tm (NN)b | Amplicon length | GenBank accession no. |
|---|---|---|---|---|---|---|
| MEL40 | Melampsora spp. | Melampsora40Fm | CCTGGTACTCCAACTATCATCTTA | 60.3 °C | 131 bp | MH171821-MH171846 |
| Melampsora40Rm | GAAWGTGCACGCGATTGACG | 59.3 °C | ||||
| Melampsora40T (40–66) | CCTCTGAGTGATGGCGAAGACCCTTTA | 68.1 °C | ||||
| MEL100 | Melampsora spp. | Melampsora100Fm | CACGAAAGTCBCAAGTGGC | 60.6 °C | 148 bp | MH171847-MH171875 |
| Melampsora100Rm | GTRCAGTCATGAGGTACGATA | 59.3 °C | ||||
| Melampsora100T (66–85) | CCTCCTCCTCCGCTCTACGC | 68.8 °C | ||||
| MEL176 | Melampsora spp. | Melampsora176Fm | GCCCTTGCCGTTGCTAT | 60.7 °C | 112 bp | MH171876-MH171903 |
| Melampsora176Rm | GRCTCGTGCTGATCAGTC | 60.2 °C | ||||
| Melampsora176Td (37–62) | ACCACTCACCATCCGTAYATCAACGC | 68.2 °C | ||||
| MM53 | Melampsora medusae | Mm53F | ACAACCAGGTGACGGAAATC | 59.1 °C | 129 bp | MH171799-MH171810 |
| Mm53R | GAATCGTCCGAGGAGTCATT | 58.1 °C | ||||
| Mm53Trev (52–28) | CTTAGGTCGTTCACCCTCTGATTCG | 64.6 °C | ||||
| MM74 | Melampsora medusae | Mm74Fm | CACCATGCAAATCACCAATCAC | 58.4 °C | 118 bp | MH171811-MH171820 |
| Mm74R | TTTGGCTCAGCCTCAGTTTT | 58.5 °C | ||||
| Mm74T (29–57) | TGGACCTTCTTTGCAAATCAACACAGTAT | 63.4 °C | ||||
| MLP104 | Melampsora larici-populina | Mlp104Fm | CGGCCAGAAATTGTGATGGAT | 59.4 °C | 104 bp | MH171781-MH171789 |
| Mlp104R | TGCATAGCCTTTGTGGACAG | 59.9 °C | ||||
| Mlp104T(28–56) | CGTATGAACCATTGATTGAAGGCTTGGAC | 65.1 °C | ||||
| MLP133 | Melampsora larici-populina | Mlp133Fm | ATGGACCGGGAATATGAAC | 56.7 °C | 126 bp | MH171790-MH171798 |
| Mlp133R | TCGTTGATCGTATCGTGGAA | 56.0 °C | ||||
| Mlp133T(33–55) | CGCATCAGGTGGATTTGGTGAAG | 62.9 °C |
a In parentheses: probe position within amplicon.
b Values estimated by Oligo Analyzer V1.5.
Specificity of the real-time PCR assays
Mono-uredinial cultures and environmental material from 11 Cronartium and 18 Melampsora rust species were used to assess the specificity of the assays. Cross-reaction of the assays with DNA from non-target species was also evaluated as well as the performance of the assays in detecting the presence of targeted taxonomic groups on environmental material (Fig 1). For Cronartium spp., environmental samples included aeciospores of C. ribicola produced on naturally infected white pines (Pinus strobus), urediospores and single telia produced on blackcurrants (Ribes nigrum) and symptomatic white pine stem tissue and chlorotic needle infection spots of white pine seedlings artificially inoculated with C. ribicola. Insects intercepted in a white pine plantation infected with blister rust were also used as some feed on the nectar of spermogonia produced on the surface of the pine tree bark during the initial spore stage infection on pine [28] (S3 Table). It has been postulated that Megaselia spp. and Paracacoxenus guttatus are the main insects responsible for cross-fertilization of C. ribicola [28]. Megaselia spp. accounted for seven out of nine insects tested in this study. Melampsora spp. environmental samples included aeciospores and urediospores of both M. medusae f. sp. deltoidae and M. larici-populina produced on naturally infected larch (Larix laricina) and poplar (Populus spp. and interspecific hybrids), respectively (S4 Table).
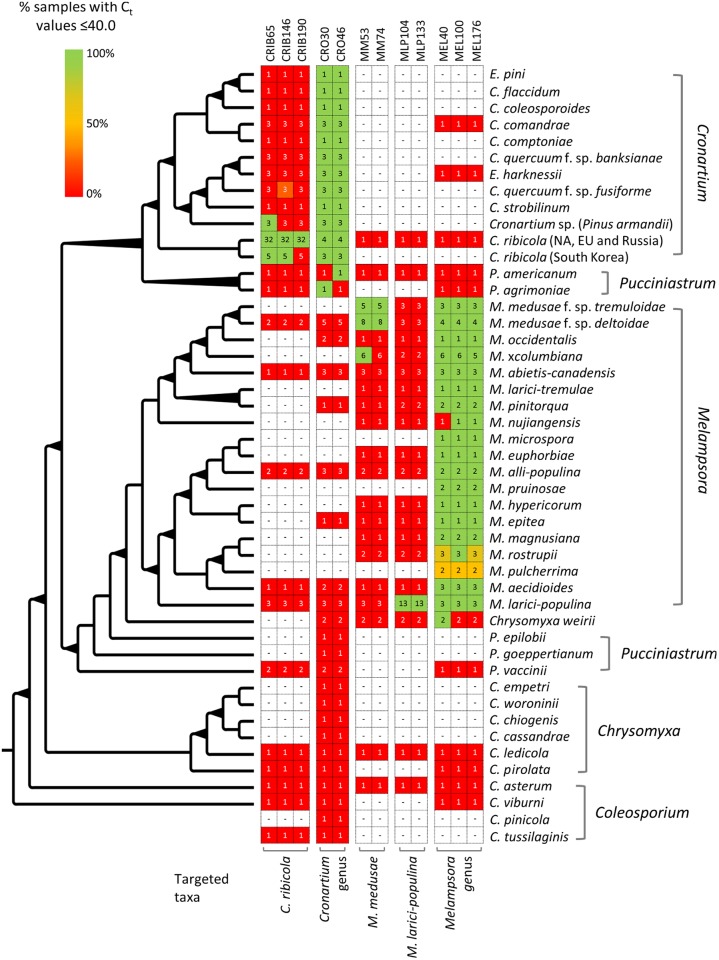
Fig 1. Specificity of the pine and poplar rust assays.
The NJ phylogenetic tree represents evolutionary relationships among the rust samples used, inferred from an alignment of ITS sequences; bolded nodes received bootstrap values ≥ 80%. Rows represent the number of rust samples used within each taxon and columns stand for the different assays. For each taxon, the relative abundance of samples with Ct values ≤ 40.0, as determined by real-time PCR, is depicted by color scale with the legend at the upper left side.
Sensitivity of the real-time PCR assays on a known number of spores
Sensitivity of the real-time PCR assays was determined by testing seven different amounts of spores for C. ribicola, M. medusae f. sp. deltoidae and M. larici-populina: 30,000, 500, 100, 25, 10, 2 and 1 spore(s). DNA was isolated from three biological replicates for each amount of spores. Three technical replicates were performed for all reactions, using the same real-time PCR conditions as those described above. Graphs were generated by plotting the number of spores on the x-axis against the mean Ct (cycle threshold) values determined by real-time PCR on the y-axis. Each dot represents a biological independent replicate (corresponding to a known amount of spores), which was obtained by averaging the Ct values of three technical real-time PCR replicates. Amplification reaction efficiency was calculated from the slope of the lines, using the following formula:
Data analysis and interpretation
Naïve Bayes classifier
We built a naïve Bayes classifier [29] to i. select the optimal combination of real-time PCR assays producing the most accurate detection of the targeted species and ii. interpret assay outcomes. Using a training set of Ct values obtained from the collection of positive and negative samples for each targeted taxon (see below), the classifier estimates for each assay the parameters of probability density of the Ct values obtained for two given classes X|i (positive samples) and X|j (negative samples). Then, for any new sample tested with a Ct value X, the method computes the posterior probability P(i|X) of that sample belonging to the class i and the posterior probability P(j|X) to belong to j according to the naïve Bayes model [29–31]:
Probability densities P(X|i) and P(X|j) have to be integrated into the Bayes model through a function of probability density calculated over the training set. Such a function f(x) represents the normal distribution of a numerical attribute x as a function of a mean μ and its standard deviation σ (assuming a normal distribution of Ct values in the experiment):
with and
Finally, assuming independence between each assay, the Bayes model is applied recursively for each selected assay; thus, it is possible to use every a posteriori probability to be i P(i|Xn) as a priori probability for the next assay. P(i|Xn) becomes P(i) to calculate P(i|Xn+1) on the next Bayes iteration as follows:
The method then classifies the sample data according to the largest posterior probability (D positive predicts i else predicts j).
Training sets
Training sets of positive and negative samples were built with Ct values obtained from the tests made on mono-uredinial cultures and environmental rust samples (See Specificity of the real-time PCR assays). Only samples whose species identity was verified by DNA barcoding of the ITS region were retained (S1–S5 Tables). DNA barcodes were obtained from previous studies for Melampsora and Chrysomyxa spp. [32–34]; for the other rust fungi, the ITS region was amplified/sequenced using ITS-1f [35] and ITS-4 [36] (or ITS-4BR [37] or ITS-4BR2: 5’- GGATTATCACCCTCAATGAT -3’) primers, under the same thermochemical conditions as described above, except for the primer annealing temperature which was set at 54–55°C.
Assay combinations
For each targeted taxonomic group, we tested each single assay and all assay combinations with the naïve Bayes classifier to reassign samples of the corresponding training set to be either positive or negative (Python script available upon request). Samples were then identified as being true positive (TP), true negative (TN), false positive (FP) and false negative (FN), depending on the correspondence between their DNA barcode and the result of the naïve Bayes classifier. TP samples were those with a DNA barcode corresponding to the targeted species and identified as being positive with the naïve Bayes classifier; TN were samples with a DNA barcode other than the targeted species and identified as being negative with the naïve Bayes classifier; FP were samples with a DNA barcode other than the targeted species and identified as being positive with the naïve Bayes classifier; finally, FN were samples with a DNA barcode of the targeted species that returned a negative identification with the naïve Bayes classifier. The accuracy of a detection assay is then defined as the rate of correct positive (defined as sensitivity of detection) and correct negative (specificity of detection) identifications:
The best minimal assay combination is the one that provides the highest accuracy over the entire reference population and includes the lowest number of assays.
Simulation of new samples
To circumvent potential issues with unbalanced sets of positive and negative samples and obtain higher precision on the estimates of TP, TN, FP and FN and accuracy rates, we simulated new sets of samples. For each targeted taxonomic group, 1,000 random datasets of 500 positive and 500 negative samples were simulated and accuracy measurement was re-estimated by using the naïve Bayes classifier on these new datasets (Python script available upon request). For each simulated dataset, Ct values for the positive samples were obtained based on the modeling of a normal distribution of the experimental data obtained for positive samples. Ct values of the negative samples were considered as the left tail part of a distribution right-truncated at the last cycle of the real-time PCR run (i.e. 40.0) and were modeled using an exponential distribution as suggested in Chandelier et al. [38].
Results and discussion
Tree rust-specific and sensitive real-time PCR assays were developed using the GEDI (genome-enhanced detection and identification) method which identifies genome regions that are unique to targeted taxa or groups of related taxa following whole genome comparisons [21]. These assays were designed for species-specific detection of Cronartium ribicola, Melampsora medusae and Melampsora larici-populina and for the detection of taxa belonging to Cronartium and Melampsora genera.
Assay design
The GEDI bioinformatics method, previously described [21] and applied in this study, resulted in 1341, 1542 and 1519 unique genes identified as species-specific to C. ribicola, M. medusae f. sp. deltoidae and M. larici-populina, respectively. At the genus level, 34 candidate gene clusters uniquely shared among the four Cronartium/Endocronartium species analysed were identified as compared to 270 among the seven Melampsora species (Table 2 in [21]).
Assay selection, specificity and sensitivity
Pine rusts
Subsets of species- and genus-specific candidate genes were screened by PCR on a reduced panel of DNA samples from target and non-target species among the Pucciniales. Among the 20 primer pairs screened for C. ribicola, three yielded amplicons with the ten selected C. ribicola samples collected in North America, Europe and Russia: CRIB65, CRIB146 and CRIB190 (Table 1 and S1 Table). None of these primer pairs produced amplicons with DNA from other rust species (n = 20 samples representative of ten Cronartium spp., other than C. ribicola, and two Melampsora spp.). Once the primer pair screening was completed, TaqMan probes were incorporated into the master mixes and the resulting assays were tested on a larger panel of DNA samples (Fig 1 and S1 Table). The specificity of CRIB65, CRIB146 and CRIB190 assays was confirmed, with the exception of the CRIB65 assay that generated a signal with Cronartium sp. collected on Pinus armandii in China. Multiple lines of evidence, based on host specialization, spore morphology and protein and DNA analyses indicate that this blister rust pathogen on P. armandii is different from C. ribicola, even though this fungus has not been recognized as a distinct taxon yet (for a review see [39]). Similarly, another Asian blister rust pathogen, collected on Pinus koraiensis in South Korea was detected with CRIB65 and CRIB146 assays, but not with the CRIB190 assay. Phylogenetic relationships of C. ribicola samples collected on different telial and aecial host species in Eurasia and North America as well as artificial inoculation studies on different telial host species suggest that this pathogen might have sub-species or ecological races [40].
At a higher taxonomic level, eight Cronartium-specific candidate genes were screened by PCR on a reduced panel of DNA samples, including 21 samples representative of 11 Cronartium/Endocronartium spp. and eight samples representative of as many non-target species among the Pucciniales. Two primer pairs yielded amplicons with the 21 selected Cronartium/Endocronartium spp. samples: CRO30 and CRO46, but not with DNA from other rust species (Table 1 and S1 Table). TaqMan probes were incorporated into the master mixes of the two selected primer pairs to design real-time PCR assays that were tested on a larger panel of DNA samples (Fig 1 and S1 Table). The specificity of CRO30 and CRO46 assays was confirmed, with the exception of positive signals for the CRO30 assay on a Pucciniastrum agrimoniae sample, and the CRO46 assay on a specimen of Pucciniastrum americanum. These two rust species are known to be closely related to the Cronartium genus [41], as confirmed in our phylogenetic reconstruction (Fig 1).
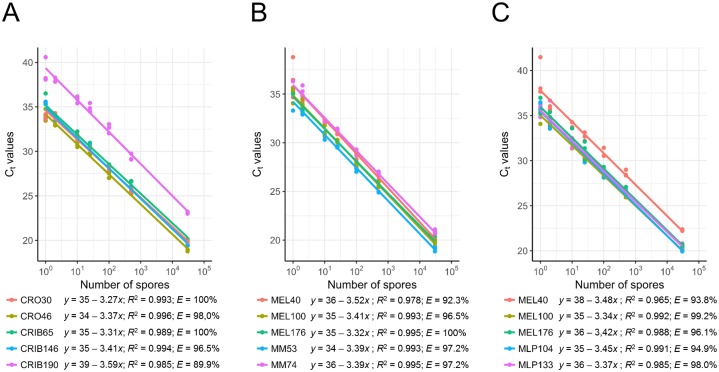
Using as few as one spore per reaction resulted in successful detection, as indicated by mean Ct values between 33.44 and 40.58, with the five TaqMan assays specific to C. ribicola or Cronartium spp. (S6 Table). Sensitivity in spore detection with the genus-specific assay CRO46 was slightly higher than sensitivity with the other genus-specific assay CRO30 or the species-specific assays CRIB65 and CRIB146 (Fig 2A and S6 Table). The species-specific assay CRIB190 was the least sensitive, with detection at higher Ct values for all spore quantities tested (average difference of 3.7 to 4.1 cycles with CRIB65 and CRIB146 assays, respectively). Linearity was observed across the range of spores counted, and from which DNA was isolated, and the mean Ct values determined by real-time PCR for each assay (Fig 2A). The high coefficients of determination (R2 = 0.985–0.996) indicated low variability between the three independent DNA isolations performed for each amount of spores of C. ribicola. Estimates of the efficiency of the amplification reaction were high, with values ranging from 96.5% (CRIB146) to 100% (CRO30 and CRIB65), except for the CRIB190 assay which had an efficiency of 89.9% (Fig 2A).
Fig 2. Relationship between the number of Cronartium ribicola (A), Melampsora medusae f. sp. deltoidae (B) and Melampsora larici-populina (C) spores from which DNA was isolated and Ct values, as determined by real-time PCR.
Each dot represents a biological independent replicate (corresponding to a known amount of spores), which was obtained by averaging the Ct values of three technical real-time PCR replicates.
Poplar rusts
Reduced panels of DNA samples from target and non-target species among the Pucciniales were used in PCR screening of subsets of species-specific candidate genes for both M. medusae f. sp. deltoidae and M. larici-populina and a subset of Melampsora-specific candidate genes. Among the ten primer pairs screened for M. medusae f. sp. deltoidae, two yielded amplicons with the eight M. medusae f. sp. deltoidae samples collected in North America and South Africa: MM53 and MM74 (Table 2 and S2 Table). No other rust taxa produced amplicons (n = 20 samples representative of 12 Melampsora spp., other than M. medusae f. sp. deltoidae), except for M. medusae f. sp. tremuloidae and M. ×columbiana. There are two host-specific formae speciales within M. medusae, named M. medusae f. sp. deltoidae (pathogenic on Populus deltoides and poplars in section Aigeiros) and M. medusae f. sp. tremuloidae (pathogenic on Populus tremuloides and poplars in section Populus) [42, 43]. A recent phylogenetic study supported the split of these formae speciales into two distinct and independent evolutionary lineages i.e. species [33]. Sequencing of the M. medusae f. sp. tremuloidae genome and comparison with the one of M. medusae f. sp. deltoidae should help to identify species-specific genome regions that would allow for the design of real-time PCR assays unique to each of these two rust taxa. The MM53 primer pair produced a faint amplification with some of the M. ×columbiana samples tested. This result was expected as M. ×columbiana is an interspecific hybrid between the two North American poplar rusts M. medusae f. sp. deltoidae and M. occidentalis [44, 45]. When the TaqMan probe was incorporated into the master mix, the resulting MM53 assay generated a signal for all six M. ×columbiana samples tested. The other assay targeting M. medusae, MM74, did not generate a signal for any M. ×columbiana samples tested nor did it generate a signal with the additional species of the expanded panel (Fig 1 and S2 Table). Melampsora ×columbiana hybrids exhibit various morphological features of both parents, sometimes mixed with the diagnostic alleles of only one of the parental species, indicating different degrees of hybridization from F1 hybrids to back-crosses with one of the parental species [44]. The ability to detect this hybrid with the MM53 and MM74 assays likely depends on the degree of introgression that will determine which of the M. medusae f. sp. deltoidae or M. occidentalis alleles are present at these two targeted loci in the M. ×columbiana samples tested.
For M. larici-populina-specific candidate genes, two out of ten primer pairs screened yielded amplicons with all 11 selected M. larici-populina samples collected in North America, New Zealand and China: MLP104 and MLP133 (Table 2 and S2 Table). Neither primer pair produced amplicons with DNA isolated from other rust species (n = four samples representative of as many non-target Melampsora spp.). Once this primer pair screening was completed, TaqMan probes were incorporated into the master mixes and the resulting assays were tested on a larger panel of DNA samples (Fig 1 and S2 Table). The specificity of the MLP104 and MLP133 assays was confirmed.
At a higher taxonomic level, five Melampsora-specific candidate genes were screened by PCR on a reduced panel of DNA samples, including 26 samples representative of 14 Melampsora spp. and ten samples representative of as many non-target species among the Pucciniales. Three primer pairs produced amplicons with 24 out of 26 selected Melampsora spp. samples: MEL40, MEL100 and MEL176 (Table 2 and S2 Table). No other rust taxa were amplified with these primer pairs. Once the TaqMan probes were incorporated into the master mixes, the resulting assays were tested on the expanded panel. The specificity of the MEL40, MEL100 and MEL176 assays was confirmed, except for the MEL40 assay which generated a signal with the spruce needle rust species Chrysomyxa weirii (Fig 1 and S2 Table). This result was not unexpected as C. weirii is known to share similar teliospore morphology and basidium germination characteristics with representative species of the genus Melampsora [46]. Recent multi-gene phylogenies have confirmed C. weirii as being more closely related to Melampsora than the Chrysomyxa genus [34], which is corroborated by our phylogenetic reconstruction, based on the ITS region (Fig 1).
Sensitivity of spore detection with the poplar leaf rust assays was similar to that obtained with the pine rust assays, with as few as one spore detected (Fig 2B and 2C and S7 and S8 Tables). Linearity was observed across the range of spores tested, and from which DNA was isolated, and the mean Ct values determined by real-time PCR for each assay (Fig 2B and 2C). Low variability was also observed between the three independent DNA isolations performed for each amount of spores of M. medusae f. sp. deltoidae or M. larici-populina (R2 = 0.965–0.995). Estimates of the efficiency of the amplification reactions were high, with values ranging from 92.3% to 100% (Fig 2B and 2C).
Tests on environmental samples
Pine rusts
DNA obtained from five types of environmental samples was tested with the pine rust assays. Three of the five assays yielded 100% detection success (measured as Ct values < 40.0) with inoculated and naturally infected material, regardless of the tissue infected or symptom type (white pine needle and stem tissues, blackcurrant leaves or single telia; S3 Table). One C. ribicola-specific assay (CRIB190) failed to detect C. ribicola in three inoculated samples and two naturally infected samples. One Cronartium-specific assay (CRO30) failed to detect the pathogen in a single telium sample (S3 Table). When DNA samples associated with Ct values > 40.0 were retained, positive results increased to between 90.9% and 100%. Assays CRO30, CRO46, CRIB65 and CRIB146 detected comparable amounts of C. ribicola DNA from DNA isolated from insects captured in a white pine plantation diseased with blister rust: eight out of the nine isolates produced positive results (88.9%), with Ct values < 40.0 (S3 Table). In contrast, positive results decreased to 33.3% with the CRIB190 assay when isolates with Ct values > 40.0 were excluded. However, when the Ct value threshold was raised to 41.5, all nine DNA isolates produced positive results, which were confirmed by fungal DNA barcoding. Insects associated with spore dispersal might facilitate spermogonia fertilization by carrying C. ribicola spermatia to the spermogonia, which produce a nectar that attracts the insects. Once fertilization has occurred, a dikaryotic mycelium develops, which later organizes to form aecidia on pines [47].
Poplar rusts
All poplar rust assays have the ability to detect M. medusae f. sp. deltoidae and/or M. larici-populina DNA on naturally infected larch needles and poplar leaves (S4 Table). One hundred percent of DNA isolates from naturally infected larch samples produced positive results with the MEL40, MEL100 and MEL176 assays. The species-specific assays showed that M. medusae f. sp. deltoidae was detected almost four times more frequently than M. larici-populina on the larch needles analysed. Furthermore, all 33 samples from naturally infected poplar leaves produced positive results with the three genus-specific assays. Co-infection by both M. medusae f. sp. deltoidae and M. larici-populina was detected in 32 out of the 33 poplar leave samples tested with the species-specific assays. All of these results were corroborated by previous results achieved through real-time PCR assays, using species-specific primers targeting the ITS region [48]. Moreover, the variable abundance, based on Ct values of both Melampsora species detected on these leaves with our poplar rust assays, targeting single- or low-copy genes, is consistent with observations using an assay targeting the multi-copy region of the internal transcribed spacers 1 and 2 ([49] and unpublished data).
Accuracy of the assays
Detection of pathogens by real-time PCR usually requires setting a Ct value threshold to declare a positive reaction. As originally proposed by Liu and Saint [50], the fluorescent signal of a real-time PCR reaction fits the sigmoidal mathematical model. In theory, under optimal and standardized conditions, the fluorescent threshold value for detecting one copy of the target sequence should be consistent across samples and assays. However, in practice, many factors impact PCR efficiency (e.g. the quantity and quality of the template DNA, the PCR reagents, the thermocycler). As demonstrated by Grosdidier et al. [51], specific Ct cutoff values need to be calculated for any given target*matrix*thermocycler combination since a single change of target, matrix (physicochemical nature of the reaction mixture) or thermocycler will affect this cutoff value. This approach is empirical and generally based on theoretical, unproven assumptions [51]. Assuming that the Ct values obtained from reference collections of positive and negative samples were independent variables [30, 31], we anticipate that the overlap could be interpreted in a Bayesian context. We employed naïve Bayes classifier learning on these distributions to determine the probability that an unknown sample is a true positive. For each real-time PCR assay, the classifier was trained on the Ct values obtained on an extensive set of positive and negative samples (mono-uredinial cultures and environmental rust samples) that were previously identified as true positive and true negative samples by using a DNA barcoding approach [32]. Following this learning stage, the classifier assigned unknown samples as being either positive or negative.
We first used the classifier to re-evaluate each sample of the training sets and generate measurements of performance (TP and FP rates, accuracy) for each assay and for each combination of two and three assays. For the three targeted species and the two genera, we obtained an accuracy of detection of 100% (i.e. TP = 100% and FP = 0.0%; Table 3). A single assay was sufficient to reach this level of accuracy for the targets of C. ribicola with CRIB146, M. larici-populina with MLP104 or MLP133 and Cronartium spp. with CRO46, whereas a minimum of two combined assays were needed for M. medusae f. sp. deltoidae with MM53 + MM74 and Melampsora spp. with MEL100 + MEL176 (Table 3). Interestingly, the combination of the two assays CRO30 and CRO46 for detecting Cronartium spp. resulted in a lower accuracy than using CRO46 alone; the low accuracy obtained with the assay combination CRO30 + CRO46 likely resulted from the low TP rate (92.7%) and the substantial FP rate (3.5%) yielded by CRO30. This assay failed to detect C. ribicola on one out of the nine insect vectors, positive with our DNA barcoding approach, as well as one out of 14 single telia harvested on blackcurrant leaves (S3 Table). CRO30 also cross-reacted with Pucciniastrum agrimoniae (Fig 1).
Table 3. Assignment predictions made with the naïve Bayes classifier.
| Pine and poplar rust assays | % TP a | % FP a | Accuracy (%) |
|---|---|---|---|
| Cronartium spp.—41/29b | |||
| CRO30 | 92.7 (94.59) | 3.5 (11.72) | 94.3 (91.44) |
| CRO46 | 100.0 (99.11) | 0.0 (0.45) | 100.0 (99.33) |
| CRO30 + CRO46 | 100.0 (99.86) | 3.5 (2.47) | 98.6 (98.70) |
| Cronartium ribicola—64/34b | |||
| CRIB65 | 96.9 (99.43) | 5.9 (2.32) | 95.9 (98.56) |
| CRIB146 | 100.0 (100.0) | 0.0 (0.69) | 100.0 (99.66) |
| CRIB190 | 90.6 (97.95) | 0.0 (0.64) | 93.9 (98.65) |
| CRIB65 + CRIB46 | 100.0 (100.0) | 0.0 (0.21) | 100.0 (99.90) |
| CRIB65 + CRIB190 | 95.3 (99.86) | 0.0 (0.11) | 96.9 (99.88) |
| CRIB146 + CRIB190 | 100.0 (100.0) | 0.0 (0.40) | 100.0 (99.80) |
| CRIB65 + CRIB146 + CRIB190 | 100.0 (100.0) | 0.0 (0.14) | 100.0 (99.93) |
| Melampsora spp.—105/12b | |||
| MEL40 | 92.4 (91.21) | 8.3 (7.85) | 92.3 (91.68) |
| MEL100 | 99.1 (99.61) | 0.0 (0.46) | 99.2 (99.57) |
| MEL176 | 99.1 (99.12) | 0.0 (0.23) | 99.2 (99.45) |
| MEL40 + MEL100 | 99.1 (99.85) | 0.0 (0.69) | 99.2 (99.58) |
| MEL40 + MEL176 | 99.1 (99.77) | 0.0 (0.86) | 99.2 (99.45) |
| MEL100 + MEL176 | 100.0 (99.99) | 0.0 (0.11) | 100.0 (99.94) |
| MEL40 + MEL100 + MEL176 | 100.0 (100.0) | 0.0 (0.39) | 100.0 (99.80) |
| Melampsora medusae—67/31b | |||
| MM53 | 98.5 (99.10) | 0.0 (0.39) | 99.0 (99.36) |
| MM74 | 92.5 (95.04) | 3.2 (3.11) | 93.9 (95.97) |
| MM53 + MM74 | 100.0 (99.93) | 0.0 (0.88) | 100.0 (99.52) |
| Melampsora larici-populina—91/46b | |||
| MLP104 | 100.0 (99.33) | 0.0 (0.39) | 100.0 (99.47) |
| MLP133 | 100.0 (98.90) | 0.0 (0.27) | 100.0 (99.31) |
| MLP104 + MLP133 | 100.0 (99.98) | 0.0 (0.14) | 100.0 (99.92) |
Values on the left are estimates obtained from the training set and values on the right, in parentheses, are those obtained from resampling. Lines highlighted in bold characters correspond to the best assay or assay combinations, as predicted after the resampling procedure.
aTP: true positive; FP: false positive.
bNumber of samples tested (positives/negatives).
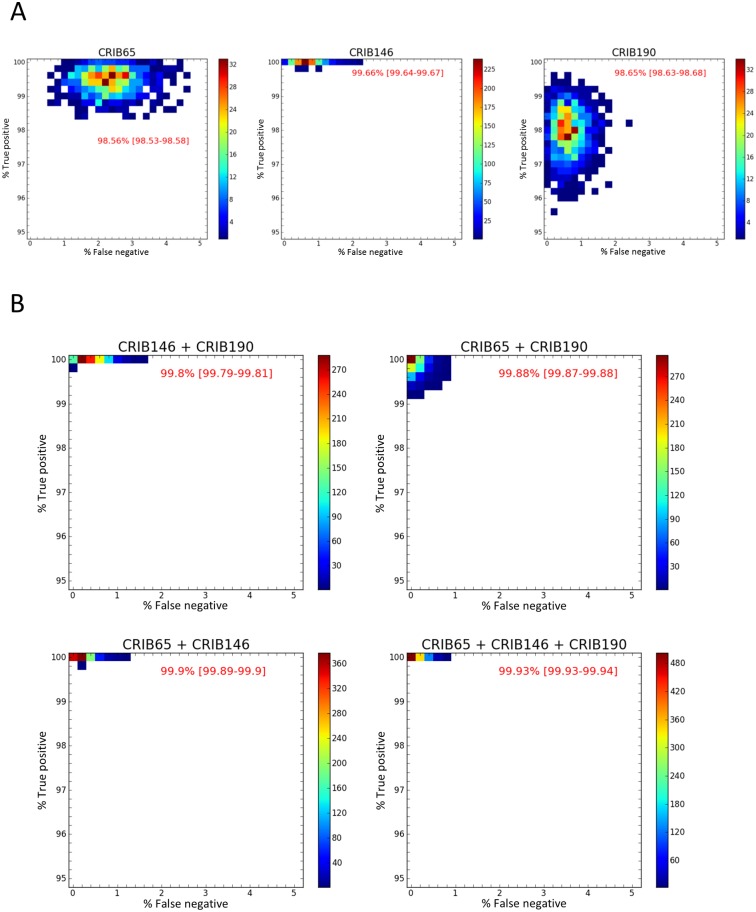
To obtain a higher accuracy, we generated a new dataset of random positive and negative samples for each targeted taxonomic group (500 for each class) based on the Ct value distributions of the training sets. Results obtained with the naïve Bayes classifier on the five simulated datasets confirmed those previously obtained for Cronartium spp. (highest accuracy for CRO46; 99.33%), M. medusae f. sp. deltoidae (MM53 + MM74; 99.52%) and Melampsora spp. (MEL100 + MEL176; 99.94%) (Table 3). In the two other cases (C. ribicola and M. larici-populina), analysis of the simulated datasets resulted in improved precision and allowed us to draw some slightly different conclusions about the assay combinations. Previous accuracy estimates (see above) indicated that using CRIB146 alone was sufficient to obtain a detection accuracy of 100% (Table 3). New estimates obtained from the simulations showed that this assay produced a lower detection accuracy of 99.66% (resulting from a FP rate of 0.69% i.e. one false positive every 145 samples tested). Combining the two additional C. ribicola-specific assays decreased the FP rate to one false positive every 714 samples tested and raised the accuracy to 99.93% (Fig 3). Similarly, we also improved precision for the assays targeting M. larici-populina. The combination of the two available assays was required to reach a maximum accuracy of 99.92% for this species (Table 3). However, from a practical point of view, such a slight increase in accuracy likely would not justify the time and resources required to run additional assays.
Fig 3. Distribution of true positive (TP) and false positive (FP) rates estimated with a naïve Bayes classifier, over 1,000 simulations of 500 positive and 500 negative samples, for the three specific assays targeting Cronartium ribicola.
Results are presented for each single assay (A) and for all possible combinations of two or three assays (B). Color scale represents the number of simulations that were assigned into each TP rate X FP rate category. Values in red represent the percentage of accuracy with 95% confidence intervals.
Conclusions
The advantage of using the GEDI approach is that unique genome regions are more likely to provide highly specific assays that can discriminate closely related taxa and are amenable to multiplexing. The disadvantage is that currently the number of sequenced genomes available is still far smaller than the number of DNA sequences available in public databases for the well conserved genes that are used for DNA barcoding and DNA detection. We do foresee, however, a dramatic increase in the number of pathogen genomes that will be available in the future, making the GEDI approach extremely relevant. In addition, future uses of this approach could combine the front portion of the GEDI method with target-enrichment of the target genome regions, followed by high-throughput sequencing, instead of real-time PCR. This approach, already used for human pathogens in clinical settings [52], will likely become the standard in the future for biosurveillance of plant pathogens.
As the proposed assays target some of the most important tree rust pathogens, the adoption of these valuable tools for the monitoring of these fungi should contribute to forest health protection. On a broader perspective, the methodology used in this study provides a framework for the development and validation of robust molecular detection assays that should be widely applicable across different species.
Supporting information
(TIFF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Values indicate the number of positive (+) and negative (-) samples per species that were tested for each targeted taxonomic group.
(DOCX)
The replicate numbers in the first column correspond to the three independent extractions from the same number of spores.
(DOCX)
The replicate numbers in the first column correspond to the three independent extractions from the same number of spores.
(DOCX)
The replicate numbers in the first column correspond to the three independent extractions from the same number of spores.
(DOCX)
Acknowledgments
Authors gratefully acknowledge people and institutions for kindly providing DNA samples, fungal isolates, herbarium specimens or environmental samples. They would also like to thank M. Blais, E. Dussault, P. Labrie and D. Plourde (Canadian Forest Service, Natural Resources Canada, Québec) for technical advice in evaluating spore count and artificial inoculation of white pine seedlings with C. ribicola spores. Finally, the authors would like to acknowledge C. Morin for the design of the ITS-4BR2 primer as well as C. Coulombe and M.-C. Nicole for the trapping of insect vectors.
Data Availability
Data are available at GenBank (accession numbers: MH171727-MH171903).
Funding Statement
This research was funded by Genome Canada, Genome British Columbia and Génome Québec Large-Scale Applied Research Program, projects #2112 and #10106 to RCH; https://www.genomecanada.ca/en/genomics-based-forest-health-diagnostics-and-monitoring; https://www.genomecanada.ca/en/biosurveillance-alien-forest-enemies-biosafe. Author PT received funding from the Government of Canada from the Genomic Research and Development Initiative program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Santini A, Ghelardini L, De Pace C, Desprez-Loustau M-L, Capretti P, Chandelier A, et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013;197(1):238–250. 10.1111/j.1469-8137.2012.04364.x [DOI] [PubMed] [Google Scholar]
- 2.Pautasso M, Schlegel M, Holdenrieder O. Forest health in a changing world. Microbial Ecology. 2015;69(4):826–842. 10.1007/s00248-014-0545-8 [DOI] [PubMed] [Google Scholar]
- 3.Loo JA. Ecological impacts of non-indigenous invasive fungi as forest pathogens. Biological Invasions. 2009;11(1):81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484(7393):186 10.1038/nature10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung B, Lodge DM, Finnoff D, Shogren JF, Lewis MA, Lamberti G. An ounce of prevention or a pound of cure: bioeconomic risk analysis of invasive species. Proceedings of the Royal Society of London B: Biological Sciences. 2002;269(1508):2407–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geils BW, Hummer KE, Hunt RS. White pines, Ribes, and blister rust: a review and synthesis. Forest Pathol. 2010;40(3–4):147–185. [Google Scholar]
- 7.Kim M-S, Klopfenstein NB, Ota Y, Lee SK, Woo K-S, Kaneko S. White pine blister rust in Korea, Japan and other Asian regions: comparisons and implications for North America. Forest Pathol. 2010;40(3–4):382–401. [Google Scholar]
- 8.Sniezko RA, Smith J, Liu J-J, Hamelin RC. Genetic resistance to fusiform rust in southern pines and white pine blister rust in white pines—a contrasting tale of two rust pathosystems—current status and future prospects. Forests. 2014;5(9):2050–2083. [Google Scholar]
- 9.Ostry ME, Ramstedt M, Newcombe G, Steenackers M. Diseases of poplars and willows In: Isebrands JG, Richardson J, editors. Poplars and willows: trees for society and the environment. Rome, Italy: CAB International and Food and Agriculture Organization of the United Nations; 2014. pp. 443–458. [Google Scholar]
- 10.Carnegie AJ, Kathuria A, Pegg GS, Entwistle P, Nagel M, Giblin FR. Impact of the invasive rust Puccinia psidii (myrtle rust) on native Myrtaceae in natural ecosystems in Australia. Biological Invasions. 2016;18(1):127–144. [Google Scholar]
- 11.Jalkanen R. Synthesis and new observations on needle pathogens of larch in Northern Finland. Forests. 2016;7(1):25. [Google Scholar]
- 12.Sinclair WA, Lyon HH. Diseases of trees and shrubs. Second ed Ithaca, NY, USA: Comstock Publishing Associates, a division of Cornell University Press; 2005. 660 p. [Google Scholar]
- 13.Organization EaMPP. EPPO global database—Distribution of Melampsora medusae. 2018 [updated 2018-05-29]; https://gd.eppo.int/taxon/MELMME/distribution.
- 14.Organization EaMPP. EPPO A2 List of pests recommended for regulation as quarantine pests. 2018 [updated September 2018]; https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list.
- 15.Innes L, Marchand L, Frey P, Bourassa M, Hamelin RC. First report of Melampsora larici-populina on Populus spp. in eastern North America. Plant Dis. 2004;88(1):85–85. [DOI] [PubMed] [Google Scholar]
- 16.Grondin J, Bourassa M, Hamelin RC. First report of the aecial state of Melampsora larici-populina on Larix spp. in North America. Plant Dis. 2005;89(11):1242–1242. [DOI] [PubMed] [Google Scholar]
- 17.Husson C, Ioos R, Andrieux A, Frey P. Development and use of new sensitive molecular tools for diagnosis and detection of Melampsora rusts on cultivated poplar. Forest Pathol. 2013;43(1):1–11. [Google Scholar]
- 18.Boutigny A-L, Guinet C, Vialle A, Hamelin RC, Andrieux A, Frey P, et al. Optimization of a real-time PCR assay for the detection of the quarantine pathogen Melampsora medusae f. sp. deltoidae. Fungal Biology. 2013;117(6):389–398. 10.1016/j.funbio.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 19.Boutigny A-L, Guinet C, Vialle A, Hamelin R, Frey P, Ioos R. A sensitive real-time PCR assay for the detection of the two Melampsora medusae formae speciales on infected poplar leaves. Eur J Plant Pathol. 2013;136(3):433–441. [Google Scholar]
- 20.Guinet C, Boutigny A-L, Vialle A, Hamelin RC, Frey P, Ioos R. Simultaneous monitoring and quantification of Melampsora allii-populina and Melampsora larici-populina on infected poplar leaves using a duplex real-time PCR assay. Plant Pathol. 2016;65(3):380–391. [Google Scholar]
- 21.Feau N, Beauseigle S, Bergeron M-J, Bilodeau GJ, Birol I, Cervantes-Arango S, et al. Genome-Enhanced Detection and Identification (GEDI) of plant pathogens. PeerJ. 2018;6:e4392 10.7717/peerj.4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Withers S, Gongora-Castillo E, Gent D, Thomas A, Ojiambo PS, Quesada-Ocampo LM. Using next-generation sequencing to develop molecular diagnostics for Pseudoperonospora cubensis, the cucurbit downy mildew pathogen. Phytopathology. 2016;106(10):1105–1116. 10.1094/PHYTO-10-15-0260-FI [DOI] [PubMed] [Google Scholar]
- 23.Duplessis S, Cuomo CA, Lin Y-C, Aerts A, Tisserant E, Veneault-Fourrey C, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proceedings of the National Academy of Sciences. 2011;108(22):9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toome M, Ohm RA, Riley RW, James TY, Lazarus KL, Henrissat B, et al. Genome sequencing provides insight into the reproductive biology, nutritional mode and ploidy of the fern pathogen Mixia osmundae. New Phytol. 2014;202(2):554–564. 10.1111/nph.12653 [DOI] [PubMed] [Google Scholar]
- 25.Firrincieli A, Otillar R, Salamov A, Schmutz J, Khan Z, Redman RS, et al. Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Frontiers in Microbiology. 2015;6:978 10.3389/fmicb.2015.00978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13(9):2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Research. 2012;40(15):e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunt RS. Experimental evidence of heterothallism in Cronartium ribicola. Canadian Journal of Botany. 1985;63(6):1086–1088. [Google Scholar]
- 29.Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Machine Learning. 1997;29(2–3):103–130. [Google Scholar]
- 30.Lowd D, Domingos P, editors. Naive Bayes models for probability estimation. Proceedings of the 22nd International Conference on Machine Learning; 2005; Bonn, Germany. New York, USA: ACM; 2005.
- 31.Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: a review of classification and combining techniques. Artificial Intelligence Review. 2006;26(3):159–190. [Google Scholar]
- 32.Feau N, Vialle A, Allaire M, Tanguay P, Joly DL, Frey P, et al. Fungal pathogen (mis-) identifications: a case study with DNA barcodes on Melampsora rusts of aspen and white poplar. Mycol Res. 2009;113(6–7):713–724. [DOI] [PubMed] [Google Scholar]
- 33.Vialle A, Feau N, Frey P, Bernier L, Hamelin RC. Phylogenetic species recognition reveals host-specific lineages among poplar rust fungi. Mol Phylogenet Evol. 2013;66(3):628–644. 10.1016/j.ympev.2012.10.021 [DOI] [PubMed] [Google Scholar]
- 34.Feau N, Vialle A, Allaire M, Maier W, Hamelin RC. DNA barcoding in the rust genus Chrysomyxa and its implications for the phylogeny of the genus. Mycologia. 2011;103(6):1250–1266. 10.3852/10-426 [DOI] [PubMed] [Google Scholar]
- 35.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts Mol Ecol. 1993;2(2):113–118. [DOI] [PubMed] [Google Scholar]
- 36.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego, CA, USA: Academic Press, Inc; 1990. pp. 315–322. [Google Scholar]
- 37.Vialle A, Feau N, Allaire M, Didukh M, Martin F, Moncalvo J-M, et al. Evaluation of mitochondrial genes as DNA barcode for Basidiomycota. Molecular Ecology Resources. 2009;9(s1):99–113. [DOI] [PubMed] [Google Scholar]
- 38.Chandelier A, Planchon V, Oger R. Determination of cycle cut off in real-time PCR for the detection of regulated plant pathogens. EPPO Bulletin. 2010;40(1):52–58. [Google Scholar]
- 39.Zhang XY, Lu Q, Sniezko RA, Song RQ, Man G. Blister rusts in China: hosts, pathogens, and management. Forest Pathol. 2010;40:369–381. [Google Scholar]
- 40.Richardson BA, Kim M-S, Klopfenstein NB, Ota Y, Woo KS, Hamelin RC, editors. Tracking the footsteps of an invasive plant-pathogen: intercontinental phylogeographic structure of the white-pine-blister-rust fungus, Cronartium ribicola Breeding and Genetic Resources of Five-Needle Pines: Ecophysiology, Disease Resistance and Developmental Biology; 2008; Yangyang, Korea. Seoul, Korea: Korea Forest Research Institute; 2009. [Google Scholar]
- 41.Maier W, Begerow D, Weiss M, Oberwinkler F. Phylogeny of the rust fungi: an approach using nuclear large subunit ribosomal DNA sequences. Canadian Journal of Botany. 2003;81(1):12–23. [Google Scholar]
- 42.Feau N, Joly DL, Hamelin RC. Poplar leaf rusts: model pathogens for a model tree. Botany. 2007;85(12):1127–1135. [Google Scholar]
- 43.Shain L. Evidence for formae speciales in the poplar leaf rust fungus, Melampsora medusae. Mycologia. 1988;80(5):729–732. [Google Scholar]
- 44.Newcombe G, Stirling B, McDonald S, Bradshaw HD. Melampsora xcolumbiana, a natural hybrid of M. medusae and M. occidentalis. Mycol Res. 2000;104:261–274. [Google Scholar]
- 45.Frey P, Gérard P, Feau N, Husson C, Pinon J. Variability and population biology of Melampsora rusts on poplars In: Pei MH, McCracken AR, editors. Rust diseases of willow and poplar. Cambridge, MA, USA: CABI Publishing; 2005. pp. 63–72. [Google Scholar]
- 46.Maier WFA, Crane PE, Wingfield BD. Chrysomyxa weirii is more closely related to Melampsora than to Chrysomyxa. Abstracts of Papers Presented at the Meeting of the Mycological Society of Japan. 2005;49:156. [Google Scholar]
- 47.Hamelin RC. Genetic diversity between and within cankers of the white pine blister rust. Phytopathology. 1996;86(8):875–879. [Google Scholar]
- 48.Boyle B, Hamelin RC, Seguin A. In vivo monitoring of obligate biotrophic pathogen growth by kinetic PCR. Appl Environ Microbiol. 2005;71(3):1546–1552. 10.1128/AEM.71.3.1546-1552.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyle B, Grondin J, Feau N, Innes L, Hamelin RC. Real-time PCR to study the distribution of two major rust species in Eastern North America. Phytopathology. 2006;96(6 (Supplement)):S15. [Google Scholar]
- 50.Liu W, Saint DA. Validation of a quantitative method for real time PCR kinetics. Biochemical and Biophysical Research Communications. 2002;294(2):347–353. 10.1016/S0006-291X(02)00478-3 [DOI] [PubMed] [Google Scholar]
- 51.Grosdidier M, Aguayo J, Marcais B, Ioos R. Detection of plant pathogens using real-time PCR: how reliable are late Ct values? Plant Pathol. 2017;66(3):359–367. [Google Scholar]
- 52.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. 2017;12(6):1261–1276. 10.1038/nprot.2017.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Values indicate the number of positive (+) and negative (-) samples per species that were tested for each targeted taxonomic group.
(DOCX)
The replicate numbers in the first column correspond to the three independent extractions from the same number of spores.
(DOCX)
The replicate numbers in the first column correspond to the three independent extractions from the same number of spores.
(DOCX)
The replicate numbers in the first column correspond to the three independent extractions from the same number of spores.
(DOCX)
Data Availability Statement
Data are available at GenBank (accession numbers: MH171727-MH171903).