Abstract
In the 15 years following the release of the first complete human genome sequences, our understanding of the ability of rare and common genetic variation to determine cardiovascular disease susceptibility, prognosis, and therapeutic response has grown exponentially. As such, the use of genomics to enhance the care of patients with cardiovascular diseases (CVDs) has garnered increased attention from clinicians, researchers, and regulatory agencies eager to capitalize on the promise of precision genomic medicine. However, owing to a large burden of “complex” common diseases, an unrelenting desire for evidence-based practice, and a degree of unfamiliarity/discomfort with the language of genomic medicine, the development and implementation of genomics-guided approaches designed to further individualize the clinical management of a variety of cardiovascular disorders remains a challenge. In this Review, we detail a practical approach to genetic testing initiation and interpretation as well as review the current state of cardiovascular genetic and pharmacogenomics testing in the context of relevant society and regulatory agency recommendations/guidelines.
INTRODUCTION
Since the sentinel discovery of the first heritable monogenic cardiovascular disease (CVD)-susceptibility genes in the early-to-mid 1990’s, genetic testing for familial aortopathies,1, 2 cardiomyopathies,3, 4 cardiac channelopathies,5, 6 and hypercholesterolemia7, 8 has transitioned rapidly from early research-based endeavors to a full complement of reimbursable commercially-available genetic tests. Furthermore, following the release of the first complete human genome sequences in 2001,9, 10 ensuing genome-wide association studies (GWAS) have identified a plethora of common genetic variants that underlie risk of developing common CVDs such as coronary heart disease (CHD)11 and atrial fibrillation (AF)12 as well as inter-individual variability in cardiovascular drug-response. Collectively, genetic testing for rare monogenic CVDs, ongoing development of genetic risk scores (GRS) for common polygenic CVDs, and the implementation of pharmacogenomics testing to predict individual response to cardiovascular drugs represent the spectrum of genetic tests that currently impact the diagnosis, risk-stratification, and clinical management of patients with rare and common CVDs.
With the announcement of the Precision Medicine Initiative in early 2015, interest in precision genomic medicine has intensified, and the stage has been set for an unprecedented proliferation of genetics- and genomics-guided approaches. Although cardiovascular providers stand to benefit immensely from these advances, the rapid pace of genomic discoveries, gaps in genomics education/literacy, and paucity of data from randomized controlled trials (RCTs) designed to determine the clinical utility of genomic-aided approaches have left many overwhelmed and thereby ill prepared to deliver high-quality, genomics/genetics-guided care. As such, this Review aims to summarize the current clinical utility, commonly encountered pitfalls, and areas of emerging interest pertaining to the use of genetic and pharmacogenomic testing to individualize the clinical management of an array of CVDs.
Basic Principles Governing the Initiation and Interpretation of Cardiovascular Genetic Tests
With each passing year, cost-lowering technological advances, improved payer reimbursement, and legislation aimed at eliminating genetic discrimination make genetic testing increasingly accessible and appealing. However, as the pendulum has swung from inaccessible to more readily available, the increased, and at times, inappropriate utilization of genetic testing has brought with it a new set of obstacles.13 As such, the ensuing paragraphs aim to help providers avoid common pitfalls associated with the inappropriate use of genetic testing, namely poor phenotyping, inappropriate genetic test selection, and misinterpretation of results, by outlining common indications, expected results, and basic interpretative strategies when considering CVD genetic testing.
At present, CVD genetic testing is reserved typically for one of three clinical indications: 1) comprehensive genetic testing to aid in or confirm the diagnosis of a heritable CVD in which there is a strong index of clinical suspicion (class I recommendation for many, but not all monogenic CVDs),6 2) mutation-specific cascade screening of appropriate relatives (class I recommendation for all monogenic CVDs),6 and 3) the selected use of pharmacogenomics testing to aid in the selection and or dosing of certain cardiovascular medications (variable society and regulatory agency recommendations). It is important to note that due to variable expressivity and incomplete penetrance of monogenic CVDs coupled with significant background genetic variation in many monogenic CVD-causative genes, diagnostic genetic testing should be viewed as probabilistic rather than binary/deterministic in nature.5, 14, 15
As such, the clinical utility of a given genetic test is dependent highly on the pre-test probability of disease (i.e. strength of clinical phenotype/diagnosis) and disease-specific genetic test performance metrics (diagnostic yield, signal-to-noise ratio, etc.). In other words, in patients with weak/non-equivocal clinical phenotypes, the diagnostic yield of genetic testing declines, the signal-to-noise ratio rises, and the risk of encountering false-positives increases exponentially. Therefore, a “one-size-fits-all” mentality to genetic testing is ill-advised and genetic testing should only be undertaken if significant suspicion for an underlying genetic CVD remains following a thorough clinical evaluation, including but not limited to a detailed family history, comprehensive cardiovascular work-up, and assessment for multisystem syndromes. Given these nuances, the initiation and interpretation of cardiovascular genetic tests requires a multidisciplinary approach involving the coordinated efforts of general practitioners/general cardiologists, genetic counselors, medical geneticists, and cardiovascular sub-specialists with expertise in the CVD of interest. When feasible, the ordering, interpretation, and communication of monogenic CVD genetic test results should be done under the guidance of a genetically-oriented cardiologist and/or medical geneticist with expertise in heritable CVDs in conjunction with a cardiovascular-oriented genetic counselor. Regardless of the responsible provider, the patient and his or her family should receive genetic counseling. By assisting the multidisciplinary team with 1) the generation of multigenerational pedigrees and identification of the most appropriate individual(s) to initially test, 2) the selection of appropriate genetic tests, 3) the accurate interpretation and continued reassessment of genetic test results, and 4) assuring that psychosocial ramifications of genetic testing are adequately addressed, such genetic counseling provides additional assurance that genetic testing will be utilized appropriately and that high-quality, cost-effective care is delivered.16
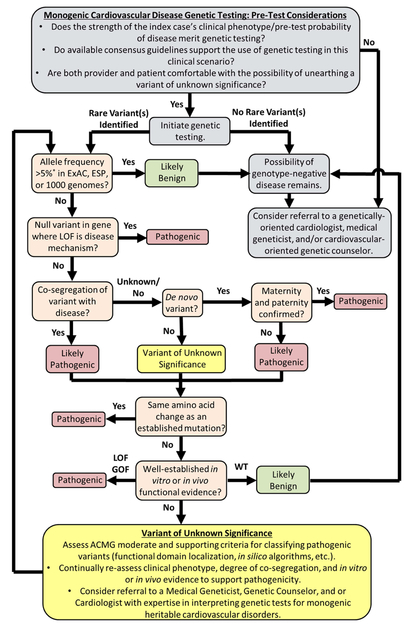
Once a decision is made to pursue genetic testing and the patient apprised of the potential risks, benefits, and limitations of genetic testing (ideally by a genetic counselor), including the fact that a negative test cannot definitively rule out disease (except for the case of mutation-specific cascade screening) and variants of unknown/uncertain significance (VUS) without sufficient supporting evidence to be deemed pathogenic may be encountered, the work is far from over. First, the genetic test results should be interpreted in light of established criteria set forth by the American College of Medical Genetics (ACMG)17 and other organizations as summarized in Figure 1. In some instances, a bona fide pathogenic mutation meeting one if not all of the ACMG’s very strong/strong evidence of pathogenicity (i.e. null, established, and de novo variants as outlined in Figure 1)17 is unearthed making interpretation straightforward.
Figure 1 |.
A rational approach to genetic test initiation, rare variant interpretation, and the fluid re-assessment of variants of unknown/uncertain significance. Gray boxes denote basic considerations pertaining to the initiation of genetic testing. Beige boxes denote key steps in the classification of rare genetic variation based on widely acceptable American College of Medical Genetics criteria. Yellow boxes denote basic considerations pertaining to the identification of a rare variant of unknown/uncertain significance that currently lacks sufficient evidence to either up or down grade its probability of pathogenicity. *Allele frequency > 5% or greater than widely accepted estimates of disease prevalence in the Exome Aggregation Consortium, Exome Sequencing Project, or 1000 genomes serve as stand-alone or strong evidence of pathogenicity, respectively. ACMG = American College of Medical Genetics; ESP = Exome Sequencing Project; ExAC = Exome Aggregation Consortium; GOF = gain-of-function; LOF = loss-of-function; VUS = variant of unknown/uncertain significance; WT = wild-type.
However, in many cases, the commercial genetic test reports will return “positive” results with cryptic language such as “possible deleterious mutation” or “VUS” next to the identified variant(s). In this scenario, it is important to realize that “positive” is not synonymous with “disease-causative” as the identified VUS, in many circumstances, has a nearly equal chance of being a pathogenic mutation as it does a rare innocuous variant. When there is insufficient evidence to tip the scale in either direction, an agonizing situation for patients and providers develops recently referred to as “genetic purgatory”.13, 18 Although genetic purgatory is a situation all providers hope to avoid, once there, it is imperative to resist the temptation to act on a VUS by 1) escalating clinical management of the index case and/or 2) initiating mutation/variant-specific cascade screening as these can lead to potentially harmful diagnostic miscues. Rather, providers need to closely scrutinize available clinical data and intermittently reassess the potential pathogenicity of the VUS in light of any new clinical, molecular, or computational data that may elevate or downgrade its status based on the ACMG’s fluid evidence of pathogenicity framework (Figure 1).17
Genetic Testing for Commonly Encountered Monogenic (Mendelian) Cardiovascular Disorders (CVDs)
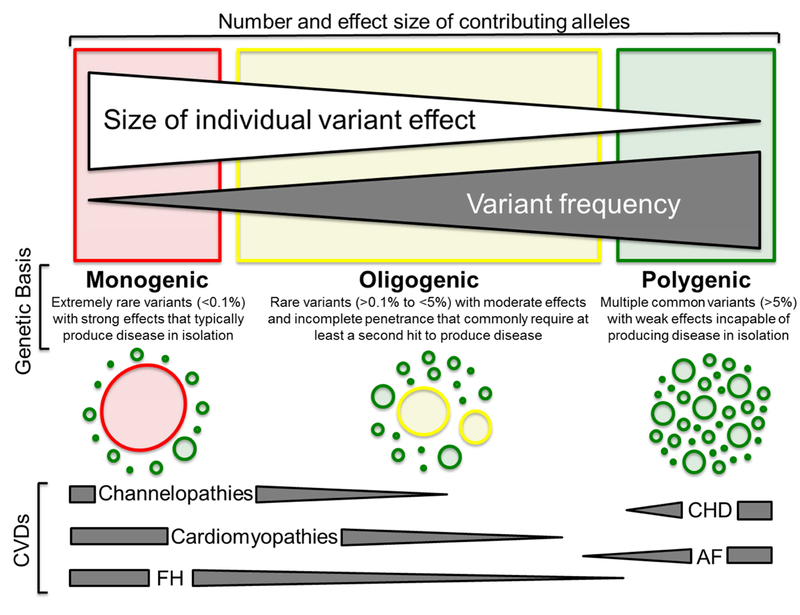
Classically, monogenic or Mendelian disorders arise from a rare mutation(s), passed from generation-to-generation in defined inheritance patterns (autosomal dominant, autosomal recessive, X-linked, etc.), that perturb the intended biological function of a single, disease-causative, gene-encoded protein (Figure 2).19 In the subsequent sections, we summarize the current state of diagnostic clinical genetic testing for four commonly encountered classes of monogenic CVDs (aortopathies, cardiomyopathies, cardiac channelopathies, and familial hypercholesteremia) and how the judicious utilization of genetic testing can enhance and in some cases individualize the diagnosis, risk-stratification, and/or clinical management of patients afflicted by these potentially life-threatening disorders.
Figure 2 |.
The spectrum of genetic variation underlying the heritable component of commonly encountered cardiovascular disorders (CVDs). At the severe (red) end of the spectrum are extremely rare disease-causative mutations with strong effects on gene function that typically result in monogenic disorders such as long QT syndrome, hypertrophic cardiomyopathy, and familial hypercholesterolemia. In the middle of the spectrum (yellow) are rare variants with moderate effects on gene function that rarely produce disease in isolation, but in the presence of one or more second hits result in disease as seen in some instances of arrhythmogenic cardiomyopathy. Lastly, at the benign (green) end of the spectrum are common variants with weak effects on gene function that are incapable of producing disease in isolation, but may confer disease risk when multiple risk-associated common variants are present within the genome of an individual with environmental risk factors for disorders such as coronary heart disease and atrial fibrillation. In recognition that the genetic basis of most heritable CVDs is variable, dark gray triangles denote the spectrum of genetic variation underlying each CVD or class of CVDs. ACM = arrhythmogenic cardiomyopathy; AF = atrial fibrillation; CHD = coronary heart disease; CVD = cardiovascular disease; FH = familial hypercholesterolemia; HCM = hypertrophic cardiomyopathy; LQTS = long QT syndrome. Adapted from Giudicessi, J.R., and Ackerman, M.J. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Translational Research 161(1), 1–14 (2013) with permission from Elsevier.19
Aortopathies
The thoracic aortopathies include a spectrum of heritable connective tissue disorders such as Marfan syndrome (MFS), Loeys-Dietz syndrome, and vascular Ehlers-Danlos syndrome, that largely arise secondary to dysregulated transforming growth factor-beta signaling and predispose affected individuals to aortic dilatation/aneurysm, premature death secondary to aortic dissection/rupture, and a host of variable overlapping cardiac (arrhythmia, valvular dysfunction, etc.) and extra-cardiac (ophthalmologic, orthopedic, etc.) manifestations.1, 2 In addition, thoracic aortic aneurysm and dissection without evident systemic connective tissue abnormalities can occur in families often in an autosomal dominant fashion. Mutations in several genes have been implicated in such syndromes of familial thoracic aortic aneurysm and dissection (FTAAD). Current commercially-available genetic testing panels cover ~16 aortopathy-susceptibility genes responsible for at least six distinct clinical entities as detailed in Table 1.
Table 1:
Current Recommendations and Clinical Utility of Commercially-Available Genetic Tests for Commonly Encountered Heritable Aortopathies and Cardiomyopathies
| Class | Disorder | Genes | Clinical Impact | Society Recommendationsb | Ref. |
|---|---|---|---|---|---|
| Aortopathies (systemic) |
vEDS | COL3A1c | Diagnosis | ACC/AHA/AATS and ESC guidelines: 1) Patients with suspected vEDS based on clinical criteria should be referred to a geneticist to facilitate diagnostic genetic testing for COL3A1 mutations (class I). 2) Mutation-specific cascade screening recommended if a bona fide disease-causative mutation identified (class I). |
20, 21 |
| LDS |
TGFBRIc, TGFBR2c, SMAD3, TGFB2, and TGFB3 |
Diagnosis | ACC/AHA/AATS and ESC guidelines: 1) Patients with suspected LDS based on clinical criteria should be referred to a geneticist to facilitate diagnostic genetic testing (class I). 2) Mutation-specific cascade screening recommended if a bona fide disease-causative mutation identified (class I). |
20, 21 | |
| MFS | FBNlc | Diagnosis Risk-stratification Management |
ACC/AHA/AATS and ESC guidelines: 1) Patients with suspected MFS based on revised Ghent nosology should be referred to a geneticist to facilitate diagnostic genetic testing (class I). 2) Mutation-specific cascade screening recommended if a bona fide disease-causative mutation identified (class I). |
20, 21 | |
| Aortopathies (non-systemic) |
FTAAD |
ACTA2, MAT2A, MYH11, MYLK, PRKG1, and TGFB2 |
Diagnosis | ACC/AHA/AATS and ESC guidelines:break 1) Known syndromic FTAADs (vascular EDS, LDS, and MFS) should be ruled out and the index case referred to a geneticist for consideration of diagnostic genetic testing (class I). 2) Mutation-specific cascade screening recommended if a bona fide disease-causative mutation identified (class I). |
20, 21 |
| Cardiomyopathies | ACM |
Major:
DSC2c, DSG2c, DSPc, and PKP2c Minor: JUP, RYR2, TGFB3, and TMEM43 |
Diagnosis Risk-stratification |
HRS/EHRA guidelines: 1) Genetic testing can be useful in patients who satisfy 2010 ESC task force diagnostic criteria (class IIa). 2) Mutation-specific cascade screening recommended (class I). 3) Genetic testing can be considered in patients with possible ACM based on 2010 ESC task force diagnostic criteria (class IIb).d |
6, 22 |
| DCM |
Autosomal DCM:~60+ genes identified to date. DCM with conduction disease: LMNAc and SCN5A X-linked DCM: DMD and TAZ |
Diagnosis Management |
HRS/EHRA guidelines: 1)Comprehensive or LMNA/SCN5A targeted genetic testing is recommended for all patients with DCM and significant cardiac conduction disease or family history of SCD (class I) 2) Mutation-specific cascade screening recommended (class I). 3) Comprehensive genetic testing can be useful in confirming the diagnosis of familial DCM and establishing a molecular target for cascade screening (class IIa).d |
6 | |
| HCMe |
Major:
MYBPC3c and MYH7c Minor: ACTC, ACTN2, ANKRD1, CSRP3, JPH2, LBD3, MYH6, MYL2, MYL3, MYOZ2, PLN, TNNC1, TNNI3, TNNT2c, TPM1, TTN, TCAP, and VCL |
Diagnosis Risk-stratification Management |
ACC/AHA. HRS/EHRA. and ESC guidelines: 1) Genetic testing recommended in patients fulfilling diagnostic criteria for or with signs/symptoms suggestive of HCM (class I). 2) Mutation-specific cascade screening recommended (class I). 3) Genetic testing should be considered in borderline cases after assessment by HCM specialist (class IIa).d 4) Genetic testing should be considered in deceased patients with pathologically confirmed HCM to facilitate cascade screening (class IIa). |
6,23,24 |
AATS = American Academy of Thoracic Surgery; ACC = American College of Cardiology; AHA = American Heart Association; ACM, arrhythmogenic cardiomyopathy; DCM = dilated cardiomyopathy; EHRA = European Heart Rhythm Association; ESC = European Society of Cardiology; FTAAD = familial thoracic aortic aneurysm and dissection; HCM = hypertrophic cardiomyopathy; LDS = Loeys-Dietz syndrome; MFS = Marfan syndrome; and vEDS = vascular Ehlers-Danlos syndrome.
Class of evidence indicated when available.
Denotes commonly encountered genes with strong evidence that disease-causative mutations within these genes contribute directly to disease pathogenesis.
Use of large gene panels in weak/borderline clinical cases substantially increases the risk of encountering a non-diagnostic VUS.
Denotes the most clinically useful monogenic cardiovascular genetic tests.
Although a relative paucity of clinically relevant genotype-phenotype correlations exist for the aortopathies, mutations within exons 24-32 of the MFS-causative FBN1 gene that encodes fibrillin-1 gene are associated with a form of atypically severe, early onset MFS classically referred to as neonatal MFS.25 Emerging evidence also suggests that MFS patients with haploinsufficient FBN1 mutations (truncating/frameshift mutations that fail to produce a protein product) are at greater risk for premature aortic events/cardiovascular death26, 27 and may be more responsive to angiotensin-receptor blockers28 than counterparts with dominant-negative FBN1 mutations (missense/exon-skipping mutations that yield an aberrantly functioning protein). As such, the clinical utility of genetic testing for these disorders is confined to 1) diagnostic confirmation in patients with a high pretest probability of disease (e.g. FBN1 testing in MFS patients that meet revised Ghent criteria), 2) cascade genetic screening starting with first-degree relatives to determine those who would benefit from imaging surveillance and those who can be dismissed potentially, and 3) to allow for differentiation between clinical entities given the degree of phenotypic overlap particularly since the current American College of Cardiology (ACC)/American Heart Association (AHA) Thoracic Aortic Disease recommendations regarding surveillance imaging and timing of surgical intervention differ between MFS, Loeys-Dietz, and vascular Ehlers-Danlos.20 An overview of available society recommendations and clinical utility of genetic testing for the heritable thoracic aortopathies are summarized succinctly in Table 1.
Cardiomyopathies
The inherited cardiomyopathies are a group of phenotypically and genetically heterogeneous heart failure- and sudden cardiac death (SCD)-predisposing CVDs that arise secondary to mutations in genes that encode key cardiomyocyte structural components (myofilaments, Z-disc, desmosome, etc.) and are classified by functional and morphologic features as arrhythmogenic cardiomyopathy (ACM; previously referred to as arrhythmogenic right ventricular cardiomyopathy/dysplasia), dilated cardiomyopathy (DCM), hypertrophic cardiomyopathy (HCM), left ventricular non-compaction (LVNC), or restrictive cardiomyopathy (RCM).3 Given the considerable phenotypic and genetic overlap between these cardiomyopathies, the use of commercial pan-cardiomyopathy gene panels has gained favor, particularly in cases where the addition of molecular insights afford an opportunity to further refine the clinical diagnosis. However, the use of large gene panels is likely best reserved for those individuals who have exhausted conventional cardiomyopathy-specific genetic testing as 1) nearly half of the > 60 cardiomyopathy-susceptibility genes identified to date lack sufficient evidence to be considered as bona fide disease-susceptibility genes and are considered “limited evidence genes”,29, 30 2) next-generation sequencing (NGS)-based panels further complicate the already difficult task of differentiating rare benign genetic variation from disease-causative mutations by enhancing the detection of low frequency benign variants, and 3) current society guidelines recommend comprehensive/targeted diagnostic screening of only those genes (e.g. myofilament-only for HCM, desmosome-only for ACM, etc.) commonly and or very strongly associated with the clinically suspected cardiomyopathy (Table 1).6, 23, 31
In general, the lack of disease-modifying therapies limits the clinical utility of cardiomyopathy genetic testing to diagnosis and prognostication/risk-stratification. However, there are some situations where genetic testing for patients with suspected cardiomyopathies does have direct and immediate therapeutic implications. For example, in HCM, phenocopies such as Fabry disease, a lysosomal storage disease responsible for up to ~3% of familial HCM in some populations, 32 are amenable to treatment with enzyme replacement therapy. In addition, patients with DCM secondary to mutations in either LMNA-encoded lamin A/C or DES-encoded desmin are at much higher risk for conduction disease and/or malignant arrhythmias/SCD.33-35 Here, genetic testing may guide the use of implantable cardioverter-defibrillators as primary prevention.
Unfortunately, due to a relative paucity of clinically relevant genotype-phenotype correlations, the clinical impact of genetic testing for ACM, DCM, LVNC, and RCM is confined largely to diagnostic confirmation for the proband and facilitation of cascade testing for the relatives at this time (Table 1). Although few clinically relevant gene- or mutation-specific genotype-phenotype correlations exist in HCM, multiple studies have demonstrated that those individuals with genotype-positive HCM (i.e. a positive genetic test), particularly those with mutations in the cardiac myofilaments, have a more severe clinical phenotype (younger age at diagnoses, more hypertrophy, higher rate of progression to New York Heart Association class III/IV heart failure, and risk of cardiovascular death) compared to patients with HCM but a negative genetic test (i.e. heretofore genotype-negative HCM).36–39 Furthermore, in comparison to individuals with thin-filament HCM (ACTC, TNNI3, TNNT2 and TPM1), those with thick myofilament HCM (MYH7 and MYBPC3) have more severe hypertrophy as well as higher rates of left ventricular outflow tract obstruction and progression to New York Heart Association class III/IV heart failure, but no difference in the risk of arrhythmia/SCD.40 Lastly, ~5–10% of individuals with HCM and ACM harbor >1 disease-causative mutation resulting in so-called compound or digenic heterozygosity that is associated typically with a more severe clinical phenotype.41–44
These observations, coupled with the massive discordance between the genotypic and phenotypic prevalence of many inherited cardiomyopathies unearthed by analysis of recent large-scale exome sequencing studies,45 suggest that 1) many genetic variants in cardiomyopathy-susceptibility genes, previously believed to be pathogenic, may be merely disease-modifiers or non-pathogenic altogether and/or 2) the classic monogenic/Mendelian autosomal-dominant mode of inheritance may be an oversimplification, particularly for ACM where a significant number of patients harbor >1 putative ACM-causative mutation,41, 42 suggesting that an oligogenic model, whereby > 2 hits in genes encoding the same functional unit (i.e. desmosome, sarcomere, etc.) are required to produce overt disease, may prove ultimately to be the best fit for even these “monogenic” forms of genetic heart disease (Figure 2).
In summary, mutation-specific cascade testing of appropriate relatives remains a class I recommendation for all inherited cardiomyopathies once a bona fide disease-causative mutation is identified in an index case. In contrast, owing to the genetic complexity of these disorders, the challenges associated with evaluating the pathogenicity of rare variants, and the paucity of clinically useful genotype-phenotype correlations at present, diagnostic genetic testing of the index case is a class I recommendation only for patients with either HCM or those with DCM and significant cardiac conduction disease (Table 1).6 It is anticipated that ongoing and future longitudinal studies that couple NGS-aided genotyping with in-depth clinical phenotyping may yield the additional insights necessary to further define the optimum role of genetic testing for patients with suspected ACM, DCM, LVNC, and RCM. Until then, recommendations for genetic testing for patients with these cardiomyopathies are class IIa/IIb (Table 1).6
Cardiac Channelopathies
The term cardiac channelopathies is used colloquially to describe a set of clinically and genetically diverse heritable cardiac arrhythmia syndromes, including Brugada syndrome (BrS), catecholaminergic polymorphic ventricular tachycardia (CPVT), and long QT syndrome (LQTS). These channelopathies collectively arise from defects in either critical cardiac ion channel macromolecular complexes or proteins critical for intracellular calcium-handling. Patients with a channelopathy typically have a structurally normal heart but are predisposed to arrhythmic syncope/seizures and SCD.5, 46 In excess of 40 channelopathy-susceptibility genes have been described to date with the majority of commercially-available genetic tests covering at least 20 of these genes through the use of disease-specific tests or comprehensive pan-channelopathy gene panels as outlined in Table 2.
Table 2:
Current Recommendations and Clinical Utility of Commercially-Available Genetic Tests for Commonly Encountered Heritable Cardiac Channelopathies and Familial Hypercholesterolemia
| Class | Disorder | Genes | Clinical Impact | Society Recommendationsb | Ref. | |
|---|---|---|---|---|---|---|
| Channelopathies | BrS |
Major:
SCN5A(BrS1)c Minor: ABCC9, CACNA1Cc, CACNA2D1, CACNB2, FGF12, GPD1L, HCN4, KCND2, KCND3, KCNE3, KCNE5, KCNJ8, PKP2 MOG1, SCN1B, SCN2B, SCN3B, SCN10A, SLMAP, SEMA3A, and TRPM4 |
Diagnosis Risk-stratification |
HRS/ERHA guidelines: 1) Mutation-specific cascade screening recommended (class I). 2) Genetic testing can be useful when there is clinical suspicion (class IIa).d |
6 | |
| CPVTe |
Major:
RYR2(CPVT1)c and CASQ2 (CPVT2)c Minor: CALM1, CALM2, CALM3, KCNJ2, and TRDN |
Diagnosis Risk-stratification |
HRS/EHRA guidelines: 1) Genetic testing recommended when there is clinical suspicion (class I). 2) Mutation-specific cascade screening recommended (class I). |
6 | ||
| LQTSe |
Major:
KCNQ1(LQT1)c, KNCH2(LQT2)c, and SCN5A (LQT3)c Minor: AKAP9, ANKB, CACNA1C, CALM1, CALM2, CALM3, CAV3, KCNE1, KCNE2, KCNJ2, KCNJ5, SCN4B, SNTA1 and TRDN |
Diagnosis Risk-stratification Management |
HRS/EHRA guidelines: 1)Genetic testingrecommended when there is either clinical suspicion or in asymptomatic patients with unexplained QT prolongation (QTc > 480 ms pre-puberty or QTc > 500 ms post-puberty, class I). 2) Mutation-specific cascade screening recommended (class I). 3) Genetic testing may be considered in asymptomatic patients with otherwise unexplained QT prolongation (QTc > 460 ms pre-puberty or QTc >480 ms post-puberty, class IIb)d |
6 | ||
| Familial Hypercholesterole mia |
FHe |
Major:
LDLRc, APOBc, and PCSK9c Minor: APOE, LDLRAPc, and STAP1 |
Diagnosis Risk-stratification |
AHA, EAS, and NLA statements/guidelines: 1) Genetic testing strongly recommended for individuals with a definite or probable diagnosis of FH based on validated clinical scorecards (DLCN, Simon-Broome, etc.). 2) Mutation-specific cascade screening provides a diagnostic gold-standard in families with an identified disease-causative mutation(s). |
8,47, 48 | |
AHA = American Heart Association; BrS = Brugada syndrome; CPVT = catecholaminergic polymorphic ventricular tachycardia; DLCN = Dutch Lipid Clinic Network; EAS = European Atherosclerosis Society; EHRA = European Heart Rhythm Association; FH = familial hypercholesterolemia; HRS = Heart Rhythm Society; LQTS = long QT syndrome; and NLA = National Lipid Association.
Class of evidence indicated when available.
Denotes commonly encountered genes with strong evidence that disease-causative mutations within these genes contribute directly to disease pathogenesis.
Use of large gene panels in weak/borderline clinical cases substantially increases the risk of encountering a non-diagnostic VUS.
Denotes the most clinically useful monogenic cardiovascular genetic tests.
Similar to the cardiomyopathies, the first-line use of commercial channelopathy panels should be approached with great caution as the inclusion of channelopathy-susceptibility genes without definitive clinical association and the enhanced detection of low frequency benign variants can confound the already difficult task of rare variant interpretation. Although the clinical utility of genetic testing in the diagnosis of cardiac channelopathies is well established and evidenced by recent Heart Rhythm Society (HRS)/European Heart Rhythm Association (EHRA) recommendations,6 LQTS, with its particularly robust genotype-phenotype correlations, represents one of the few monogenic CVDs where genetic testing facilitates a genomic/genetic-guided approach to both risk-stratification and treatment and is therefore the focus of the ensuing paragraphs.
Amongst clinically definitive LQTS cases (i.e. heart rate-correct QT interval > 480 msec and Schwartz score > 3.5),49 ~75% have a mutation in one of the three canonical LQTS-susceptibility genes, the KCNQl-encoded Kv7.1 potassium channel (LQT1, ~35%), the KCNH2-encoded Kv11.1/hERG potassium channel (LQT2, ~30%), or the SCN5A-encoded Nav1.5 sodium channel (LQT3, ~10%), with an additional ~5–10% expected to harbor mutations in the remaining 14 “minor” LQTS-susceptibility genes.50, 51 Even among the three major LQTS-susceptibility genes, there is a significant rate of background genetic noise (~2.7% of 60,000-plus individuals in Exome Aggregation Consortium cohort harbor rare amino acid-altering genetic variation in the major LQTS genes).13, 14 This complicates LQTS genetic test interpretation to the extent that occasional calls for universal LQTS genetic testing must be deemed ill-informed. However, when viewed in the context of an individual’s entire clinical picture (e.g. non-genetic risk factors such as age, gender, and degree of QT prolongation) and the established genotype-phenotype correlations (e.g. genotype-specific triggers and genotype-dependent responsiveness to primary therapy, i.e. beta blockers), the identification of a putative mutation in one of the major LQTS-susceptibility genes enables genotype-specific approaches to risk-stratification and clinical management.49 Unfortunately, a seemingly positive genetic test for a minor LQTS gene, with the notable exception of exceedingly rare multisystem forms of LQTS such as Timothy syndrome (CACNA1C), Andersen-Tawil Syndrome (KCNJ2), the Calmodulinopathies (CALM1–3), and Triadin Knockout Syndrome (TRDN),49‘ 52 does not carry the same weight as the major LQTS subtypes and thus contributes little-to-no significance on risk-stratification and clinical management.13
Current HRS/EHRA guidelines recommend (class I) LQTS genetic testing for any individual with a strong clinical suspicion of LQTS based on clinical/family history and electrocardiographic phenotype OR an asymptomatic individual with unexplained, serial QT prolongation (> 480 msec before puberty and > 500 msec after puberty.).6 Similarly, under current HRS/EHRA guidelines, comprehensive or targeted genetic testing for individuals with a strong clinical suspicion for CPVT is recommended, whereas targeted screening of the SCN5A-encoded Nav1.5 sodium channel in BrS can be useful in establishing a diagnosis in individuals with a strong clinical suspicion of disease based on clinical/family history and electrocardiographic phenotype.6 Lastly, given that many genotype-positive LQTS, BrS, and CPVT patients fail to manifest clinical/electrographic evidence of disease at baseline, mutation/variant-specific cascade screening of all at-risk relatives is recommended by HRS/EHRA following the identification of a bona fide channelopathy-susceptibility mutation in an index case.6
Familial Hypercholesterolemia
Familial hypercholesterolemia (FH) is a relatively common47, 53, 54 predominantly autosomal dominant disorder of lipid/lipoprotein metabolism characterized clinically by elevated low-density lipoprotein cholesterol levels (LDL-C), tendon (xanthoma) and corneal (corneal arcus) cholesterol deposition, and if untreated a high risk for premature atherosclerotic CVD.7 Due to the relatively high prevalence of FH across all racial/ethnic groups and devastating complications of unrecognized/untreated disease, FH is the only monogenic CVD that currently meets World Health Organization criteria for universal, population-based screening.8, 55 Although current National Heart Lung and Blood Institute (NHLBI) expert guidelines strongly recommend universal lipid screening for children ages 9 to 11 56, the rate of pre-pubertal lipid screening in the United States is low (~10%)57 and the optimal approach to universal FH screening remains undefined (i.e. lipid screening alone vs. lipid screening followed by family-based cascade genetic screening).8,57
In recent years, the recognition of a significant overlap in LDL-C levels between mutation-positive and mutation-negative relatives 58 as well as between individuals with heterozygous FH (single mutation in a FH-susceptibility gene)47, 53 and homozygous/compound heterozygous FH [>1 mutation in FH-susceptibility gene(s)]59 has led to an increased reliance on genetic testing. Specifically, the World Health Organization 60 and European Atherosclerosis Society47 both recommend family-based, genetic cascade screening to enhance diagnostic precision in FH once a mutation-positive index case is identified. The role of genetic testing in FH is further highlighted by the incorporation of genetic test results into the two most commonly employed sets of validated FH diagnostic clinical criteria, the Dutch Lipid Clinic Network 61 and Simon Broome Registry criteria.58
Although six FH-susceptibility genes have been discovered to date (Table 2), most commercially-available panels and expert recommendations focus on initial sequencing and deletion/duplication analysis for three key genes (LDLR, APOB, and PCSK9) that account for ~>90% of mutation-positive, clinically definite FH cases.47 Similar to other genetic disorders, the overall diagnostic yield of FH is dependent on the pre-test probability of disease (e.g. 63% for definite, 35% for probable, and 22% for possible FH based on Dutch Lipid Clinic Network criteria)62 and the major FH-susceptibility genes are also subject to an inherent rate of background genetic variation. Thus, it is imperative that FH genetic test results are cautiously interpreted in light of the aforementioned ACMG guidelines.17 As such, the primary clinical utility of FH genetic testing is 1) to confirm diagnosis/identify FH-causative mutation in those individuals with a definite/probable clinical diagnosis of FH (e.g. adults with LDL-C >190 mg/dL and children with LDL-C >160 mg/dL and personal/family history of premature atherosclerotic CVD or tendinous xanthomas) and 2) to facilitate cascade screening of first-, second-, and third-degree relatives of mutation-positive FH index cases.
Lastly, despite the identification of several potentially clinically relevant genotype-phenotype correlations shown to influence phenotypic severity 59, 63and statin-responsiveness,64 risk-stratification and clinical management decisions in FH are driven currently by LDL-C levels and therapeutic response. As our understanding of the genetic architecture of FH continues to evolve, particularly with regards to the ability of cholesterol-raising and cholesterol-lowering genetic variants with modest effect sizes to modulate the phenotypic expression of primary FH-causative mutations, it is anticipated that the role of genetics in tailoring individualized approaches to the risk-stratification and management of patients with FH will continue to grow.
Genetic Testing for Cardiovascular Diagnostic Odysseys
The term “diagnostic odyssey” refers to any patient or family with a suspected genetic disorder whereby the precise underlying clinical entity remains undifferentiated following standard genetic testing.65 With its ability to detect changes throughout the coding regions of the human genome and increasingly cost-effective nature, whole exome sequencing (WES) is proving to be an indispensable diagnostic clinical tool for elucidating the genetic etiology of diagnostic odysseys.66–68 In fact, WES has been successful in elucidating an underlying genetic basis for ~25% of diagnostic odysseys65, 69–71 and in numerous cases has led to the identification of novel monogenic/Mendelian CVD-susceptibility genes/genetic loci in patients with seemingly genotype-negative disease.66–68
In addition to the use of clinical WES to investigate cardiovascular diagnostic odysseys in the living, several groups have explored the utility of WES-based molecular autopsies (WEMA)72–77as a cost-effective means of screening cases of sudden unexplained death in the young (SUDY) for previously undiagnosed SCD-predisposing monogenic/Mendelian CVDs, such as the cardiac channelopathies and cardiomyopathies, that are often undetectable on autopsy and known to underlie ~25%-35% of SUDY cases.73, 76, 78 Although the majority of these studies, including a recent population-based prospective trial that utilized a 55 cardiac gene panel,76 identified clinically actionable variants and increased the overall diagnostic yield in SUDY,73-76the use of large cardiac gene panels to probe ambiguous phenotypes such as SUDY needs to be approached with caution. Not only do these gene panels contain a significant number of polymorphic genes that collectively have the ability to produce an overwhelming amount of background genetic noise, but SUDY phenotypes are often poorly defined making it next to impossible to interpret suspected SUDY-causative variants in the context of the pre-test probability of any single heritable CVD. As a result, the use of WEMAs is likely to unearth many ultra-rare VUS in potential SUDY-susceptibility genes and the bulk of these variants will lack the supporting evidence needed to definitively ascertain pathogenicity. One can envision how the results of a 50-100 gene WEMA could be misinterpreted easily by well-intentioned clinicians triggering the misguided cascade screening of at-risk relatives that ultimately may result in harmful diagnostic miscues. As such, a tiered approach to WEMA starting with those genes (KCNQ1, KCNH2, SCN5A, RYR2, PKP2, CALM1–3, etc.) most likely to harbor clinically actionable variants based on prior SUDY studies 74, 75 is advisable and consistent with current HRS/EHRA recommendations when performed in conjunction with a comprehensive postmortem examination and clinical/cardiologic evaluation of first degree relatives.6
Genetic Risk Scores (GRS) in the Prediction and Prevention of Polygenic (Non-Mendelian) Cardiovascular Disorders (CVDs)
In contrast to monogenic/oligogenic CVDs that typically arise from a rare, single gene mutation(s), many common CVDs, including AF, CHD, and hypertension, have a clear heritable component attributable to the collective contribution of multiple independent or interacting variants that in isolation account for a miniscule fraction of the complex trait or disease in question resulting in a so-called polygenic inheritance pattern (Figure 2).79, 80 Although no commercial genetic tests are available currently for so-called “polygenic” CVDs, the following paragraphs briefly examine ongoing efforts to translate bona fide CVD-associated single nucleotide polymorphisms (SNPs)/genetic loci discovered through GWAS into a clinically useful aggregate genetic risk score (GRS) that, in conjunction with traditional clinical risk factors, can enhance risk-stratification and prevention of polygenic CVDs.
Over the past decade, multi-cohort GWAS meta-analyses led by transatlantic consortia such as the AF Genetics (AFGen),12 Global Blood Pressure Genetics (BPgen),81 and Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM)11 consortia, have yielded a large number of CVD-associated SNPs/genetic loci that reach genome-wide significance. However, owing to the small effect size of each individual SNP, the clinical utility of individual SNPs to predict disease likelihood is modest.82 As a result, the concept of a “genetic risk score” (GRS) was conceived.83 Here, a panel of weighted or unweighted diseasemodifier SNPs is used to generate a single aggregate score that undergoes subsequent predictive modelling (i.e. area under the receiver operator curve or net reclassification improvement index) to determine if the GRS improves predictive capacity, and therefore may have clinical utility, incremental to traditional clinical risk factors.83
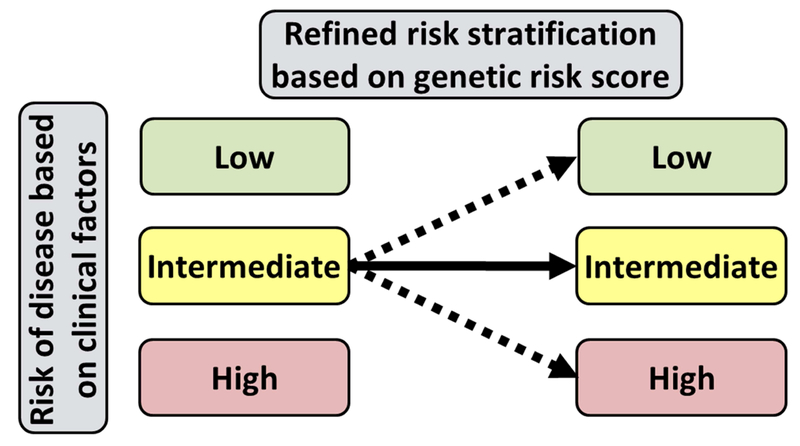
To date, several GRS studies have demonstrated a relatively modest, but statistically significant incremental predictive ability for incident AF 84, 85 and adverse CHD events.86-91 Furthermore, the recent Myocardial Infarction Genes (MI-GENES) clinical trial demonstrated that the incorporation of a CHD-GRS into a conventional risk prediction algorithm and subsequent disclosure of genetic risk for CHD to study participants led to lower LDL-C levels in comparison to disclosure of clinical risk factors alone.92 Furthermore, disclosure of CHD genetic risk did not induce significant patient anxiety.92 As such, the knowledge of an underlying genetic predisposition to common polygenic CVDs may lower the overall burden of CVD by prompting providers and patients to more aggressively address modifiable risk factors before disease onset (Figure 3). However, as the majority of CVD GWAS meta-analyses were conducted in cohorts of European ancestry, ongoing studies are needed to ascertain whether the current CVD-GRSs can be generalized to other racial/ethnic groups. Lastly, large prospective clinical trials are needed to determine if the use of a CVD-GRS can improve clinical outcomes, decrease disease burden, and lower healthcare costs.
Figure 3 |.
Refinement of clinical disease risk estimates with the use of genetic risk scores (GRS). Generalized schematic displaying how the use of an aggregate GRS can be used to refine the risk assessment in individuals at intermediate risk for disease based on clinical risk factors leading to earlier identification of individuals at increased risk of developing disease. GRS = genetic risk score.
Cardiovascular Pharmacogenomics
Drugs used in the prevention and treatment of CVD, including β-blockers, flecainide, clopidogrel, statins, and warfarin, account for manyof the most widely prescribed pharmacologic agents worldwide. Although these drugs are highly effective and safe at the population-level, individual patients occasionally display variability in the degree of efficacy and/or rate of adverse drug reactions. This clinical conundrum birthed the field of Pharmacogenomics, which aims to enhance the utility of available pharmacologic agents by linking variation in genes that govern a drug’s pharmacokinetic (effect of the body on drug concentration/tissue distribution) and pharmacodynamics (effect of the drug on body molecular/cellular/organ function) properties to inter-individual variability in drug response. Furthermore, the concept of pre-emptive pharmacogenomics, wherein individuals are genotyped for relevant pharmacogenomic genes/variants and actionable results are then placed in the electronic health record, with linkage to clinical decision support, is being pursued actively.93
Due to a relative paucity of RCTs aimed at defining the clinical utility of pharmacogenomics testing and the availability of alternative agents with decreased or no known pharmacogenomic liability, current society and regulatory agency recommendations/guidelines are variable in regards to the use of pharmacogenomics testing in clinical practice (Table 3). Although a thorough discussion of cardiovascular pharmacogenomics is outside the scope of this Review, current professional society and regulatory agency recommendations/guidelines pertaining to the use of pharmacogenomics testing when prescribing β-blockers, clopidogrel, statins, and warfarin are outlined in Table 3 and an expanded discussion of how pharmacogenomics data may be used to individualize cardiovascular drug and dosage selectionis contained within the online supplement.94-139
Table 3:
Expert Guidelines Regarding Utilization of Genotype-Guided Drug/Dose Selection in Cardiovascular Disease
| Class | Medication(s) | Gene(s) | Clinical Concern | Relevant Society/Regulatory Agency Recommendations |
CPIC Strength |
Ref. |
|---|---|---|---|---|---|---|
| Anti-platelet | Clopidogrel | CYP2C19 | Clinical efficacy in ACS patients undergoing PCI |
AHA/ACC recommendations: Due to lack of RCTs, the evidence base is insufficient to recommend routine CYP2C19 genotyping, but can be considered on a case-by-case basis for individuals at high-risk for poor outcomes. CPIC Dosing Guidelines: Ultra-rapid - Standard dosing Extensive - Standard dosing. Intermediate - Alternative agent. Poor - Alternative agent. FDA recommendations: Due to the estimated 2–14% of individuals who are poor metabolizers alternative dosing or anti-platelet agents should be considered. Clinical CYP2C19 genotyping is available to identify poor metabolizers |
Moderate -to- strong |
113, 141 |
| β-blockers | Carvedilol Metoprolol Propranolol |
ADRB1 CYP2D6 GRK4 GRK5 |
Therapeutic response |
No recommendations have been made by CPIC, AHA/ACC, or regulatory agencies. |
N/A | N/A |
| Statins | Simvastatin | SLCO1B1 | Statin-induced myopathy |
AHA/ACC recommendations: No formal recommendations. yCPIC Dosing Guidelines: Normal function - Standard starting dose. Intermediate function - Consider alternative. Low function - Consider alternative; CK surveillance. FDA recommendations: Simvastatin 80 mg should not be started in patients with a new clinical indication for a statin. |
Strong | 123 |
| Vitamin K antagonist |
Warfarin | CYP2C9 and VKORC1 |
Clinical efficacy and toxicity |
AHA/ACC recommendations: No formal recommendations. CPIC Dosing Guidelines: Recommend use of pharmacogenetic-guided warfarin dosing algorithms that account for CYP2C9 and VKORC1 genetic variation (e.g. http://www.warfarindosing.org). FDA recommendations: Recommend use of electronic (e.g. http://www.warfarindosing.org) or table genetic-guided algorithms that account for CYP2C9 and VKORC1 genetic variation to establish initial dosing when possible. |
Strong | 142 |
ACC = American College of Cardiology; ACS = acute coronary syndrome; AHA = American Heart Association; CK = creatinine kinase; CPIC = Clinical Pharmacogenomics Implementation Consortium; FDA = Food and Drug Administration; and PCI = percutaneous coronary intervention.
CONCLUSION
Although this Review details many examples of how genetic and pharmacogenomics testing has already led or will one day lead to genotype-guided approaches to the diagnosis, risk-stratification, and management of patients with an array of CVDs, the full promise of precision genomic medicine is far from being realized. As echoed throughout this Review, a number of significant barriers currently limit and complicate the use of genetic and pharmacogenomics testing in clinical practice. As such, it is imperative that innovative, well-designed, and adequately powered studies are undertaken to 1) elucidate genotype-phenotype correlations utilized in the development of individualized genotype-guided approaches to the risk-stratification and management of monogenic CVDs, 2) enhance the ability to distinguish pathogenic mutations from rare, benign, background genetic variants, 3) better define the genomic architecture of common CVDs, inter-individual response to common cardiovascular drugs, and the phenomena of incomplete penetrance and variable expressivity in monogenic CVDs, 4) determine the clinical utility of established GRSs and pharmacogenomics tests through prospective RCTs, and 5) implement precision cardiovascular genomic medicine at the point of care by integrating genetic/genomic testing results into the electronic health record with linkage to clinical decision support.140 When coupled with the development of educational resources to ensure that current and future cardiovascular providers are prepared to utilize evidence-based genetic/genomic approaches and ongoing support from broad proposals such as Precision Medicine Initiative, tackling these barriers should assure the future of precision cardiovascular medicine remains bright.
Supplementary Material
Acknowledgments
FUNDING SOURCES
This work was supported by the Windland Smith Rice Sudden Comprehensive Sudden Cardiac Death Program (to Dr. Ackerman). Dr. Giudicessi thanks the Mayo Clinic Internal Medicine Residency and Clinician Investigator Training Programs for fostering an outstanding environment for physician-scientist training. Dr. Kullo is supported by grant U01 HG06379 from the National Human Genome Research Institute (NHGRI).
Financial support and conflict of interest disclosure: M.J.A. is a consultant for Boston Scientific, Gilead Sciences, Invitae, Medtronic, MyoKardia, and St. Jude Medical. From 2004 through 2016, M.J.A. and Mayo Clinic received sales-based royalties from Transgenomic for their FAMILION-LQTS and FAMILION-CPVT genetic tests. However, none of these entities participated in this study. I.J.K. has received honoraria from Amgen. J.R.G. has nothing to disclose.
Abbreviations and Acronyms:
- ACC
American College of Cardiology
- ACMG
American College of Medical Genetics
- AF
atrial fibrillation
- AHA
American Heart Association
- ACM
arrhythmogenic cardiomyopathy
- BrS
Brugada syndrome
- CHD
coronary heart disease
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- CVD
cardiovascular disease
- DCM
dilated cardiomyopathy
- EHRA
European Heart Rhythm Association
- FH
familial hypercholesterolemia
- GRS
genetic risk score
- GWAS
genome-wide association study
- HCM
hypertrophic cardiomyopathy
- HRS
Heart Rhythm Society
- LDL-C
low-density lipoprotein C
- LQTS
long QT syndrome
- LVNC
left ventricular non-compaction
- MFS
Marfan syndrome
- NGS
next-generation sequencing
- PCI
percutaneous coronary intervention
- RCM
restrictive cardiomyopathy
- SCD
sudden cardiac death
- SNPs
single nucleotide polymorphisms
- SUDY
sudden unexplained death in the young
- VUS
variant of unknown significance
- WEMA
whole exome molecular autopsy.
Footnotes
SUPPLEMENTAL ONLINE MATERIAL Supplemental material can be found online at:
http://www.mavoclinicproceedings.org.Supplemental material attached to journal articles has not been edited, and the authors take responsibility for the accuracy of all data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Loeys BL, Schwarze U, Holm T, et al. Aneurysm Syndromes Caused By Mutations In The Tgf-Beta Receptor. NEngl JMed 2006;355(8):788–798. [DOI] [PubMed] [Google Scholar]
- 2.Paterick TE, Humphries JA, Ammar KA, et al. Aortopathies: Etiologies, Genetics, Differential Diagnosis, Prognosis And Management. Am J Med 2013;126(8):670–678. [DOI] [PubMed] [Google Scholar]
- 3.Watkins H, Ashrafian H, Redwood C. Inherited Cardiomyopathies. The New England journal of medicine. 2011;364(17):1643–1656. [DOI] [PubMed] [Google Scholar]
- 4.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 Accf/Aha Guideline For The Diagnosis And Treatment Of Hypertrophic Cardiomyopathy: Executive Summary: A Report Of The American College Of Cardiology Foundation/American Heart Association Task Force On Practice Guidelines. J Am Coll Cardiol 2011;58(25):2703–2738. [DOI] [PubMed] [Google Scholar]
- 5.Giudicessi JR, Ackerman MJ. Genetic Testing In Heritable Cardiac Arrhythmia Syndromes: Differentiating Pathogenic Mutations From Background Genetic Noise. Curr Opin Cardiol 2013;28(1):63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackerman MJ, Priori SG, Willems S, et al. Hrs/Ehra Expert Consensus Statement On The State Of Genetic Testing For The Channelopathies And Cardiomyopathies This Document Was Developed As A Partnership Between The Heart Rhythm Society (Hrs) And The European Heart Rhythm Association (Ehra). Heart Rhythm. 2011;8(8): 1308–1339. [DOI] [PubMed] [Google Scholar]
- 7.Marks D, Thorogood M, Neil HA, Humphries SE. A Review On The Diagnosis, Natural History, And Treatment Of Familial Hypercholesterolaemia. Atherosclerosis. 2003; 168(1): 1–14. [DOI] [PubMed] [Google Scholar]
- 8.Gidding SS, Champagne MA, de Ferranti SD, et al. The Agenda For Familial Hypercholesterolemia: A Scientific Statement From The American Heart Association.Circulation. 2015;132(22):2167–2192. [DOI] [PubMed] [Google Scholar]
- 9.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291(5507): 1304–1351. [DOI] [PubMed] [Google Scholar]
- 10.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- 11.Nikpay M, Goel A, Won HH, et al. A Comprehensive 1,000 Genomes-Based Genome-Wide Association Meta-Analysis Of Coronary Artery Disease. Nat Genet 2015;47(10): 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamin EJ, Rice KM, Arking DE, et al. Variants In Zfhx3 Are Associated With Atrial Fibrillation In Individuals Of European Ancestry. Nat Genet 2009;41(8):879–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ackerman MJ. Genetic Purgatory And The Cardiac Channelopathies: Exposing The Variants Of Uncertain/Unknown Significance Issue. Heart Rhythm. 2015; 12(11):2325–2331. [DOI] [PubMed] [Google Scholar]
- 14.Kapa S, Tester DJ, Salisbury BA, et al. Genetic Testing For Long-Qt Syndrome: Distinguishing Pathogenic Mutations From Benign Variants. Circulation. 2009;120(18):1752–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingles J, Semsarian C. Conveying a probabilistic genetic test result to families with an inherited heart disease. Heart Rhythm. 2014; 11(6): 1073–1078. [DOI] [PubMed] [Google Scholar]
- 16.Arscott P, Caleshu C, Kotzer K, et al. A Case for Inclusion of Genetic Counselors in Cardiac Care. Cardiol Rev 2016;24(2):49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards And Guidelines For The Interpretation Of Sequence Variants: A Joint Consensus Recommendation Of The American College Of Medical Genetics And Genomics And The Association For Molecular Pathology. Genet Med 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapplinger JD, Giudicessi JR, Ye D, et al. Enhanced Classification Of Brugada Syndrome-Associated And Long-Qt Syndrome-Associated Genetic Variants In The Scn5a-Encoded Na(V)1.5 Cardiac Sodium Channel. Circ Cardiovasc Genet 2015;8(4):582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giudicessi JR, Ackerman MJ. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res 2013; 161(1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiratzka LF, Bakris Gl, Beckman JA, et al. 2010 Accf/Aha/Aats/Acr/Asa/Sca/Scai/Sir/Sts/Svm Guidelines For The Diagnosis And Management Of Patients With Thoracic Aortic Disease. A Report Of The American College Of Cardiology Foundation/American Heart Association Task Force On Practice Guidelines, American Association For Thoracic Surgery, American College Of Radiology, American Stroke Association, Society Of Cardiovascular Anesthesiologists, Society For Cardiovascular Angiography And Interventions, Society Of Interventional Radiology, Society Of Thoracic Surgeons, And Society For Vascular Medicine. JAm Coll Cardiol 2010;55(14):e27–el29. [DOI] [PubMed] [Google Scholar]
- 21.Erbel R, Aboyans V, Boileau C, et al. 2014 Esc Guidelines On The Diagnosis And Treatment Of Aortic Diseases: Document Covering Acute And Chronic Aortic Diseases Of The Thoracic And Abdominal Aorta Of The Adult. The Task Force For The Diagnosis And Treatment Of Aortic Diseases Of The European Society Of Cardiology (Esc). European heart journal. 2014;35(41):2873–2926. [DOI] [PubMed] [Google Scholar]
- 22.Marcus FI, McKenna WJ, Sherrill D, et al. Diagnosis Of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia: Proposed Modification Of The Task Force Criteria. Eur Heart J. 2010;31(7):806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Authors/Task Force m, Elliott PM, Anastasakis A, et al. 2014 Esc Guidelines On Diagnosis And Management Of Hypertrophic Cardiomyopathy: The Task Force For The Diagnosis And Management Of Hypertrophic Cardiomyopathy Of The European Society Of Cardiology (Esc). Eur Heart J. 2014;35(39):2733–2779. [DOI] [PubMed] [Google Scholar]
- 24.American College of Cardiology Foundation/American Heart Association Task Force on Practice G, American Association for Thoracic S, American Society of E, et al. 2011 Accf/Aha Guideline For The Diagnosis And Treatment Of Hypertrophic Cardiomyopathy: Executive Summary: A Report Of The American College Of Cardiology Foundation/American Heart Association Task Force On Practice Guidelines. JThorac Cardiovasc Surg 2011;142(6): 1303–1338. [DOI] [PubMed] [Google Scholar]
- 25.Tiecke F, Katzke S, Booms P, et al. Classic, Atypically Severe And Neonatal Marfan Syndrome: Twelve Mutations And Genotype-Phenotype Correlations In Fbn1 Exons 24–40. Eur J Hum Genet 2001;9(1): 13–21. [DOI] [PubMed] [Google Scholar]
- 26.Baudhuin LM, Kotzer KE, Lagerstedt SA. Increased Frequency Of Fbn1 Truncating And Splicing Variants In Marfan Syndrome Patients With Aortic Events. Genet Med 2015;17(3): 177–187. [DOI] [PubMed] [Google Scholar]
- 27.Franken R, Groenink M, de Waard V, et al. Genotype Impacts Survival In Marfan Syndrome. Eur Heart J. 2016. [DOI] [PubMed] [Google Scholar]
- 28.Franken R, den Hartog AW, Radonic T, et al. Beneficial Outcome Of Losartan Therapy Depends On Type Of Fbn1 Mutation In Marfan Syndrome. Circ Cardiovasc Genet 2015;8(2):383–388. [DOI] [PubMed] [Google Scholar]
- 29.Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited Cardiomyopathies: Molecular Genetics And Clinical Genetic Testing In The Postgenomic Era. J Mol Diagn 2013; 15(2): 158–170. [DOI] [PubMed] [Google Scholar]
- 30.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-Based Variation In Cardiomyopathy Genes. Circ Cardiovasc Genet 2012;5(4):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 Accf/Aha Guideline For The Diagnosis And Treatment Of Hypertrophic Cardiomyopathy: A Report Of The American College Of Cardiology Foundation/American Heart Association Task Force On Practice Guidelines. Circulation. 2011;124(24):e783–831. [DOI] [PubMed] [Google Scholar]
- 32.Havndrup O, Christiansen M, Stoevring B, et al. Fabry Disease Mimicking Hypertrophic Cardiomyopathy: Genetic Screening Needed For Establishing The Diagnosis In Women. Eur J Heart Fail. 2010;12(6):535–540. [DOI] [PubMed] [Google Scholar]
- 33.Meune C, Van Berlo JH, Anselme F, Bonne G, Pinto YM, Duboc D. Primary Prevention Of Sudden Death In Patients With Lamin A/C Gene Mutations. NEngl J Med 2006;354(2):209–210. [DOI] [PubMed] [Google Scholar]
- 34.van Rijsingen IA, Arbustini E, Elliott PM, et al. Risk Factors For Malignant Ventricular Arrhythmias In Lamin A/C Mutation Carriers A European Cohort Study. J Am Coll Cardiol 2012;59(5):493–500. [DOI] [PubMed] [Google Scholar]
- 35.van Tintelen JP, Van Gelder IC, Asimaki A, et al. Severe Cardiac Phenotype With Right Ventricular Predominance In A Large Cohort Of Patients With A Single Missense Mutation In The Des Gene. Heart Rhythm. 2009;6(11):1574–1583. [DOI] [PubMed] [Google Scholar]
- 36.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament Protein Gene Mutation Screening And Outcome Of Patients With Hypertrophic Cardiomyopathy. Mayo Clin Proc 2008;83(6):630–638. [DOI] [PubMed] [Google Scholar]
- 37.Andersen PS, Havndrup O, Hougs L, et al. Diagnostic Yield, Interpretation, And Clinical Utility Of Mutation Screening Of Sarcomere Encoding Genes In Danish Hypertrophic Cardiomyopathy Patients And Relatives. HumMutat 2009;30(3):363–370. [DOI] [PubMed] [Google Scholar]
- 38.Millat G, Bouvagnet P, Chevalier P, et al. Prevalence And Spectrum Of Mutations In A Cohort Of 192 Unrelated Patients With Hypertrophic Cardiomyopathy. Eur J Med Genet 2010;53(5):261–267. [DOI] [PubMed] [Google Scholar]
- 39.Bos JM, Will ML, Gersh BJ, Kruisselbrink TM, Ommen SR, Ackerman MJ. Characterization Of A Phenotype-Based Genetic Test Prediction Score For Unrelated Patients With Hypertrophic Cardiomyopathy. Mayo Clin Proc 2014;89(6):727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppini R, Ho CY, Ashley E, et al. Clinical Phenotype And Outcome Of Hypertrophic Cardiomyopathy Associated With Thin-Filament Gene Mutations. J Am Coll Cardiol 2014;64(24):25 89–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christensen AH, Benn M, Bundgaard H, Tybjaerg-Hansen A, Haunso S, Svendsen JH. Wide Spectrum Of Desmosomal Mutations In Danish Patients With Arrhythmogenic Right Ventricular Cardiomyopathy. J Med Genet 2010;47(11):736–744. [DOI] [PubMed] [Google Scholar]
- 42.Bhonsale A, Groeneweg JA, James CA, et al. Impact Of Genotype On Clinical Course In Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy-Associated Mutation Carriers. Eur Heart J. 2015;36(14):847–855. [DOI] [PubMed] [Google Scholar]
- 43.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin Binding Protein C Mutations And Compound Heterozygosity In Hypertrophic Cardiomyopathy. J Am Coll Cardiol 2004;44(9): 1903–1910. [DOI] [PubMed] [Google Scholar]
- 44.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound And Double Mutations In Patients With Hypertrophic Cardiomyopathy: Implications For Genetic Testing And Counselling. J Med Genet 2005;42(10):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andreasen C, Nielsen JB, Refsgaard L, et al. New Population-Based Exome Data Are Questioning The Pathogenicity Of Previously Cardiomyopathy-Associated Genetic Variants. Eur J Hum Genet 2013;21(9):918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giudicessi JR, Ackerman MJ. Potassium-Channel Mutations And Cardiac Arrhythmias--Diagnosis And Therapy. Nat Rev Cardiol 2012;9(6):319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordestgaard BG, Chapman MJ, Humphries SE, et al. Familial Hypercholesterolaemia Is Underdiagnosed And Undertreated In The General Population: Guidance For Clinicians To Prevent Coronary Heart Disease: Consensus Statement Of The European Atherosclerosis Society. Eur Heart J. 2013;34(45):3478–3490a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg AC, Hopkins PN, Toth PP, et al. Familial Hypercholesterolemia: Screening, Diagnosis And Management Of Pediatric And Adult Patients: Clinical Guidance From The National Lipid Association Expert Panel On Familial Hypercholesterolemia. J Clin Lipidol 2011;5(3 Suppl):S1–8. [DOI] [PubMed] [Google Scholar]
- 49.Giudicessi JR, Ackerman MJ. Genotype- And Phenotype-Guided Management Of Congenital Long Qt Syndrome. Curr Probl Cardiol 2013;38(10):417–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Splawski I, Shen J, Timothy KW, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102(10):1178–1185. [DOI] [PubMed] [Google Scholar]
- 51.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2(5):507–517. [DOI] [PubMed] [Google Scholar]
- 52.Giudicessi JR, Ackerman MJ. Calcium Revisited: New Insights Into The Molecular Basis Of Long-Qt Syndrome. Circ Arrhythm Electrophysiol 2016;9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neil HA, Hammond T, Huxley R, Matthews DR, Humphries SE. Extent Of Underdiagnosis Of Familial Hypercholesterolaemia In Routine Practice: Prospective Registry Study. BMJ. 2000;321(7254): 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Safarova MS, Liu H., and Kullo IJ Rapid Identification Of Familial Hypercholesterolemia From Electronic Health Records: The Search Study J Clin Lipidol 2016;in press( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Safarova MS, Kullo IJ. My Approach To The Patient With Familial Hypercholesterolemia. Mayo Clin Proc 2016;91(6):770–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Expert Panel on Integrated Guidelines for Cardiovascular H, Risk Reduction in C, Adolescents, National Heart L, Blood I. Expert Panel On Integrated Guidelines For Cardiovascular Health And Risk Reduction In Children And Adolescents: Summary Report. Pediatrics. 2011; 128 Suppl 5(S213–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margolis KL, Greenspan LC, Trower NK, et al. Lipid Screening In Children And Adolescents In Community Practice: 2007 To 2010. Circ Cardiovasc Qual Outcomes. 2014;7(5):718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starr B, Hadfield SG, Hutten BA, et al. Development Of Sensitive And Specific Age- And Gender-Specific Low-Density Lipoprotein Cholesterol Cutoffs For Diagnosis Of First-Degree Relatives With Familial Hypercholesterolaemia In Cascade Testing. Clin Chem Lab Med 2008;46(6):791–803. [DOI] [PubMed] [Google Scholar]
- 59.Sjouke B, Kusters DM, Kindt I, et al. Homozygous Autosomal Dominant Hypercholesterolaemia In The Netherlands: Prevalence, Genotype-Phenotype Relationship, And Clinical Outcome. Eur Heart J. 2015;36(9):560–565. [DOI] [PubMed] [Google Scholar]
- 60.Green RF, Dotson WD, Bowen S, Kolor K, Khoury MJ. Genomics In Public Health: Perspective From The Office Of Public Health Genomics At The Centers For Disease Control And Prevention (Cdc). Healthcare (Basel). 2015;3(3):830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umans-Eckenhausen MA, Defesche JC, Scheerder RL, Cline F, Kastelein JJ. [Tracing Of Patients With Familial Hypercholesterolemia In The Netherlands]. Ned Tijdschr Geneeskd. 1999;143(22): 1157–1161. [PubMed] [Google Scholar]
- 62.Damgaard D, Larsen ML, Nissen PH, et al. The Relationship Of Molecular Genetic To Clinical Diagnosis Of Familial Hypercholesterolemia In A Danish Population. Atherosclerosis. 2005;180(1): 155–160. [DOI] [PubMed] [Google Scholar]
- 63.Guardamagna O, Restagno G, Rolfo E, et al. The Type Of Ldlr Gene Mutation Predicts Cardiovascular Risk In Children With Familial Hypercholesterolemia. J Pediatr 2009;155(2): 199–204 e192. [DOI] [PubMed] [Google Scholar]
- 64.Choumerianou DM, Dedoussis GV. Familial Hypercholesterolemia And Response To Statin Therapy According To Ldlr Genetic Background. Clin Chem Lab Med 2005;43(8):793–801. [DOI] [PubMed] [Google Scholar]
- 65.Lazaridis KN, Schahl KA, Cousin MA, et al. Outcome Of Whole Exome Sequencing For Diagnostic Odyssey Cases Of An Individualized Medicine Clinic: The Mayo Clinic Experience. Mayo Clin Proc 2016;91(3):297–307. [DOI] [PubMed] [Google Scholar]
- 66.Boczek NJ, Best JM, Tester DJ, et al. Exome Sequencing And Systems Biology Converge To Identify Novel Mutations In The L-Type Calcium Channel, Cacna1c, Linked To Autosomal Dominant Long Qt Syndrome. Circ Cardiovasc Genet 2013;6(3):279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shameer K, Klee EW, Dalenberg AK, Kullo IJ. Whole Exome Sequencing Implicates An Ino80d Mutation In A Syndrome Of Aortic Hypoplasia, Premature Atherosclerosis, And Arterial Stiffness. Circ Cardiovasc Genet 2014;7(5):607–614. [DOI] [PubMed] [Google Scholar]
- 68.Altmann HM, Tester DJ, Will ML, et al. Homozygous/Compound Heterozygous Triadin Mutations Associated With Autosomal-Recessive Long-Qt Syndrome And Pediatric Sudden Cardiac Arrest: Elucidation Of The Triadin Knockout Syndrome. Circulation. 2015;131(23):2051–2060. [DOI] [PubMed] [Google Scholar]
- 69.Yang Y, Muzny DM, Xia F, et al. Molecular Findings Among Patients Referred For Clinical Whole-Exome Sequencing. JAMA. 2014;312(18):1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee H, Deignan JL, Dorrani N, et al. Clinical Exome Sequencing For Genetic Identification Of Rare Mendelian Disorders. JAMA. 2014;312(18): 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y, Muzny DM, Reid JG, et al. Clinical Whole-Exome Sequencing For The Diagnosis Of Mendelian Disorders. NEngl J Med 2013;369(16): 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loporcaro CG, Tester DJ, Maleszewski JJ, Kruisselbrink T, Ackerman MJ. Confirmation Of Cause And Manner Of Death Via A Comprehensive Cardiac Autopsy Including Whole Exome Next-Generation Sequencing. Arch Pathol Lab Med 2014;138(8): 1083–1089. [DOI] [PubMed] [Google Scholar]
- 73.Bagnall RD, Das KJ, Duflou J, Semsarian C. Exome Analysis-Based Molecular Autopsy In Cases Of Sudden Unexplained Death In The Young. Heart Rhythm. 2014;11(4):655–662. [DOI] [PubMed] [Google Scholar]
- 74.Narula N, Tester DJ, Paulmichl A, Maleszewski JJ, Ackerman MJ. Post-Mortem Whole Exome Sequencing With Gene-Specific Analysis For Autopsy-Negative Sudden Unexplained Death In The Young: A Case Series. Pediatr Cardiol 2015;36(4):768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson JH, Tester DJ, Will ML, Ackerman MJ. Whole-Exome Molecular Autopsy After Exertion-Related Sudden Unexplained Death In The Young. Circ Cardiovasc Genet 2016;9(3):259–265. [DOI] [PubMed] [Google Scholar]
- 76.Bagnall RD, Weintraub RG, Ingles J, et al. A Prospective Study Of Sudden Cardiac Death Among Children And Young Adults. NEngl J Med 2016;374(25):2441–2452. [DOI] [PubMed] [Google Scholar]
- 77.Torkamani A, Muse ED, Spencer EG, et al. Molecular Autopsy For Sudden Unexpected Death. JAMA. 2016;316(14): 1492–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tester DJ, Medeiros-Domingo A, Will ML, Haglund CM, Ackerman MJ. Cardiac Channel Molecular Autopsy: Insights From 173 Consecutive Cases Of Autopsy-Negative Sudden Unexplained Death Referred For Postmortem Genetic Testing. Mayo Clin Proc 2012;87(6):524–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding K, Kullo IJ. Genome-Wide Association Studies For Atherosclerotic Vascular Disease And Its Risk Factors. Circ Cardiovasc Genet 2009;2(1):63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kullo IJ, Cooper LT. Early Identification Of Cardiovascular Risk Using Genomics And Proteomics. Nat Rev Cardiol 2010;7(6):309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-Wide Association Study Identifies Eight Loci Associated With Blood Pressure. Nat Genet 2009;41(6):666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wray NR, Goddard ME, Visscher PM. Prediction Of Individual Genetic Risk To Disease From Genome-Wide Association Studies. Genome Res 2007;17(10):1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith JA, Ware EB, Middha P, Beacher L, Kardia SL. Current Applications Of Genetic Risk Scores To Cardiovascular Outcomes And Subclinical Phenotypes. Curr Epidemiol Rep 2015;2(3): 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.j/>Tada H, Shiffman D, Smith JG, et al. Twelve-Single Nucleotide Polymorphism Genetic Risk Score Identifies Individuals At Increased Risk For Future Atrial Fibrillation And Stroke. Stroke. 2014;45(10):2856–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel Genetic Markers Improve Measures Of Atrial Fibrillation Risk Prediction. Eur Heart J. 2013;34(29):2243–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morrison AC, Bare LA, Chambless LE, et al. Prediction Of Coronary Heart Disease Risk Using A Genetic Risk Score: The Atherosclerosis Risk In Communities Study. Am J Epidemiol 2007;166(1):28–35. [DOI] [PubMed] [Google Scholar]
- 87.Ripatti S, Tikkanen E, Orho-Melander M, et al. A Multilocus Genetic Risk Score For Coronary Heart Disease: Case-Control And Prospective Cohort Analyses. Lancet. 2010;376(9750): 13931400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thanassoulis G, Peloso GM, Pencina MJ, et al. A Genetic Risk Score Is Associated With Incident Cardiovascular Disease And Coronary Artery Calcium: The Framingham Heart Study. Circ Cardiovasc Genet 2012;5(1): 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ganna A, Magnusson PK, Pedersen NL, et al. Multilocus Genetic Risk Scores For Coronary Heart Disease Prediction. Arterioscler Thromb Vasc Biol 2013;33(9):2267–2272. [DOI] [PubMed] [Google Scholar]
- 90.Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic Risk Prediction And A 2-Stage Risk Screening Strategy For Coronary Heart Disease. Arterioscler Thromb Vasc Biol 2013;33(9):2261–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mega JL, Stitziel NO, Smith JG, et al. Genetic Risk, Coronary Heart Disease Events, And The Clinical Benefit Of Statin Therapy: An Analysis Of Primary And Secondary Prevention Trials. Lancet. 2015;385(9984):2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kullo IJ, Jouni H, Austin EE, et al. Incorporating A Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect On Low-Density Lipoprotein Cholesterol Levels (The Mi-Genes Clinical Trial). Circulation. 2016;133(12):1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive Genotyping For Personalized Medicine:Design Of The Right Drug, Right Dose, Right Time-Using Genomic Data To Individualize Treatment Protocol. Mayo Clin Proc 2014;89(1):25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ismail R, Teh LK. The Relevance Of Cyp2d6 Genetic Polymorphism On Chronic Metoprolol Therapy In Cardiovascular Patients. J Clin Pharm Ther 2006;31(1):99–109. [DOI] [PubMed] [Google Scholar]
- 95.Goryachkina K, Burbello A, Boldueva S, Babak S, Bergman U, Bertilsson L. Cyp2d6 Is A Major Determinant Of Metoprolol Disposition And Effects In Hospitalized Russian Patients Treated For Acute Myocardial Infarction. Eur J Clin Pharmacol 2008;64(12): 1163–1173. [DOI] [PubMed] [Google Scholar]
- 96.Giessmann T, Modess C, Hecker U, et al. Cyp2d6 Genotype And Induction Of Intestinal Drug Transporters By Rifampin Predict Presystemic Clearance Of Carvedilol In Healthy Subjects. Clin Pharmacol Ther 2004;75(3):213–222. [DOI] [PubMed] [Google Scholar]
- 97.Baudhuin LM, Miller WL, Train L, et al. Relation Of Adrb1, Cyp2d6, And Ugt1a1 Polymorphisms With Dose Of, And Response To, Carvedilol Or Metoprolol Therapy In Patients With Chronic Heart Failure. Am J Cardiol 2010;106(3):402–408. [DOI] [PubMed] [Google Scholar]
- 98.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, et al. A Polymorphism Within A Conserved Beta(1)-Adrenergic Receptor Motif Alters Cardiac Function And Beta-Blocker Response In Human Heart Failure. Proc Natl Acad Sci U S A. 2006;103(30): 11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pacanowski MA, Gong Y, Cooper-DeHoff RM, et al. Beta-Adrenergic Receptor Gene Polymorphisms And Beta-Blocker Treatment Outcomes In Hypertension. Clinical Pharmacology & Therapeutics. 2008;84(6):715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aleong RG, Sauer WH, Sauer WH, et al. Prevention Of Atrial Fibrillation By Bucindolol Is Dependent On The Beta(1)389 Arg/Gly Adrenergic Receptor Polymorphism. JACCHeart Fail. 2013;1(4):338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-Adrenergic Receptor Polymorphisms And Antihypertensive Response To Metoprolol. Clin Pharmacol Ther 2003;74(1):44–52. [DOI] [PubMed] [Google Scholar]
- 102.Biolo A, Clausell N, Santos KG, et al. Impact Of Beta1-Adrenergic Receptor Polymorphisms On Susceptibility To Heart Failure, Arrhythmogenesis, Prognosis, And Response To Beta-Blocker Therapy. Am J Cardiol 2008;102(6):726–732. [DOI] [PubMed] [Google Scholar]
- 103.Bhatnagar V, O’Connor DT, Brophy VH, et al. G-Protein-Coupled Receptor Kinase 4 Polymorphisms And Blood Pressure Response To Metoprolol Among African Americans: Sex-Specificity And Interactions. Am JHypertens. 2009;22(3):332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vandell AG, Lobmeyer MT, Gawronski BE, et al. G Protein Receptor Kinase 4 Polymorphisms: Beta-Blocker Pharmacogenetics And Treatment-Related Outcomes In Hypertension. Hypertension. 2012;60(4):957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muskalla AM, Suter PM, Saur M, Nowak A, Hersberger M, Krayenbuehl PA. G-Protein Receptor Kinase 4 Polymorphism And Response To Antihypertensive Therapy. Clin Chem 2014;60(12): 1543–1548. [DOI] [PubMed] [Google Scholar]
- 106.Liggett SB, Cresci S, Kelly RJ, et al. A Grk5 Polymorphism That Inhibits Beta-Adrenergic Receptor Signaling Is Protective In Heart Failure. Nat Med 2008; 14(5):510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cresci S, Kelly RJ, Cappola TP, et al. Clinical And Genetic Modifiers Of Long-Term Survival In Heart Failure. J Am Coll Cardiol 2009;54(5):432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hulot JS, Bura A, Villard E, et al. Cytochrome P450 2c19 Loss-Of-Function Polymorphism Is A Major Determinant Of Clopidogrel Responsiveness In Healthy Subjects. Blood.2006; 108(7): 2244–2247. [DOI] [PubMed] [Google Scholar]
- 109.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines For Cyp2c19 Genotype And Clopidogrel Therapy: 2013 Update. Clin Pharmacol Ther 2013;94(3):317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mega JL, Simon T, Collet JP, et al. Reduced-Function Cyp2c19 Genotype And Risk Of Adverse Clinical Outcomes Among Patients Treated With Clopidogrel Predominantly For Pci: A MetaAnalysis. JAMA. 2010;304(16): 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. Cyp2c19 Genotype, Clopidogrel Metabolism, Platelet Function, And Cardiovascular Events: A Systematic Review And Meta Analysis. JAMA. 2011;306(24):2704–2714. [DOI] [PubMed] [Google Scholar]
- 112.Jneid H, Anderson JL, Wright RS, et al. 2012 Accf/Aha Focused Update Of The Guideline For The Management Of Patients With Unstable Angina/Non-St-Elevation Myocardial Infarction (Updating The 2007 Guideline And Replacing The 2011 Focused Update): A Report Of The American College Of Cardiology Foundation/American Heart Association Task Force On Practice Guidelines. JAm Coll Cardiol 2012;60(7):645–681. [DOI] [PubMed] [Google Scholar]
- 113.Holmes DR Jr., Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol 2010;56(4):321–341. [DOI] [PubMed] [Google Scholar]
- 114.Tailored Antiplatelet Therapy Following PCI (TAILOR-PCI). https://clinicaltrials.gov/ct2/show/NCT01742117: http://clinicaltrials.gov; 2013.
- 115.Assessment of Prospective CYP2C19 Genotype Guided Dosing of Anti-Platelet Therapy in Percutaneous Coronary Intervention (ADAPT). https://clinicaltrials.gov/ct2/show/NCT02508116: http://clinicaltrials.gov; 2015.
- 116.PRAsugrel or clopIdogrel in Acute Coronary SyndromE Patients With CYP2C19 Polymorphism Before Percutaneous Coronary Intervention (PRAISE-GENE).https://clinicaltrials.gov/ct2/show/NCT01641510: http://clinicaltrials.gov; 2012.
- 117.Ticagrelor or Prasugrel Versus Clopidogrel in Elderly Patients With an Acute Coronary Syndrome and a High Bleeding Risk: Optimization of Antiplatelet Treatment in High-risk Elderly (POPular AGE). https://clinicaltrials.gov/ct2/show/NCT02317198: http://clinicaltrials.gov; 2014.
- 118.Pereira NL, Sargent DJ, Farkouh ME, Rihal CS. Genotype-Based Clinical Trials In Cardiovascular Disease. Nat Rev Cardiol 2015;12(8):475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wiviott SD, Braunwald E, McCabe CH, et al. Prasugrel Versus Clopidogrel In Patients With Acute Coronary Syndromes. NEngl J Med 2007;357(20):2001–2015. [DOI] [PubMed] [Google Scholar]
- 120.Wallentin L, Becker RC, Budaj A, et al. Ticagrelor Versus Clopidogrel In Patients With Acute Coronary Syndromes. N Engl J Med 2009;361(11): 1045–1057. [DOI] [PubMed] [Google Scholar]
- 121.DiNicolantonio JJ, D’Ascenzo F, Tomek A, Chatterjee S, Niazi AK, Biondi-Zoccai G.Clopidogrel Is Safer Than Ticagrelor In Regard To Bleeds: A Closer Look At The Plato Trial. International Journal of Cardiology. 2013;168(3): 1739–1744. [DOI] [PubMed] [Google Scholar]
- 122.Buettner C, Rippberger MJ, Smith JK, Leveille SG, Davis RB, Mittleman MA. Statin Use And Musculoskeletal Pain Among Adults With And Without Arthritis. The American journal of medicine. 2012;125(2): 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ramsey LB, Johnson SG, Caudle KE, et al. The Clinical Pharmacogenetics Implementation Consortium Guideline For Slco1b1 And Simvastatin-Induced Myopathy: 2014 Update. Clinical pharmacology and therapeutics. 2014;96(4):423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Link E, Parish S, Armitage J, et al. Slco1b1 Variants And Statin-Induced Myopathy--A Genomewide Study. The New England journal of medicine. 2008;359(8):789–799. [DOI] [PubMed] [Google Scholar]
- 125.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms In Oatp-C: Identification Of Multiple Allelic Variants Associated With Altered Transport Activity Among European- And African-Americans. J Biol Chem 2001;276(38):35669–35675. [DOI] [PubMed] [Google Scholar]
- 126.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. Slcolbl Polymorphism Markedly Affects The Pharmacokinetics Of Simvastatin Acid. Pharmacogenet Genomics. 2006;16(12):873–879. [DOI] [PubMed] [Google Scholar]
- 127.Niemi M Transporter Pharmacogenetics And Statin Toxicity. Clin Pharmacol Ther 2010;87(1):130–133. [DOI] [PubMed] [Google Scholar]
- 128.Voora D, Shah SH, Spasojevic I, et al. The Slco1b1*5 Genetic Variant Is Associated With Statin-Induced Side Effects. JAm Coll Cardiol 2009;54(17):1609–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Danik JS, Chasman DI, MacFadyen JG, Nyberg F, Barratt BJ, Ridker PM. Lack Of Association Between Slco1b1 Polymorphisms And Clinical Myalgia Following Rosuvastatin Therapy. Am Heart J. 2013;165(6): 1008–1014. [DOI] [PubMed] [Google Scholar]
- 130.Genetically Guided Statin Therapy. https://clinicaltrials.gov/ct2/show/NCT018942302013.
- 131.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-Warfarin Metabolism Catalysed By The R144c Allelic Variant Of Cyp2c9. Pharmacogenetics. 1994;4(1):39–42. [DOI] [PubMed] [Google Scholar]
- 132.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association Of Polymorphisms In The Cytochrome P450 Cyp2c9 With Warfarin Dose Requirement And Risk Of Bleeding Complications. Lancet. 1999;353(9154):717–719. [DOI] [PubMed] [Google Scholar]
- 133.Rost S, Fregin A, Ivaskevicius V, et al. Mutations In Vkorc1 Cause Warfarin Resistance And Multiple Coagulation Factor Deficiency Type 2. Nature. 2004;427(6974):537–541. [DOI] [PubMed] [Google Scholar]
- 134.Rieder MJ, Reiner AP, Gage BF, et al. Effect Of Vkorc1 Haplotypes On Transcriptional Regulation And Warfarin Dose. NEngl J Med 2005;352(22):2285–2293. [DOI] [PubMed] [Google Scholar]
- 135.Cooper GM, Johnson JA, Langaee TY, et al. A Genome-Wide Scan For Common Genetic Variants With A Large Influence On Warfarin Maintenance Dose. Blood. 2008;112(4): 1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mega JL, Walker JR, Ruff CT, et al. Genetics and the clinical response to warfarin and edoxaban: findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet. 2015;385(9984):2280–2287. [DOI] [PubMed] [Google Scholar]
- 137.Verhoef TI, Ragia G, de Boer A, et al. A Randomized Trial Of Genotype-Guided Dosing Of Acenocoumarol And Phenprocoumon. N Engl J Med 2013;369(24):2304–2312. [DOI] [PubMed] [Google Scholar]
- 138.Pirmohamed M, Burnside G, Eriksson N, et al. A Randomized Trial Of Genotype-Guided Dosing Of Warfarin. NEngl J Med 2013;369(24):2294–2303. [DOI] [PubMed] [Google Scholar]
- 139.Kimmel SE, French B, Kasner SE, et al. A Pharmacogenetic Versus A Clinical Algorithm For Warfarin Dosing. NEngl J Med 2013;369(24):2283–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kullo IJ, Jarvik GP, Manolio TA, Williams MS, Roden DM. Leveraging The Electronic Health Record To Implement Genomic Medicine. Genet Med 2013;15(4):270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clinical pharmacology and therapeutics. 2013;94(3):317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clinical pharmacology and therapeutics. 2011;90(4):625–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.