SUMMARY
Extracellular RNAs (exRNAs) can be released by numerous cell types in vitro, are often protected within vesicles, and can modify recipient cell function. To determine how the composition and cellular sources of exRNAs and the extracellular vesicles (EVs) that carry them change in vivo during tissue inflammation, we analyzed bronchoalveolar lavage fluid (BALF) from mice before and after lung allergen challenge. In the lung, extracellular microRNAs (ex-miRNAs) had a composition that was highly correlated with airway-lining epithelium. Using cell type-specific membrane tagging and single vesicle flow, we also found that 80% of detected vesicles were of epithelial origin. After the induction of allergic airway inflammation, miRNAs selectively expressed by immune cells, including miR-223 and miR-142a, increased and hematopoietic-cell-derived EVs also increased >2-fold. These data demonstrate that infiltrating immune cells release ex-miRNAs and EVs in inflamed tissues to alter the local extracellular environment.
Graphical Abstract
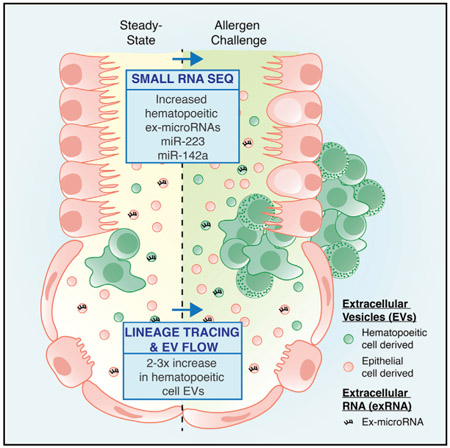
In Brief
Using small RNA sequencing and cell type-specific membrane tracing coupled with single vesicle flow, Pua et al. demonstrate that immune cell miRNAs and extracellular vesicles increase in airway fluid during allergic inflammation. This provides insight into how infiltrating immune cells alter the local extracellular environment during tissue immune responses.
INTRODUCTION
Body fluids, including serum/plasma, urine, saliva, bronchoalveolar lining fluid, and cerebrospinal fluid, are routine clinical specimens. These biofluids, which are rich in metabolites, proteins, and lipids, are also now recognized to carry extracellular nucleic acids, including microRNAs (miRNAs). Cellular miRNAs are non-coding ~22 nucleotide RNAs that function within cells as post-transcriptional inhibitors of gene expression, modulating networks of downstream target genes (Bartel, 2018). Extracellular miRNAs (ex-miRNAs) are produced by a wide range of cells and have proposed roles in normal physiology as well as disease states (Mateescu et al., 2017). They can be transferred between cells, including cells of the immune system (Alexander et al., 2015; Momen-Heravi et al., 2015; Montecalvo et al., 2012). The export of miRNAs can be selective (Cha et al., 2015; Santangelo et al., 2016; Shurtleff et al., 2017; Villarroya-Beltri et al., 2013), and recent studies provide experimental evidence that miRNA transfer may affect target cell behavior in immunologic processes in vivo, including sepsis (Alexander et al., 2015) and atherosclerosis (Li et al., 2015).
In body fluids, ex-miRNAs have been most extensively studied within the fluid compartment of the blood (serum and plasma). Ex-miRNAs in the blood are stably packaged, occurring in extracellular vesicles (EVs) (Hunter et al., 2008), in protein complexes (Arroyo et al., 2011), and/or in association with lipoprotein particles (Vickers et al., 2011). They are apparently derived from circulating hematopoietic cells (Pritchard et al., 2012) and from tissues such as the placenta during pregnancy (Williams et al., 2013) and adipose tissue (Thomou et al., 2017). A distinct set of ex-miRNAs has also been identified as being associated with the respiratory tract in induced sputum and bronchoalveolar lavage fluid (BALF) (Maes et al., 2016; Seumois et al., 2012), as well as the EV-enriched 100,000 g pelleted fraction from BALF (Levänen et al., 2013). However, for disease processes in the lung, many fundamental questions remain to be answered. Determining the cellular sources and composition of EVs and exRNAs in vivo are essential first steps in understanding their function in pathologic processes in this organ. Additional work on exRNA packaging and target cell delivery will be necessary to harness their clinical potential.
In the present study, we show that ex-miRNAs are abundant in airway lining fluid (as measured through bronchoalveolar washes), but they have a composition that is distinct from that of serum. We find that the miRNA composition of BALF largely mirrors that of the local tissue, in particular, the mucosal epithelium lining the airways. We demonstrate that EVs are present within BALF and that ex-miRNAs are associated with vesicleenriched fractions. Coupling a membrane fluorescent protein reporter system with EV flow, we characterize the sources and heterogeneity of EVs in the lung. Finally, we identify increases in ex-miRNAs selectively expressed as intracellular miRNAs in hematopoietic cells and a corresponding increase in the number of hematopoietic cell-derived EVs in a mouse model of allergic airway disease.
RESULTS
Abundant Ex-miRNAs in Airway Lining Fluid
To determine the composition of ex-miRNAs in the airways, BALF was collected as a single 1-mL wash from mice. miRNA sequencing libraries were generated from cell-free BALF based on previous literature with minor modifications (Williams et al., 2013), and the data were processed using the exceRpt small RNA-sequencing (RNA-seq) pipeline for alignment, annotation, and counting (http://www.genboree.org/site/, developed by the Data Integration and Analysis Component of the Extracellular RNA Communication Consortium). Modification to the published sequencing protocol included the addition of 5 random nucleotides at the ligating ends of both the 5′ and 3′ adapters. Addition of random nucleotides can reduce ligation bias, which is particularly problematic with short invariant sequences such as miRNAs (Jayaprakash et al., 2011; Zhuang et al., 2012).
In addition to BALF, 3 more specimens were collected and submitted for sequencing (Figure 1A). Serum was collected and sequenced as a second biofluid containing ex-miRNAs. We also sequenced miRNAs from the cellular sources we hypothesized as being the most likely to contribute to BALF ex-miRNAs: lung epithelial cells and hematopoietic cells in the airways. Bronchial epithelial cells were collected from airway brushings, and hematopoietic cells from the airways were collected as 300-g cellular pellets from BALF washes. An average of 550,000 miRNA reads were obtained from each sample (Table S1). In most cases, miRNAs constituted >80% (range, 69%–95%) of all mapped sequencing reads, with the remainder of the reads being composed predominantly of mRNA (average of 5%), rRNA (average of 4%), and tRNA (average of 3%) fragments. Circular RNAs and PIWI-interacting RNAs (piRNAs) each accounted for <0.5% of the total aligned reads. Overall, no striking qualitative differences in the distribution of small RNA types were observed between the sequenced samples (Figure 1B).
Figure 1. Ex-miRNAs Are Abundant in BALF and Reflect the Composition of the Local Tissue Environment.
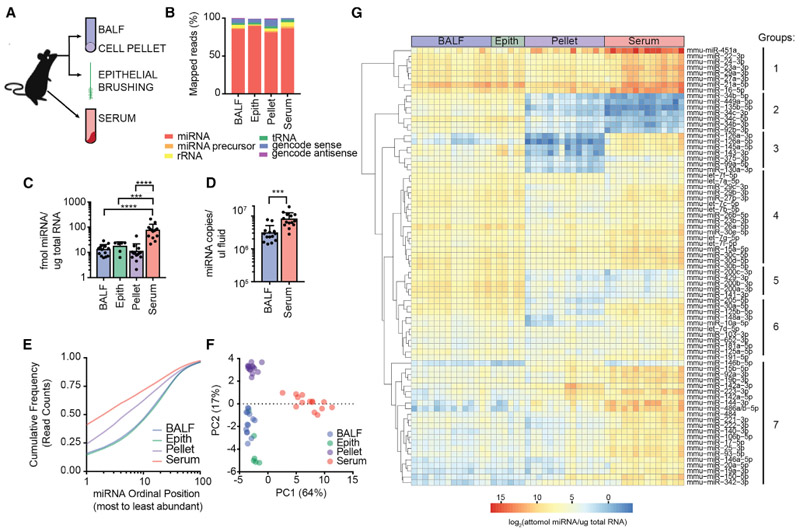
(A) Schematic of extracellular and cellular specimens collected for miRNA sequencing.
(B) Average percentage of mapped reads by RNA class.
(C) Calibrator-calculated amount of miRNAs per total input RNA for each specimen type (n = 6–14, 1-way ANOVA with Tukey’s multiple comparison test).
(D) Calibrator-calculated molecules of miRNA per input volume of biofluids (n = 14, unpaired t test).
(E) Cumulative distribution frequency of reads for top 100 expressed miRNAs for each sample type.
(F) Principle component analysis of the 100 most variant miRNAs in sequenced samples.
(G) Heatmap of calibrator-calculated miRNA amount per total input RNA for top 50 expressed miRNAs from each sample type. Clustering of sample types was supervised and clustering of miRNAs was unsupervised.
Error bars are mean + SD; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
See also Figures S1, S2, and S3 and Table S1.
During library preparation, a mixture of 5 calibrator sequences were spiked into the ligation reaction at a known concentration. These calibrators are miRNA-like synthetic oligoribonucleotides with 5′-monophosphates and no matches to the mouse genome (Williams et al., 2013). The total number of miRNA reads from each sample could then be estimated as a molar concentration of miRNAs sequenced per input of total RNA using mapped calibrator reads (Figure S1). In BALF, epithelial brushings and hematopoietic cell-rich pellets on the order of 10 fmol of miRNAs were sequenced per microgram of input total RNA (Figure 1C). This was significantly less than the amount that was sequenced from serum, which averaged closer to 100 fmol of miRNAs per microgram of input total RNA. Bioanalyzer tracings of total RNA isolated from both biofluids showed similar size distributions and a predominance of small RNA species (Figure S2). In contrast, cellular RNA contained intact large ribosomal RNA subunits. Therefore, despite the similar miRNA content per microgram of total RNA in BALF and cellular samples, these data indicate that BALF contains little or no intact cellular RNA. Rather, it suggests that blood and airway biofluids may contain different relative amounts of other small RNAs or RNA fragments that were not sequenced by this library preparation protocol.
Using our calibrators and average input volumes for RNA purification, we also calculated the number of ex-miRNAs per unit volume in the 2 biofluids examined. We calculate an average of 3 million total miRNAs per microliter of BALF and 8 million total miRNAs per microliter of serum (Figure 1D). Estimates of the volume of epithelial lining fluid in the lung suggest that the process of bronchoalveolar lavage dilutes this biofluid 10- to 100- fold (Dargaville et al., 1999; Rennard et al., 1986). Therefore, it seems likely that on a per-volume basis, the airway lining fluid has at least as many miRNAs as the blood.
A relatively small number of unique miRNAs contributed the majority of sequenced reads in the miRNAome of each specimen type (Figure 1E). The most highly expressed miRNA in each specimen type (miR-21a-5p in BALF and epithelial brushings, and miR-451a in serum and pellet) constituted 15%–41% of all reads, and the top 30 miRNAs made up 76%–83% of all of the reads sequenced. Thus, although sequenced reads mapped to >400 individual annotated miRNAs in all of the samples, there were a limited number of abundant miRNAs in both body fluids and cells.
Epithelial Cell miRNAs Dominate BALF Ex-miRNA Composition in Lungs
Principal-component analysis of the top 100 most variant miRNAs showed strong clustering of individual samples by specimen type (Figure 1F). Three clearly distinct clusters emerged from this analysis: a serum cluster, a hematopoietic cell-rich pellet cluster, and a mixed BALF and epithelial brushing cluster. Principal-component 1, which accounted for 64% of variance, separated serum from all of the other specimen types. Principal-component 2, which accounted for 17% of variance, separated the cell pellet from the mixed BALF and epithelial group. Consistent with the principal-component analysis, BALF and epithelial brushings had the strongest correlation coefficients (Pearson’s r = 0.97) when the top 200 most abundant miRNAs were used for comparison (Figure S3). The BALF ex-miRNA profile was much more poorly correlated with that of serum (Pearson’s r = 0.52) and intermediately correlated with miRNAs in the hematopoietic cell-rich pellets from bronchial washings (Pearson’s r = 0.72). This analysis shows that miRNAs expressed in BALF are more similar to cells present in and lining the airways than the other biofluid sequenced in this study—serum.
To visualize the absolute expression of the most abundant miRNAs sequenced, clustering was performed for only the top 50 expressed miRNAs from each specimen type. Based on cumulative frequency calculations (Figure 1E), this constituted ≥80% of the total reads sequenced in each sample. After supervising the clustering of samples by specimen type, unsupervised hierarchical clustering of miRNAs was performed using calibrator normalized attomols of miRNA per microgram of input total RNA. Among all of the specimen types, there were shared abundant (group 1), intermediate (group 4), and lower (group 6) expressed groups of miRNAs. Notable among these shared abundant miRNAs (group 1) were miR-21a-5p, all 3 members of the Mirc11 cluster of miRNAs (miR-23a, miR-27a, and miR-24-3p), miR-22-3p, and miR-29a-3p (Figure 1G). There were also 2 miRNAs known to be highly expressed in red blood cells that were the most abundant in serum, miR-451a and miR-16-5p. Visual inspection of the heatmap showed a coordinated intensity of miR-451a, miR-16-5p, and miR144-3p within select individual samples, and likely represents a signature of red blood cell lysis (Kirschner et al., 2011; Pritchard et al., 2012).
This analysis also highlighted a subset of miRNAs that were present at roughly equivalent levels in BALF and epithelial brushings, but were absent (group 2) to low (group 5) in both serum and hematopoietic cell-rich bronchoalveolar lavage cell pellets (Figure 1G). Notable among these were multiple members of the miR-34/449 family of miRNAs, which are highly expressed in ciliated epithelium, including lung epithelium, and are required for proper cilia formation and function (Marcet et al., 2011; Song et al., 2014). Multiple members of the miR-141/200/429 family were also present in this group. These miRNAs are highly expressed in the lung (Hasuwa et al., 2013), and deficiency of some family members such as miR-200b result in altered lung function in the mouse (Khoshgoo et al., 2017). Similarly, there was a group of miRNAs that were more highly abundant in serum and/or hematopoietic cell-rich pellets than in BALF or epithelial brushings (group 7). Included among these were miRNAs with preferential expression in the immune system and roles in immune cell differentiation and function (Chen et al., 2004; Landgraf et al., 2007; Monticelli et al., 2005). For example, miR-150-5p and miR-142a both are important for lymphocyte development (Kramer et al., 2015; Xiao et al., 2007), and miR-223 regulates myeloid cell differentiation and activation (Johnnidis et al., 2008). These data indicate that airway epithelial cells are by far the greatest contributor to the abundant ex-miRNA pool present in the airway lining fluid of lungs.
Ex-miRNAs in the BALF Are Protected from Degradation
Free RNA molecules are notoriously unstable due to the ubiquity of RNases that induce their rapid degradation. In blood, endogenous ex-miRNAs have been shown to be surprisingly stable (Arroyo et al., 2011). To test whether endogenously produced ex-miRNAs in the BALF are protected from degradation, RNA spike in experiments were performed. Calibrators from miRNA sequencing were once again used, this time to test the stability of naked RNAs in BALF. When 60 million copies of calibrator 1 and 600,000 copies of calibrator 2 were spiked into 200 μL BALF after the addition of Trizol in preparation for RNA extraction, these calibrators could be detected at 24 and 31 cycles, respectively, in quantitative RT-PCR analyses (Figure 2A). However, when these calibrators were spiked into BALF and incubated for 5 min at 37°C before the addition of Trizol reagent, both were undetectable. The pre-addition of an RNase inhibitor significantly protected these synthetic miRNAs from degradation. These data show that endogenous ex-miRNAs are stabilized in epithelial lining fluid in the lung and protected from the activity of endogenous RNases. RNases have been documented in the BALF and may arise in part from alveolar macrophages located there (Cormier et al., 2002).
Figure 2. Ex-miRNA Are Stable and Protected from RNases in BALF.
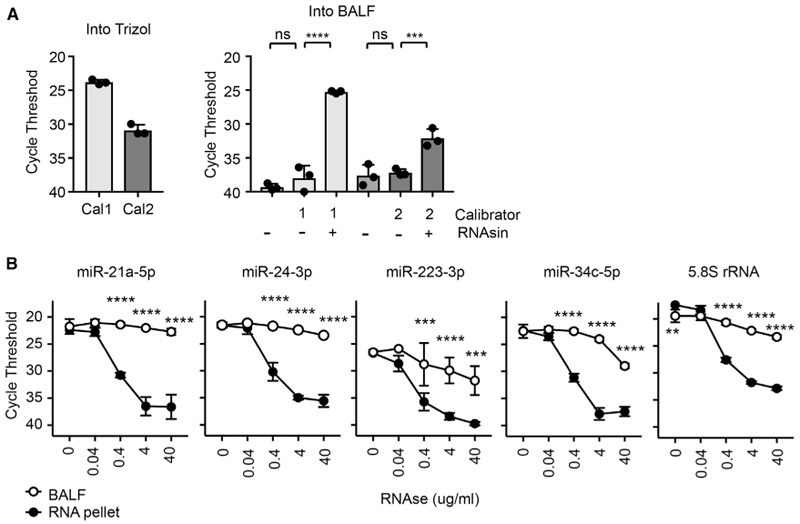
(A) qPCR of synthetic miRNAs (“calibrators” Cal1 and Cal2) spiked into Trizol or BALF with or without an RNase inhibitor (RNAsin) (n = 3 from 1–2 independent experiments, 1-way ANOVA with Bonferroni’s multiple comparison test).
(B) qPCR of analysis of miRNAs from BALF and RNA pellets treated with RNaseA (n = 3 from 3 independent experiments, 2-way ANOVA with Bonferroni’s multiple comparison test).
Error bars are mean + SD; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To confirm the stability of ex-miRNAs present in lung lining fluid, freshly isolated BALF from mice was obtained and incubated with additional RNase A. Two highly abundant miRNAs, miR-21a-5p and miR-24-3p, were protected from RNase A degradation in the BALF when compared to RNA previously extracted from the same BALF (Figure 2B). This was also true for miRNAs in the BALF that were preferentially expressed in the serum and inflammatory cell pellet (miR-223-3p) or in epithelial cells (miR-34c-5p), as well as for 5.8SrRNA (Figure 2B). In all of the cases, these RNAs were rapidly degraded when RNase A was added to RNA previously extracted from the BALF. These data confirm that ex-miRNAs in the BALF are present in a protected form.
EVs Are Present in the BALF
Given the stability of miRNAs in the BALF, we hypothesized that EVs present in lung lining fluid may carry miRNAs. To characterize EVs present in the lung, nanoparticle tracking was performed. On the order of 108 particles were present per microliter of this diluted biofluid. Although not statistically significantly different due to a single high outlier among the tested serum samples, the median number of particles in serum appeared to be higher, at close to 109 particles per microliter (Figure 3A). Nanoparticle tracking allows the assessment of particle size in addition to concentration. The size distribution of particles in the BALF was larger than that seen in the serum, with a mode of ~125 nm in BALF and ~90 nm in serum (Figure 3B). These results show that abundant sub-200-nm particles are present in both biofluids, which is consistent with the presence of small EVs.
Figure 3. EVs Are Present in BALF.
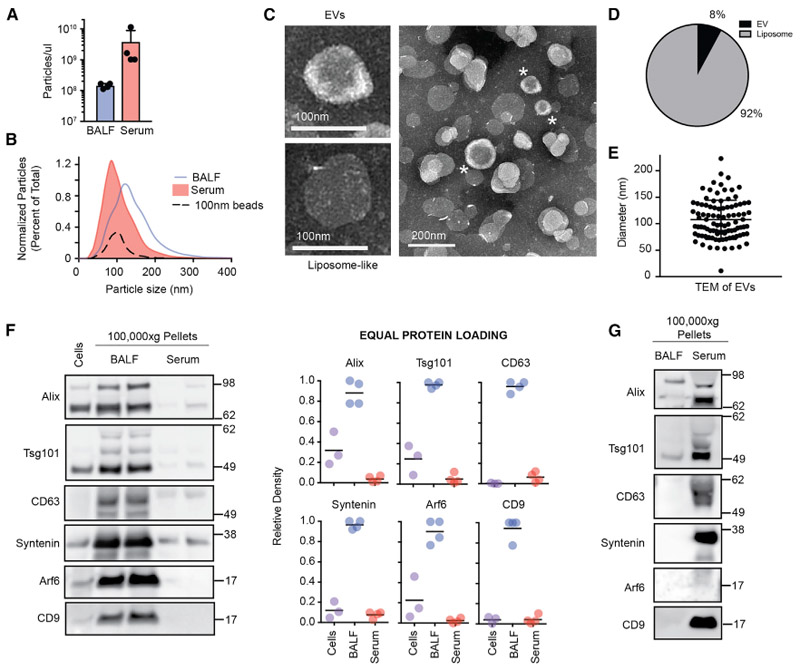
(A) Concentration of particles in BALF and serum by nanoparticle tracking assays. Each data point represents the average of 3 technical replicate readings for a single mouse (n = 4, 2-tailed t test).
(B) Size distribution of particles in BALF and serum in the same mice as (A); 100 nm beads are included as a positive control.
(C) Example transmission electron micrograph of EV-like and liposome-like vesicles observed in BALF.
(D) Percentage of morphologically distinct structures in BALF (n > 400 vesicles from 5 images).
(E) Size distribution of EV-like vesicles identified by transmission electron microscopy (TEM) from BALF (n = 96 vesicles).
(F) Western blot analysis and quantification of EV markers after loading 10 μg of cell lysate, 100,000 g pellet from BALF, and 100,000 g pellet from serum (n = 4 from 2 independent experiments).
(G) Western blot analysis after loading 1 × 109 particles from 100,000 g pellets of BALF and serum. Data representative of 2 independent experiments.
To visualize EVs present in the BALF, we performed transmission electron microscopy of negatively stained 100,000 g BALF pellets that had been pre-cleared of debris, as well as largersized EVs and aggregates by prior serial centrifugation at 1,200 and 10,000 × g. We observed 2 discrete populations of vesicles. By morphology, ~90% of the vesicles appeared liposome-like with a uniform electron density, variable size, and irregular cell border (Figures 3C and 3D). These structures are interpreted to be derived from the disruption of the surfactant lipid monolayer lining the lung during the process of lavage. In contrast, just under 10% of the vesicles had a morphology that was consistent with that of EVs. This second group of vesicles had a donut-shaped appearance of electron density with proteins visibly studding the surface and an average size of ~100 nm diameter (99 nm median, 108 nm mean) (Figures 3C–3E). These EV particles share similarities in morphologies to exosome-like vesicles present in human plasma (Caby et al., 2005).
To determine whether EVs present in the BALF express EV-associated protein markers, western blots of 100,000 g pellets were performed after differential centrifugation. BALF from this pelleted fraction contained EV membrane organizers, including CD9 and CD63, as well as proteins associated with EV biogenesis, including Alix, Tsg101, Syntenin, and Arf6 (Figure 3F). When equal total protein was loaded, markers were more highly enriched in 100,000 g BALF pellets than lysates either collected from hematopoietic cells in the airway or 100,000 g pellets collected from serum. However, when equal numbers of particles from BALF and serum 100,000 g pellets were compared, EV markers were more highly expressed in the serum (Figure 3G). These differences suggest that EVs present in these 2 biofluids exist amid different compositions of free proteins and lipid particles in airways and the circulation. In summary, data from nanoparticle tracking, electron microscopy, and western blot analysis demonstrate that among particles recovered from cell-free BALF, small EVs are present.
Select Ex-miRNAs Are Changed in the BALF during Allergic Inflammation
Asthma is a prevalent disease that affects >8% of the US population (Moorman et al., 2012) and causes dramatic changes in the lungs of affected individuals. In the Th2-high endotype of asthma, these changes include the recruitment of a mixed type 2 inflammatory infiltrate with eosinophils to the lung parenchyma and bronchoalveolar space, as well as metaplasia of bronchial epithelium and stromal remodeling (Fahy, 2015). Given these changes in cell composition and cell function, we hypothesized that ex-miRNAs present in the lung lining fluid would change during asthmatic inflammation. To model allergic-type asthmatic responses with recruitment of type 2 inflammatory cells to the lung, we used an antigen sensitization and challenge model. Mice were sensitized to the model antigen ovalbumin (OVA) via intraperitoneal immunization with the adjuvant alum and then challenged by aspiration of OVA. We and others have shown that after 3 days of OVA challenge, a robust type 2 immune response is induced in the lungs with recruitment of Th2 cells and eosinophils to the airways and mucus metaplasia of bronchial epithelium (Nials and Uddin, 2008; Pua et al., 2016).
The miRNA sequencing performed on each specimen type (see Figure 1) included samples from 3 experimental groups (Figure 4A). One cohort of mice (CTR) was neither sensitized by intraperitoneal injection nor challenged in the lung. One cohort of mice (IMM) was immunized intraperitoneally with OVA and alum but challenged in the lung with PBS only. The last cohort of mice (AA) was immunized with both OVA and alum intraperitoneally and then challenged in the lung with OVA to recruit a local type 2 lung inflammatory response. When the calibrator-calculated amount of total miRNAs sequenced was compared between CTR, IMM, and AA mice for each sample type, no statistically significant difference in the miRNA content was observed (Figure 4B). In addition, no differences in the concentration of ex-miRNAs per volume of biofluid were detected in BALF between the 3 experiment groups, and there was overall only a trend toward slightly increased ex-miRNA in the serum of IMM or AA mice when compared with CTR (Figure 4C).
Figure 4. Select Ex-miRNAs Are Increased in Lung after the Induction of Allergic Airway Inflammation.
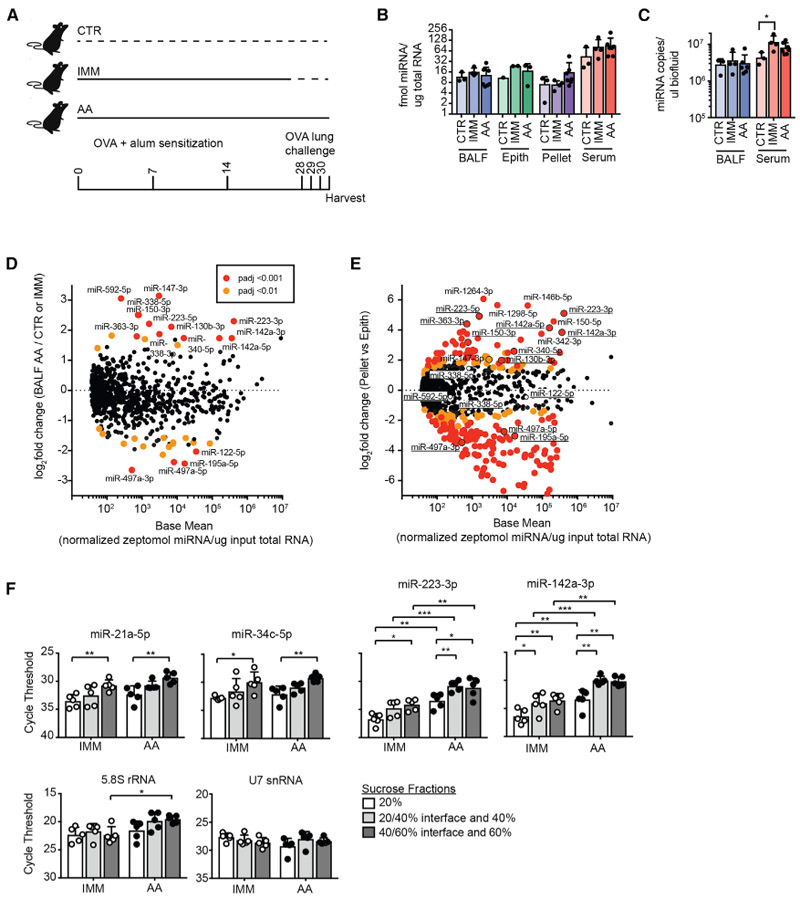
(A) Schematic of 3 experimental groups sequenced: CTR, IMM, and AA.
(B) Calibrator-calculated amount of miRNA per total input RNA for each specimen type (n = 1–7, 2-way ANOVA with Tukey’s multiple comparison test for comparing between CTR, IMM, and AA in each specimen type).
(C) Calibrator-calculated molecules of ex-miRNAs per input volume of biofluids (n = 3–7, 2-way ANOVA with Tukey’s multiple comparison test for comparing between CTR, IMM, and AA in each specimen type).
(D and E) MA plots comparing base mean expression of miRNAs across all specimen and sample types to log2 fold change of ex-miRNA in BAL in AA versus CTR/IMM (D) or in AA pellet versus AA epithelium (E). Outlined points with underlined miRNAs in (E) were significantly upregulated or downregulated in (D) (padj < 0.001).
(F) qPCR for ex-miRNAs and other extracellular small RNAs in 20%, 40%, and 60% sucrose fractions after flotation of 100,000 g pellets from BALF isolated from IMM and AA mice (n = 5 from 2 independent experiments, 2-way ANOVA with Tukey’s multiple comparison test for comparing among fractions and Sidak’s multiple comparison test for comparing between IMM and AA in 1 fraction).
Error bars are mean + SD; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. See also Tables S2, S3, and S4.
However, allergic airway inflammation induced major changes in the abundance of specific miRNAs in airway-lining fluid. To identify changed miRNAs after the induction of a type 2 inflammatory response in the lungs, differential expression analysis was performed to compare the BALF of AA to CTR and IMM mice (Table S2). Since no differentially expressed miRNAs were observed between IMM and CTR BALF (Table S3), these samples were grouped for comparison to AA samples. A total of 12 miRNAs were upregulated in the BALF of mice with allergic airway inflammation having high statistical significance (padj < 0.001) (Figure 4D; Table S4). Among the most abundant of these miRNAs were miR-223-3p and miR-142a (both 5p and 3p). Since these miRNAs are highly and preferentially expressed in hematopoietic cells (Chen et al., 2004; Landgraf et al., 2007; Monticelli et al., 2005), we next performed a differential expression analysis between hematopoietic cell pellets and epithelial brushings collected from AA mice. Almost all of the miRNAs upregulated in AA BALF were also preferentially expressed in hematopoietic cell pellets relative to epithelial brushings (Figure 4E; Table S4).
Not all of the abundant miRNAs preferentially expressed in hematopoietic cell-rich pellets were increased in AA BALF. Notable among those absent from the list were miR-150-5p (log2 fold change AA versus CTR/IMM BALF = 0.9, padj = 0.16), miR-146b-5p (log2 fold change AA versus CTR/IMM BALF = −0.3, padj = 0.65), and miR-342-3p (log2 fold change AA versus CTR/IMM BALF = −0.4, padj = 0.64). This is particularly striking for miR-150, as prior studies have shown that lymphocyte activation is associated with decreased cellular levels of select miRNAs, including miR-150, with accumulation in exosomes, and correlates with enhanced extracellular levels in serum after immunization in mice and humans (de Candia et al., 2013). To determine whether increases in select ex-miRNAs in the allergic lung may result from sequence-specific secretion, we performed motif analysis. Although sequence-specific trafficking of miRNAs into EVs has been documented in immune cells (Villarroya-Beltri et al., 2013), we did not identify significantly enriched motifs among ex-miRNAs upregulated in AA BALF. These data suggest that upregulation of select ex-miRNAs in AA BALF may result from differences in the immune cell populations present and/or export mechanism that depend on proteins but are independent of primary miRNA sequence, as has been suggested for miR-223 and the RNA binding protein YBX1 (Shurtleff et al., 2016). Overall, our findings support the hypothesis that the influx of inflammatory cells during an allergic response contribute a select subpopulation of ex-miRNAs in the airways.
To determine whether ex-miRNAs selectively expressed in hematopoietic cells and increased in allergic airways were present in vesicle-enriched fractions, we performed a qPCR analysis of 100,000 g pellets that were floated into sucrose density gradients. Prior work has demonstrated that in a 20%/40%/60% sucrose gradient, EVs will be enriched at the interface of the 20% and 40% fractions (Shurtleff et al., 2016). Across all of the miRNAs tested, we saw the fewest ex-miRNAs in the 20% fraction (Figure 4F). When fractions were compared between experimental groups, no differences in the amounts of miR-21a-5p or miR-34c-5p were found between IMM and AA mice (Figure 4F). These data are consistent with differential expression analyses of RNA-seq data, in which no difference was observed in these abundant miRNAs between groups. In contrast, ex-miRNAs whose expression was increased in AA BALF were also increased by qPCR analysis. Differences of 3–4 cycles (estimated 6- to 8-fold) were seen across all of the fractions collected. Of note, the amount of other small RNA species (e.g., 5.8S rRNA and U7 small nuclear RNA [snRNA]) was largely unchanged between fractions and after the induction of allergic airway inflammation (Figure 4F). These data support the hypothesis that a subset of EV-packaged miRNAs are upregulated during allergic airway inflammation.
EV Flow with Conditionally Expressed Membrane Fluorescent Proteins
Pioneering work on EV flow has enabled the measurement of single vesicles from complex biofluids. This has allowed for the characterization of EV size and surface markers expression (Higginbotham et al., 2016; Inglis et al., 2015; Morales-Kastresana et al., 2017; Nolte-’t Hoen et al., 2013; Stoner et al., 2016). Given our data showing that hematopoietic miRNAs increase in the BALF with allergic inflammation, we combined a lineage-specific fluorescent labeling system with flow analysis to trace the origin of EVs in our mouse model of asthma. We used a CytoFLEX flow cytometer (Beckman Coulter), which was optimized for the detection of EVs and could detect beads down to 100 nm (Figure 5A), and we obtained transgenic mice that express membrane-anchored fluorescent proteins. These mice (hereafter referred to as mTmG mice) express a membrane-bound tandem dimer Tomato (mT) that is genetically replaced with membrane-bound GFP (mGFP) in cells expressing Cre recombinase (Muzumdar et al., 2007). EV flow of BALF collected from mTmG mice showed the presence of red Tomato+ EVs (Figure 5B). When mTmG mice were crossed with the ubiquitously expressed Actin-Cre transgene, all of the fluorescent EVs detected in the BALF were green and not red. The number of fluorescent EVs detected after a single 1-mL BAL wash in each mouse was on the order of 105 vesicles/μL (Figure 5C). Using beads to calculate a linear regression of violet side scatter (SSC) to bead size, the median vesicle size detected in both was equivalent (Figure 5D). EV flow was also performed on serum; however, no reliable mT signal could be detected above background (Figure S4A). Nevertheless, these results permit single vesicle analysis of BALF EVs using membrane-associated fluorescent proteins expressed in the cells of origin.
Figure 5. EV Flow with Conditionally Expressed Membrane Fluorescent Proteins.
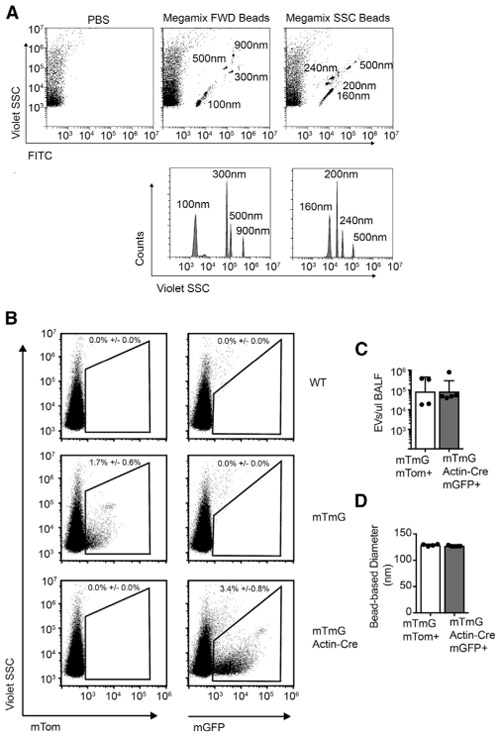
(A) Flow cytometry of 100–900 nm fluorescent beads. Size is detected by SSC using the 405 laser (violet SSC) (representative of >10 independent experiments).
(B) Flow cytometry of BALF from wild-type (WT) control, mTmG, and mTmG Actin-Cre mice for the detection of EVs positive for membrane-bound Tomato (mTom) and mGFP (membrane-bound GFP).
(C and D) mGFP+ vesicle counts (C) and size (D) are also quantified (n = 4–5 from 2–3 independent experiments, 2-tailed t test).
Error bars are mean + SD; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. See also Figures S4 and S6.
Inflammatory Cell-Derived Airway EVs Increase with Allergic Inflammation
Given the strong correlation between the miRNA profiles of epithelial cell brushings and the BALF, we wanted to determine the relative contribution of epithelial and hematopoietic cells to the EV pool in this biofluid. To specifically label lung epithelial cells with membrane-targeted GFP, mTmG mice were crossed to Nkx2.1-Cre mice (Xu et al., 2008). This transgene exhibits expression in the lung epithelium as well as the thyroid, pituitary, and subdomains of the brain, but not in immune cells or other structural cells in the lung. To specifically label immune cells, the mTmG line was also crossed to the pan-hematopoietic expressed Vav-Cre line (de Boer et al., 2003). Fluorescent microscopy of lung sections demonstrated mG labeling of bronchial and alveolar epithelium in mTmG Nkx2.1-Cre and alveolar macrophage and scattered hematopoietic cell labeling in mTmG Vav-Cre mice, as expected (Figure 6A).
Figure 6. EVs of Hematopoietic Origin Are Increased in Allergic Airway Inflammation.
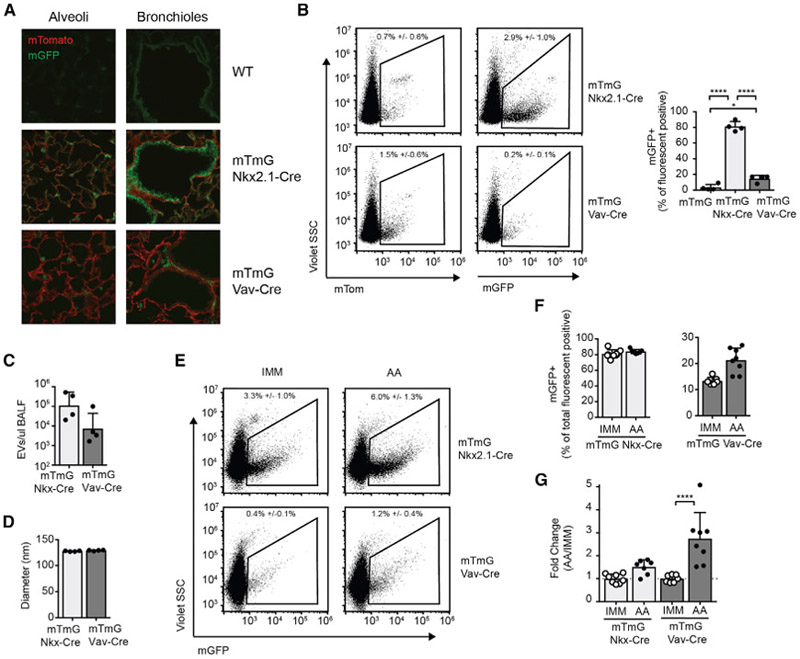
(A) Fluorescence microscopy of lungs from WT, mTmG Nkx2.1-Cre, and mTmG Vav-Cre mice.
(B) EV flow of BALF from mTmG Nkx2.1-Cre and mTmG Vav-Cre mice for the detection of EVs positive for mTom and mGFP. Relative percentage of mGFP+ among all fluorescent positive EVs (mGFP+ and mTom+) is graphed (n = 4 mice from 3 independent experiments, 1-way ANOVA with Tukey’s multiple comparison test).
(C and D) mGFP+ EV counts (C) and size (D) are also quantified in BALF (n = 4–5 from 3 independent experiments, 2-tailed t test).
(E) EVs among all fluorescent positive particles in IMM or AA mTmG Nkx2.1-Cre and mTmG Vav-Cre mice.
(F and G) Quantification of percentage (F) and fold change (G) in mGFP+ EVs in IMM and AA BALF. Fold change is normalized to IMM average in each experiment (n = 7–9 mice from 4 independent experiments, 1-way ANOVA with comparing IMM versus AA with Bonferroni’s multiple comparison test).
Error bars are mean + SD; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. See also Figures S4 and S5.
EV flow analysis of BALF showed that ~80% of fluorescent positive EVs in mTmG Nkx2.1-Cre mice were mG+ and therefore of epithelial origin in the lung (Figure 6B). No mG+ EVs were detected above baseline in the serum of these mice (Figure S4B). In BALF from mTmG Vav-Cre mice, ~15% of EVs detected by EV flow were of hematopoietic cell origin (Figure 6B). In contrast, the vast majority of EVs detected in the serum were of hematopoietic origin (Figure S4B). Overall, we could visualize on the order of 100 million fluorescent positive epithelial and 10 million fluorescent positive hematopoietic-derived EVs in a 1-mL BALF wash (Figure 6C). No difference in the median size of epithelial or hematopoietic EVs was identified (Figure 6D).
To determine whether there was a change in epithelial or hematopoietic origin EVs in the setting of a local type 2 inflammatory response in the lung, EV flow was performed on BALF from IMM and AA mTmG Nkx2.1-Cre and mTmG Vav-Cre mice. The total number and mode of the diameter of EVs detected in the BALF by nanoparticle tracking was unchanged in mice with allergic airway inflammation (Figure S5). However, mTmG Vav-Cre mice did exhibit a statistically significant increase in the percentage of mG+ hematopoietic-derived EVs among all of the fluorescent positive EVs, from 13% in IMM mice to 21% in AA mice (Figures 6E and 6F). This correlated with a 2.7-fold (range, 1.5- to 5.1-fold) increase in the absolute number of hematopoietic-derived EVs during allergic inflammation (Figure 6G). There was also a trend toward increased epithelial EVs in mTmG Nkx2.1-Cre in the inflamed lung; however, this did not achieve statistical significance, and no difference in the percentage of mG+ EVs in these mice was observed. Our data indicate that airway inflammation increases the contribution of infiltrating immune cells to airway-lining fluid EVs and ex-miRNAs.
DISCUSSION
Body fluids harbor a surprisingly high concentration of small exRNAs stabilized in association with EVs. Active secretion of EVs and ex-miRNAs may influence the function of both source and recipient cells and provides an opportunity for biomarker discovery in accessible biospecimens. In this study, we set out to determine how the composition and cellular sources of ex-miRNAs and the EVs that carry them change during tissue inflammation. Studying allergic lung inflammation using mouse models of asthma, we found that hematopoietic miRNAs and vesicles specifically increased in BALF. This occurred in the context of a biofluid that contained predominantly epithelial ex-miRNAs and EVs arising from the most abundant cells lining the airways. These findings matched our expectation that immune cell secretion of RNA and vesicles into the extracellular space will alter the local tissue environment and the biofluids associated with it.
In our studies, we performed both qualitative and quantitative studies of ex-miRNAs and EVs in BALF. Although our EV flow cytometry studies successfully discriminated EVs of epithelial and immune cell origin, the total number of particles detected using this method was substantially lower than the number counted by nanoparticle tracking assays (0.1% on average). This likely reflects a combination of limitations in the detection of small and dimly fluorescent vesicles by EV flow cytometry, and highlights both the need for and great potential of continued technical innovation in the field. Still, our work allows us to begin to address important quantitative questions such as how many molecules of RNA are packed into an individual EV in vivo. For both BALF and serum, we detected ~50–100 times more EV-sized particles by nanoparticle tracking than ex-miRNAs per microliter of fluid. Studies on purified EVs isolated from glioma stem cell cultures have estimated approximately 1 miRNA per EV generated in conditioned media in vitro (Wei et al., 2017). These results suggest that BALF is likely to contain a subpopulation of EVs that carry ex-miRNAs, which is not an altogether surprising finding given that BALF contains a complex mixture of surfactant, proteins, and EVs from different cellular sources. Important and difficult-to-address questions remain as to the number and distribution of RNA within heterogeneous populations of individual EVs, particularly in vivo.
While our results supported our overall hypothesis that miRNAs and EVs would change with mouse models of asthma, there were a number of surprising findings. We did not identify striking changes in any epithelial miRNAs, despite a prior study identifying the downregulation of miR-34/449 and miR-141/200/429 families in EV-enriched fractions from asthmatic patients that predicted lung function (Levänen et al., 2013). Perhaps the investigation of mouse models that better mimic a mild, chronic form of the disease will identify a role for epithelial EVs and/or ex-miRNAs in defining and contributing mechanistically to disease pathogenesis. Also of interest was the fact that although the induction of systemic inflammation dramatically changed the ex-miRNA profile in serum, we could detect neither a specific ex-miRNA signature nor epithelial EVs in circulation in AA mice that reflected the induction of local inflammation in the lung. It will be interesting to determine whether inflammation induced in and restricted to the lung can produce a detectible inflammatory signal in the circulation.
Perhaps some of the most exciting long-term questions raised by this work are what may be the functional consequences of alterations in ex-miRNAs and EVs derived from different cell sources in vivo in pathologic inflammation. Two of the ex-miRNAs identified as upregulated in allergic BALF may be particularly attractive for future study, as both miR-223-3p and miR-142a-3p are upregulated in the sputum of patients with a severe form of the neutrophilic subtype of asthma, and their levels correlate with disease severity (Maes et al., 2016). miR-223 is highly expressed in myeloid linage cells and limits neutrophil production, maturation, and activation (Johnnidis et al., 2008). This function is particularly important in the lung, as mice deficient in this miRNA develop spontaneous lung inflammation and have enhanced susceptibility to acute lung injury (Johnnidis et al., 2008; Neudecker et al., 2017). miR-142 contributes to the development and function of multiple hematopoietic cell subsets, including megakaryocytes, dendritic cells, mast cells, and lymphocytes (Chapnik et al., 2014; Kramer et al., 2015; Mildner et al., 2013; Yamada et al., 2014). In addition, interleukin-4 (IL-4) and IL-13 can induce mir-142a in macrophages to promote a profibrogenic pathway in the lung (Su et al., 2015). Studying the sources and function of these 2 ex-miRNAs may contribute to our understanding of asthma, especially given that extracellular miR-223 can be transferred to lung epithelial cells in vitro (Neudecker et al., 2017).
In this study, we set out to begin to answer a number of fundamental questions about ex-miRNAs and EVs in airway-lining fluid. Our findings allowed us to assign cellular sources in both healthy lungs and lungs with allergic airway inflammation. They also highlight that changes in extracellular fluids reflect changes in the composition and/or function of cells in the environment during local tissue inflammation. Our study sits amid an emerging literature on the biogenesis and delivery of exRNA (Patton et al., 2015) and the EVs that can carry them (Tkach and Tháry, 2016). It is a field that holds promise for the discovery of basic principles of cell biology and molecular physiology, as well as strategies that can be leveraged for diagnostic and therapeutic advancement.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mark Ansel (mark.ansel@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Wild-type, mTmG, mTmG Actin-Cre, mTmG Vav-Cre, and mTmG Nkx2.1-Cre male and female mice were used at greater than 5 weeks of age. mTmG mice (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, RRID:IMSR_JAX:007676) were crossed to Actin-Cre mice (B6N.FVB-Tmem163Tg(ACTB-cre)2Mrt/CjDswJ), RRID:IMSR_JAX:019099), Vav-Cre mice (B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J, RRID:IMSR_JAX:008610) or Nkx2.1-Cre mice (C57BL/6J-Tg(Nkx2-1-cre)2Sand/J, RRID:IMSR_JAX:008661, The Jackson Laboratory) to generate mice with membrane GFP labeling in all cells, hematopoietic cells or lung epithelial cells, respectively. All mice were housed and bred in specific pathogen-free conditions at the University of California, San Francisco, or at Vanderbilt University Medical Center. Animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco, or Vanderbilt University Medical Center.
METHOD DETAILS
Allergic airways and specimen collection
Bronchoalveolar lavages were performed with 1 mL of cold PBS. BAL fluid was centrifuged at 500 g for 5 minutes to separate cell free BALF supernatant from hematopoietic cell rich cell pellets. To perform epithelial brushings, brushes were made from 60-grit sandpaper–polished polyethylene PE-10 tube (BD Biosciences) (Sugimoto et al., 2012). The brush was inserted postmortem through the trachea to the mid mediastinum (past the bifurcation of the right and left mainstem bronchus). The epithelium was gently brushed and collected cells washed directly into Trizol. Blood was drawn slowly from the inferior vena cava immediately postmortem into a 1 or 3ml syringe using a 23G needle to limit hemolysis and collected into a microtainer serum separator tube (Beckton Dickinson). Serum was isolated by centrifugation at 6000 rpm for 3 minutes, followed by centrifugation at 1200 g for 20 minutes to remove residual red blood cells. Visibly hemolyzed specimens were not used for study. For allergic airway experiments, mice were injected intraperitoneally weekly for 3-4 weeks with 50 ug ovalbumin (OVA, Sigma-Aldrich) in 100 ul of PBS plus 100 ul of Imject alum (Thermo Scientific). Starting 7 days after the last injection, mice were challenged daily by oropharyngeal aspiration of 50 ug OVA in 20 ul of PBS or 20 ul of PBS alone for three consecutive days. Specimens were collected on day 4 after the first lung challenge.
Small RNA sequencing and data analysis
Total RNA was isolated using Trizol LS (for fluids, Invitrogen) and Trizol (for cell pellets, Invitrogen) from 500ul of cell free BALF, bronchoalveolar lavage cell pellets, epithelial brushings or ~200ul of serum. Each biologic replicate was obtained from a single mouse, except for epithelial brushings, which were pools of 1-3 mice. Total RNA was quantified using the RNA 6000 Pico Kit on a Bioanalyzer (Agilent). 20-65ng of total RNA was used for small RNA sequencing library generation as previously described (Williams et al., 2013). The following modifications were made to the published small RNA sequencing protocol. Five random nucleotides were added to the 5′ end of the 3′ oligodeoxynucleotide adaptor with the resulting modified adaptor sequence: 5Phos/NNNNNXXXXXTGGA ATTCTCGGGTGCCAAGG/3AmMO (where “N” are random nucleotides and “X” are pentamer indexing barcodes as outlined in the original protocol). Five random nucleotides were also added to the 3′ end of the 5′ oligoribonculeotide adaptor with the resulting modified adaptor sequence: rGrUrUrCrArGrArGrUrUrCrUrArCrArGrUrCrCrGrArCrGrArUrCrNrNrNrNrN (where “rN” are random nucleotides). 3′ adapters were adenylated enzymatically using the 5′ DNA Adenylation Kit (New England Biolabs) for 80min at 65oC. Adenylated adapters were subsequently ethanol precipitated. A mix of calibrators 1, 2, 3, 4 and 7 were used at a final combined concentration of 0.025nM in the 3′ ligation reaction. For RT of the library, the following primer was used: GCCTTGGCACCCGAGAATTCCA. The library was amplified by qPCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs). The 5′ PCR primer was as specified in the original protocol. The 3′ primer used was CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA. Amplification was stopped before the reaction reached plateau phase, and the product purified by phenol:chloroform extraction. Libraries were sequenced on a HiSeq2500 (Illumina). Data was demultiplexed and adapters trimmed using FastX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) and cutadapt (Martin, 2011). Sequence alignment and small RNA read counts were obtained using the exceRpt small RNA-seq Pipeline (v4) (http://www.genboree.org/site/). Comparative expression analysis was performed with DESeq2 (Love et al., 2014). Motif analysis of mature miRNA sequences upregulated in BALF was performed using DREAM (Discriminative Regular Expression Motif Elicitation), (http://meme-suite.org/doc/dreme.html) (Bailey, 2011) using ex-miRNAs that were unchanged or downregulated as a control and returned no results with an E-value of < 0.05. Motif analysis was also performed using Improbizer (https://users.soe.ucsc.edu/~kent/improbizer/improbizer.html) with a Markov 0 background model, an initial minimum motif size of 4 nucleotides, and randomly generated control data. The significance score for the top motif in miRNA sequences upregulated in AA BALF (3.63) was similar to the average value obtained in 10 control runs with random sequences (3.67 ± 0.17).
exRNA stability
To test whether synthetic miRNA-like oligoribonucleotide sequences were intrinsically stable in BALF, 22 nucleotide RNA calibrators (calibrators 1-4 (Williams et al., 2013)) were spiked into BALF and incubated for 5min at 37°C with or without RNAsin ribonuclease inhibitor (Promega) before RNA extraction. To test whether endogenous ex-miRNAs were stable to exogenous RNases, RNase A (Thermo Scientific) was added to BALF or total RNA previously isolated from BALF at 0.04-40ug/ml and incubated for 37°C for 15min before RNA extraction.
qPCR
Total RNA was isolated from fluids using Trizol LS (Invitrogen) and cells or vesicle pellets using Trizol (Invitrogen). Small RNA reverse transcription was performed using the miR-X First Strand Synthesis kit (Clonetech). Forward primers for miR-21a-5p, miR-24-3p, miR-34c-5p, miR-142a-3p, miR-200b-3p, and miR-223-3p were the full mature miRNA sequence (www.mirbase.org). Forward primers for other small RNAs included the 5.8S ribosomal RNA ATCGTAGGCACCGCTACGCCTGTCTG and U7 small nuclear RNA GTTACAGCTCTTTTAGAATTTGTCTAGC. The miR-X First Strand Synthesis kit (Clonetech) common reverse primer was used for all reactions. qPCR was performed on a CFX96 Quantitative PCR Thermocycler (Biorad) or a QuantStudio 3 (Applied Bio-systems) in technical duplicate using Roche FastStart Universal SYBR Green Master mix [Rox] (Sigma-Aldrich). Technical replicates with > 1 cycle s.d. were excluded from analysis.
Extracellular vesicle fractionation
After BALF was centrifuged at 300-500 g for 10 minutes to remove cells, fluid was serially centrifuged at 1,200 g for 20min, 10,000 g for 30min and 100,000 g for 2hrs all at 4oC in a Microfuge 20R (Beckman Coulter) and an Optima L-90K Utracentrifuge (Beckman Coulter). For density gradient flotation experiments, the 100,000 g pellet was collected in a 60% sucrose solution followed by layering of 40% and 20% sucrose. Samples were spun overnight (16-18hrs) at 150,000 g then particles recovered from three fractions by centrifugation at 150,000 g for 1hr. All fractions were finally resuspended in PBS.
Nanoparticle tracking and electron microscopy
Nanoparticle tracking as performed using a NanoSight LM10 (Malvern Instruments) or a ZetaView (Particle Metrix). A serially centrifuged 100,000 g pellet or 40% sucrose float fraction from the BALF of 6 pooled mice was resuspended in 50 ul of PBS and submitted for negative staining using uranyl acetate on copper coated grids and visualized with a JEOL 100CX transmission electron microscope (Gladstone Institutes, University of California, San Francisco). The fraction of liposome-like and extracellular vesicle-like particles was assessed by manual counting. The diameter of the vesicles were determined using ImageJ, RRID:SCR_003070 (Schneider et al., 2012).
Western blot
Cells and 100,000 g cell pellets were lysed in RIPA buffer (10mM Tris-HCL, 1mM EDTA, 0.5mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140mM NaCl) with 1mM PMSF for 15min on ice. Protein was quantified using a Pierce BCA Protein Assay (Thermo Scientific). Lysates were resolved on a 4%–12% BisTris Bolt mini gel (Invitrogen) and transferred to nitrocellulose membrane. Membranes were blocked with 5% powdered milk or 5% BSA and washed with TBS-T (0.1% Tween-20, 137mM NaCl, 20mM Tris Base). Primary antibodies were used at manufacturer recommended dilutions and included anti-Alix (Abcam, EPR15314 ab186429, RRID: AB_2754981), anti-Tsg101 (Abcam, ab30871, RRID: AB_2208084), anti-CD63 (Abcam, EPR21151, ab217345, RRID: AB_2754982), anti-Syntenin (Abcam, ab19903, RRID: AB_445200), anti-Arf6 (Abcam, ab77581, RRID: AB_2058475), and anti-CD9 (Santa Cruz, KMC8.8, sc-18869, RRID: AB_2076043). Secondary HRP antibodies were goat anti-rabbit (Invitrogen, A16110, RRID: AB_2534782) and donkey anti-rat (Invitrogen, A18745, RRID: AB_2535522), and blots detected with Pierce ELC Western Substrate (Thermo) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo) on an AI600 imager (GE Healthcare). Bands were quantified using ImageJ, RRID:SCR_003070.
EV flow
A Cytoflex Flow Cytometer (Beckman Coulter) was configured according to the manufacturer’s instructions for vesicle flow cytometry with the SSC detected using the violet 405 laser. BALF and serum samples were diluted in PBS and collected for 2min on low or medium speed for a target event rate of ~4,000 events/second (and no more than ~8,000 events/second) to avoid swarm effects due to coincident particle detection (Figure S6). Event collection was thresholded on violet SSC such that average particle counts for PBS alone were on average ≥ 400 events/ second (or approximately 10% of the total event rate of sample being run).
Immunofluorescent microscopy
Lungs were inflated with 0.75ml of 75% OCT (Tissue-Tek) and 2% paraformaldehyde, then placed in a 4% paraformaldehyde solution in PBS for 1hr at room temperature. The lungs were washed with 20ml of PBS for 1hr then incubated overnight in 30% sucrose in PBS with 0.02% sodium azide. The following day, they were incubated for 2hrs with 50% OCT and 15% sucrose in PBS, placed in a cryomold with OCT for 1 hour on ice, fixed in dry ice and sectioned on a cryostat. Sections were covered by ProLong Gold Antifade reagent (Invitrogen) and imaged on a Zeiss Axio upright fluorescent microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Excel (Microsoft), Prism (GraphPad), FlowJo (TreeStar) and R Studio (RStudio Team, 2017) were used for data analysis. Individual statistical tests performed are included in the data legends. All data was assumed to be normally distributed.
DATA AND SOFTWARE AVAILABILITY
The accession number for the miRNA sequencing data reported in this paper is GEO: GSE121818.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Alix [EPR15314] N-terminal | Abcam | Cat# ab186429; RRID:AB_2754981 |
| Rabbit polyclonal anti-Tsg101 | Abcam | Cat# ab30871; RRID:AB_2208084 |
| Rabbit monoclonal anti-CD63 [EPR21151] | Abcam | Cat# ab217345; RRID:AB_2754982 |
| Rabbit polyclonal anti-Syntenin | Abcam | Cat# ab19903; RRID:AB_445200 |
| Rabbit polyclonal anti-Arf6 | Abcam | Cat# ab77581; RRID:AB_2058475 |
| Rat monoclonal anti-CD9 (KMC8.8) | Santa Cruz | Cat# sc-18869; RRID:AB_2076043 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, HRP | Invitrogen | Cat# A16110; RRID:AB_2534782 |
| Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, HRP | Invitrogen | Cat# A18745; RRID:AB_2535522 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BD Microtainer SST | Becton Dickinson | Cat# 365967 |
| Imject Alum | Thermo Scientific | Cat# 77161 |
| Ovalbumin | Sigma-Aldrich | Cat# A5503; CAS Number 9006-59-1 |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0530 |
| RNase A | Thermo Scientific | Cat# EN0531 |
| RNasin | Promega | Cat# N2511 |
| Roche FastStart Universal SYBR Green Master mix (Rox) | Sigma-Aldrich | Cat# 04913850001 |
| TRIzol | Invitrogen | Cat# 15596018 |
| TRIzol LS | Invitrogen | Cat# 10296028 |
| Critical Commercial Assays | ||
| DNA Adenylation Kit | New England BioLabs | Cat# E2610 |
| miR-X First Strand Synthesis Kit | Clonetech | Cat# 638315 |
| Pierce BCA Protein Assay Kit | Thermo Scientific | Cat# 23227 |
| RNA 6000 Pico Kit | Agilent | Cat# 5067-1513 |
| Deposited Data | ||
| miRNA sequencing data | GEO | GSE121818 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Actin-cre (B6N.FVB-Tmem163Tg(ACTB-cre)2Mrt/CjDswJ) | The Jackson Laboratory | RRID:IMSR_JAX:019099 |
| Mouse: mTmG (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J) | The Jackson Laboratory | RRID:IMSR_JAX:007676 |
| Mouse: Nkx2.1-cre (C57BL/6J-Tg(Nkx2-1-cre)2Sand/J) | The Jackson Laboratory | RRID:IMSR_JAX:008661 |
| Mouse: Vav-cre (B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J) | The Jackson Laboratory | RRID:IMSR_JAX:008610 |
| Mouse: Wild type (C57BL/6J) | NCI Mouse Repository | Cat# 01B96 |
| Oligonucleotides | ||
| See Table S5 | N/A | |
| Software and Algorithms | ||
| cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/installation.html |
| DESeq2 | Love et al., 2014 | https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| DREAM | Bailey, 2011 | http://meme-suite.org/doc/dreme.html |
| exceRpt small RNA-seq Pipeline v4 | Bioinformatics Research Laboratory, Baylor College of Medicine | http://www.genboree.org/site/ |
| FastX Toolkit | Cold Spring Harbor Laboratory | http://hannonlab.cshl.edu/fastx_toolkit/index.html |
| FlowJo v9 and v10 | TreeStar | FlowJo, RRID:SCR_008520; https://www.flowjo.com/solutions/flowjo |
| ImageJ | Schneider et al., 2012 | ImageJ, RRID:SCR_003070; https://imagej.net/Welcome |
| Improbizer | University of California, Santa Cruz | https://users.soe.ucsc.edu/~kent/improbizer/improbizer.html |
| Prism 7 | GraphPad Software | GraphPad Prism, RRID:SCR_002798; https://www.graphpad.com/ |
| R Studio | RStudio Team, 2017 | https://www.rstudio.com |
Highlights.
Lung extracellular miRNAs have a composition similar to lung epithelium
Extracellular vesicles (EVs) in airway fluid contain miRNAs
Genetic membrane tagging and single vesicle flow identify cell sources of EVs in vivo
Immune extracellular miRNAs and EVs increase during allergic lung inflammation
ACKNOWLEDGMENTS
This publication is part of the NIH Extracellular RNA Communication Consortium paper package and was supported by the NIH Common Fund’s exRNA Communication Program. We would like to thank Matt Spitzer for use of the CytoFLEX instrument. We would like to thank Andy Vaughan and Harold Chapman for lung fixation protocols and the use of their fluorescent microscope. We would like to thank the Gladstone Electron Microscopy Core, and especially Jinny S. Wong, for processing and imaging BALF vesicle pellets. We would like to thank Shinya Sato and Alissa Weaver for training and use of their ZetaView for nanoparticle tracking. We would also like to thank Jeff Rathmell’s lab (in particular Melissa Wolf) and Peggy Kendall’s lab (in particular Christina Thurman) for sharing mice, reagents, and/or equipment with us. This work was supported by NIH U19CA179512 (K.M.A.), NIH K08AI116949 (H.H.P.), and the Department of Pathology, Microbiology and Immunology at Vanderbilt University Medical Center (H.H.P.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 figures and 5 tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2019.01.002.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPORTING CITATIONS
The following reference appears in the Supplemental Information: Bronevetsky et al., 2013.
REFERENCES
- Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, and O’Connell RM (2015). Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun 6, 7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 108, 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL (2011). DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, et al. (2013). T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med 210, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caby M-P, Lankar D, Vincendeau-Scherrer C, Raposo G, and Bonnerot C (2005). Exosomal-like vesicles are present in human blood plasma. Int. Immunol 17, 879–887. [DOI] [PubMed] [Google Scholar]
- Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, et al. (2015). KRAS-dependent sorting of miRNA to exosomes. eLife 4, e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapnik E, Rivkin N, Mildner A, Beck G, Pasvolsky R, Metzl-Raz E, Birger Y, Amir G, Tirosh I, Porat Z, et al. (2014). miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. eLife 3, e01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li L, Lodish HF, and Bartel DP (2004). MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science 303, 83–86. [DOI] [PubMed] [Google Scholar]
- Cormier SA, Yuan S, Crosby JR, Protheroe CA, Dimina DM, Hines EM, Lee NA, and Lee JJ (2002). T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol 27, 678–687. [DOI] [PubMed] [Google Scholar]
- Dargaville PA, South M, Vervaart P, and McDougall PN (1999). Validity of markers of dilution in small volume lung lavage. Am. J. Respir. Crit. Care Med 160, 778–784. [DOI] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, and Kioussis D (2003). Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol 33, 314–325. [DOI] [PubMed] [Google Scholar]
- de Candia P, Torri A, Gorletta T, Fedeli M, Bulgheroni E, Cheroni C, Marabita F, Crosti M, Moro M, Pariani E, et al. (2013). Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PLoS One 8, e75348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy JV (2015). Type 2 inflammation in asthma–present in most, absent in many. Nat. Rev. Immunol 15, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasuwa H, Ueda J, Ikawa M, and Okabe M (2013). MiR-200b and miR-429 Function in Mouse Ovulation and Are Essential for Female Fertility. Science 341, 71–73. [DOI] [PubMed] [Google Scholar]
- Higginbotham JN, Zhang Q, Jeppesen DK, Scott AM, Manning HC, Ochieng J, Franklin JL, and Coffey RJ (2016). Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J. Extracell. Vesicles 5, 29254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee M-LT, Schmittgen TD, et al. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 3, e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis HC, Danesh A, Shah A, Lacroix J, Spinella PC, and Norris PJ (2015). Techniques to improve detection and analysis of extracellular vesicles using flow cytometry. Cytometry A 87, 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash AD, Jabado O, Brown BD, and Sachidanandam R (2011). Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 39, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, and Camargo FD (2008). Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129. [DOI] [PubMed] [Google Scholar]
- Khoshgoo N, Visser R, Falk L, Day CA, Ameis D, Iwasiow BM, Zhu F, Öztürk A, Basu S, Pind M, et al. (2017). MicroRNA-200b regulates distal airway development by maintaining epithelial integrity. Sci. Rep 7, 6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, and Reid G (2011). Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 6, e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer NJ, Wang W-L, Reyes EY, Kumar B, Chen C-C, Ramakrishna C, Cantin EM, Vonderfecht SL, Taganov KD, Chau N, and Boldin MP (2015). Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 125, 3720–3730. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. (2007). A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levänen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, Sköld CM, Svartengren M, Grunewald J, Gabrielsson S, et al. (2013). Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol 131, 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Ching D, Luk FS, and Raffai RL (2015). Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-κB-driven inflammation and atherosclerosis. Circ. Res 117, e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, De Preter K, Bracke KR, Conickx G, Mesnil C, Vandesompele J, et al. (2016). Asthma inflammatory phenotypes show differential microRNA expression in sputum. J. Allergy Clin. Immunol 137, 1433–1446. [DOI] [PubMed] [Google Scholar]
- Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi L-E, Cibois M, Robbe-Sermesant K, Jolly T, Cardinaud B, Moreilhon C, et al. (2011). Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol 13, 693–699. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. [Google Scholar]
- Mateescu B, Kowal EJK, van Balkom BWM, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FWN, Das S, et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles 6, 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A, Chapnik E, Manor O, Yona S, Kim K, Aychek T, Varol D, Beck G, Itzhaki ZB, Feldmesser E, et al. (2013). Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood 121, 1016–1027. [DOI] [PubMed] [Google Scholar]
- Momen-Heravi F, Bala S, Kodys K, and Szabo G (2015). Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci. Rep 5, 9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan MLG, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. (2005). MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6, R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman JE, Akinbami LJ, Bailey CM, Zahran HS, King ME, Johnson CA, and Liu X (2012). National surveillance of asthma: United States, 2001-2010. Vital Health Stat. 3 35, 1–58. [PubMed] [Google Scholar]
- Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, Rosner A, Demberg T, Watson DC, Karpova TS, et al. (2017). Labeling Extracellular Vesicles for Nanoscale Flow Cytometry. Sci. Rep 7, 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Neudecker V, Brodsky KS, Clambey ET, Schmidt EP, Packard TA, Davenport B, Standiford TJ, Weng T, Fletcher AA, Barthel L, et al. (2017). Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci. Transl. Med 9, eaah5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nials AT, and Uddin S (2008). Mouse models of allergic asthma: acute and chronic allergen challenge. Dis. Model. Mech 1, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-’t Hoen ENM, van der Vlist EJ, de Boer-Brouwer M, Arkesteijn GJA, Stoorvogel W, and Wauben MHM (2013). Dynamics of dendritic cell-derived vesicles: high-resolution flow cytometric analysis of extracellular vesicle quantity and quality. J. Leukoc. Biol 93, 395–402. [DOI] [PubMed] [Google Scholar]
- Patton JG, Franklin JL, Weaver AM, Vickers K, Zhang B, Coffey RJ, Ansel KM, Blelloch R, Goga A, Huang B, et al. (2015). Biogenesis, delivery, and function of extracellular RNA. J. Extracell. Vesicles 4, 27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, and Tewari M (2012). Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila.) 5, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, Chiou N-T, Gallman A, de Kouchkovsky D, Jeker LT, et al. (2016). MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production. Immunity 44, 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennard SI, Basset G, Lecossier D, O’Donnell KM, Pinkston P, Martin PG, and Crystal RG (1986). Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of dilution. J. Appl. Physiol 60, 532–538. [DOI] [PubMed] [Google Scholar]
- RStudio Team (2017). RStudio: Integrated Development for R (RStudio). [Google Scholar]
- Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, and Tripodi M (2016). The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 17, 799–808. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seumois G, Vijayanand P, Eisley CJ, Omran N, Kalinke L, North M, Ganesan AP, Simpson LJ, Hunkapiller N, Moltzahn F, et al. (2012). An integrated nano-scale approach to profile miRNAs in limited clinical samples. Am. J. Clin. Exp. Immunol 1, 70–89. [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, and Lambowitz AM (2017). Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 114, E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, et al. (2014). miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner SA, Duggan E, Condello D, Guerrero A, Turk JR, Narayanan PK, and Nolan JP (2016). High sensitivity flow cytometry of membrane vesicles. Cytometry A 89, 196–206. [DOI] [PubMed] [Google Scholar]
- Su S, Zhao Q, He C, Huang D, Liu J, Chen F, Chen J, Liao JY, Cui X, Zeng Y, et al. (2015). miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat. Commun 6, 8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Kudo M, Sundaram A, Ren X, Huang K, Bernstein X, Wang Y, Raymond WW, Erle DJ, Åbrink M, et al. (2012). The αvβ6 integrin modulates airway hyperresponsiveness in mice by regulating intraepithelial mast cells. J. Clin. Invest 122, 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, and Théry, C. (2016). Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, and Remaley AT (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol 13, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernán-dez D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, and Sánchez-Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, et al. (2017). Coding and non-coding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, and Tuschl T (2013). Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 110, 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, and Rajewsky K (2007). MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159. [DOI] [PubMed] [Google Scholar]
- Xu Q, Tam M, and Anderson SA (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol 506, 16–29. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Kosaka K, Miyazawa T, Kurata-Miura K, and Yoshida T (2014). miR-142-3p enhances FcεRI-mediated degranulation in mast cells. Biochem. Biophys. Res. Commun 443, 980–986. [DOI] [PubMed] [Google Scholar]
- Zhuang F, Fuchs RT, Sun Z, Zheng Y, and Robb GB (2012). Structural bias in T4 RNA ligase-mediated ′3-adapter ligation. Nucleic Acids Res. 40, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the miRNA sequencing data reported in this paper is GEO: GSE121818.


