Abstract
Heterokonts, Alveolata protists, green algae from Charophyta and Chlorophyta divisions, and all Embryophyta plants possess an aldehyde dehydrogenase (ALDH) gene named ALDH12. Here, we provide a biochemical characterization of two ALDH12 family members from the lower plant Physcomitrella patens and higher plant Zea mays. We show ALDH12 encodes an NAD+-dependent glutamate γ-semialdehyde dehydrogenase (GSALDH), which irreversibly converts glutamate γ -semialdehyde (GSAL), a mitochondrial intermediate of the proline and arginine catabolism, to glutamate. Sedimentation equilibrium and small-angle X-ray scattering analyses reveal that in solution both plant GSALDHs exist as equilibrium between a domain-swapped dimer and the dimer-of-dimers tetramer. Plant GSALDHs share very low sequence identity with bacterial, fungal, and animal GSALDHs (classified as ALDH4), which are the closest related ALDH superfamily members. Nevertheless, the crystal structure of ZmALDH12 at 2.2 Å resolution shows that nearly all key residues involved in the recognition of GSAL are identical to those in ALDH4, indicating a close functional relationship with ALDH4. Phylogenetic analysis suggests that the transition from ALDH4 to ALDH12 occurred during the evolution of the endosymbiotic plant ancestor, prior to the evolution of green algae and land plants. Finally, ALDH12 expression in maize and moss is downregulated in response to salt and drought stresses, possibly to maintain proline levels. Taken together, these results provide molecular insight into the biological roles of the plant ALDH12 family.
Keywords: aldehyde dehydrogenase, ALDH12, gene expression, glutamate γ-semialdehyde, Physcomitrella patens, proline, site-directed mutagenesis, X-ray crystallography, Zea mays
Introduction
The mitochondrial enzyme Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (P5CDH; E.C.1.2.1.88) is often annotated as ALDH4 in reference to its membership in family 4 of the aldehyde dehydrogenase (ALDH) superfamily. Early works on human ALDH4 identified P5C/glutamate-γ-semialdehyde (GSAL) and NAD+ as physiological substrates and L-glutamate as the product1,2. P5C, which is the cyclic Schiff base of GSAL, is in non-enzymatic equilibrium with GSAL in solution3. More recently, P5CDH has been annotated as GSAL dehydrogenase (GSALDH) to reflect the true substrate and recognize that the enzyme belongs to the ALDH structural superfamily4.
GSAL is an intermediate in L-proline catabolism and is generated by the flavoenzyme proline dehydrogenase (PRODH) (Figure 1)5,6. Proline acts both as a compatible osmolyte, which accumulates during drought, salinity, or cold stresses, and an energy source during pollen maturation and germination in plants 7,8. Proline catabolism has also been shown to impact cellular signaling processes that combat pathogens and delay senescence in different plant tissues9,10. Therefore, a careful regulation of proline levels is necessary for cellular homeostasis and conferring tolerance to both abiotic and biotic stresses. GSAL is also an intermediate in the mitochondrial L-arginine catabolic pathway11 (Figure 1). L-Arginine is hydrolyzed via arginase to L-ornithine12, which is reversibly metabolized by the 2-oxoglutarate-dependent ornithine-δ-aminotransferase (δ-OAT, E.C. 2.6.1.13) to GSAL and glutamate. GSAL is further oxidized to glutamate by GSALDH.
Figure 1. Reaction scheme of proline and arginine catabolism.
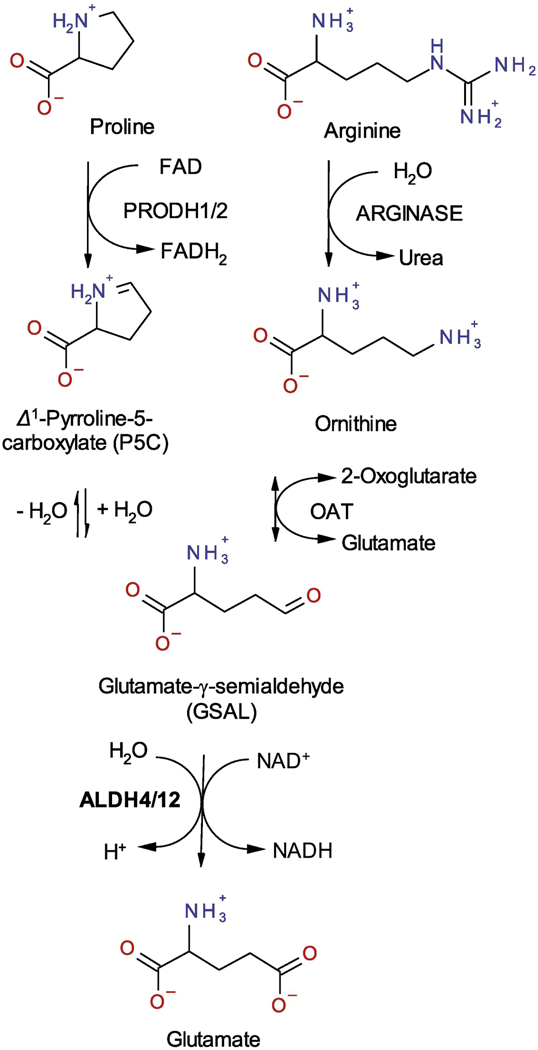
Degradation of proline and arginine in plant mitochondria leads to GSAL, which is oxidized by ALDH12 into glutamate using NAD+ a coenzyme. The same reaction in catalyzed by ALDH4 in other species.
Available plant genomes display the presence of two PRODH genes, one GSALDH gene, and one δ-OAT gene13,14. δ-OAT has been studied in Arabidopsis and pea13,15. In plants, PRODH and GSALDH enzymatic activities were first described in maize mitochondria16, and kinetic parameters were also determined for potato GSALDH17. The GSALDH gene, originally named flax inducible sequence 1 (FIS1), was up-regulated in flax (Linum usitatissimum) in response to infection with Melampsora lini, the causal agent of flax rust18. The Arabidopsis GSALDH gene (At5g62530) has also been identified and studied using reverse genetics14,19. The highest AtGSALDH transcript levels were detected in buds and flowers. Arabidopsis gsaldh mutants displayed reduced thermo-tolerance in response to heat stress, consistent with previously identified proline toxicity at high temperatures in gsaldh mutant plants20,21. The mutants also displayed three-fold higher proline levels compared to wild-type under salt stress, whereas no difference in gene expression was observed compared to wild-type under normal conditions19.
The amino acid sequence identity of plant to non-plant GSALDHs is below 24%. Because ALDHs have to share ≥40% amino acid sequence identity to be considered members of the same ALDH family22, plant GSALDHs are grouped into the ALDH12 rather than the ALDH4 family23. Unlike ALDH12, the ALDH4 family has been extensively characterized. Crystal structures and kinetic data are available for ALDH4 from several organisms: Thermus thermophilus24,25, Saccharomyces cerevisiae26, Mus musculus21 and Homo sapiens27,28. Additionally, in some prokaryotes, ALDH4 is fused to PRODH into the bifunctional enzyme proline utilization A (PutA)29, which oxidizes L-proline to glutamate using spatially-separated PRODH and GSALDH active sites.
Herein we report biochemical and structural studies of ALDH12 from Zea mays (ZmALDH12) and Physcomitrella patens (PpALDH12). Kinetic parameters and substrate preference were determined for both enzymes. Further, in-solution oligomeric state was investigated by analytical ultracentrifugation and small-angle X-ray scattering. To better understand substrate specificity, we determined the crystal structure of ZmALDH12 in complex with NAD+ to 2.20 Å resolution. Mutagenesis and structural comparisons to members of the ALDH4 family were used to analyze the substrate and coenzyme binding sites. These analyses revealed amino acid residues crucial for coenzyme specificity (Asp226) and GSAL binding (Ser331, Lys329 and Glu205). The spatial and temporal expression of ALDH12 in maize seedlings and full-grown plants was also analyzed. Finally, the expression of maize and moss ALDH12 is downregulated under salt and drought stresses possibly to maintain proline levels.
RESULTS AND DISCUSSION
Cloning and heterologous expression of two plant ALDH12 family members.
The complementary DNA (cDNA) of ALDH12 from moss and maize were cloned to identify the correct gene models and to obtain recombinant proteins (primers are shown in Table S1). The unique PpALDH12 gene (Phytozome ID: Pp3c5_4940V3) encodes a 571 amino acid protein23. The ZmALDH12 gene (Phytozome ID: GRMZM2G090087) encodes a 549 amino acid protein. Because the maize cultivar Cellux (Morseva, Czech Rep.) was used in this study, the cDNA sequence differs in 15 bases leading to five amino acid substitutions (P86S, V88M, A127T, V468I and V532I) compared to the sequenced B73 cultivar. The moss and maize ALDH12 sequences are 71% identical (88% similar). Both enzymes also have putative N-terminal mitochondrial targeting sequences. Heterologous expression of both complete cDNAs cloned into pCDFDuet, pETDuet, or pET32b expression vectors in E. coli resulted in protein sequestered in inclusion bodies. Therefore, pCDFDuet constructs lacking the putative mitochondrial targeting sequence (Δ1–36PpALDH12 and Δ1–16ZmALDH12) were prepared and yielded large quantities of active, soluble enzyme. Although the protein yield in pET32b vectors was more than 6-fold higher, the specific activities after purification were lower than those expressed in the pCDFDuet vector. The specific activities of ZmALDH12 and PpALDH12 with 1 mM GSAL were 32 nkat mg−1 and 1.6 nkat mg−1, respectively. The reason for such a low activity of the moss enzyme could be attributed to lower stability as identified by nano differential scanning fluorimetry (nanoDSF). Whereas PpALDH12 had the highest Tm of 53.0°C in 150 mM sodium pyrophosphate, pH 7.5 (containing 5% glycerol), ZmALDH12 displayed a much higher Tm of 75.7°C in 150 mM Tris-HCl buffer, pH 7.5 (containing 100 mM NaCl and 5% glycerol) (Figure 2a and 2b). In addition, the maize enzyme is greatly stabilized in presence of NaCl, which is not the case of the moss enzyme. Because the maize enzyme displayed higher activity and stability, it was chosen for further kinetic and structural analyses.
Figure 2. Thermal stability of ALDH12 from maize (Zea mays) and moss (Physcomitrella patens).
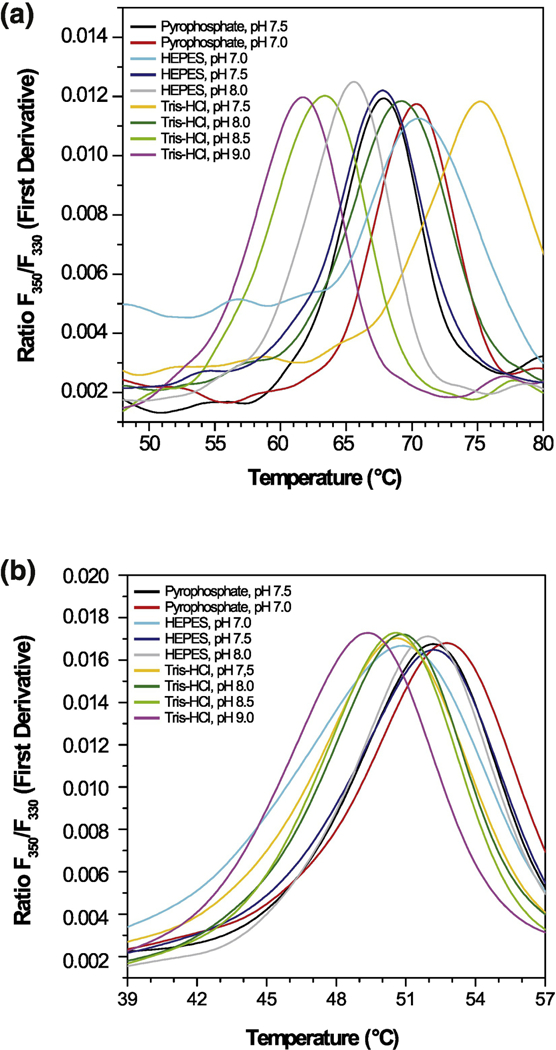
Effect of pH and buffer composition on the thermal unfolding of ZmALDH12 (a) and PpALDH12 (b) measured by nanoDSF. All buffers were at 150 mM concentration with 100 mM NaCl and 5% glycerol.
Phylogenetic analysis of the ALDH4 and ALDH12 families.
The ALDH4 gene coding for GSALDH is present in bacterial (as rocA)30, archaeal, fungal (as Put2p), and Chordata genomes. For example, human and mouse ALDH4A1 (HsALDH4, MmALDH4) share only 23.7% and 23.9% sequence identity with ZmALDH12, respectively. The ALDH4 gene is present in genomes of Cyanidioschyzon merolae (M1V4V6) and Galdieria sulphuraria (NCBI ID: XP_005705265). In some prokaryotes, ALDH4 is fused to PRODH to form the bifunctional enzyme PutA. Predecessors of chloroplasts such as photosynthetic cyanobacteria Synechococcus (Uniprot ID: K9SVM7) and Nostoc (D7E1V8) carry ALDH4 (but not ALDH12) fused into PutA.
Protists are considered the oldest eukaryotes, and the ALDH4 sequence is found also among amoeboid protists such as Dictyostelium or Polysphondylium (Uniprot IDs: Q54RA2 and D3BP41). However, the ALDH12 gene occurs among Alveolata protists, including endosymbiotic dinoflagellates, such as unicellular alga Symbiodinium microadriaticum; the oyster pathogen Perkinsus marinus; the mammalian pathogen Toxoplasma gondii and others. Among the Excavata clade of protists, ALDH4 is present, for example, in Leishmania and Trypanosoma species (S6EYD1 and E9ACG7) while ALDH12 is present in Stygiella incarcerata (A0A192ZIT0) from a Jakobea class of Excavata, which are flagellated protists with aerobic mitochondria.
Conversely, the complete ALDH12 sequence is found in the genomes of green algae and all studied Embryophyta, including non-vascular, vascular, and higher flowering plants (listed in Table S2). The only exception is one of the oldest groups of eukaryotic algae - Rhodophyta (red algae). ALDH12 is thus present in green algae from both Chlorophyta and Charophyta divisions, in Cryptomonads carrying a red-algal derived chloroplast such as Guillardia theta, in Chlorarachniophytes carrying green-algal derived chloroplasts such as Bigelowiella natans, in Haptophytes such as Emiliania huxleyi and also among heterokonts comprising both algae diatoms, such as Thalassiosira and Phaeodactylum, and colorless plant pathogens including Phytophthora and others (Figure S1). Taken together, these findings indicate that the ALDH4 to ALDH12 switch appeared among the oldest eukaryotes during the evolution of the endosymbiotic plant ancestor, prior to the evolution of green algae and land plants.
Plant ALDH12 is usually a single copy gene with 15 or 16 exons. For example, the ZmALDH12 gene studied in this work is composed of 15 exons and lies on maize chromosome 6, and the PpALDH12 gene is composed of 16 exons and lies on chromosome 5 of Physcomitrella patens. The ZmALDH12 and PpALDH12 amino acid sequences are >60% identical and thus belong to a single subfamily, ALDH12A23. Alveolata ALDH12 sequences exhibit only 52 – 59% amino acid sequence identity to ALDH12A sequences and therefore are sorted into the ALDH12B subfamily.
Substrate specificity of the plant ALDH12 family.
Several aldehydes were screened as potential substrates using NAD+ as the coenzyme (Figure 3a). Both moss and maize enzymes display a narrow substrate preference for GSAL and glutaric semialdehyde (GRSAL) at an optimal pH of 7.5 (Figure 3b). GSAL and GRSAL are 5-carbon semialdehydes differing only in the presence of a positively-charged amino group in GSAL. α-Aminoadipic semialdehyde (AASAL), a substrate analog of GSAL with a 6-carbon chain, is the major substrate of ALDH731 and is oxidized at a rate of ~6% compared to GSAL. Succinic semialdehyde, a substrate analog of GRSAL with a 4-carbon chain, is the major substrate of ALDH5 and ALDH2132. Succinic semialdehyde is oxidized at ~2% rate compared to GSAL. Other aliphatic and aromatic aldehydes (substrates of ALDH2)31, D- and L-glyceraldehyde 3-phosphates (substrates of ALDH11), and ω-aminoaldehydes (substrates of ALDH10)33 such as 4-aminobutanal or 3-aminopropanal were tested. Each of these substrates exhibited rates substantially lower than the preferred substrate GSAL. For example, D-glyceraldehyde 3-phosphate was oxidized at 7.5% the rate of GSAL, and the rates observed for 4-aminobutanal, phenylacetaldehyde and phenylpropionaldehyde were ~ 5% the rate of GSAL.
Figure 3. Screening of substrate specificity of maize and moss ALDH12.
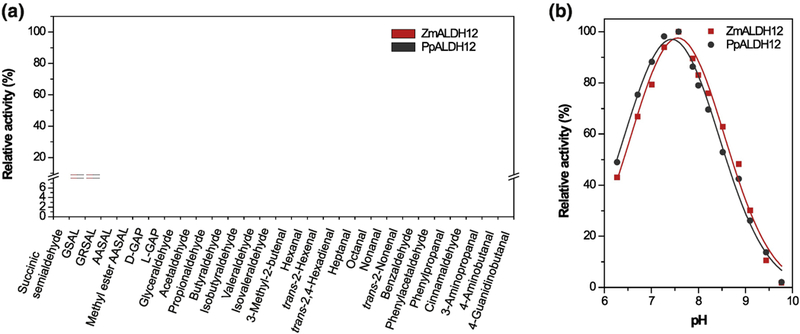
(a) Measurements were performed with 1 mM substrates in 100 mM pyrophosphate buffer, pH 7.5 containing 3.0 mM NAD+. Specific activity values with GSAL and NAD+ of 32 nkat mg−1 for ZmALDH12 and 1.6 nkat mg−1 for PpALDH12 were arbitrarily taken as 100% for each isoform. Error bars stand for S.D. from four measurements. (b) Influence of pH on the catalytic activity of ZmALDH12 and PpALDH12. Measurements were performed with 1 mM GSAL and 3.0 mM NAD+ in Britton-Robinson buffers in the pH range of 6 – 10.
Kinetic properties of ZmALDH12 with the preferred substrates GSAL and GRSAL were explored further. The apparent catalytic efficiency (kcat/Km) for NAD+ is ~ 60-fold higher (1.7 × 104 s−1M−1) than for NADP+ (2.7 × 102 s−1 M−1, Table 1) using 0.3 mM GSAL. The enzyme displays the highest catalytic efficiency (3.4 × 104 s−1M−1) for GSAL with Km of 198 ± 21 μM and kcat of 6.8 ± 0.5 s−1. The catalytic efficiency of ZmALDH12 for GSAL is thus similar to that of Oryza sativa ALDH12 (3.3 × 104 s−1M−1)34, higher than that of Saccharomyces cerevisiae ALDH4 (kcat/Km = 1.4 × 104 s−1M−1, kcat = 1.5 s−1, and Km = 104 μM)26 but lower than of HsALDH4 (kcat/Km = 3.1 × 106 s−1 M−1, Km = 32 μM, and kcat = 10.0 s−1)35. Although the maximal rate of GRSAL oxidation is slower that GSAL, the Km value of 84 ± 5 μM is also lower compared to GSAL making it the second best substrate of the maize enzyme (kcat/Km = 2.3 104 s−1M−1). Because GSAL causes cell death in human tumor cell lines, it has been hypothesized that GSALDH plays a crucial role in protecting the cell from GSAL hyperaccumulation36. It has likewise been suggested that GSAL levels are tightly controlled by the P5C-Pro cycle in which P5C is transported (as P5C) into the cytosol and converted to proline by P5C reductase20.
Table 1. Kinetic parameters for ZmALDH12 and PpALDH12.
Saturation curves for NAD+ and NADP+ were measured in 100 mM pyrophosphate buffer (pH 7.5) using 0.3 mM GSAL, saturation curves for aldehyde substrates were measured with 3.0 mM NAD+. Kinetic constants Km and kcat including their standard error values were determined using GraphPad Prism 5.0 software. The lower kcat values for NAD+ (indicated by asterisks) compared to those for GSAL result from using a fixed sub-saturating GSAL concentration in the saturation of the enzymes by NAD+. n.d. - not determined
| Substrate | ZmALDH12 |
PpALDH12 |
ZmALDH12 D226A |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| (μM) | (s−1) | (s−1M1) | μM) | (s−1) | (s−1M−1) | μM) | (s−1) | (s−1M1) | |
| NAD+ | 185 ± 14 | *3.1 ± 0.1 | 1.7 ± 0.2 ×104 | 227 ± 14 | *0.17 ± 0.01 | 7.4 ± 0.8 × 102 | 119 ± 7 | *1.7 ± 0.1 | 1.4 ± 0.1 × 104 |
| NADP+ | 2872 ± 360 | 0.8 ± 0.1 | 2.7 ± 0.5 × 102 | 3066±186 | 0.05 ± 0.01 | 1.4 ± 0.1 × 101 | 71 ± 6 | 1.5 ± 0.1 | 2.1 ± 0.1 × 104 |
| GSAL | 198 ± 21 | 6.8 ± 0.5 | 3.4 ± 0.6 × 104 | 228 ± 40 | 0.44 ± 0.05 | 1.9 ± 0.6 × 103 | 266 ± 34 | 3.6 ± 0.3 | 1.3 ± 0.3 × 104 |
| GRSAL | 84 ± 5 | 1.9 ± 0.2 | 2.3 ± 0.4 × 104 | 82 ± 6 | 0.30 ± 0.02 | 3.7 ± 0.6 × 103 | n.d. | n.d. | n.d. |
| AASAL | 173 ± 12 | 0.04 ± 0.02 | 2.0 ± 0.2 × 102 | 211 ± 13 | 0.005 ± 0.001 | 2.6 ± 0.2 × 101 | n.d. | n.d. | n.d. |
| D-GAP | 1860±99 | 0.4 ± 0.1 | 2.1 ± 0.2 × 102 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
Dissociation constants (Kd) of 166 ± 14 μM for GSAL and 34 ± 11 μM for GRSAL were measured by microscale thermophoresis (MST) in the absence of the coenzyme. These values correlate well with the respective Km values for GSAL and GRSAL. The catalytic efficiency for AASAL is only 2.1 × 102 s−1M−1. This decreased value is attributed to low turnover rate because GSAL and AASAL have similar Km values. The enzyme follows substrate inhibition kinetics with both GSAL and GRSAL. As anticipated, the wild-type enzyme affinity for the reaction product glutamate is much lower (Kd = 3.1 ± 0.3 mM, Figure S3). By comparison, a Ki value of 12 mM for glutamate was reported for MmALDH428.
Structure and domain architecture of ZmALDH12.
The structure of ZmALDH12 co-crystallized with NAD+ was determined to 2.20 Å resolution. The overall fold of ZmALDH12 resembles that of other ALDH superfamily members including ALDH4. There are three domains22 within ZmALDH12 (Figure 4a): an NAD+-binding fold (residues 28 – 170, 190 – 295, and 519 – 527), a catalytic α/β fold (residues 296 – 497), and an oligomerization flap (residues 171 – 189 and 528 – 549). Like other GSALDHs and ALDH superfamily members, the NAD+-binding domain exhibits an abbreviated Rossmann fold consisting of a parallel five-stranded β-sheet. The catalytic α/β fold contains the catalytic Cys330 on a loop that sits in the crevice between the NAD+-binding and catalytic domains. The C-terminal oligomerization flap is a three-stranded antiparallel β-sheet that mediates formation of the classic ALDH domain swapped dimer. Finally, residues 498 – 518 form an inter-domain linker that provides both a lid for the active site and an aldehyde anchor loop within the active site. In other ALDHs, including GSALDHs, this anchor loop provides stabilizing contacts for the aldehyde substrate27,28.
Figure 4. Structure of ZmALDH12.
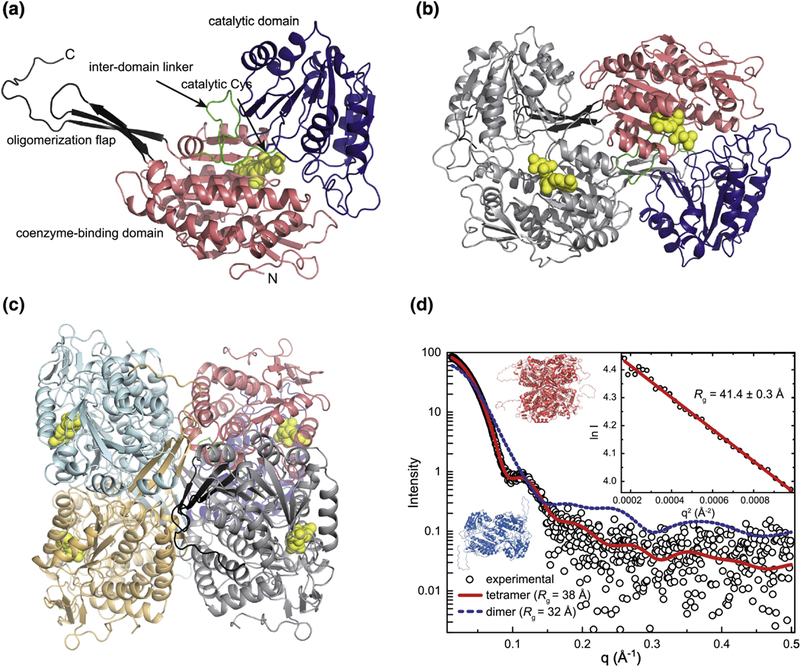
(a) A subunit of ZmALDH12 highlighting domain architecture. (b) The domain-swapped dimer. One subunit is colored according to domains as in the first panel. The other subunit has a single color. The coenzyme molecule is in yellow. (c) The ZmALDH12 tetramer formed in solution. One subunit is colored according to domains as in panel a. The other subunits have single colors. (d) An experimental SAXS curve for ZmALDH12 (open circles) extrapolated to infinite dilution. The inset shows the Guinier plot. The red curve was calculated from a complete model generated by AllosMod-FoXS using the crystallographic dimer-of-dimers tetramer as a template (χ2 = 0.6). The blue dashed curve was calculated from a ZmALDH12 crystallographic dimer (Figure 4b).
Quaternary structure and in solution oligomeric state.
The asymmetric unit of the ZmALDH12 crystal structure contains four monomers arranged as two domain-swapped dimers (Figure 4b). Analysis of crystal packing with PDBePISA37 identified a potentially stable dimer-of-dimers tetramer formed by the application of a crystallographic 2-fold operator either of the dimers in the asymmetric unit (Figure 4c). We note this tetramer has also been observed in other ALDHs, such as ALDH7A131,38, but never before in GSALDHs. Because previous studies of GSALDHs from human, mouse, yeast, and bacteria revealed dimeric and hexameric assemblies35,26,39, we determined the oligomeric state of His-tagged ZmALDH12 in solution using sedimentation equilibrium (Figure S2). Data were fit to a single-body model to determine the average molecular mass (Mr) of 206 ± 7 kDa, which is between the expected mass of a dimer (121 kDa) and a tetramer (242 kDa). This result suggests a dimer-tetramer equilibrium. Therefore, the data were reanalyzed using a dimer-tetramer equilibrium model revealing a dimer-tetramer association constant (K2–4) of 910,000 ± 60,000 M−1, corresponding to a Kd of 1.1 μM (relative to the dimer Mr). Similar analysis of His-tagged PpALDH12 yielded-a dimer-tetramer Kd of 1.3 μM (Figure S2). Thus, both plant ALDH12 enzymes form dimer-tetramer equilibria at low concentration in solution.
To further explore the quaternary structure of ZmALDH12 in solution, SAXS analysis was performed at higher protein concentrations where the tetramer is expected to predominate. SAXS intensities were collected at three nominal protein concentrations (0.8 – 2.6 mg mL−1, 6 – 20x Kd), then extrapolated to infinite dilution to account for mild sample aggregation apparent in the two highest protein concentrations (Figure 4d). The Guinier radius of gyration (Rg) from the extrapolated SAXS data is 41.4 ± 0.3 Å, compared to 36.5 Å for the crystallographic tetramer. We considered the possibility that the 5 Å discrepancy in Rg could be due in part to the incompleteness of the crystal structure, which lacks the N-terminal 27 residues because of disorder. Tetrameric models that included the missing residues were generated from the AllosMod server. The models not only have larger Rg (37.8 – 38.6 Å), but also agree very well with the experimental SAXS curve (χ2 = 0.6 – 0.8). The SAXS curve calculated from one of the tetramer models is shown in Figure 4d (χ2 = 0.6). In contrast, AllosMod models for the ZmALDH12 dimer bearing the disordered N-terminal residues have a much lower Rg and have very poor fit to the experimental scattering profile (Rg = 32.0 – 32.8 Å; χ2 = 57.4 – 60.4, Figure 4d). Attempts to fit a mixture of the AllosMod dimer and tetramer with MultiFoXS did not statistically improve the fit to the experimental data suggesting the tetramer predominates in the SAXS samples, consistent with the relatively high enzyme concentration used. The implications of our results for the in vivo enzyme are unclear because the cellular concentration of ALDH12, and thus oligomeric state, are unknown. It is possible that molecular crowding in the cell may shift the equilibrium observed in sedimentation equilibrium study to favor the tetrameric assembly in planta.
Structural analysis of the ZmALDH12 coenzyme binding site.
ZmALDH12 was co-crystallized with NAD+, and electron density consistent with NAD+ was observed in all four chains of the asymmetric unit (Figure 5a). However, the electron density for the nicotinamide riboside moiety was diffuse and weak, so the NAD+ was conservatively modeled as an ADP fragment. NAD+ adopts the canonical binding pose expected for Rossmann dinucleotide binding proteins41. The coenzyme binds at the C-termini of a 5-stranded β-sheet in an extended conformation (Figure 5b). Several of the interactions with NAD+ in ZmALDH12 are also observed in ALDH4 structures35,42 (Figure 5b and 5c) including a lysine residue Lys224 that interacts with the adenosine ribose and a serine residue Ser277 that hydrogen bonds to the pyrophosphate (Lys233 and Ser287 in MmALDH4). Modeling of the nicotinamide riboside into the active site based on the weak electron density and the pose seen in other ALDHs predicts bidentate hydrogen bonding between a conserved glutamate residue (Glu434) and the nicotinamide ribose.
Figure 5. Coenzyme binding in ZmALDH12.
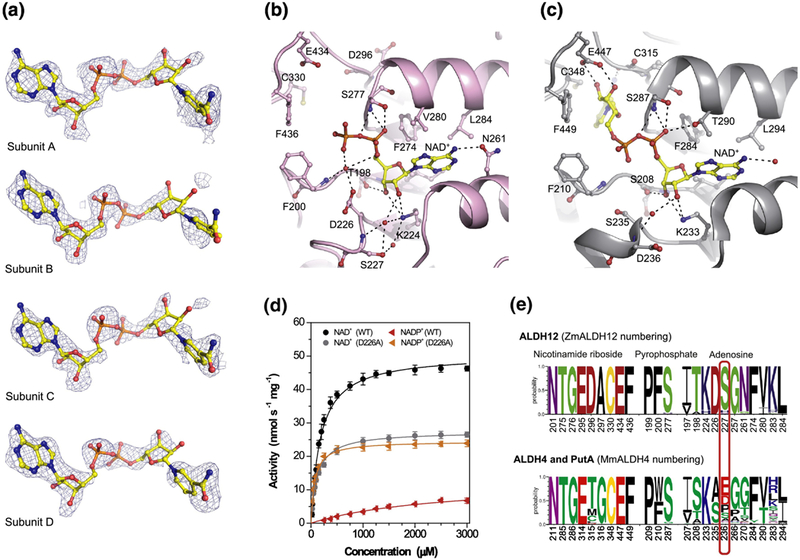
(a) Omit electron density evidence for NAD+. The cage represents a simulated annealing Fo-Fc omit map contoured at 3σ. The nicotinamide riboside moiety does not appear in the PDB deposition but is included here to guide the eye. (b) Binding of NAD+ in ZmALDH12 (pink color, PDB ID: 6D97). The NAD+ molecule is shown in yellow and atom-coded color sticks. Important residues are labeled. (c) NAD+ binding site in the mouse ALDH4 (grey color, PDB ID: 3V9L)35. (d) Saturation curves for NAD+ and NADP+ with ZmALDH12 and D226A variant. The data were measured in 100 mM sodium pyrophosphate buffer, pH 7.5 and 0.3 mM GSAL (sub-saturating concentration). (e) An overview of the conservation of amino acid residues forming the coenzyme-binding site in enzymes from the ALDH12 family, which is compared with those found ALDH4 and PutA isoforms. Sequence logos were made using WebLogo 3 (http://weblogo.threeplusone.com)
Additional structural and biochemical analysis explains the preference for NAD+ over NADP+. Specifically, the Km value for NADP+ is 2872 ± 360 μM, which is 15-times higher than that of NAD+ (Km = 185 ± 14 μM) (Figure 5d and Table 1). The Kd of 186 ± 25 μM for NAD+ was determined by MST, in good agreement with the Km value (Figure S3). Recently published data on rice ALDH1234 revealed an even higher Km of 644 ± 30 μM for NAD+. ZmALDH12 is NAD+-specific due to presence of the Asp226, whose negatively charged carboxylate side chain likely repels the 2’-phosphate group of NADP+. For comparison, the plant ALDH21 family members, which are NADP+-dependent, have alanine at this position32, suggesting that the identity of the residue at this position drives coenzyme preference. To test this hypothesis, the coenzyme preference of the ZmALDH12 D226A mutant was analyzed. The D226A variant displayed comparable catalytic efficiencies for both NAD+ (Km = 119 ± 7 μM) and NADP+ (Km = 71 ± 6 μM) (Figure 5d; Table 1). These results suggest the presence of Asp226 results in a preference of ZmALDH12 to bind NAD+ over NADP+.
Other ALDH superfamily members bearing a glutamate residue at equivalent position of Ser227, such as ALDH2 and ALDH10, display even lower Km values for NAD+ 43,31,44. At the same time, this glutamate also repels the 2’-phosphate group of NADP+. Serine at the equivalent position of Ser227 is strictly conserved in ALDH12 of all Embryophyta plants and heterokonts. This observation is not true for green algae, which have either a histidine, glutamine, serine, or threonine residue at this position. The residue is also not conserved among PutAs and ALDH4s. Many bacterial ALDH4s have a glutamate residue at this position, and ALDH4 isoforms of Chordata phylum, including MmALDH4 and HsALDH4, have an aspartate residue at this position (Figure 5e). Consequently, HsALDH435 displays a lower Km value of 100 μM and higher affinity Kd value of 15 ± 1 μM for NAD+. Likewise, there are certain cases when rearrangement of the glutamate side chain permits NADP+ binding as in the case of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa45 or ALDH4 from Thermus thermophilus24,25 In the latter case, the bacterial ALDH4 displays turnover rate with NADP+ only four times lower than with NAD+.
Analysis of the ZmALDH12 active site.
Attempts at generating a ZmALDH12 structure with the product glutamate bound in the active site either by co-crystallization or soaking experiments were not successful. Nevertheless, the substrate cavity of ZmALDH12 can be analyzed by comparison to that of mouse ALDH4 (MmALDH4) with the product glutamate bound (PDB ID: 3V9K)35. The RMSD between the two active site structures is 0.69 Å and almost all key residues involved is GSAL binding are identical (Figure 6a, 6b). Thus, the MmALDH4 complexes with glutamate or glutarate (PDB ID: 4LH3)28 provide a basis for performing structural analysis of the ZmALDH12 active site combined with site-directed mutagenesis.
Figure 6. Substrate binding site and kinetics of ZmALDH12.
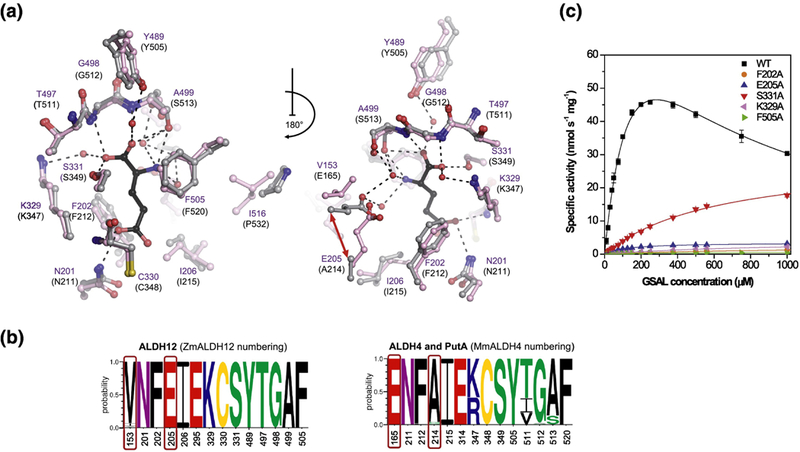
(a) The substrate cavity and surrounding residues in ZmALDH12 colored in pink (PDB ID: 6D97, this work). Residues of MmALDH4 (gray, PDB ID: 3V9K)35 involved in binding of the reaction product glutamate (colored in black) are shown for comparison. A repositioning of active-site glutamate E165 in MmALDH4 and E205 in ZmALDH12 is indicated by the red arrow. (b) An overview of the conservation of amino acid residues forming the substrate-binding site in enzymes from the ALDH12 family, which is compared with those found in ALDH4 used as a reference and numbered as MmALDH4. Sequence logos were made using WebLogo 3. (c) Saturation curves for active-site variants of ZmALDH12 with GSAL. The data were measured in 100 mM pyrophosphate buffer, pH 7.5 using 3.0 mM NAD+ as a coenzyme. Error bars stand for S.D. from four measurements.
The catalytic Cys330 is poised on a loop flanked on either side by conserved Lys329 and Ser331, which likely function to coordinate the carboxylate group of GSAL. The oxyanion hole residue Asn201, which donates a hydrogen bond to the O atom of the aldehyde group, is positioned on a loop adjacent to the catalytic loop. Phe202 combined with Phe505 form an aromatic box that presumably stabilizes the aliphatic chain of GSAL and is conserved in all ALDH4 and ALDH12 members. The aldehyde anchor loop comprising residues 497–499 seals the substrate cavity over both carboxylate and amino groups of substrate/product. Importantly, the loop is in nearly identical conformation as that in MmALDH4 (residues 511–513), suggesting that it stabilizes the amino acid backbone of the substrate/product through hydrogen bonding interactions. The remaining portion of the active site is filled in with two hydrophobic amino acids Ile206 and Ile516. MmALDH4 has the smaller residue proline at this position.
There are two noteworthy differences between the ZmALDH12 and MmALDH4 active sites (Figure 6a, 6b). First, Ser513 of MmALDH4 is replaced by Ala499 in ZmALDH12. This variation is notable because the side chain of Ser513 hydrogen bonds to the α-amino group of GSAL in MmALDH4. This serine side-chain, however, may not be essential: an alanine is present at this position in many bacterial ALDH4 modules of PutAs46,47. Second, the carboxylate of Glu165 in MmALDH4 forms a water-mediated hydrogen bond with the α-amino group of GSAL. The conformation of this glutamate is maintained by Phe169 located on the same helix in the second substrate interaction sphere. We note that both of these residues are absent in ZmALDH12. Instead, the corresponding glutamate Glu205 is located on a neighboring helix and the bulky Phe169 of MmALDH4 corresponds with Leu209. We note that superposition of the ZmALDH12 and MmALDH4 active sites reveals similar positioning of the δ-carboxylate of Glu205, consistent with the capability of this residue to form a water-mediated hydrogen bond with the α-amino group of GSAL. Taken together, the structure and composition of the ZmALDH12 active site is consistent with the enzyme functioning as a GSALDH.
We performed site-directed mutagenesis of ZmALDH12 to assess the role of six structurally conserved active residues and determined kinetic constants for each variant with GSAL and GRSAL (Table 2, Figure 6c and Figure S4). Previous structural analyses of MmALDH4 revealed no conformational differences between glutamate and glutarate binding (GSAL and GRSAL products)28,35. All active-site mutants studied in this work followed Michaelis-Menten kinetics with GSAL (Table 2) except for the coenzyme-site variant D226A. Conversely, all variants except F202A and S331A followed substrate inhibition kinetics when GRSAL was used as the substrate. Circular dichroism (CD) spectroscopy was used to determine whether mutant variants were folded in a similar fashion to the wildtype enzyme. Further, their thermal stability, determined by nanoDSF analysis, was found similar to wildtype (Figure S5).
Table 2. Kinetic parameters for ZmALDH12 active-site mutant variants.
Saturation curves for substrates GSAL and GRSAL were measured in 100 mM pyrophosphate buffer (pH 7.5) containing 3.0 mM NAD+. Kinetic constants Km and kcat including their standard error values were determined using GraphPad Prism 5.0 software. n.d.- not determined, *only specific activity measured with 1 mM substrate.
| Enzyme |
Km (μM) |
kcat (s−1) |
kcat/Km (s−1M−1) |
Km (μM) |
kcat (s−1) |
kcat/Km (s−1M−1) |
|
|---|---|---|---|---|---|---|---|
| GSAL | GRSAL | ||||||
| ZmALDH12 | 198 ± 21 | 6.76 ± 0.47 | 3.4 ± 0.6 × 104 | 84 ± 5 | 1.91 ± 0.21 | 2.3 ± 0.4 × 104 | |
| F202A | 969 ± 45 | 0.15 ± 0.01 | 1.5 ± 0.1 × 102 | 480 ± 28 | 0.21 ± 0.01 | 4.4 ± 0.4 × 102 | |
| E205A | 120 ± 5 | 0.21 ± 0.01 | 1.8 ± 0.1 × 103 | 32 ± 4 | 0.53 ± 0.02 | 1.6 ± 0.3 × 104 | |
| C330A | n.d. | 6.8 ± 0.9 × 10−4* | n.d. | n.d. | 1.7 ± 0.3 × 10−4* | n.d. | |
| S331A | 624 ± 27 | 1.83 ± 0.04 | 2.8 ± 0.2 × 103 | 227 ± 16 | 0.89 ± 0.03 | 3.9 ± 0.4 × 103 | |
| K329A | 958 ± 47 | 0.26 ± 0.01 | 2.7 ± 0.2 × 102 | 442 ± 70 | 0.31 ± 0.04 | 7.0 ± 1.9 × 102 | |
| F505A | 170 ± 18 | 0.03 ± 0.001 | 1.9 ± 0.3 × 102 | 379 ± 41 | 0.52 ± 0.04 | 1.4 ± 0.3 × 103 | |
Mutational analysis identified key residues that are crucial for catalysis. For example, the predicted catalytic Cys330 plays a crucial role in catalysis, as expected; the C330A mutation renders ZmALDH12 nearly inactive (kcat = 6.8 × 10−4 s−1). Importantly, the mutant still binds GSAL with similar affinity as the wild-type enzyme (Kd = 315 μM ± 48 μM), suggesting the defect in catalysis is due to the loss of the β-thiol of Cys330. In addition, both aromatic box residues - Phe202 and Phe505 - also strongly contribute to catalysis. The F202A and F505A variants display 50 and 220-fold lower kcat values for GSAL corresponding to reduced catalytic efficiencies of 0.4% and 0.6%, respectively, compared to wild-type. Further, of the two conserved residues Lys329 and Ser331 that flank the catalytic Cys330 and are predicted to interact with the carboxylate of GSAL, the lysine residue was found more important for catalysis. The K329A variant displays five-fold higher Km value and 26-fold lower kcat value for GSAL strongly reducing the overall catalytic efficiency to 0.8% of the wild-type enzyme. The S331A variant displays only 3.7-fold lower kcat value compared to wild-type. The F505A mutation reveals the interaction of positively charged amino group of GSAL with the negatively charged π-electron system of Phe505. The kcat value is reduced to 0.4% of the wild-type enzyme, while the kcat value for GRSAL is reduced to only 27%. In the case of the E205A variant, the catalytic efficiency value for GSAL is reduced to 5% of wild-type, while that for GRSAL reaches 70% compared to wild-type. Because GRSAL lacks the α-amino group present in GSAL, these results are consistent with the predicted role of Glu205 in the potential water-mediated interaction with the α-amino group of GSAL. Overall, these results reveal that despite low global sequence identity to ALDH4, conservation of key amino acid residues within the active site of ALDH12 results in retention of substrate specificity and GSALDH activity.
ALDH12 gene expression analysis.
We analyzedZmALDH12 expression pattern in maize seedlings during the first two weeks germination. Results show that ZmALDH12 is consistently expressed at higher levels in leaf tissue than in root tissue (Figure 7a). In 3-month old maize, leaves, shoot and main root tissues displayed comparable transcript levels. In reproductive organs, ZmALDH12 expression was higher in silks than in tassels. Transcript accumulation increased in silks after pollination suggesting a possible role for ZmALDH12 in pollen grain germination. This trend is also observed during kernel development. The highest ZmALDH12 transcript levels were observed in kernel samples more than two weeks after pollination. This finding is in agreement with high ALDH12 expression observed in the inflorescence of Arabidopsis and rice19,48. Available transcriptome data show variations among species for expression in root and shoot (leaf) tissues (http://bar.utoronto.ca/). Although ALDH12 transcripts are most abundant in reproductive organs and inflorescences in some species, higher transcript accumulation in roots than in leaves is observed for example in Brachypodium, tomato, poplar and soybean. Because the enzyme connects the proline and arginine metabolic pathways, it is possible these differences reflect differential fine-tuning of both pathways in various plants.
Figure 7. Expression profile of ZmALDH12 gene in maize.
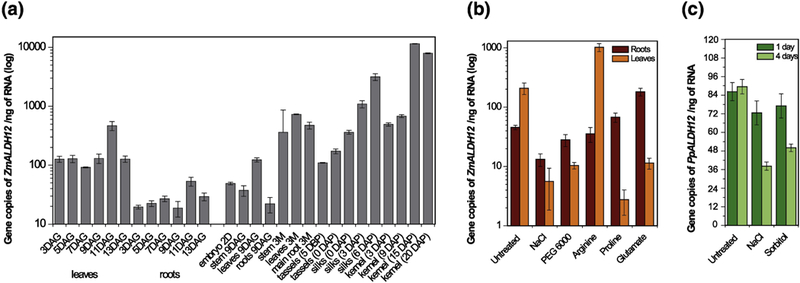
(a) Gene expression of ZmALDH12 studied in leaves and roots during 2 weeks after germination (left side) and in various organs during the lifespan of maize plants (right side). The graph shows transcript levels (in log-scale) detected in 1 ng of total RNA. DAG, days after germination; DAP, days after pollination; DBP, days before pollination. (b) Gene expression of ZmALDH12 in plants exposed to salt (NaCl) and dehydration (PEG600) for 4 days and exposed to exogenous arginine, proline and glutamate for 1 day. Seedlings were grown at 25°C on a hydroponic Hoagland solution with a light/dark photoperiod of 12/12 hours and photon flux density 280 μmol m−2 s−1. Concentrations used: 200 mM NaCl, 20% PEG 6000, 2 mM proline, 2 mM arginine and 50 mM glutamate. Cycle threshold values were normalized with respect to maize elongation factor 1α and β-actin genes and amplification efficiency. (c) Gene expression of PpALDH12 in moss grown in Knop medium exposed to salt (NaCl) and dehydration (sorbitol) for 1 and 4 days.
In many plant species, abiotic stresses, particularly salt and drought stress, induce proline accumulation by increasing biosynthesis and reducing catabolism. Therefore, we exposed 4 day-old maize seedlings to salt and drought stress using 200 mM NaCl or 20% (w/v) PEG 6000 and monitored ZmALDH12 expression in root and leaf tissue every third day for two weeks. During this period, ZmALDH12 transcript accumulated nearly 20-fold lower in leaves compared to the untreated control (Figure 7b). Similar experiments were also performed with moss using 200 mM NaCl or 400 mM sorbitol. Again, reduced PpALDH12 transcript levels were observed (Figure 7c). Thus, our qPCR data suggest proline degradation may be down-regulated under both stresses in moss and maize. Previous studies indicated that AtGSALDH was upregulated during drought stress to maintain high proline levels for osmoprotection, whereas arginine and ornithine levels decreased20. Likewise, rice ALDH12 was upregulated by drought but not by salinity48, and foxtail millet ALDH12 was upregulated by cold, drought, salinity, and abscisic acid49. This differential regulation reveals complex fine-tuning of proline and arginine metabolism among various plant species.
Maize seedlings were also exposed to exogenous proline and arginine, and ZmALDH12 expression was analyzed after 24 hours (Figure 7b). Although ZmALDH12 transcript levels in roots did not significantly differ, ZmALDH12 levels in maize leaves were significantly altered: transcript levels were 5-fold higher in response to arginine, but nearly 100-fold lower in response to proline. This result may indicate an increased contribution of arginine catabolism to cellular glutamate in leaves. Addition of exogenous glutamate resulted in decreased ZmALDH12 expression in the shoot and increased expression in roots. Results of other studies that analyzed ALDH12 expression after treatment with exogenous proline or arginine are conflicting. An earlier study on AtGSALDH showed induction of the ALDH12 transcription 24 hours after addition of exogenous proline14, whereas a study of OsGSALDH showed no changes of ALDH12 transcripts upon treatment with exogenous arginine or proline34.
A recent study of transgenic alfalfa (Medicago sativa) expressing the ALDH12 gene from Cleistogenes songorica (CsALDH12) revealed that CsALDH12 upregulation resulted in increased salt and drought stress tolerance50. Additionally, these transgenic plants also showed an increased K+:Na+ ratio and reduced malondialdehyde (a marker of oxidative stress) accumulation compared to wild-type plants, which are both signs of abiotic stress tolerance. Surprisingly, these plants accumulated 3-fold higher proline levels. The latter result is surprising because Arabidopsis gsaldh mutants also accumulated 3-fold more proline upon salt stress compared to wild-type while no difference was observed under normal conditions19,20. In agreement with the observed results in Arabidopsis, the human autosomal recessive disorder type II hyperprolinemia, which is defined by a deficiency in ALDH4 activity, results in elevated proline levels51. A possible explanation for these seemingly contradictory results is that the plant genes involved in proline biosynthesis and catabolism are simultaneously induced under certain stress conditions20,52.
Conclusions
This work provides the first structural and biochemical analyses of NAD+-dependent GSALDHs (ALDH12 family members) from two plant species - maize and Physcomitrella. Despite the very low sequence identity of plant GSALDHs to non-plant GSALDHs (ALDH4 family members), all key residues in the active-site are conserved among both families except for the position of glutamate residue near the amino group of GSAL. Along with the observed down-regulation of ALDH12 in moss and maize upon salt and drought stress, the structure highlights a similar functionality for ALDH12 and ALDH4 across species in proline metabolism.
Because the transition from ALDH4 to ALDH12 occurred among eukaryotic protists upon acquisition of a prokaryotic endosymbiont that later became the chloroplast, we speculate that the selective pressure for the switch could have arisen from altered proline/ornithine (arginine) homeostasis resulting in a new GSALDH, i.e., ALDH12, having expanded substrate selectivity for aldehydes such as N-acetyl-GSAL, an intermediate of Arg biosysnthesis. This aldehyde, which has an acetyl group attached to the amino group of GSAL, is synthesized from glutamate via several acetylated intermediates and further metabolized to the ornithine in the chloroplast. Future studies could focus on understanding any contribution to N-acetyl-GSAL oxidation that may occur due to the repositioning of this ALDH12 active-site glutamate. An oxygen atom of the acetyl group of N-acetyl-GSAL may interact with sidechain of the glutamate (Glu205 in ZmALDH12). These results could provide insight into the role of ALDH12 in modulating ornithine, and therefore arginine, metabolism.
Materials and methods
Cloning of ALDH12 from Zea mays and Physcomitrella patens
The total RNA from various maize organs (Zea mays cv. Cellux, Morseva) and from Physcomitrella (‘Gransden 2004’ strain) at the protonema stage was extracted using the RNAqueous kit and plant RNA isolation aid (Thermo Fisher Scientific). The cDNA was synthesized using Superscript III RT and oligo dT primers (Thermo Fisher Scientific). The sequences coding for ZmALDH12 (1650 bp) and PpALDH12 (1716 bp, Phytozome accession Pp3c5_4940V3.1) were amplified with the Accuprime Pfx polymerase (Thermo Fisher Scientific) using gene-specific primers with SacI and XhoI sites for the maize gene and BamHI and KpnI sites for the moss gene (listed in Table S1). The genes were cloned into pCDFDuet, pETDuet and pET32b vectors (Merck). These constructs were used as templates to prepare Δ1–16ZmALDH12 and Δ1–36PpALDH12 variants devoid of putative mitochondrial signal sequences by PCR using phosphorylated primers (Table S1). The PCR products were treated with the DpnI, gel purified and ligated using the T4 DNA ligase. All constructs were transformed into T7 Iq express E. coli cells (New England Biolabs).
qPCR analysis
RNA from four biological replicates was transcribed in two independent reactions, and PCR was performed in triplicate. Diluted cDNA samples were used as templates in qPCR with TaqMan Gene Expression Master Mix, 300 nM primers, and 250 nM TaqMan 6-FAM TAMRA probe on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). The primers and TaqMan probes (listed in Table S3) were designed using Primer Express 3.0 software. Plasmid constructs carrying ORFs of maize and moss ALDH12 genes were used both as a template for determination of PCR efficiencies of designed probes and primer pairs as well as to verify their specificity. Cycle threshold values were normalized with respect to maize elongation factor 1α and β-actin genes and amplification efficiency31. For stress experiments, 3 day-old germinated Zea mays seedlings were transferred to a hydroponic Hoagland solution and grown at 25°C with a light/dark photoperiod of 12/12 h and photon flux density 280 μmol m−2 s−1 in the absence or presence of 200 mM NaCl or 20% PEG 6000 for 4 – 14 d. The effect of presence of 2 mM L-proline, 2 mM L-arginine or 50 mM L-glutamate was determined after 24 h incubation. Moss was grown in liquid Knop medium and in the absence or presence of 200 mM NaCl and 400 mM sorbitol for 4 days.
Site-directed mutagenesis of ZmALDH12
All mutants were generated by PCR in 30 cycles using tail-to-tail oriented phosphorylated primers, with the mutation at the 5’ end of one of the primers (Table S1). The following ZmALDH12 mutants were generated: F202A, E205A, K329A, C330A, S331A and F505A. PCR products were treated with Dpnl, gel purified, and ligated using T4 DNA ligase. Clones were transformed into T7 Iq express competent cells (New England Biolabs).
CD and thermostability measurements
The far-UV CD spectra of wild-type ZmALDH12 and its variants were recorded on a J-815 spectropolarimeter (JASCO) using a 0.1 cm quartz cell and 0.4 mg mL−1 enzyme. Thermostability was measured by nanoDSF on a Prometheus NT.48 instrument (NanoTemper Technologies) with a back-reflection aggregation detection at a range of 20 – 95°C and with a heating rate of 1°C min−1. Protein unfolding was followed by tryptophan fluorescence intensity at 330 and 350 nm in various buffers covering pH range of 7.0 – 9.0 in the presence or absence of 100 mM NaCl and 5% (v/v) glycerol. Data analysis was performed using NT melting control software. The melting temperature (Tm) was determined by detecting the maximum of the first derivative of the fluorescence ratios (F350/F330) after fitting experimental data with a polynomial function. Data were measured in triplicate.
Protein production and enzyme kinetics
Protein production was induced with 0.5 mM isopropyl-β-thiogalactopyranoside (IPTG) and cells were grown at 18°C overnight. Enzyme variants were purified using a HisPur Cobalt column (Thermo Fisher Scientific) and by size-exclusion chromatography (SEC) using a HiLoad 26/60 Superdex 200 column (GE Healthcare Scientific). The final buffer for PpALDH12 contained 150 mM sodium pyrophosphate pH 7.5 and 5% (w/v) glycerol. The final buffer for ZmALDH12 and its variants contained 150 mM Tris-HCl pH 7.5, 100 mM NaCl, and 5% (w/v) glycerol. Protein content was measured using Coomassie plus protein assay kit with BSA as a standard (Thermo Fisher Scientific).
Enzyme activity was measured by monitoring the NAD(P)H formation (ɛ340 = 6.22 mM−1 cm−1) on an Agilent UV-Vis spectrophotometer 8453 and by monitoring fluorescence emission of NADH at 460 nm upon excitation at 365 nm on FluoroLog-3 spectrofluorometer (Horiba) at 30°C. Britton-Robinson buffers in the pH range of 6.0 – 10.0 and adjusted to a constant ionic strength of 0.15 M were used to determine pH optimum with 0.5 mM GSAL. The reaction mixture (2 mL) contained ten microliters of the diluted enzyme, which was mixed with 1.9 mL of Britton Robinson buffer directly in the cuvette and followed by addition of NAD+ (3 mM final).
Substrate screening was done upon addition of various aldehydes at a final concentration of 1 mM in 100 mM sodium pyrophosphate pH 7.5 and 3.0 mM NAD+. Saturating curves for GSAL, GRSAL, and AASAL were measured with 3.0 mM NAD+. Kinetic constants Km and kcat were determined using GraphPad Prism 5.0 (Table 1 and Table 2). Data were fit to the Michaelis-Menten equation. When substrate inhibition was observed, data were analyzed by nonlinear regression using Michaelis-Menten equation that accounts for partial substrate inhibition: v = Vmax[S]/(Km+ [S](1+[S]/Ki)), where v is the determined initial velocity, Vmax is the maximal velocity, [S] is the concentration of the substrate, Km is the substrate concentration at half-maximal velocity, Ki is the substrate inhibition constant. Kinetic curves for coenzymes were measured in the same buffer and in the presence of 0.3 mM GSAL, which is a sub-saturating concentration providing the maximal experimentally attainable activity and is not affected by a substrate inhibition. Therefore, the kinetic constants calculated for the coenzymes are only apparent.
α-Aminoadipic semialdehyde ethylene acetal was purchased from Chiralix. GRSAL was synthesized by oxidation of the D,L-2-aminoadipic acid with chloramine-T and further purified on Dowex 5 053. Elementary aliphatic and aromatic aldehydes, D- and L-glyceraldehyde 3-phosphate together with 4-aminobutyraldehyde and 3-aminopropionaldehyde diethylacetals were purchased from Sigma-Aldrich. Free aldehydes were freshly prepared by heating their acetals in a plugged test tube with 0.2 M HCl for 10 min and then kept on ice44. GSAL was synthesized by oxidation of D,L-5-hydroxylysine with sodium periodate54, purified on Dowex 50, and stored in 1 M HCl at 4°C. On the day of experiment, it was neutralized with 6 M NaOH and kept on ice.
Determination of substrate and coenzyme affinity
MST was used to determine binding affinities of wild-type ZmALDH12 and C330A variant for GSAL, GRSAL and NAD+. Both proteins were fluorescently labeled RED-tris-NTA dye (NanoTemper Technologies) using a 1:1 dye/protein molar ratio. Labeled protein was adjusted to 50 nM with MST buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 10 mM MgCl2) supplemented with 1 mg mL−1 BSA. Sixteen ligand dilutions covering a concentration range from 20 mM - 610 nM for NAD+ and 5 mM - 150 nM for GSAL and GRSAL were prepared. Measurements were performed on a Monolith NT.115 instrument (NanoTemper Technologies) at 25°C with 5 s/30 s/5 s laser off/on/off times and continuous sample fluorescence recording. Glutamate binding affinity was determined on a Monolith NT.LabelFree instrument using 500 nM unlabeled ZmALDH12 in MST buffer. Data analysis was conducted on three independently pipetted experiments.
Phylogenetic analysis
Amino acid alignment was performed with MUSCLE55, treated with Gblocks56, and the maximum likelihood phylogeny with bootstrap analysis was performed with PhyML v3.0 using LG amino acid replacement matrix57. Sequences were retrieved from UniProt (http://www.uniprot.org/), Phytozome 12 (https://phytozome.jgi.doe.gov/), and NCBI (https://www.ncbi.nlm.nih.gov/). Further, ALDH12 sequences were retrieved from the JGI genome portal (https://genome.jgi.doe.gov/) and ESTs from “Green algal transcriptomes for phylogenetics and comparative genomics” (https://dx.doi.org/10.6084/m9.figshare.1604778). The following sequences were retrieved: Klebsormidium flaccidum (comp6544_c0_seq4), Spirogyra pratensis (comp7165_c0_seq2), Mesostigma viride (comp42030_c0_seq1) and Chaetosphaeridium globosum (comp32236_c0_seq1). All sequence accession numbers are listed in Table S2.
Crystallization of ZmALDH12
ZmALDH12 for crystallization was purified using Ni-NTA followed by anion exchange chromatography (HiTrap Q) and SEC on Superdex 200 13/300 column equilibrated with 50 mM Tris pH 7.8, 50 mM NaCl, 0.5 mM tris(2-caboxyethyl)phosphine (TCEP), and 5% (v/v) glycerol. ZmALDH12 was concentrated to 2 mg mL−1, supplemented with 1 mM NAD+, quick-cooled in liquid nitrogen, and stored at −80°C. The crystallization condition was obtained using the Morpheus crystal screen (Molecular Dimensions) in a sitting drop format using an Oryx8 crystallization robot (Douglas Instruments). Crystals were improved by a combination of seeding and switching to a microbatch crystallization method using Al’s oil. The crystallization condition contained 0.09 M sodium nitrate, sodium phosphate, and ammonium sulfate; 50% v/v of a precipitant mixture containing 25% v/v 2-methyl-2,4-pentanediol, 25% w/v polyethylene glycol 1000, and 25% w/v polyethylene glycol 3350; and 0.1 M Tris/bicine pH 8.5. Crystals were prepared for low temperature data collection by direct plunge into liquid nitrogen without the need for additional cryoprotection.
Data collection, phasing, refinement, and model building
X-ray diffraction data were collected on Advanced Light Source beamline 4.2.2 using a Taurus-1 CMOS detector in shutterless mode. The dataset consisted of 900 images spanning 180°. The dataset was integrated and scaled with XDS58. Intensities were merged and converted to amplitudes with Aimless59. Initial phases were generated using the BALBES automated molecular replacement pipeline server60. The amino acid sequence of His-tagged ZmALDH12 and the structure factor amplitudes from Aimless were input. The best solution generated by BALBES was obtained with a search model derived from a betaine ALDH (PDB ID: 4MPY)61 and had a Q-factor of 0.49 and 68% probability of being correct. Notably, the sequence identity between the search model and ZmALDH12 was only 21% over 490 residues.
The initial phases from molecular replacement were improved by density modification and automated ab initio model building using PHENIX62. The map from BALBES/REFMAC, the experimental structure factor amplitudes, and the ZmALDH12 sequence were input to phenix.autobuild for automated ab initio model building with density modification. The resulting model from phenix.autobuild included 1,978 residues out of an expected 2,188 (90% complete) and had Rwork of 0.20 and Rfree of 0.27. The model from phenix.autobuild was used as the starting point for iterative rounds of model building in COOT63 and refinement in PHENIX64 and BUSTER65. Structure quality was validated using MolProbity66. The electron density for the nicotinamide riboside moiety of NAD+ was weak and diffuse, so only the ADP portion of the coenzyme was refined in the final model. Data processing and refinement statistics for the final model are listed in Table 3.
Table 3.
Data collection and refinement statistics.
| ZmALDH12 | |
|---|---|
| Space group | C2 |
| Asymmetric unit | 2 dimers |
| Unit cell parameters | a = 160.6 |
| (Å, °) | b = 123.7 |
| c = 103.7 | |
| β =105.6 | |
| Wavelength | 1.000 |
| Resolution (Å) | 61.87–2.20 (2.24–2.20) |
| Observations | 345979 (12672) |
| Unique reflections | 98196 (4545) |
| Rmerge(I) | 0.092 (1.007) |
| Rmeas(I) | 0.109 (1.241) |
| Rpim(I) | 0.057 (0.712) |
| Mean I/σ | 11.6 (1.1) |
| Mean CC1/2 | 0.996 (0.456) |
| Completeness (%) | 99.3 (93.4) |
| Multiplicity | 3.5 (2.8) |
| No. protein residues | 2089 |
| No. of atoms | |
| Protein | 16116 |
| NAD+ | 108 |
| Water | 578 |
| Rwork | 0.1646 (0.2521) |
| Rfreeb | 0.2100 (0.2959) |
| RMSD bond lengths (Å) | 0.003 |
| RMSD bond angles (°) | 0.676 |
| Ramachandran plotc | |
| Favored (%) | 97.31 |
| Allowed (%) | 2.69 |
| Outliers (%) | 0.00 |
| Clashscore (PR)c | 2.35 (100%) |
| MolProbity score (PR)c | 1.34 (99%) |
| Average B-factor (Å2) | |
| All atoms | 36.7 |
| Protein (chains A/B/C/D) | 35.2/37.4/36.9/37.6 |
| NAD+ | 38.5 |
| Water | 35.4 |
| NAD occupancy | 0.7 |
| Coordinate error (Å)d | 0.29 |
| PDB ID | 6D97 |
Values for the outer resolution shell of data are given in parentheses.
5 % test set.
From MolProbity. The percentile ranks (PR) for Clashscore and MolProbity score are given in parentheses.
Maximum likelihood-based coordinate error estimate reported by phenix.refine.
Analytical ultracentrifugation
Sedimentation equilibrium data were collected for ZmALDH12 and PpALDH12 at three protein concentrations (0.2, 0.4, and 0.8 mg mL−1) and three rotor speeds (6000, 9000, and 12000 rpm). Protein and reference buffer were loaded into a sedimentation equilibrium cell equipped with a six-sector charcoal-Epon centerpiece. Initially, the cell was allowed to equilibrate for 16 h at 6000 rpm, after which the absorbance (280 nm) was measured as a function of radial position. For measurements at 9000 rpm and 12,000 rpm, the samples were equilibrated for eight hours prior to acquisition of the first scan. At each rotor speed, data were collected at hourly intervals for six hours (six scans). Data were analyzed as previously described40.
SAXS data collection and analysis
Purified ZmALDH12 was dialyzed overnight against a buffer containing 50 mM Tris-HCl pH 7.8, 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol. A sample of the dialysate was kept for the SAXS background measurement. SAXS data were collected at the Advanced Light Source beamline 12.3.1 using the SAXS mail-in program67. Data were collected in shutterless mode using a Pilatus detector. Scattering intensities were measured at three protein concentrations: 0.9 mg mL−1, 1.7 mg mL−1, and 2.6 mg mL−1. For each protein concentration, 33 evenly spaced images were acquired over 10.2 s (0.3 s/frame).
Composite scattering curves were calculated using PRIMUS68. For the two lowest concentrations, 33 scattering curves (10.2 s) were averaged and used in analysis. For the high concentration sample, the first three images (0.9 s) were averaged due to radiation damage apparent after the third image. The three composite scattering curves were extrapolated to infinite dilution to account for mild aggregation apparent in the high concentration sample. PRIMUS was used to perform Guinier analysis, and GNOM69 was used to calculate the distance distribution function of the extrapolated SAXS curve. AllosMod-FoXS70,71 was used to both generate modified atomic models including the N-terminal residues that are disordered in the crystal structure, and calculate theoretical SAXS curves from these models. The experimental SAXS data along with the FoXS fits for the AllosMod tetramer and dimer were deposited in the SASBDB72 (PMID: 25352555) under the accession code SASDE96.
Accession numbers
A sequence of ZmALDH12 was submitted to GenBank (MF663524). The atomic coordinates and structure factors for ZmALDH12 have been deposited in the Protein Data Bank (PDB ID 6D97). The SAXS data have been deposited in the SASBDB (SASDE96).
Supplementary Material
Highlights.
ALDH12 is an NAD+-dependent glutamate γ-semialdehyde dehydrogenase in plants
The first crystal structure of ALDH12 is reported
Key active-site residues of ALDH12 involved in substrate binding were identified
ALDH12 displays the closest functional and sequence relationship to ALDH4
ALDH12 emerged from ALDH4 during the evolution of the endosymbiotic plant ancestorALDH12 gene expression in maize and moss is downregulated by salinity and drought
Acknowledgements
This work was supported by the NIGMS of the National Institutes of Health (NIH) under award numbers R01GM065546 and R01GM093123, grant 18–07563S from the Czech Science Foundation, grant L01204 from the National Program of Sustainability I (the Ministry of Education, Youth and Sports, Czech Republic) and grant IGA_PrF_2017_016 from Palacky University. This research also used resources of the Advanced Light Source (ALS), which is a DOE Office of Science User Facility under contract no. DE-AC02–05CH11231. Additional support for the SYBILS beamline at the ALS came from the NIH project MINOS (R01GM105404). We thank Jay Nix for help with X-ray diffraction data collection at beamline 4.2.2 of the Advanced Light Source and Katherine Burnett for collecting SAXS data through the SIBYLS mail-in program
Abbreviations:
- ALDH
aldehyde dehydrogenase
- AASAL
α-aminoadipate-semialdehyde
- BCA
bicinchoninic acid assay
- CD
circular dichroism
- GSAL
L-glutamate-γ-semialdehyde
- GSALDH
L-glutamate-γ-semialdehyde dehydrogenase
- GRSAL
glutaric-γ-semialdehyde
- HsALDH4
human ALDH4A1
- MST
microscale thermophoresis
- nanoDSF
nano differential scanning fluorimetry
- P5C
Δ1-pyrroline-5-carboxylate
- PDB
Protein Data Bank
- MmALDH4
Mus musculus ALDH4A1
- PpALDH12
Physcomitrella patens ALDH12
- SAXS
small-angle X-ray scattering
- TCEP
tris(2-caboxyethyl)phosphine
- ZmALDH12
Zea mays ALDH 12
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Forte-McRobbie CM, Pietruszko R (1986) Purification and characterization of human liver “high Km” aldehyde dehydrogenase and its identification as glutamic gamma-semialdehyde dehydrogenase. J Biol Chem 261, 2154–63. [PubMed] [Google Scholar]
- 2.Farrés J, Julià P, Parés X (1988) Aldehyde oxidation in human placenta. Purification and properties of 1-pyrroline-5-carboxylate dehydrogenase. Biochem J 256, 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearne SL, Wolfenden R (1995) Glutamate gamma-semialdehyde as a natural transition state analogue inhibitor of Escherichia coli glucosamine-6-phosphate synthase. Biochemistry 34, 11515–20. [DOI] [PubMed] [Google Scholar]
- 4.Tanner JJ (2017) Structural Biology of Proline Catabolic Enzymes. Antioxid Redox Signal. doi: 10.1089/ars.2017.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol 118, 1233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funck D, Eckard S, Muller G (2010) Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol 10, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hare PD, Cress WA (1997) Metabolic implications of stress induced proline accumulation in plants. Plant Growth Regul 21, 79–102. [Google Scholar]
- 8.Schwacke R, Grallath S, Breitkreuz KE, Stransky E, Stransky H, Frommer WB, Rentsch D (1999) LeProT1, a transporter for proline, glycine betaine, and gamma-amino butyric acid in tomato pollen. Plant Cell 11, 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cecchini NM, Monteoliva MI, Alvarez ME (2011) Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol 155, 1947–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Becker DF (2015) Connecting proline metabolism and signaling pathways in plant senescence. Front Plant Sci 6, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter G, Todd CD, Trovato M, Forlani G, Funck D (2015) Physiological implications of arginine metabolism in plants. Front Plant Sci 6, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores T, Todd CD, Tovar-Mendez A, Dhanoa PK, Correa-Aragunde N, Hoyos ME, Brownfield DM, Mullen RT, Lamattina L, Polacco JC (2008) Arginase-negative mutants of Arabidopsis exhibit increased nitric oxide signaling in root development. Plant Physiol 147, 1936–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funck D, Stadelhofer B, Koch W (2008) Ornithine-S-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol 8, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deuschle K, Funck D, Hellmann H, Däschner K, Binder S, Frommer WB (2001) A nuclear gene encoding mitochondrial Δ1-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J 27, 345–356. [DOI] [PubMed] [Google Scholar]
- 15.Stránská J, Tylichová M, Kopečný D, Snégaroff J, Šebela M (2010) Biochemical characterization of pea ornithine-delta-aminotransferase: substrate specificity and inhibition by di- and polyamines. Biochimie 92, 940–8. [DOI] [PubMed] [Google Scholar]
- 16.Elthon TE, Stewart CR (1981) Submitochondrial location and electron transport characteristics of enzymes involved in proline oxidation. Plant Physiol 67, 780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forlani G, Scainelli D, Nielsen E (1997) Δ1-pyrroline-5-carboxylate dehydrogenase from cultured cells of potato: purification and properties. Plant Physiol 113, 1413–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayliffe MA, Roberts JK, Mitchell HJ, Zhang R, Lawrence GJ, Ellis JG, Pryor TJ (2002) A plant gene up-regulated at rust infection sites. Plant Physiol 129, 169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB (2004) The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16, 3413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A (2009) Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem 284, 26482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134, 1683–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasiliou V, Bairoch A, Tipton KF, Nebert DW (1999) Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 9, 421–34. [PubMed] [Google Scholar]
- 23.Brocker C, Vasiliou M, Carpenter S, Carpenter C, Zhang Y, Wang X, Kotchoni SO, Wood AJ, Kirch HH, Kopečný D, Nebert DW, Vasiliou V (2013) Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237, 189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inagaki E, Ohshima N, Takahashi H, Kuroishi C, Yokoyama S, Tahirov TH (2006) Crystal structure of Thermus thermophilus Δ1-pyrroline-5-carboxylate dehydrogenase. J Mol Biol 362, 490–501. [DOI] [PubMed] [Google Scholar]
- 25.Inagaki E, Ohshima N, Sakamoto K, Babayeva ND, Kato H, Yokoyama S, Tahirov TH (2007) New insights into the binding mode of coenzymes: structure of Thermus thermophilus Delta1-pyrroline-5-carboxylate dehydrogenase complexed with NADP+. Acta Crystallogr Sect F Struct Biol Cryst Commun 63 462–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pemberton TA, Srivastava D, Sanyal N, Henzl MT, Becker DF, Tanner JJ (2014) Structural studies of yeast delta(1)-Pyrroline-5-carboxylate dehydrogenase (ALDH4A1): active site flexibility and oligomeric state. Biochemistry 53, 1350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, Tanner JJ (2012) The three-dimensional structural basis of type II hyperprolinemia. J Mol Biol 420, 176–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pemberton TA, Tanner JJ (2013) Structural basis of substrate selectivity of Delta(1)-pyrroline-5-carboxylate dehydrogenase (ALDH4A1): Semialdehyde chain length. Arch Biochem Biophys 538, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu LK, Becker DF, Tanner JJ (2017) Structure, function, and mechanism of proline utilization A (PutA). Arch Biochem Biophys 632, 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali NO, Jeusset J, Larquet E, Le Cam E, Belitsky B, Sonenshein AL, Msadek T, Débarbouillé M (2003) Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149, 739–750. [DOI] [PubMed] [Google Scholar]
- 31.Končitiková R, Vigouroux A, Kopečná M, Andree T, Bartoš J, Šebela M, Moréra S, Kopečný D (2015) Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7. Biochem J 468, 109–123. [DOI] [PubMed] [Google Scholar]
- 32.Kopečná M, Vigouroux A, Vilim J, Končitiková R, Briozzo P, Hájková E, Jašková L, von Schwartzenberg K, Šebela M, Moréra S, Kopečný D (2017) The ALDH21 gene found in lower plants and some vascular plants codes for a NADP+-dependent succinic semialdehyde dehydrogenase. Plant J 92, 229–243. [DOI] [PubMed] [Google Scholar]
- 33.Kopecny D, Končitíková R, Tylichová M, Vigouroux A, Moskalikovâ H, Šoural M, Sebela M, Moréra S (2013) Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate. J Biol Chem 288, 9491–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forlani G, Bertazzini M, Zarattini M, Funck D (2015) Functional characterization and expression analysis of rice S(1)-pyrroline-5-carboxylate dehydrogenase provide new insight into the regulation of proline and arginine catabolism. Front Plant Sci 6, 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, Tanner JJ (2012) The three-dimensional structural basis of type II hyperprolinemia. J Mol Biol 420, 176–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell SA, Davis GE (2000). Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA 97, 13009–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–97. [DOI] [PubMed] [Google Scholar]
- 38.Tanner JJ (2015) SAXS fingerprints of aldehyde dehydrogenase oligomers. Data in Brief 5, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo M, Singh RK, Tanner JJ (2013) Structural determinants of oligomerization of delta(1)-pyrroline-5-carboxylate dehydrogenase: identification of a hexamerization hot spot. J Mol Biol 425, 3106–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korasick DA, Tanner JJ, Henzl MT (2017) Impact of disease-Linked mutations targeting the oligomerization interfaces of aldehyde dehydrogenase 7A1. Chem Biol Interact 276, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottoms CA, Smith PE, Tanner JJ (2002) A structurally conserved water molecule in Rossmann dinucleotide-binding domains. Protein Sci 11, 2125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava D, Schuermann JP, White TA, Krishnan N, Sanyal N, Hura GL, Tan A, Henzl MT, Becker DF, Tanner JJ (2010) Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc Natl Acad Sci USA 107, 2878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrés J, Wang X, Takahashi K, Cunningham SJ, Wang TT, Weiner H (1994) Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem 269, 13854–13860. [PubMed] [Google Scholar]
- 44.Tylichová M, Kopečný D, Moréra S, Briozzo P, Lenobel R, Snégaroff J, Šebela M (2010) Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes. J Mol Biol 396, 870–882. [DOI] [PubMed] [Google Scholar]
- 45.González-Segura L, Rudiño-Piñera E, Muñoz-Clares RA, Horjales E (2009) The crystal structure of a ternary complex of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa provides new insight into the reaction mechanism and shows a novel binding mode of the 2’-phosphate of NADP+ and a novel cation binding site. J Mol Biol 385, 542–557. [DOI] [PubMed] [Google Scholar]
- 46.Singh H, Arentson BW, Becker DF, Tanner JJ (2014) Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc Nat Acad Sci USA 111, 3389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korasick DA, Gamage TT, Christgen S, Stiers KM, Beamer LJ, Henzl MT, Becker DF, Tanner JJ (2017) Structure and characterization of a class 3B proline utilization A: Ligand-induced dimerization and importance of the C-terminal domain for catalysis. J Biol Chem 292, 9652–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao C, Han B (2009) Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 431, 86–94. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z, Chen M, Xu ZS, Li LC, Chen XP, Ma YZ (2014) Characteristics and expression patterns of the aldehyde dehydrogenase (ALDH) gene superfamily of foxtail millet (Setaria italica L.). PLoS One 9, e101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan Z, Zhang D, Zhang J, Di H, Wu F, Hu X, Meng X, Luo K, Zhang J, Wang Y (2015) Co-transforming bar and CsALDH genes enhanced resistance to herbicide and drought and salt stress in transgenic Alfalfa (Medicago sativa L.). Front Plant Sci 6, 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valle DL, Phang JM, Goodman SI (1974) Type 2 hyperprolinemia: absence of delta1-pyrroline-5-carboxylic acid dehydrogenase activity. Science 185, 1053–4. [DOI] [PubMed] [Google Scholar]
- 52.Sharma S, Verslues PE (2010) Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ 33, 1838–51. [DOI] [PubMed] [Google Scholar]
- 53.Adams E Chan Y-F (1971) Preparation and assay of glutaric semialdehyde. Methods Enzymol 17B, 171–173. [Google Scholar]
- 54.Williams I, Frank L (1975) Improved chemical synthesis and enzymatic assay of delta-1-pyrroline-5-carboxylic acid. Anal Biochem 64, 85–97. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res, 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castresana J (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17, 540–552. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol, 59, 307–321. [DOI] [PubMed] [Google Scholar]
- 58.Kabsch W (2010) XDS. Acta Crystallogr. D Biol. Crystallogr, 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans PR, Murshudov GN (2013) How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69, 1204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long F, Vagin AA, Young P, Murshudov GN (2008) BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr 64, 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C, Joo JC, Brown G, Stolnikova E, Halavaty AS, Savchenko A, Anderson WF, Yakunin AF (2014) Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus. Appl Environ Microbiol 80, 3992–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO (2011) BUSTER version 2.1.0 Cambridge, United Kingdom: Global Phasing Ltd. [Google Scholar]
- 66.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL 2nd, Tsutakawa SE, Jenney FE Jr., Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6, 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. JAppl Crystallogr 36, 1277–1282. [Google Scholar]
- 69.Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25, 495–503. [Google Scholar]
- 70.Schneidman-Duhovny D, Hammel M, Sali A (2010) FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38, W540–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weinkam P, Pons J, Sali A (2012) Structure-based model of allostery predicts coupling between distant sites. Proc Natl Acad Sci USA 109, 4875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI (2015) SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res 43, D357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


