Abstract
Accumulating evidence has indicated that the dysregulation of microRNAs (miRNAs) is involved in the pathogenesis o retinoblastoma (RB); however, the potential role of miR-98 in RB remains elusive. In the present study, it was demonstrated that miR-98 is downregulated in RB tissues and cell lines, and its expression significantly associated with clinicopathological features, including differentiation, N classification and largest tumor base; patients with low miR-98 expression levels exhibited significantly poorer overall survival. Overexpression of miR-98 was suggested to suppress RB cell growth, migration and invasion. In addition, insulin-like growth factor-1 receptor (IGF1R), a well-reported oncogene, was identified as a potential target of miR-98 via a luciferase assay, reverse transcription-quantitative polymerase chain reaction and western blotting. Correlation analysis revealed a significantly negative correlation between miR-98 and IGF1R expression in tumor tissues (n=60). In addition, the results of the present study demonstrated that IGF1R function as an oncogene by promoting RB cell viability, migration and invasion. Furthermore, restoration of IGF1R was observed to reverse the anticancer effects of miR-98 on RB cell viability, migration and invasion. Importantly, the findings of the present study indicated that miR-98 suppressed RB cell growth and metastasis by inhibiting the IGF1R/k-Ras/Raf/mitogen activated protein kinase kinase/extracellular signal-regulated kinase signaling pathway. Collectively, the present study proposed that miR-98 may serve as a novel prognostic biomarker and therapeutic target in the treatment of RB.
Keywords: retinoblastoma, microRNA-98, insulin-like growth factor-1 receptor/k-Ras/Raf/mitogen activated protein kinase kinase/extracellular signal-regulated kinase signaling pathway, prognostic biomarker
Introduction
Retinoblastoma (RB) is the most common intraocular malignancy usually occurring in childhood worldwide (1). Retinoblastoma affects the eyes of children under the age of five years and accounts for 5% of blindness in children (1). In addition, RB can rapidly fill the eye, extend into the optic nerve and the central nervous system, and easily metastasizes (2). Therefore, it is urgent to identify the molecular mechanism underlying the tumorigenesis of RB; further investigation into the novel molecular targets affecting tumor growth and metastasis are required for the clinical diagnosis and treatment of patients with RB.
MicroRNAs (miRNAs) are a family of short, small, noncoding RNA molecules consisting of 21-23 nucleotides, which modulate the post-transcriptional regulation of target genes via the suppression of translation or promotion of RNA degradation (3,4). It is well reported that miRNAs have identified to act as oncogenes or tumor suppressor genes in numerous types of cancers; miRNAs may target cell signaling pathways to regulate a variety of biological processes, including cell proliferation, apoptosis, differentiation and migration (5,6). Numerous studies have demonstrated that the ectopic expression of miRNAs can regulate cell growth, apoptosis, migration, or invasion in RB (7-9). Huang et al (10) revealed that inhibition of miR-182 may suppress cell viability, invasion and angiogenesis in RB through inactivation of the PI3K/AKT pathway. miR-145 has been identified to be downregulated in RB tissues and cell lines, and suppressed RB cell proliferation, migration and invasion by targeting ADAM metallopeptidase domain 19 in vitro (11). Previously, increasing evidence reported that miR-98 may be associated with various cancers, including prostate cancer, head and neck squamous cell carcinoma and breast cancer (12-14). miR-98 has been demonstrated to suppress prostate cancer growth, and tumor angiogenesis and invasion by targeting matrix metalloproteinase-11 and activating receptor-like kinase-4 (12,14); however, the molecular mechanism underlying the role of miR-98 in the development and progression of RB is unknown.
In the present study, the miRNA expression profiles associated with RB tumorigenesis were determined and the molecular mechanism underlying the biological function of miRNAs in the development of RB was investigated. The results of the present study demonstrated that miR-98 was downregulated in RB tissues and its expression may be considered as a predictor of poor prognosis in RB. In addition, the findings of the present study revealed that miR-98 inhibits RB cell growth and metastasis by suppressing the insulin like growth factor-1 receptor (IGF1R)/k-Ras/Raf/mitogen activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway, which suggested the potential value of miR-98 in the clinical diagnosis and treatment of patients with RB.
Materials and methods
Patients and specimens
Human RB samples were obtained from 60 patients from the Department of Ophthalmology, The First People's Hospital of Shangqiu (Shangqiu, China), between February 2014 and November 2016. All of the 60 RB patients received enucleation or enucleation + chemotherapy ± radiation therapy. Of the 60 RB patients, there were 24 females and 36 males. The age of the patients ranged from 0-7 years, with an average age of 2.6 years. All 60 RB patients were confirmed histopathologically using the based on the American Joint Commission for Cancer (AJCC) staging system (15) and all tumors were classified based on the International Retinoblastoma Staging System (16). The clinicopathological features of patients with RB were summarized in Table I. A total of 9 normal retinal samples from patients who had succumbed to mortality due to conditions other than ophthalmologic diseases were collected in the First People's Hospital of Shangqiu. Of the 9 patients with normal retinas, there were 5 females and 4 males. The age of the patients ranged from 0-8 years, with an average age of 2.7 years. All patients provided written informed consent for the use of human specimens for clinical research. The present study was approved by the Institute Research Ethics Committee of The First People's Hospital of Shangqiu.
Table I.
Association between miR-98 and clinicopathological features of patients with retinoblastoma.
| Feature | Total n=60 | miR-98
|
P-value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Sex | 0.8251 | |||
| Male | 36 | 13 | 23 | |
| Female | 24 | 8 | 16 | |
| Age at presentation (years) | 0.4211 | |||
| ≤5 | 49 | 16 | 33 | |
| >5 | 11 | 5 | 6 | |
| Tumor enucleated location | 0.9352 | |||
| Right | 31 | 11 | 20 | |
| Left | 29 | 10 | 19 | |
| T classification | 0.1964 | |||
| T1-2 | 38 | 11 | 27 | |
| T3-4 | 22 | 10 | 12 | |
| Clinical stage | 0.2415 | |||
| I-II | 17 | 4 | 13 | |
| III-IV | 43 | 17 | 26 | |
| N classification | 0.0251a | |||
| N0 | 26 | 5 | 21 | |
| N1 + 2 | 34 | 16 | 18 | |
| Differentiation | 0.0136a | |||
| Well and moderate | 21 | 3 | 18 | |
| Poorly | 39 | 18 | 21 | |
| Largest tumor base (mm) | 0.0242a | |||
| ≤15 | 37 | 17 | 20 | |
| >15 | 23 | 4 | 19 | |
P<0.05.
Cell culture
The human RB cell lines, including WERI-Rb-1, Y79 and SO-RB50, and the normal retinal pigmented epithelium cell line ARPE-19 were employed in the present study. ARPE-19, Y79 and SO-RB50 cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The WERI-Rb-1 cell line was obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 100 IU/ml penicillin (Thermo Fisher Scientific, Inc.), and 100 mg/ml streptomycin (Thermo Fisher Scientific, Inc.), at 37°C in a humidified atmosphere containing 5% CO2. Based on our preliminary experiment, the two RB cell lines, SO-RB50 and Y79, with the lowest expression levels of miR-98, were selected for subsequent analysis in the present study.
miRNA microarray analysis
miRNA microarray analysis was used to determine the miRNA expression profiles in clinical samples. Total RNAs were isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and the miRNA fraction was further purified by a mirVana miRNA isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The isolated miRNAs were labeled with Hy3 using the miRCURY array labeling kit (Exiqon; Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocols, and hybridized with miRCURY locked nucleic acid (LNA) microRNA arrays (v8.0; Exiqon; Qiagen, Inc.). Microarray analysis was conducted using a Genepix 4000B scanner (Molecular Devices, LLC, Sunnyvale, CA, USA) and Genepix Pro 6.0 software (Molecular Devices, LLC).
Vector constructs and cell transfection
The specific miR-98 mimics (cat. no. HMI0982) or inhibitor (cat. no. HLTUD0460), and corresponding negative control (NC; mimics NC, cat. no. HMC0002 and inhibitor NC, cat. no. NCSTUD001) were obtained from Sigma-Aldrich (Merck KGaA), as well as specific small interfering RNA for IGF1R (si-IGF1R-1, si-IGF1R-2 and si-IGF1R-3), and si-Scramble were designed and purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences of si-IGF1R were: 5′-CAA CAGUGGUCAUCAUGGAACUGUTT-3′ (sense) and 5′-AUC AGUUCCAUGAUGACCACUGUUGTT-3′ (antisense). The sequences of si-Scramble were: 5′-UUCUCCGAACGUGUC ACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGA GAATT-3′ (antisense). The IGF1R 3′-UTR was cloned into a pMIR-REPORT luciferase reporter vector (Ambion; Thermo Fisher Scientific, Inc.) using SpeI and MluI or HindIII and SpeI restriction sites for luciferase assays. For plasmid construction, the IGF1R 3′-UTR was cloned into the XhoI and KpnI sites of the pcDNA3.1 expression vector (Invitrogen Thermo Fisher Scientific, Inc.). For the transfection of siRNAs or miRNA mimics/inhibitors, cells that reached 50% confluence were transfected with siRNAs or miRNA mimics/inhibitors at a final concentration of 50 nM using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in 6-well plates according to the manufacturer's protocols. For the luciferase assay, the cells were cultured in 96-well plates and co-transfected with 400 ng of pMIR-REPORT luciferase reporter vector and 50 ng miR-98 mimic, inhibitor or corresponding NC using Lipofectamine® 2000 reagent. IGF1R ectopic expression was achieved through pcDNA3.1-IGF1R (500 ng) transfection using Lipofectamine® 2000 reagent, with an empty pCDNA3.1 vector used as a control. Untransfected cells were employed as the blank group. At 48 h after transfection, the transfection efficiency was evaluated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) or western blot analysis, and these cells were employed for subsequent analysis.
RT-qPCR
Total RNA from tissues and cultured cells were extracted using TRIzol reagent according to the manufacturer's protocols. For RT, 10 ng of RNA, 50 nM of stem-loop RT primer, 1X RT buffer, 0.25 mM each of dNTPs, 3.33 U/μl MultiScribe reverse transcriptase and 0.25 U/μl RNase inhibitor (all from TaqMan MicroRNA Reverse Transcription kit, Applied Biosystems; Thermo Fisher Scientific, Inc.) were used. Reaction mixtures (15 μl) were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. qPCR was performed using the Applied Biosystems 7500 instrument; 20 μl PCR reaction mixture included 1.33 μl of RT product, 1X TaqMan (AmpErase UNG) Universal PCR Master Mix and 1 μl of primer and probe mix of the TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Reactions were incubated in a 96-well optical plate at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 10 min. The primers used for miR-98 were 5′-TGTATGACTGCCGTATGTTTCCTATT-3′ (forward) and 5′-AATTCTTAAAGTATGCTTTCCATTCC-3′ (reverse). The primers used for IGF1R were 5′-GCGGTTCTGTTGATAGTGG-3′ (forward) and 5′-GCCTCGTTCACC GTCTTA-3′ (reverse). The primers for U6 5′-CTC GCT TCG GCA GCA CA-3′ (forward) and 5′-AAC GCT TCA CGA ATT TGC GT-3′ (reverse). The primers for GAPDH were 5′-AAC GTG TCA GTG GTG GAC CTG-3′ (forward) and 5′-AGT GGG TGT CGC TGT TGA AGT-3′ (reverse). The expression of miR-98 and IGF1R in tissues or cells were normalized to that of expression of U6 and GAPDH, respectively. Experiments were performed in triplicate, and data were calculated using the relative quantification (2−∆∆Cq) method as previously described (17).
Cell viability analysis
Cell viability was evaluated with an MTT assay according to the manufacturer's protocols. Briefly, cells were seeded into 96-well plates at a density of 1×104 cells and incubated overnight at 37°C. Then, 5 μl MTT solution (5 mg/ml, Sigma-Aldrich; Merck KGaA) was added to each well, incubated at 37°C for 24 h at 1, 2, 3 and 4 days. Subsequently, the reaction was terminated with 200 μl dimethyl sulfoxide. The blank control wells contained medium only (RPMI-1640 medium). Following incubation with MTT, the absorbance of the samples was measured with an ELISA microplate reader at 490 nm (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate.
Cell apoptosis assay
Cell apoptosis was measured via flow cytometry analysis. The cells (2×105) were seeded on 6-well plates and were incubated at 37°C in a humidified chamber containing 5% CO2 for 48 h following transfection. The cells then were collected and washed twice with pre-chilled PBS. Following treatment with 0.25% trypsin at room temperature for 5 min, cells were harvested and washed with pre-cooled PBS once. Subsequently, the cells were resuspended in binding buffer (BD Pharmingen; BD Biosciences, Franklin Lakes, NJ, USA) and stained with 5 μl Annexin V-fluorescein isothiocyanate (FITC) and 1 μl propidium iodide (PI, 50 μg/ml) (BD Biosciences). The stained cells were analyzed using a flow cytometer (Cytomics FC-500, Beckman Coulter, Brea, CA, USA), which demonstrated the living cells as scatter diagram at Q4 (FITC-/PI-). The results of apoptotic cells in the early stage were presented in Q3 (FITC+/PI-), and Q2 presented the apoptotic cells in the advanced stage (FITC+/PI+). The portion of apoptosis was determined as the apoptosis rate (apoptotic cell number/total cell number) in the early stage (%; Q3) plus apoptosis rate in the advanced stage (%; Q2). MultiCycle Software version 5.0 (Phoenix Flow Systems, San Diego, CA, USA) for Windows 7 32-bit (Microsoft Corporation, Redmond, WA, USA) was used to analyze the experimental data. Each experiment was performed at least three times.
Western blot analysis
Cells were lysed as previously described (18). A Bicinchoninic Acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) was used to determine protein concentration. Protein samples (60 μg) were separated via 10% SDS-PAGE (Sigma-Aldrich; Merck KGaA) and transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.). Following blocking with 5% skimmed milk at room temperature for 1 h, the membranes were incubated primary antibodies against proliferating cell nuclear antigen (PCNA; 1:1,000; cat. no. SC-56; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), Ki-67 (1:1,000; cat. no. SC-7846; Santa Cruz Biotechnology, Inc.), cleaved-caspase-3 (1:1,000; cat. no. 9661; Cell Signaling Technology, Inc., Danvers, MA, USA), B-cell lymphoma-2 (Bcl-2)-associated x (Bax; 1:500; cat. no. SC-526; Santa Cruz Biotechnology, Inc.), Bcl-2 (1:1,000; cat. no. SC-492; Santa Cruz Biotechnology, Inc.), E-cadherin (1:1,000; cat. no. SC-7870; Santa Cruz Biotechnology, Inc.), α-catenin (1:1,000; cat. no. SC-59890; Santa Cruz Biotechnology, Inc.), fibronectin (1:1,000; cat. no. SC-18825; Santa Cruz Biotechnology, Inc.), vimentin (1:1,000; cat. no. SC-5565; Santa Cruz Biotechnology, Inc.), IGF1R (1:1,000; cat. no. SC-81464; Santa Cruz Biotechnology, Inc.), k-Ras (1:1,000; cat. no. SC-30; Santa Cruz Biotechnology, Inc.), phosphorylated (p)-Raf/1 (Ser338, 1:1,000; cat. no. 9427; Cell Signaling Technology, Inc.), Raf/1 (1:1,000; cat. no. ab137435; Abcam), MEK1/2 (1:1,000; cat. no. SC-436; Santa Cruz Biotechnology, Inc.), p-MEK1/2 (1:1,000; cat. no. 9154S; Cell Signaling Technology, Inc.), p-ERK1/2 (1:1,000; cat. no. SC-81492; Santa Cruz Biotechnology, Inc.) and ERK1/2 (1:1,000; cat. no. SC-514302; Santa Cruz Biotechnology, Inc.). β-actin (1:750; cat. no. A5060; Sigma-Aldrich; Merck KGaA) served as an internal control. A horseradish peroxidase-conjugated antibody (1:1,000; cat. no. SC-2060; Santa Cruz Biotechnology, Inc.) was employed. Subsequently, the protein bands were scanned on an X-ray film using an enhanced chemiluminescence detection system (PerkinElmer, Inc., Waltham, MA, USA). The band intensity was quantified with 3.2 AlphaImager 2200 software (Alpha Innotech Corporation, San Leandro, CA, USA). Analysis was performed at least three times.
Cell invasion assay
A total of 1×105 cells were plated onto 25 μg Matrigel-coated 24-well FluoroBlok cell culture inserts with an 8 μm-pore membrane (Corning Incorporated, Corning, NY, USA). After 48 h of incubation at 37°C, cells were resuspended in 200 μl serum-free RPMI-1640 medium and placed into the upper chamber of the insert without Matrigel. Medium-supplemented with 5% FBS was used as a chemoattractant in the lower chamber. The cells were incubated at 37°C for 24 h and cells that did not invade via the pores were removed with a cotton swab. Cells on the lower surface of the membranes were fixed with 20% methanol at 4°C for 30 min and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 20 min. The number of cells on the lower surface of the membrane was calculated in six random fields using an IX71 inverted microscope (Olympus Corporation, Tokyo, Japan) and then photographed (magnification, ×200). Experiments were performed in triplicate.
Cell migration assay
The wound healing assay was used to measure cell migration. The Y79 or SO-RB50 cells were plated into a 6-well plate and cultured at 37°C for 24 h until 80-90% confluence was attained. Wounds were created in monolayers of cells using a 200 μl sterilized tip. Following two washes with PBS, the cells were incubated in RPMI-1640 medium (10% FBS). After 48 h of incubation at 37°C, the wounds were analyzed in six random fields using an IX71 inverted microscope (Olympus Corporation) (magnification, ×200). The wound area was determined and the percentage of closure of denuded area was calculated using ImageJ software (version 1.44, National Institutes of Health, Bethesda, MD, USA).
Luciferase assay
The miR-98 mimics, inhibitor and corresponding negative control (NC) were obtained from Shanghai GenePharma Co., Ltd. The potential binding sequence between IGF1R and miR-98 was investigated using TargetScan (Version 7.1, http://www.targetscan.org) and miRanda (Version 3.3a, http://www.microrna.org/microrna/home.do/). The wild-type (wt) and mutant (mut) human IGF1R 3′-untranslated regions (UTRs) containing the putative binding site of miR-98 were constructed, respectively and were subsequently cloned into a pMIR-REPORT luciferase reporter vector (Ambion; Thermo Fisher Scientific, Inc.). Site-directed mutagenesis of the miR-98 target-site in the IGF1R 3′-UTR was performed using a QuikChange kit (Qiagen, Inc.). For the luciferase assay, the Y79 cells were cultured in 96-well plates and co-transfected with 400 ng of pMIR-IGF1R-wt-3′-UTR or pMIR-IGF1R-mut-3′-UTR, and 50 ng miR-98 mimic, inhibitor or corresponding NC using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h following transfection, the relative firefly luciferase activity was normalized with that of Renilla luciferase as measured using a Dual-Light luminescent reporter gene assay (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Immunohistochemistry
Immunohistochemistry was performed using paraformaldehyde-fixed (ice-cold 4% paraformaldehyde for 24 h) paraffin sections. k-Ras (1:1,000; cat. no. SC-30; Santa Cruz Biotechnology, Inc.), p-ERK1/2 (1:1,000; cat. no. SC-81492; Santa Cruz Biotechnology, Inc.) and p-MEK1/2 (1:1,000; cat. no. 9154S; Cell Signaling Technology, Inc.) antibodies were used in immunohistochemistry followed by a streptavidin peroxidase-conjugated method (19). Following washing with PBS, the slides were incubated with horseradish peroxide-conjugated secondary antibody (1:2,000, cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) for 30 min at 37°C; detection was conducted using 3,3′-diaminobenzidine (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) and stained with hematoxylin for 15 sec at room temperature. The slides were examined under an inverted light microscope (Nikon Eclipse TS 100, Nikon Corporation, Tokyo, Japan). The percentage of positive tumor cells was graded according to the following criteria: 0, <10%; 1, 10-30%; 2, 31-50%; 3, >50%. Patients with different k-Ras, p-ERK1/2 and p-ERK1/2 expression levels in RB tissues were defined as the low-expression group (0 or 1) and the high-expression group (2 or 3).
Statistical analysis
All statistical analyses were performed using SPSS software (version 18.0, SPSS, Inc., Chicago, IL, USA). Experiments were conducted in triplicate and data were presented as the mean ± standard deviation. The clinicopathological features of RB patients that were associated with miR-98 low and high expression groups were determined by the χ2 test. The median value of miR-98 expression level (1.932) was selected as the cut-off value for the miR-98 high and low expression groups. The correlation between IGF1R and miR-98 expressions was tested with two-tailed Pearson's correlation analysis. A Student's t-test or one way analysis of variance with a Tukey's post hoc test were also performed. Survival analysis was determined using the Kaplan-Meier method followed by a log-rank test. P<0.05 and P<0.01 were considered to indicate a significant difference and a highly significant difference, respectively.
Results
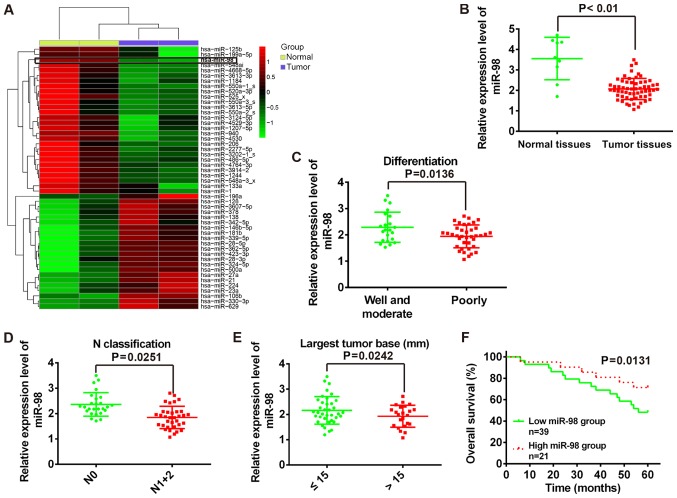
miR-98 is downregulated in RB tissues and is associated with poor patient survival rate
To investigate miRNA expression associated with the tumorigenesis of RB, miRNA microarray analysis was performed to determine miRNA expression profiles in RB tissues and normal retinas. As presented in Fig. 1A, numerous miRNAs were downregulated; and miR-98 was observed as one of the most notably downregulated in human RB tissues compared with in normal tissues. With primers specific to miR-98, qPCR analysis was performed to further verify the results of microarray analysis. The present study reported that the expression levels of miR-98 were significantly downregulated in RB tissues (n=60) compared with in normal tissues (n=9) (P<0.01; Fig. 1B). In addition, the association between miR-98 expression and the clinicopathological features of patients with RB were analyzed (Table I). The results demonstrated that low miR-98 expression levels were significantly associated with differentiation (P=0.0136; Fig. 1C), N classification (P=0.0251; Fig. 1D) and largest tumor base (P=0.0242; Fig. 1E); however, no significant association between miR-98 expression levels and patient sex, age, tumor enucleated location, T classification or clinical stage were observed (Table I). To investigate the association between miR-98 expression and the prognosis of patients with RB, Kaplan-Meier survival analysis was performed to plot survival curves of RB, which revealed that patients with low miR-98 expression levels (n=39) exhibited significantly poorer overall survival compared with in patients with high miR-98 expression (n=21) (P=0.0131; Fig. 1F). Collectively, the data indicated that miR-98 is downregulated in RB tissues and low miR-98 expression may be a predictor of poor prognosis in patients with RB. The findings of the present study suggest that miR-98 may function as a potential biomarker in the clinical diagnosis of RB.
Figure 1.
Downregulation of miR-98 in human RB tissues is associated with poor patient survival. (A) Microarray analysis was used to determine the miRNA expression profile in RB tissues and normal retinas. Red and green colors indicate high and low expression in the heatmap, respectively. (B) Relative expression of miR-98 was determined using RT-qPCR analysis in tumor tissues (n=60) and normal tissues (n=9). (C-E) RT-qPCR was used to detect the miR-98 expression levels in tumor tissues which were grouped according to clinicopathological features of patients with RB: Differentiation, N classification and largest tumor base. (F) Kaplan-Meier survival analysis of patients with RB (n=60) with low and high miR-98 expression levels. miR, microRNA; RB, retinoblastoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
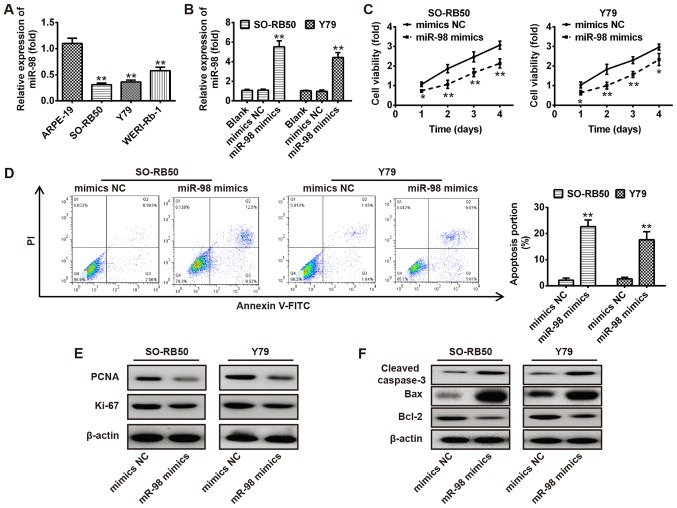
Overexpression of miR-98 suppresses RB cell proliferation and promotes apoptosis
To investigate the functions of miR-98 in RB cells, the expression levels of miR-98 were investigated in human RB cells, including WERI-Rb-1, Y79 and SO-RB50, and normal retinal pigmented epithelium cells ARPE-19 via RT-qPCR. As presented in Fig. 2A, the expression levels of miR-98 were significantly downregulated in all RB cells compared with in the normal retinal pigmented epithelium cells ARPE-19 (P<0.01). Then, SO-RB50 and Y79 cells were transfected with miR-98 mimics or NC; RT-qPCR was used to evaluate the overexpression efficiency of miR-98. As presented in Fig. 2B, miR-98 was significantly overexpressed in SO-RB50 and Y79 cells compared with the corresponding NC and blank groups (P<0.01). Subsequently, an MTT assay and flow cytometry analysis were used to measure cell viability and apoptosis of SO-RB50 and Y79 cells transfected with miR-98 mimics or NC. The results revealed that overexpression of miR-98 significantly reduced cell viability and promoted apoptosis compared with in the control (P<0.01; Fig. 2C and D). To further investigate the molecular mechanisms underlying the effects of miR-98 on cell growth, western blot analysis was conducted to detect the expression of proliferation markers (PCNA and Ki-67) and apoptosis-associated proteins (cleaved-caspase-3, Bax and Bcl-2) in SO-RB50 and Y79 cells transfected with miR-98 mimics or mimics NC, respectively. The results revealed that overexpression of miR-98 markedly decreased the expression of proliferation markers (PCNA and Ki-67) and anti-apoptotic protein (Bcl-2), and increased that of the pro-apoptotic proteins (cleaved-caspase-3 and Bax) (Fig. 2E and F). Collectively, the results suggested that overexpression of miR-98 inhibited RB cell viability and induced cell apoptosis.
Figure 2.
Overexpression of miR-98 suppresses RB cell growth. (A) RT-qPCR was conducted to determine the expression levels of miR-98 in human RB cell lines, including WERI-Rb-1, Y79 and SO-RB50, and normal retinal pigmented epithelium cells, ARPE-19. (B) miR-98 expression levels in SO-RB50 and Y79 cells following transfection with miR-98 mimics or NC were measured via RT-qPCR. (C) An MTT assay was used to analyze the cell viability of SO-RB50 and Y79 cells transfected with miR-98 mimics or NC. (D) Cell apoptosis was measured via flow cytometry analysis in SO-RB50 and Y79 cells transfected with miR-98 mimics or NC. (E and F) SO-RB50 and Y79 cells were transfected as aforementioned (C), and western blot analysis was performed to detect the expression of proliferation markers (PCNA and Ki-67) and apoptosis-associated proteins (cleaved-caspase-3, Bax and Bcl-2) in SO-RB50 and Y79 cells, respectively. β-actin was used as an internal control for protein loading. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 vs. ARPE-19 cells, blank or NC. Bcl-2, B cell lymphoma-2; Bax, Bcl-2-associated X; FITC, fluorescein isothiocyanate; miR, microRNA; NC, negative control; PCNA, proliferating cell nuclear antigen; PI, propidium iodide.
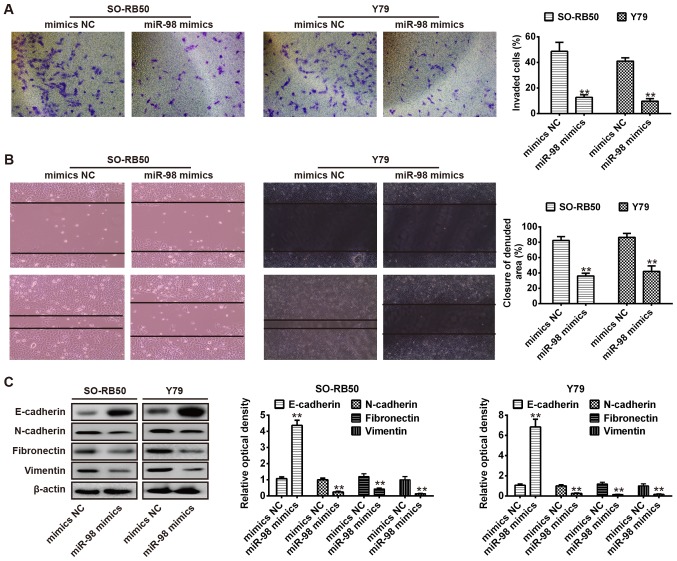
Overexpression of miR-98 suppresses cell migration and invasion
The present study further investigated the effects of miR-98 overexpression on cell invasion and migration, key factors associated with malignant progression and metastasis (20). As presented in Fig. 3A, overexpression of miR-98 significantly inhibited the invasion of SO-RB50 and Y79 cells compared with the NC group (P<0.01). Additionally, a wound healing assay was performed to determine cell migration following transfection with miR-98 mimics or NC. The results demonstrated that overexpression of miR-98 also significantly inhibited the migration of SO-RB50 and Y79 cells compared with in the NC group (P<0.01; Fig. 3B). The data indicated that overexpression of miR-98 suppressed cell migration and invasion; however, the underlying molecular mechanism requires further investigation.
Figure 3.
Overexpression of miR-98 suppresses cell migration and invasion via inhibiting cellular epithelial-mesenchymal transition. (A) SO-RB50 and Y79 cells were transfected with miR-98 mimics or NC, and cell invasion was evaluated using a Transwell invasion assay (magnification, ×200). (B) Wound healing assay was conducted to determine cell migration in SO-RB50 and Y79 cells transfected with miR-98 mimics or NC (magnification, ×200). (C) Expression of epithelial marker E-cadherin, and the mesenchymal markers N-cadherin, fibronectin and vimentin in SO-RB50 and Y79 cells following transfection with miR-98 mimics or NC were analyzed. Data are presented as the mean ± standard error of three independent experiments. **P<0.01 vs. NC. miR, microRNA; NC, negative control.
Previous studies revealed that tumor cell invasion and metastasis are closely associated with numerous different processes, including an epithelial to mesenchymal transition (EMT) in cancer cells. The progression of epithelial tumors to more aggressive metastatic forms was reported during EMT, which was associated with the upregulation of mesenchymal protein markers, including N-cadherin and vimentin, and loss of the epithelial protein marker E-cadherin (21-23). It is well reported that EMT in a variety of cancers is modulated by posttranscriptional mechanisms, such as that of miRNAs (24,25). Therefore, to investigate the molecular mechanism of miR-98-suppressed RB cell metastasis, SO-RB50 and Y79 cells were transfected with miR-98 mimics or NC, and western blot analysis was applied to determine the expression of the epithelial marker, E-cadherin and the mesenchymal markers, N-cadherin, fibronectin and vimentin. As presented in Fig. 3C, overexpression of miR-98 resulted in notable E-cadherin upregulation in SO-RB50 and Y79 cells; however, the expression levels of N-cadherin, fibronectin and vimentin were downregulated. Collectively, these findings suggested that miR-98 may suppress RB cell migration and invasion via the inhibition of EMT.
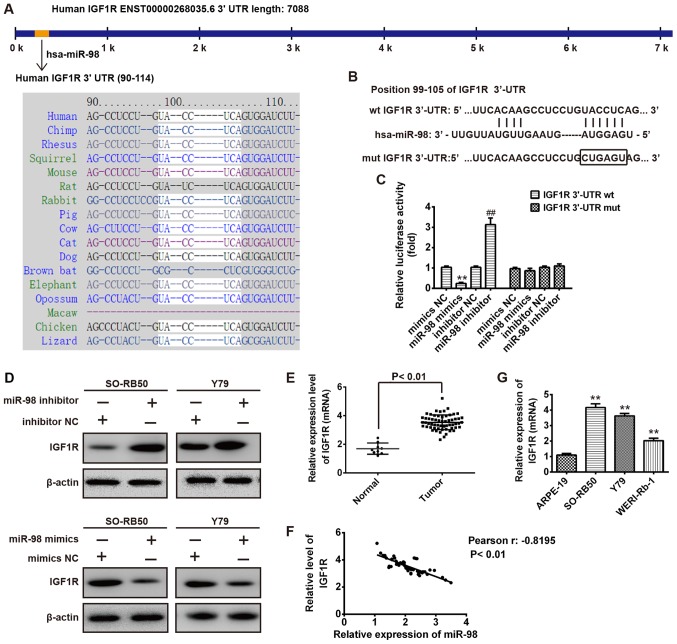
IGF1R is a target gene of miR-98 in RB cells
TargetScan and miRanda were used to predict the target genes of miR-98; IGF1R was determined as a potentials target (Fig. 4A). To confirm this bioinformatic prediction, luciferase reporter plasmids harboring the wt or mut 3′-UTR segments of IGF1R (Fig. 4B). Then, SO-RB50 cells were co-transfected with wt or mut reporter plasmid along with miR-98 mimics, inhibitor or NC, and the luciferase activity was analyzed. As presented in Fig. 4C, miR-98 mimics significantly inhibited the luciferase activity compared with the NC (P<0.01), whereas miR-98 inhibitor significantly increased the luciferase activity in the presence of the wt 3′-UTR compared with in the inhibitor NC (P<0.01). In addition, miR-98 did not notably suppress the luciferase activity of the reporter vector containing 3′-UTR of IGF1R with mutations in the miR-98-binding site (Fig. 4C). To further verify whether IGF1R expression is modulated by miR-98, SO-RB50 and Y79 cells were transfected with miR-98 mimics, inhibitor or NC, and the IGF1R protein expression levels were determined by western blotting. The present study observed that upregulation of miR-98 decreased IGF1R protein expression; however, downregulation of miR-98 increased the IGF1R expression levels compared with in the NC group (Fig. 4D). To further clarify the association between miR-98 and IGF1R in RB, RT-qPCR analysis was conducted to detect IGF1R expression in human RB tissues (n=60). The results demonstrated that the expression levels of IGF1R were significantly increased in the 60 cases of tumors compared with in normal retinas (n=9; P<0.01; Fig. 4E). More importantly, correlation analysis revealed a significantly negative correlation between miR-98 and IGF1R expression in 60 tumor tissues (r=-0.8195, P<0.01; Fig. 4F). Furthermore, IGF1R expression levels were significantly increased in the RB cell lines, WERI-Rb-1, Y79 and SO-RB50, compared with in ARPE-19 cells (P<0.01; Fig. 4G). The data indicated that miR-98 negatively modulated IGF1R expression and that a negative correlation between miR-98 an IGF1R may detected in clinical samples.
Figure 4.
IGF1R is a target gene of miR-98 in RB cells. (A) Prediction of IGF1R as a target of miR-98 in different species. (B) Schematic view of the miR-98 putative targeting site in the wt and mut 3′-UTR of IGF1R. (C) Relative luciferase activity of IGF1R wt or mut 3′-UTR in SO-RB50 cells transfected with miR-98 mimic, inhibitor or corresponding NC. **P<0.01 vs. NC, ##P<0.01 vs. inhibitor NC. (D) SO-RB50 and Y79 cells were transfected with miR-98 mimic, inhibitor or corresponding NC, and IGF1R expression was determined via western blot analysis. β-actin served as an internal control. (E) Reverse transcription-quantitative polymerase chain reaction analysis was used to determine IGF1R expression levels in human RB samples (n=60) and normal retinas (n=9). (F) Negative correlation between IGF1R and miR-98 expression levels in patients with RB (r=-0.8195, P<0.01). (G) IGF1R expression levels were measured in human RB cell lines WERI-Rb-1, Y79 and SO-RB50, and the normal retinal pigmented epithelium cell line ARPE-19. **P<0.01 vs. ARPE-19. Data are presented as the mean ± standard deviation of three individual experiments. IGF1R, insulin-like growth factor-1 receptor; mut, mutant; wt, wild-type.
IGF1R regulates RB cell viability, migration and invasion
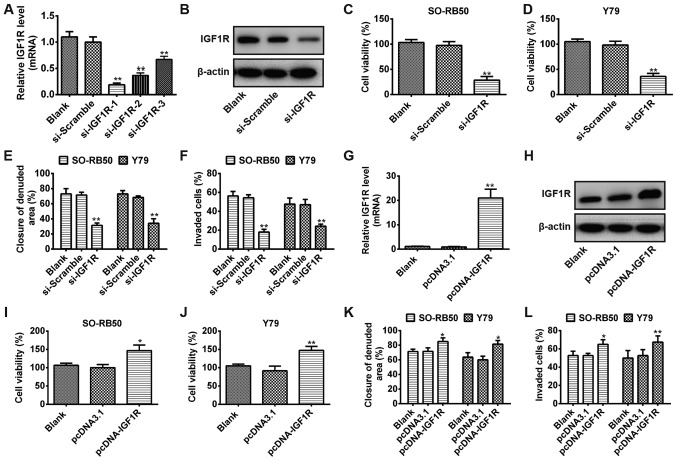
To investigate whether IGF1R act as oncogene in RB cells, the expression levels of IGF1R were downregulated via transfection with si-IGF1R. SO-RB50 cells were transfected with three si-IGF1Rs (si-IGF1R-1, si-IGF1R-2 and si-IGF1R-3), or a scrambled NC (si-Scramble). As presented in Fig. 5A, the siRNA (si-IGF1R-1) with the most significant effect compared with in the blank and si-Scramble groups was selected for subsequent analysis (P<0.01). In addition, the protein expression levels of IGF1R were notably downregulated in RB cells transfected with si-IGF1R-1 compared with in the si-Scramble group (P<0.01; Fig. 5B). The results indicated that IGF1R expression was successfully downregulated via transfection of RB cells with si-IGF1R. Subsequently, MTT, wound healing and Transwell invasion assays were performed to evaluate the viability, and migration and invasive abilities of SO-RB50 and Y79 cells transfected with si-IGF1R or si-Scramble, respectively. The results indicated that downregulation of IGF1R significantly suppressed cell viability, migration and invasion compared with in the blank and si-Scramble groups (P<0.01) (Fig. 5C-F). To further determine whether ectopic expression of IGF1R modulates cell growth and metastasis, RB cells were transfected with pcDNA-IGF1R or vector control pcDNA3.1. The mRNA expression levels of IGF1R were reported to be significantly increased as observed by RT-qPCR (Fig. 5G); western blotting revealed a notable increase in the protein expression of IGF1R (Fig. 5H). Additionally, the MTT, wound healing and Transwell invasion assays revealed that overexpression of IGF1R significantly promoted the cell viability, migration and invasion compared with in pcDNA3.1-transfected cells (P<0.01; Fig. 5I-L). The results of the present study suggested that IGF1R may act as an oncogene via the regulation of cell viability, and migration and invasive abilities of RB cells.
Figure 5.
IGF1R regulates RB cell viability, migration and invasion. (A) SO-RB50 cells were transfected with three si-IGF1Rs (si-IGF1R-1, si-IGF1R-2 and si-IGF1R-3) or si-Scramble, and the mRNA expression of IGF1R was measured by RT-qPCR. (B) Western blot analysis was used to determine IGF1R protein expression in SO-RB50 cells transfected with si-IGF1R or si-Scramble. (C-F) SO-RB50 and Y79 cells were transfected with si-IGF1R or si-Scramble; MTT, wound healing and Transwell invasion assays were used to evaluate cell viability, migration and invasion, respectively. **P<0.01 vs. blank or si-Scramble. (G and H) SO-RB50 cells were transfected with pcDNA-IGF1R or vector control pcDNA3.1, and the mRNA and protein expression of IGF1R were determined using RT-qPCR and western blot analysis, respectively. (I-L) MTT, wound healing and Transwell invasion assays were conducted to assess the cell viability, migration and invasion in SO-RB50 and Y79 cells transfected with pcDNA-IGF1R or vector control pcDNA3.1, respectively. *P<0.05, **P<0.01 vs. blank or pcDNA3.1. Data were presented as the mean ± standard deviation of three independent experiments. IGF1R, insulin-like growth factor-1 receptor; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si, small interfering RNA; si-Scramble, scrambled negative control.
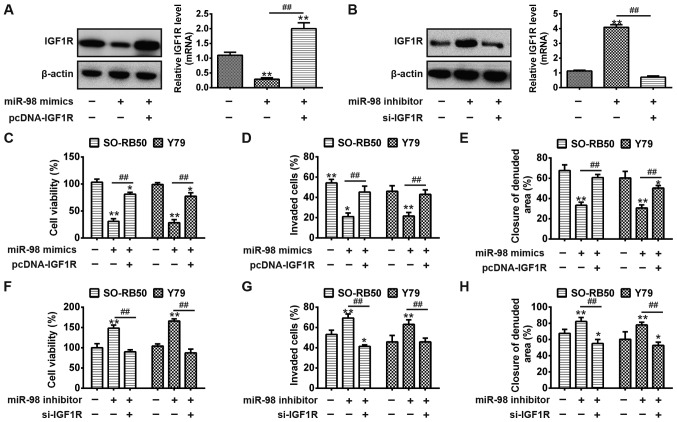
IGF1R rescues the suppressive effects of miR-98 on RB cell viability, migration, and invasion
To determine whether IGF1R is a functional target of miR-98, SO-RB50 cells were transfected miR-98 mimics, inhibitor or were co-transfected with pcDNA-IGF1R or si-IGF1R. Cells that were not transfected served as the control (blank group). As presented in Fig. 6A, overexpression of miR-98 notably inhibited the protein expression of IGF1R and the mRNA expression levels were significantly reduced compared with in the blank; however, pcDNA-IGF1R significantly rescued the suppressive effects of miR-98 on IGF1R expression (P<0.01). Conversely, miR-98 inhibitor upregulated the protein and mRNA expression of IGF1R, but was reversed via si-IGF1R in RB cells (P<0.01; Fig. 6B). Then, cell viability, migration and invasion of each group were analyzed via MTT, wound healing and Transwell invasion assays, respectively. The present study reported that overexpression of miR-98 significantly suppressed cell viability, migration, and invasion compared with in the blank group; upregulation of IGF1R resulted in a significant increase in cell viability, migration and invasion in SO-RB50 and Y79 cells following co-transfection with miR-98 mimics and pcDNA-IGF1R plasmid (P<0.01) (Fig. 6C-E). Conversely, downregulation of miR-98 significantly promoted cell viability, migration and invasion compared with in the blank group; however, these miR-98 inhibitor-mediated promoting effects on cell viability, migration and invasion were significantly reversed by si-IGF1R in SO-RB50 and Y79 cells (P<0.01; Fig. 6F-H). The data indicated that miR-98 suppressed RB cell growth and metastasis via the modulation of IGF1R expression.
Figure 6.
IGF1R rescues the effects of miR-98-mediated RB cell viability, migration, and invasion. SO-RB50 cells were transfected miR-98 mimics or inhibitor, or were co-transfected with mimics or inhibitor and pcDNA-IGF1R/si-IGF1R. (A and B) Protein and mRNA expression of IGF1R were measured via western blot analysis and reverse transcription-quantitative polymerase chain reaction, respectively. **P<0.01 vs. blank group. ##P<0.01 vs. miR-98 mimics. (C and F) MTT assay was used to evaluate cell viability. (D and G) Cell invasion was measured via a Transwell invasion assay. (E and H) Wound healing assay was performed to determine cell migration. **P<0.01 vs. blank group. ##P<0.01 vs. miR-98 mimics or inhibitor. Data are presented as the mean ± standard deviation of three individual experiments. IGF1R, insulin-like growth factor-1 receptor; miR, microRNA.
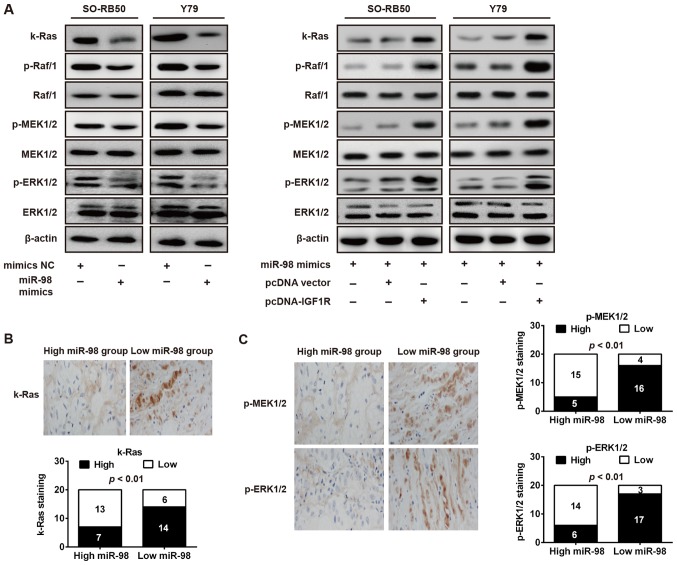
miR-98 inhibits the IGF1R/k-Ras/Raf/MEK/ERK signaling pathway
A previous study reported that IGF1R can activate numerous downstream genes are involved in malignant cell proliferation, including k-Ras, Raf1, MEK1/2 and ERK1/2, which were key components of the Raf1-MEK1/2-ERK1/2 pathway (26). The present study further investigated the potential effect of miR-98 on these molecules in RB cells. As presented in Fig. 7A, overexpression of miR-98 notably downregulated these phosphorylated molecules (k-Ras, p-Raf1, p-MEK1/2 and p-ERK1/2) in SO-RB50 and Y79 cells transfected with miR-98 mimics compared with in cells transfected with NC. Furthermore, restoration of IGF1R abrogated the suppressive effects of miR-98 on proteins of k-Ras/Raf/MEK/ERK signaling, in SO-RB50 and Y79 cells (Fig. 7A). To validate whether miR-98 expression is associated with these molecules in RB tissues, immunohistochemistry was performed to measure the expression levels of k-Ras, p-MEK1/2 and p-ERK1/2 in RB tissues. According to miR-98 expression levels, RB tissues were divided into low-expression (scores 0 and 1) and a high expression group (scores 2 and 3). As presented in Fig. 7B and C, lower miR-98 expression in RB tissues was associated with increased k-Ras, p-MEK1/2 and p-ERK1/2 expression (P<0.01). The data suggested that miR-98 may suppress RB cell growth and metastasis via the suppression of the IGF1R/k-Ras/Raf/MEK/ERK signaling pathway.
Figure 7.
miR-98 inhibits IGF1R/k-Ras/Raf/MEK/ERK signaling pathway. (A) SO-RB50 and Y79 cells were transfected with miR-98 mimics, mimics NC, pcDNA-vector or pcDNA-IGF1R and k-Ras, p-Raf/1, Raf/1, p-MEK1/2, MEK1/2, p-ERK1/2 and ERK1/2 were measured using western blot analysis. β-actin served as an internal control. (B and C) Immunohistochemistry was conducted to detect k-Ras, p-MEK1/2 and p-ERK1/2 expression in RB tissues with high or low miR-98 expression (magnification, ×200). Bar graphs demonstrated a significant inverse association between miR-98 and k-Ras, p-MEK1/2 and p-ERK1/2 expression in RB tissues (n=20). Data are presented as the mean ± standard deviation of three individual experiments. ERKK, extracellular signal-regulated kinase; IGF1R, insulin-like growth factor-1 receptor; MEK, mitogen activated protein kinase kinase; miR, microRNA; p, phosphorylated.
Discussion
Accumulating evidence has revealed that the aberrant expression of miRNAs serve a key role in cancer development (27-29). In the present study, the results demonstrated that miR-98 is downregulated in RB tissues and cells, and low miR-98 expression may be a predictor of poor prognosis in RB. In addition, overexpression of miR-98 was observed to suppress the viability and induce apoptosis in RB cells, and was suggested to inhibit migration and invasion via the suppression of EMT. Additionally, IGF1R was predicted as a target gene of miR-98 in RB cells and acts as an oncogene by modulating cell growth and metastasis in RB. Importantly, the present study verified that miR-98 may function as a tumor suppressor gene in RB via targeting the IGF1R/k-Ras/Raf/MEK/ERK signaling pathway.
Numerous miRNAs have been identified to be involved in RB development, and act as oncogene or tumor suppressor by regulating cell proliferation, apoptosis, cell cycle arrest, invasion and migration (30,31). miR-204 has been demonstrated to be downregulated in RB tissues and cell lines, and overexpression of miR-204 suppressed RB cell proliferation and invasion by targeting cyclin D2 and matrix metalloproteinase (MMP)-9 (32). Wang et al (8) reported that miR-183 inhibited RB cell proliferation, migration and invasion via downregulating low-density lipoprotein receptor-related protein 6. Previous studies indicated that miR-98 suppressed prostate cancer growth and invasion by targeting activin receptor-like kinase-4 and MMP-11 (12,14). A recent study also identified that miR-98 expression was downregulated in the serum samples of patients with RB (33). Consistent with a previous report, the results of the present study further suggested that miR-98 is downregulated in RB tissues compared with in in normal tissues. Furthermore, it was observed that the expression of miR-98 was significantly associated with clinicopathological features, including differentiation, N classification and largest tumor base; patients with low miR-98 expression exhibited significantly poorer overall survival. The data indicated that low miR-98 expression may be considered as a predictor of poor prognosis in RB, suggesting that miR-98 may serve as a potential prognostic biomarker for RB.
Increasing evidence has demonstrated the association between miR-98 expression levels and numerous types of cancer (12-14). Previously, miR-98 was identified to exert a suppressive effect on prostate cancer by inhibiting cancer cell growth, invasion and tumor angiogenesis (12,14). To further investigate the role of miR-98 in RB, the present study conducted an MTT assay and flow cytometry analysis to measure the cell viability and apoptosis in RB cells transfected with miR-98 mimics or mimics NC, respectively. It was observed that overexpression of miR-98 significantly suppressed cell viability and induced the apoptosis of RB cells. Additionally, the results of the present study further indicated that overexpression of miR-98 significantly downregulated the expression of proliferation markers (PCNA and Ki-67) and anti-apoptotic protein (Bcl-2), and upregulated that of proapoptotic proteins (cleaved-caspase-3 and Bax). These results suggested that miR-98 exhibits its suppressive effects on RB cells by inhibiting cell viability and inducing apoptosis.
Cell invasion and migration have been defined to be the crucial factors in malignant progression and metastasis (20). Furthermore, a previous study also reported that metastasis is a major factor that limits the successful treatment of RB (34). To investigate the effects of miR-98 on tumor cell metastasis, Transwell invasion and wound healing assays were performed in the present study to evaluate the invasion and migration of RB cells following overexpression of miR-98. The results revealed that overexpression of miR-98 significantly suppressed cell migration and invasion. EMT has been considered to be one of the key molecular steps in the process of distant metastasis, which induces the invasion and migration in a variety of cancers (35,36). A previous study reported that the EMT cascade can cause dissolution of cell-cell junctions and loss of apico-basolateral polarity, resulting in the formation of migratory mesenchymal cells with invasive properties (24). Zhou et al (37) revealed that miR-98 repressed non-small cell lung cancer (NSCLC) cells migration and invasion via impeding EMT. Thus, to further investigate the molecular mechanism underlying the effects of miR-98 on RB cell metastasis, western blotting was conducted in the present study to measure the expression of EMT-associated markers (E-cadherin, N-cadherin, fibronectin and vimentin) in RB cells transfected with miR-98 mimics or mimics NC. The results demonstrated that overexpression of miR-98 increased E-cadherin, and decreased N-cadherin, fibronectin and vimentin expression in SO-RB50 and Y79 cells. These results indicated that miR-98 inhibited RB cell migration and invasion via the inhibition of EMT.
IGF1R, a receptor tyrosine kinase, which modulates IGF1-induced signaling events and serves an important role in cellular processes including differentiation, proliferation and migration (38). Emerging evidence revealed that IGF1R may be associated with tumorigenesis in a variety of cancers, including breast and gastric cancers (39,40). Recently, Du et al (41) identified that miR-98 inhibits cell growth and metastasis in oral squamous cell carcinoma by targeting IGF1R. In the present study, it was reported that IGF1R is a direct target of miR-98 in RB cells, and its expression was inversely correlated with miR-98 in RB tissues. Additionally, the results demonstrated that IGF1R may function as an oncogene by modulating cell viability, migration and invasion in RB. In addition, the present study demonstrated that restoration of IGF1R may attenuate the tumor-suppressive effects of miR-98 on RB cell viability, migration and invasion. Collectively, these results suggested that miR-98 may exert inhibiting effects on RB cells via the suppression of IGF1R expression.
IGF1R has been identified as a main component in the activation of Raf1-MEK1/2-ERK1/2 signaling pathway, which is involved in tumorigenesis, apoptosis and metastasis in a variety of cancer types (42,43). To further investigate the molecular mechanisms underlying the suppressive effects of miR-98 on RB, western blotting was conducted in the present study to analyze the expression of major molecules of the Raf1-MEK1/2-ERK1/2 signaling pathway in RB cells following miR-98 overexpression. The results demonstrated that overexpression of miR-98 downregulated the levels of phosphorylated molecules, including k-Ras, p-Raf1, p-MEK1/2 and p-ERK1/2 in RB cells. In addition, the expression of three major molecules k-Ras, p-MEK1/2 and p-ERK1/2 were observed to be inversely correlated with miR-98 expression in RB tissues. Collectively, the findings of the present study indicated that miR-98 may serve as a tumor suppressor in RB via targeting the IGF1R/k-Ras/Raf/MEK/ERK signaling pathway.
A previous study revealed that miR-98 inhibits cell proliferation, migration and invasion in various types of cancers via the inhibition of numerous targets, including enhancer of zeste homolog 32, integrin β chain β 3 and IκB kinase (44-46). In the present study, it was reported that miR-98 suppressed the k-Ras/Raf/MEK/ERK signaling pathway via targeting IGF1R in RB. To further validate whether miR-98 functions as oncogene suppressor via inhibiting other targets in RB, further investigation into the different targets of miR-98 is required.
In summary, the results of the present study revealed that miR-98 is significantly downregulated in RB tissues and cell lines. Overexpression of miR-98 inhibited the growth and metastasis of RB cells by targeting IGF1R. Furthermore, it was demonstrated that miR-98 may exert the suppressive effects on RB by inhibiting the IGF1R/k-Ras/Raf/MEK/ERK signaling pathway. The findings of the present study suggested that miR-98 may be considered as a potential biomarker for the prognosis and a therapeutic target for patients with RB.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LG, YB, SJ and HM performed the experiments and data analysis, and wrote the manuscript. LG made substantial contributions to the design on the present study and acquired experimental materials. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All individuals provided informed consent for the use of human specimens for clinical research. The present study was approved by The First People's Hospital of Shangqiu Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dimaras H, Dimba EAO, Gallie BL. Challenging the global retinoblastoma survival disparity through a collaborative research effort. Br J Ophthalmol. 2010;94:1415–1416. doi: 10.1136/bjo.2009.174136. [DOI] [PubMed] [Google Scholar]
- 2.Narang S, Mashayekhi A, Rudich D, Shields CL. Predictors of long-term visual outcome after chemoreduction for management of intraocular retinoblastoma. Clin Exp Ophthalmol. 2012;40:736–742. doi: 10.1111/j.1442-9071.2012.02757.x. [DOI] [PubMed] [Google Scholar]
- 3.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang LK, Xu GP, Pan XR, Lou YJ, Zou QP, Xia D, Yan WW, Zhang YT, Jia PM, Tong JH. MicroRNA-181a-mediated downregulation of AC9 protein decreases intracellular cAMP level and inhibits ATRA-induced APL cell differentiation. Cell Death Dis. 2014;5:e1161. doi: 10.1038/cddis.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Hu C, Wang Y, Shi G, Li Y, Wu H. miR-124 inhibits proliferation and invasion of human retinoblastoma cells by targeting STAT3. Oncol Rep. 2016;36:2398–2404. doi: 10.3892/or.2016.4999. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Wang X, Li Z, Liu H, Teng Y. MicroRNA-183 suppresses retinoblastoma cell growth, invasion and migration by targeting LRP6. FEBS J. 2014;281:1355–1365. doi: 10.1111/febs.12659. [DOI] [PubMed] [Google Scholar]
- 9.Shen F, Mo M-H, Chen L, An S, Tan X, Fu Y, Rezaei K, Wang Z, Zhang L, Fu SW. MicroRNA-21 Down-regulates Rb1 expression by targeting PDCD4 in retinoblastoma. J Cancer. 2014;5:804–812. doi: 10.7150/jca.10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang YX, Nie XG, Li GD, Fan DS, Song LL, Zhang XL. Downregulation of microRNA-182 inhibits cell viability, invasion and angiogenesis in retinoblastoma through inhibition of the PI3K/AKT pathway and CADM2 upregulation. Int J Oncol. 2018;53:2615–2626. doi: 10.3892/ijo.2018.4587. [DOI] [PubMed] [Google Scholar]
- 11.Sun Z, Zhang A, Jiang T, Du Z, Che C, Wang F. miR-145 suppressed human retinoblastoma cell proliferation and invasion by targeting ADAM19. Int J Clin Exp Pathol. 2015;8:14521–14527. [PMC free article] [PubMed] [Google Scholar]
- 12.Ting H-J, Messing J, Yasmin-Karim S, Lee Y-F. Identification of microRNA-98 as a therapeutic target inhibiting prostate cancer growth and a biomarker induced by vitamin D. J Biol Chem. 2013;288:1–9. doi: 10.1074/jbc.M112.395947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siragam V, Rutnam ZJ, Yang W, Fang L, Luo L, Yang X, Li M, Deng Z, Qian J, Peng C, et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget. 2012;3:1370–1385. doi: 10.18632/oncotarget.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 16.Chantada G, Doz F, Antoneli CB, Grundy R, Clare Stannard FF, Dunkel IJ, Grabowski E, Leal-Leal C, Rodríguez-Galindo C, Schvartzman E, et al. A proposal for an international retino-blastoma staging system. Pediatr Blood Cancer. 2006;47:801–805. doi: 10.1002/pbc.20606. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-∆ ∆ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Lai YJ, Lin CI, Wang CL, Chao JI. Expression of survivin and p53 modulates honokiol-induced apoptosis in colorectal cancer cells. J Cell Biochem. 2014;115:1888–1899. doi: 10.1002/jcb.24858. [DOI] [PubMed] [Google Scholar]
- 19.Duan J, Li X, Huang S, Zeng Y, He Y, Liu H, Lin D, Lu D, Zheng M. GOLPH2, a gene downstream of ras signaling, promotes the progression of pancreatic ductal adenocarcinoma. Mol Med Rep. 2018;17:4187–4194. doi: 10.3892/mmr.2018.8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 21.Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, et al. Matrix metallopro-teinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2:737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang Y, Massagué J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Natalwala A, Spychal R, Tselepis C. Epithelial-mesenchymal transition mediated tumourigenesis in the gastrointestinal tract. World J Gastroenterol. 2008;14:3792–3797. doi: 10.3748/wjg.14.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JZ-H, Gao W, Lei W-B, Zhao J, Chan JY, Wei WI, Ho WK, Wong TS. MicroRNA 744-3p promotes MMP-9-mediated metastasis by simultaneously suppressing PDCD4 and PTEN in laryngeal squamous cell carcinoma. Oncotarget. 2016;7:58218–58233. doi: 10.18632/oncotarget.11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Wang Q, Wang GD, Wang HS, Huang Y, Liu XM, Cai XH. miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2ERK1/2 pathway in osteosarcoma. FEBS Lett. 2013;587:1366–1372. doi: 10.1016/j.febslet.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 28.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Mei Q. miRNA signature identification of retino-blastoma and the correlations between differentially expressed miRNAs during retinoblastoma progression. Mol Vis. 2015;21:1307–1317. doi: 10.3390/molecules21101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beta M, Venkatesan N, Vasudevan M, Vetrivel U, Khetan V, Krishnakumar S. Identification and insilico analysis of retino-blastoma serum microRNA profile and gene targets towards prediction of novel serum biomarkers. Bioinform Biol Insights. 2013;7:21–34. doi: 10.4137/BBI.S10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Zeng Y, Wu S, Zhong J, Wang Y, Xu J. miR-204, down-regulated in retinoblastoma, regulates proliferation and invasion of human retinoblastoma cells by targeting cyclin D2 and MMP-9. FEBS Lett. 2015;589:645–650. doi: 10.1016/j.febslet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Venkatesan N, Deepa PR, Khetan V, Krishnakumar S. Computational and in vitro investigation of miRNA-gene regulations in retinoblastoma pathogenesis: miRNA mimics strategy. Bioinform Biol Insights. 2015;9:89–101. doi: 10.4137/BBI.S21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shields CL, Shields JA. Retinoblastoma management: Advances in enucleation, intravenous chemoreduction, and intra-arterial chemotherapy. Curr Opin Ophthalmol. 2010;21:203–212. doi: 10.1097/ICU.0b013e328338676a. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Jou J, Diehl AM. Epithelial-mesenchymal transitions and hepatocarcinogenesis. J Clin Invest. 2010;120:1031–1034. doi: 10.1172/JCI42615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Huang Z, Chen X, Chen S. miR-98 inhibits expression of TWIST to prevent progression of non-small cell lung cancers. Biomed Pharmacother. 2017;89:1453–1461. doi: 10.1016/j.biopha.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Annunziata L, Li G, Pellecchia C. Group 4 metal bis(chelate) complexes of 2-anilidomethylpyridine ligands: Synthesis and catalytic activity for olefin polymerization. J Mol Catal Chem. 2011;337:1–8. doi: 10.1016/j.molcata.2011.01.004. [DOI] [Google Scholar]
- 39.Durfort T, Tkach M, Meschaninova MI, Rivas MA, Elizalde PV, Venyaminova AG, Schillaci R, François JC. Small interfering RNA targeted to IGF-IR delays tumor growth and induces proin-flammatory cytokines in a mouse breast cancer model. PLoS One. 2012;7:e29213. doi: 10.1371/journal.pone.0029213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X, Dou W, He L, Liang S, Tie J, Liu C, Li T, Lu Y, Mo P, Shi Y, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363–1372. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 41.Du Y, Li Y, Lv H, Zhou S, Sun Z, Wang M. miR-98 suppresses tumor cell growth and metastasis by targeting IGF1R in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:12252–12259. [PMC free article] [PubMed] [Google Scholar]
- 42.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Na KY, Kim YW, Park Y-K. Mitogen-activated protein kinase pathway in osteosarcoma. Pathology. 2012;44:540–546. doi: 10.1097/PAT.0b013e32835803bc. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J-J, Chen J-T, Hua L, Yao K-H, Wang C-Y. miR-98 inhibits hepatocellular carcinoma cell proliferation via targeting EZH2 and suppressing Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;85:472–478. doi: 10.1016/j.biopha.2016.11.053. [DOI] [PubMed] [Google Scholar]
- 45.Ni R, Huang Y, Wang J. miR-98 targets ITGB3 to inhibit proliferation, migration, and invasion of non-small-cell lung cancer. Onco Targets Ther. 2015;22:2689–2697. doi: 10.2147/OTT.S90998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, Xiao B, Chai Y. Overexpression of miR-98 inhibits cell invasion in glioma cell lines via downregulation of IKK ε. Eur Rev Med Pharmacol Sci. 2015;19:3593–3604. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.