Abstract
Bile acids serve a critical role in the induction of gastric intestinal metaplasia (IM) and gastric carcinogenesis. The present study investigated the effects of bile acids on the induction of gastric IM formation. The results demonstrated that the expression levels of caudal-related homeobox transcription factor 2 (CDX2), mucin 2 (MUC2) and farnesoid X receptor (FXR) were increased in vitro and in vivo following treatment with bile acids, and CDX2 transcriptionally activated MUC2 expression. Furthermore, knockdown of FXR attenuated bile acid-enhanced CDX2 promoter activity and protein expression. Conversely, the FXR agonist GW4064 synergistically enhanced bile acid-induced CDX2 promoter activity. Bile acid treatment led to an increase in nuclear factor (NF)-κB activity and protein expression. Treatment with GW4064 or the FXR antagonist Z-guggulsterone enhanced or attenuated bile acid-induced NF-κB activity, respectively. In addition, quantitative chromatin immunoprecipitation confirmed that bile acids led to enhanced binding of p50 to the CDX2 promoter, whereas this effect was not observed for p65. Treatment with GW4064 or Z-guggulsterone enhanced and attenuated the binding activity of p50 to the CDX2 promoter, respectively. These results indicated that bile acids may activate the FXR/NF-κB signalling pathway, thereby upregulating CDX2 and MUC2 expression in normal gastric epithelial cells.
Keywords: intestinal metaplasia, bile acids, caudal-related homeobox transcription factor 2, farnesoid X receptor, nuclear factor-κB
Introduction
Gastric cancer (GC) is the fifth most common type of cancer and the third leading cause of cancer-associated mortality worldwide (1). Since it is commonly diagnosed at advanced stages with lymphatic metastasis and extensive invasion, the majority of patients with GC have a poor prognosis. However, the average 5-year survival rate of patients with early GC can reach >90% (2). Gastric carcinogenesis, particularly the intestinal type, is considered to be a multistep and sequential process that includes chronic superficial gastritis, atrophic gastritis (AG), intestinal metaplasia (IM), dysplasia, intramucosal carcinoma and invasive neoplasia (3). Among these stages, IM is defined as a precancerous lesion of the gastric mucosa and is considered a risk factor in gastric tumourigenesis. However, the molecular mechanism underlying IM formation in the human stomach remains unclear. Recently, endoscopic techniques have achieved great progress in the treatment of precancerous lesions of the stomach. However, endoscopic techniques remain unsatisfactory for detecting early GC and IM due to a lack of sensitivity and high costs (4). Therefore, it is necessary to elucidate the precise mechanism underlying IM formation, and to identify valuable biomarkers for early GC and IM diagnosis and prognosis.
Caudal-related homeobox transcription factor 2 (CDX2) is an intestinal-specific transcription factor, which has been strongly implicated in the development of intestinal epithelial cells, where it regulates intestinal markers, including mucin 2 (MUC2) (5) and trefoil factor 3 (TFF3) (6). Normally, CDX2 expression is limited to the intestines, and CDX2 is absent in normal gastric mucosa. However, CDX2 exhibits a high expression level in gastric IM tissues, whereas its expression is decreased in dysplasia and GC stages (7,8). In transgenic mice expressing CDX2 in parietal cells (9), normal gastric epithelial cells are completely replaced by IM with the induction of MUC2, and these mice further develop intestinal-type adenocarcinomas, thus indicating that ectopic CDX2 expression may serve an important role in gastric tumourigenesis by inducing IM. Farnesoid X receptor (FXR) is a ligand-activated transcription factor that has a high affinity for physiological bile acids, such as chenodeoxycholic acid (CDCA) and deoxycholic acid (DCA) (10). FXR primarily regulates the homoeostasis of bile acids, including bile acid synthesis, transport and intestinal reabsorption (11,12). FXR deficiency in mice results in increased intestinal epithelial cell proliferation and intestinal tumourigenesis (13,14). In addition, upregulation of FXR has been reported in gastric IM (15); however, the function of FXR in IM formation in the stomach remains to be determined.
Clinical and experimental studies have reported that AG and IM are associated with the phenomenon of duodenogastric reflux (DGR) (16,17). Bile acids, which are the primary duodenal components of DGR, are generally considered to be carcinogens in gastric IM formation and gastric tumourigenesis (18,19). Bile acids, particularly CDCA and DCA, have been reported to be associated with the induction of CDX2 and MUC2 expression. It has previously been demonstrated that bile acid treatment can lead to an increase in CDX2 and MUC2 expression in Barrett's oesophageal cells (20). Xu et al also reported that CDCA stimulates the upregulation of CDX2 and MUC2 by activating FXR in normal rat gastric epithelial cells in a dose-dependent manner (21). However, the exact molecular mechanisms whereby bile acids promote human gastric IM formation remain unclear. Therefore, the present study aimed to investigate the effect of bile acids on molecular alterations in gastric IM formation and the molecular mechanisms involved.
Materials and methods
Clinical samples and cell culture
A total of 40 human gastric IM tissues and paired normal gastric mucosa tissues were obtained from patients (27 males and 13 females; age, 37-74 years) with DGR who had undergone endoscopic biopsy at the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) between March 2014 and July 2016. Biopsy specimens from the antrum and corpus were fixed in a solution of 4% paraformaldehyde for 24 h at room temperature and were embedded in paraffin. The 4-µm tissues were then deparaffinized using xylene and ethanol, and were stained with 0.5% hematoxylin for 5 min, followed by 0.5% eosin for 1 min at room temperature. Images were acquired using a Nikon ECLIPSE Ti-S microscope mounted with a Nikon digital camera (Nikon Corporation, Tokyo, Japan) and were assessed for the presence of intestinal metaplasia, which was recognized morphologically by the presence of goblet cells, absorptive cells and cells resembling colonocytes. Patients diagnosed with GC were excluded from this study. All tissue samples were obtained from patients that had provided informed consent, and the study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University. Normal human gastric epithelial cells (GES-1) were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were maintained at 37°C in RPMI-1640 medium supplemented with 10% foetal bovine serum (both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified incubator containing 5% CO2. Once GES-1 cells reached 70% confluence, they were serum-deprived for 24 h prior to treatment with various doses of DCA or CDCA (50, 100, 150, 200, 400 and 600 µmol/l) along with the FXR agonist GW4064 (1 µmol/l), the FXR antagonist Z-guggulsterone (20 µmol/l) or the NF-κB inhibitor pyrrolidine dithiocarbamate (PDTC; 50 µmol/l) for different durations (6, 12, 24 and 48 h). CDCA and DCA were purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). GW4064, Z-guggulsterone and PDTC were purchased from Selleck Chemicals LLC (Houston, TX, USA).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cell lines and tissue samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to a standard protocol. The PrimeScript® RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) was used to convert RNA into cDNA at 37°C for 15 min and 85°C for 5 sec, followed by maintenance at 4°C, according to the manufacturer's protocol. The PCR primer sequences were as follows: CDX2, forward, 5′-GAACCTGTGCGAGTGGATG-3′ and reverse, 5′-GGATGGTGATGTAGCGACTG-3′; MUC2, forward, 5′-CAACGATTCCTACGCTCTCC-3′ and reverse, 5′-CTTCTTCTTGTCAGCCAGCA-3′; FXR, forward, 5′-TGCAGATCAGACCGTGAATGA-3′ and reverse, 5′-TTGGTTGCCATTTCCGTCAAA-3′; and GAPDH, forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′ (all from Sangon Biotech Co., Ltd., Shanghai, China). qPCR was conducted using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.) on a CFX96™ Real-Time PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The real-time cycler conditions (two-step method) were as follows: i) Initial denaturation at 95°C for 30 sec; ii) 40 cycles of denaturation at 95°C for 5 sec and annealing/extension at 60°C for 1 min. The relative mRNA expression levels were quantified using the 2−ΔΔCq method (22). Each experiment was repeated three times.
Vector construction and transfection
Fragments of the CDX2 and MUC2 5′-flanking sequence (2,581 and 2,665 bp) were amplified using PCR from the DNA of GES-1 cells and cloned into the luciferase reporter vector pGL3.0-Basic (Promega Corporation, Madison, WI, USA). The sequences of the primers for CDX2 were forward 5′-CGGGGTACCAGAGCCACGTCTTCAGG-3′ and reverse 5′-GGAAGATCTGCACGGAGCTAGGGTAC-3′. The sequences of the primers for MUC2 were forward 5′-GAGGCTAGCCCGGGCTTCCTGGTGAGTC-3′ and reverse 5′-GAGCTCGAGCATGGTGGCTGGCAGGGGC-3′. Potential NF-κB or CDX2-binding sites in the CDX2 or MUC2 promoter were identified using bioinformatics analysis (http://jaspar.genereg.net/). Mutagenesis of the CDX2 and MUC2 promoters was performed using a site-directed mutagenesis kit (Takara Biotechnology Co., Ltd.). All constructs were verified by sequencing (data not shown). The pNF-κB-Luc plasmid was purchased from Clontech Laboratories, Inc. (Mountainview, CA, USA). The phU6-EGFP-short hairpin (sh) RNA-CDX2 lentiviral vector (target sequence, 5′-ACAAATATCGAGTGGTGTA-3′) and control vector (target sequence, 5′-TTCTCCGAACGTGTCACGT-3′) were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). Small interfering (si)RNA-FXR (target sequence, 5′-GTAGCAGAGATGCCTGTAA-3′) and scrambled control (target sequence, 5′-TTCTCCGAACGTGTCACGT-3′) were purchased from Shanghai GenePharma Co., Ltd. For siRNA transfection, cells were seeded at a density of 1×105 cells/well in 6-well plates and were transfected with 50 µM siRNA-FXR or scrambled control for 24 h using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For lentiviral infection, cells were seeded in 24-well plates overnight at a density of 5×104 cells/well and were infected with the phU6-EGFP-shRNA-CDX2 lentiviral vector and the control vector at a multiplicity of infection of 10 for 12 h, according to the manufacturer's protocol.
Dual-luciferase reporter assay
GES-1 cells were plated on 24-well plates at a density of 5×104 cells/well the day prior to transfection. The CDX2 or MUC2 promoter luciferase constructs or the pNF-κB-Luc plasmid were co-transfected with a pRL-TK construct (Promega Corporation) into the cells using Lipofectamine® 2000. A total of 24 h post-transfection, cells were incubated in medium containing 200 µmol/l bile acids for an additional 48 h and were harvested for analysis with the Dual-Luciferase Reporter Assay system (Promega Corporation), according to the manufacturer's protocol. Luciferase activity was measured using a PerkinElmer EnSpire Multilabel Reader 2300 (PerkinElmer, Inc., Waltham, MA, USA). Luciferase intensity was normalized to Renilla luciferase activity to normalize for transfection efficiency. Each experiment was repeated three times.
Cell growth assay
The cells were trypsinized and seeded into 96-well culture plates (Corning, Inc., Corning, NY, USA) at a density of 5×103 cells/well. After 24 h incubation, the cells were treated with dimethyl sulfoxide (DMSO), or 100, 200, 400 or 600 µmol/l of DCA or CDCA. The cells were then harvested at 6, 12, 24 and 48 h and cell growth was assessed using the Cell Counting Kit 8 (CCK8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. Absorbance was measured at 450 nm. Each experiment was repeated three times.
Preparation of nuclear extracts
Nuclear extracts were prepared using the Nuclear Extraction kit (Abcam, Cambridge, MA, USA). Briefly, cells were grown to 70-80% confluence on a 100-mm plate. Subsequently, culture medium was removed and cells were washed twice with PBS. After removal of the PBS, 3 ml fresh PBS was added to the plate and the cells were scraped into a 15-ml conical tube. The cells were centrifuged for 5 min at 179 × g and the supernatant was discarded; subsequently, the cell pellet was resuspended in 300 µl 1X Pre-Extraction Buffer, transferred to a microcentrifuge vial and incubated on ice for 10 min. The preparation was then vigorously vortexed for 10 sec, centrifuged for 1 min at 12,400 × g and the cytoplasmic extract was carefully removed from the nuclear pellet. Subsequently, 2 volumes of Extraction Buffer containing dithiothreitol and protease inhibitor cocktail was added to the nuclear pellet, which was incubated on ice for 15 min; the sample was vortexed for 5 sec every 3 min. Finally, the suspension was centrifuged at 16,900 × g for 10 min at 4°C and the supernatant was transferred into a new microcentrifuge vial. The nuclear protein was then quantified using Bradford reagent (Thermo Fisher Scientific, Inc.) and used for western blotting.
Western blotting
The cells were lysed with radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Shanghai, China). Protein quantification of each sample was performed using Bradford reagent (Thermo Fisher Scientific, Inc.). Subsequently, cell lysates containing 50 µg total protein were subjected to 10% SDS-PAGE (Beyotime Institute of Biotechnology) and proteins were transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). After blocking with 5% fat-free dry milk at room temperature for 2 h, the membranes were incubated overnight at 4°C with primary antibodies at 1:1,000 dilution. The membranes were then washed four times with Tris-buffered saline-0.1% Tween-20 (8 min//wash) and were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody at 1:5,000 dilution at room temperature for 2 h. Chemiluminescent HRP substrate (EMD Millipore) was used to visualize the protein bands and protein expression was semi-quantified using ImageJ version 1.46 software (National Institutes of Health, Bethesda, MD, USA). The antibodies against MUC2 (cat. no. sc-13312), FXR (cat. no. sc-13063) and GAPDH (cat. no. sc-47724) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), and the antibodies against CDX2 (cat. no. 12306), Histone H3 (cat. no. 4499), p50 (cat. no. 13586) and p65 (cat. no. 4764) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The HRP-conjugated anti-rabbit (cat. no. 7074) and anti-mouse (cat. no. 7076) secondary antibodies were purchased from Cell Signaling Technology, Inc., and the HRP-conjugated anti-goat (cat. no. sc-2354) secondary antibody was purchased from Santa Cruz Biotechnology, Inc.
Immunohistochemistry
Tissues were fixed with 10% neutral formalin at room temperature for 24 h and embedded in paraffin. Subsequently, 4-µm tissue sections were prepared. The immunohistochemical staining procedure was performed using the standard Streptavidin-Biotin Complex (SABC) staining method (23). The sections were incubated with primary antibodies against CDX2 (cat. no. 12306; Cell Signaling Technology, Inc.), MUC2 (cat. no. sc-13312) and FXR (cat. no. sc-13063) (both from Santa Cruz Biotechnology, Inc.) at 1:100 dilution overnight at 4°C, followed by incubation with biotinylated secondary antibodies (1:100 dilution) for 30 min and treatment with HRP-conjugated SABC (cat. no. SA1022 and SA1023; Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 30 min at room temperature. Images were acquired using a Nikon ECLIPSE Ti-S microscope mounted with a Nikon digital camera (Nikon Corporation). The CDX2-, MUC2- and FXR-stained sections were divided into two groups (negative and positive) based on the extent and intensity of staining. The extent of positively stained cells was categorized as follows: 0 (<5%), 1 (5-25%), 2 (25-50%), 3 (50–75%) and 4 (>75%). Staining intensity was categorized as follows: 0 (negative), 1 (weakly positive), 2 (moderately positive) and 3 (strongly positive). The immunoreactivity score (IRS) was defined as the product of the extent and intensity scores. An IRS of ≤3 was defined as negative, and a score of >3 was defined as positive. Two pathologists evaluated all specimens in a blinded manner.
Quantitative chromatin immunoprecipitation (qChIP)
qChIP assays were conducted using an EZ-ChIP™ Assay kit (EMD Millipore), as previously described (24). Chromatin-protein complexes were immunoprecipitated with 5 µg anti-CDX2, anti-p50 and anti-p65 antibodies, or 1 µg rabbit immunoglobulin G (cat. no. ab171870; Abcam) as a negative control. qPCR was performed to amplify the regions of interest or internal negative control regions. The real-time cycler conditions (two-step method) were as follows: i) Initial denaturation at 95°C for 30 sec; ii) 40 cycles of denaturation at 95°C for 5 sec and annealing/extension at 60°C for 1 min. Fold enrichment ratio was calculated as the value of the ChIP sample versus the negative control. Fold enrichment = E(Input Cq-ChIP Cq)/E(Input Cq-Negative Control Cq), where E refers to primer efficiency. The primer sequences of the CDX2 promoter region (−403 to −186 bp) were forward, 5′-TTCGAGGGGTTGTGCGTAGAGTGCG-3′ and reverse, 5′-AGGCGGTCCCTCCCTCTGGCCT-3′. The primer sequences of the MUC2 promoter region (−251 to −168 bp) were forward, 5′-CTACAGGGCTGCCTCATCCT-3′ and reverse, 5′-AATATTGATTCAGGTTATCGGAGGT-3′. Each sample was assessed in triplicate.
Animal model
A mixture of bile acids at molar concentrations, which have previously been described as near 'physiologic' (25–27), was used in this study. A total of 25 female mice (age, 7 weeks; weight, ~21 g) were purchased from Shanghai SLAC Laboratory Animal Co. Ltd. (Shanghai, China) and were housed in the Laboratory Animal Centre of Xi'an Jiaotong University. Mice had free access to water and Purina 5L79 rodent chow (Nestlé Purina PetCare Company, St. Louis, MO, USA) and were maintained under the following conditions: Temperature, 22-25°C; humidity, 50-60%; 12-h light/dark cycle. Bile acids (0.15 ml) were administered to the stomach of C57BL/6J mice via a plastic feeding tube (20 g), two times per day, for 45 days. The animals were separated into three experimental and two control groups (n=5 mice/group). The experimental groups were treated with i) DCA (10 mmol/l in 0.01 mol/l PBS), ii) CDCA (10 mmol/l in 0.01 mol/l PBS) or iii) a mixture of DCA and CDCA (10 mmol/l in 0.01 mol/l PBS). The control groups included i) a PBS-treated group (0.01 mol/l) and ii) an untreated group. After 45 days administration, the mice were sacrificed by cervical dislocation and gastric mucosa samples from each group were obtained for western blotting. The experimental protocols were evaluated and approved by the Animal Care and Use Committee of the Medical School of Xi'an Jiaotong University.
Statistical analysis
All data are presented as the means ± standard deviation. The χ2 test or one-way analysis of variance with Dunnett's post hoc test were used to analyse the differences among the groups. The correlation between CDX2 and FXR or MUC2 mRNA expression was explored using the Pearson Correlation test. All statistical analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Bile acids affect the viability of GES-1 normal human gastric epithelial cells
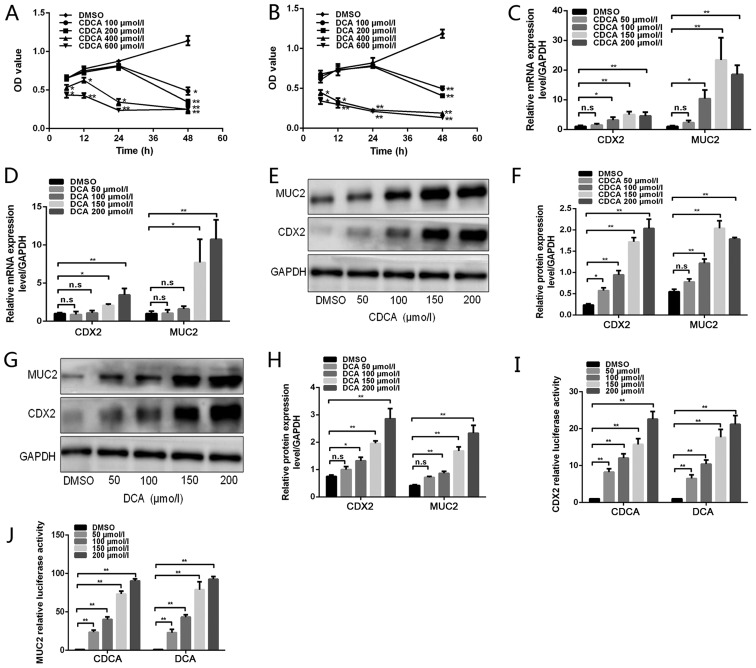
To assess the toxic effect of bile acids on GES-1 cell viability, the cells were treated with various doses of DCA or CDCA (100, 200, 400 and 600 µmol/l) for different durations (6, 12, 24 and 48 h). Cell viability was then detected using the CCK8 assay. As shown in Fig. 1A and B, compared with in the blank control group, treatment with DCA or CDCA at 100 or 200 µmol/l for 6, 12 or 24 h did not have a toxic effect. However, when cells were treated for 48 h, the viability of the cells cultured with 100 and 200 µmol/l DCA or CDCA was significantly inhibited. Treatment with 400 or 600 µmol/l DCA or CDCA, even for a short period of time, induced a severe toxic effect on cell viability. The maximum toxic effect of bile acids was detected in the group treated with 600 µmol/l for 48 h. These data demonstrated that long-term treatment with high-dose bile acids was toxic to GES-1 cells. To avoid these toxic effects, GES-1 cells were treated with physiological concentrations of bile acids (<200 µmol/l) for 24 h in subsequent experiments.
Figure 1.
Effects of bile acids on CDX2 and MUC2 expression in GES-1 cells. (A and B) GES-1 cells were treated with DMSO or various doses of bile acids (100, 200, 400 and 600 µmol/l) for different periods of time (6, 12, 24 and 48 h). Cell viability was then determined using a Cell Counting Kit 8 assay. (C and D) GES-1 cells were treated with DMSO or various concentrations of bile acids (50, 100, 150 and 200 µmol/l) for 24 h. Reverse transcription-quantitative polymerase chain reaction was performed to determine the mRNA expression levels of CDX2 and MUC2 in GES-1 cells treated with bile acids. (E) Western blot analysis of CDX2 and MUC2 protein expression in each group of GES-1 cells stimulated with CDCA. (F) Comparison of CDX2 and MUC2 protein levels in each group of cells stimulated with CDCA. (G) Western blot analysis of CDX2 and MUC2 protein expression in each group of GES-1 cells stimulated with DCA. (H) Comparison of CDX2 and MUC2 protein levels in each group of cells stimulated with DCA. (I and J) GES-1 cells treated with various concentrations of bile acids were transfected with 2.5 kb CDX2- or 2.6 kb MUC2-luc promoter constructs. A dual-luciferase reporter assay was conducted to determine (I) CDX2 and (J) MUC2 promoter activity. Data are presented as the means ± standard deviation from three independent experiments. *P<0.05, **P<0.01. CDCA, chenodeoxycholic acid; CDX2, caudal-related homeobox transcription factor 2; DCA, deoxycholic acid; DMSO, dimethyl sulfoxide; MUC2, mucin 2; n.s., not significant; OD, optical density.
Bile acids upregulate CDX2 and MUC2 expression in GES-1 cells
To determine the effects of bile acids on the formation of gastric IM, RT-qPCR and western blotting were performed to detect CDX2 and MUC2 expression levels in cells treated with bile acids (Fig. 1C–J). Treatment with CDCA at 50, 100, 150 or 200 µmol/l for 24 h increased CDX2 and MUC2 expression, at both the mRNA and protein levels, in a dose-dependent manner (Fig. 1C, E and F). Similarly, DCA treatment led to an increase in CDX2 and MUC2 expression in a dose-dependent manner (Fig. 1D, G and H). Subsequently, GES-1 cells were transfected with 2.5 kb CDX2- and 2.6 kb MUC2-luc promoter constructs and were treated with various concentrations of DCA and CDCA. As shown in Fig. 1 I and J, bile acids significantly increased CDX2 and MUC2 transcriptional activity in a dose-dependent manner. These results indicated that treatment of GES-1 cells with bile acids may lead to a dose-dependent upregulation of CDX2 and MUC2 expression.
CDX2 regulates bile acid-induced expression of MUC2
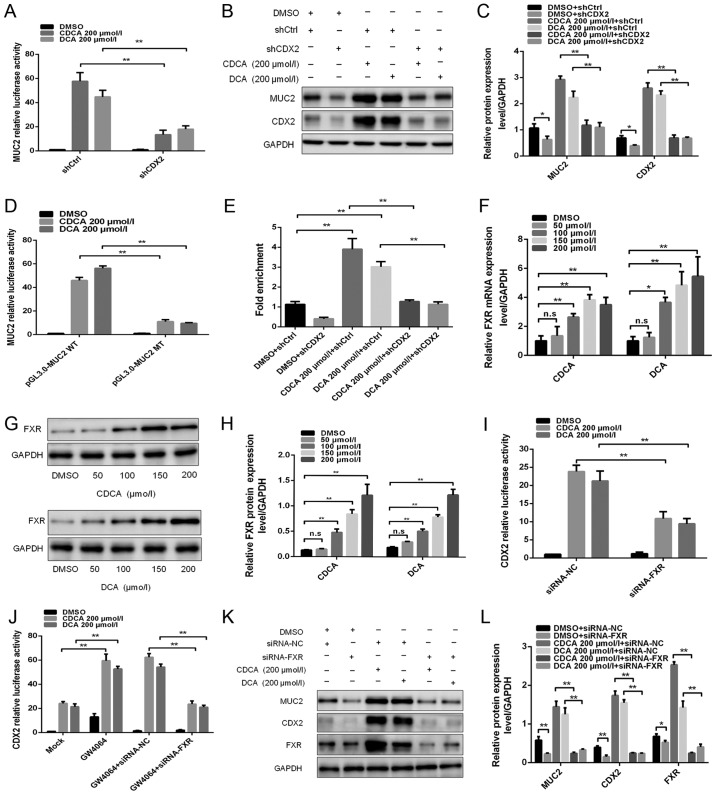
The intestinal-specific transcription factor CDX2 can directly bind to the MUC2 promoter and promote its transcription (5). This study aimed to investigate whether CDX2 has a pivotal role in bile acid-induced upregulation of MUC2 transcription. MUC2 promoter activity and protein expression levels were detected in GES-1 cells transfected with shRNA-CDX2 lentiviral vectors following treatment with 200 µmol/l bile acids (Fig. 2A–C). As shown in Fig. 2B, the shRNA-CDX2 construct successfully knocked down CDX2 expression. Knockdown of CDX2 expression attenuated bile acid-induced MUC2 promoter activity and protein expression (Fig. 2A–C).
Figure 2.
FXR is involved in the regulation of bile acid-induced CDX2 and MUC2 expression. (A) Dual-luciferase reporter assay was conducted to determine the effect of knockdown of CDX2 expression on bile acid-enhanced MUC2 promoter activity. (B) Western blotting was conducted to determine the effect of knockdown of CDX2 on bile acid-enhanced MUC2 expression. (C) Comparison of CDX2 and MUC2 protein levels in each group of GES-1 cells. (D) Mutagenesis of the MUC2 promoter was performed using a site-directed mutagenesis kit. A dual-luciferase reporter assay was conducted to detect the luciferase activities in GES-1 cells transfected with a MUC2 promoter reporter construct or mutated construct. (E) A quantitative chromatin immunopre-cipitation assay was performed to investigate the effect of bile acids on the binding of CDX2 to the MUC2 promoter. (F) Reverse transcription-quantitative polymerase chain reaction was conducted to detect the mRNA expression levels of FXR in cells treated with DMSO, or CDCA or DCA at various concentrations (0, 100, 150 and 200 µmol/l) for 24 h. (G) Western blot analysis of FXR protein expression in each group of bile acid-treated GES-1 cells. (H) Comparison of FXR protein levels in each group of GES-1 cells stimulated with CDCA or DCA. (I) siRNA-FXR was transfected into cells with the CDX2-luc promoter construct. A dual-luciferase reporter assay was conducted to determine the effect of the downregulation of FXR on bile acid-enhanced CDX2 promoter activity. (J) Effects of treatment with the FXR agonist GW4064 on bile acid-enhanced CDX2 promoter activity. (K) Western blotting was performed to investigate the effect of FXR downregulation on bile acid-induced CDX2 and MUC2 expression. (L) Comparison of CDX2, MUC2 and FXR protein expression in each group of GES-1 cells. Data are presented as the means ± standard deviation from three independent experiments. *P<0.05, **P<0.01. CDCA, chenodeoxycholic acid; CDX2, caudal-related homeobox transcription factor 2; DCA, deoxycholic acid; DMSO, dimethyl sulfoxide; FXR, farnesoid X receptor; MT, mutated type; MUC2, mucin 2; NC, negative control; n.s., not significant; sh, short hairpin RNA; si/siRNA, small interfering RNA; WT, wild-type.
Potential CDX2-binding sites were identified in the MUC2 promoter using bioinformatics analysis. To further demonstrate that CDX2 transcriptionally activated MUC2 expression by binding to the ATAAA motif in the MUC2 promoter, the MUC2 promoter luciferase construct was used to produce a mutant construct. The present study indicated that transfection with the MUC2 mutant construct led to a significantly lower level of luciferase activity in GES-1 cells after bile acids treatment compared with transfection with the wild-type construct (Fig. 2D). Furthermore, a qChIP assay was performed to investigate the effects of bile acids on the binding of CDX2 to the MUC2 promoter. The results revealed that bile acids increased the binding of CDX2 to the MUC2 promoter, whereas knockdown of CDX2 abolished enhanced binding (Fig. 2E). These results suggested that CDX2 may regulate bile acid-induced expression of MUC2 in GES-1 cells.
FXR is involved in bile acid-induced CDX2 and MUC2 expression
FXR is a bile acid nuclear receptor, which has been reported to be upregulated by bile acids in GC (10). This study aimed to investigate whether FXR is involved in bile acid-induced CDX2 and MUC2 expression in GES-1 cells. A significant increase in FXR expression at the mRNA and protein levels was observed in cells treated with bile acids (Fig. 2F–H).
The present study then assessed whether FXR participated in the regulation of bile acid-induced CDX2 transcription. siRNA-FXR was co-transfected into GES-1 cells with the CDX2-luc promoter construct. As shown in Fig. 2I, downregulation of FXR expression attenuated bile acid-enhanced CDX2 transcriptional activity. Conversely, treatment with the FXR agonist GW4064 synergistically enhanced bile acid-induced CDX2 transcriptional activity (Fig. 2J). In addition, downregulation of FXR abolished the synergistic effect of bile acids and GW4064 on bile acid-induced CDX2 promoter activity (Fig. 2J). Western blotting revealed that downregulation of FXR expression abolished the bile acid-induced upregulation of CDX2 and MUC2 expression (Fig. 2K and L). These results indicated that FXR participated in the transactivation of CDX2 induced by bile acids. Taken together, these results suggested that FXR may participate in the regulatory effects of bile acid-induced CDX2 and MUC2 expression on gastric IM formation.
NF-κB is involved in regulation of the bile acid-induced FXR/CDX2/MUC2 signalling pathway
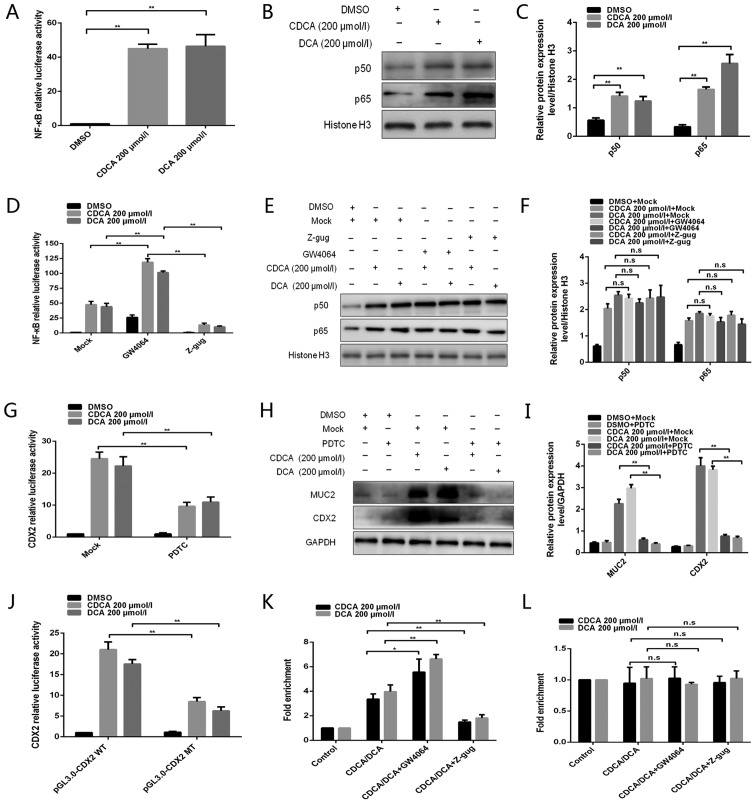
The bile acid-induced NF-κB signalling pathway serves an important role in gastric tumourigenesis (28,29). This study assessed whether NF-κB is involved in regulating the bile acid-induced FXR/CDX2/MUC2 signalling pathway in GES-1 cells. The NF-κB luciferase plasmid was transfected into GES-1 cells that were treated with 200 µmol/l bile acids. An increase in NF-κB activity was detected following treatment with bile acids (Fig. 3A). An increase in p50 and p65 protein levels was also detected in response to treatment with bile acids (Fig. 3B and C). These results indicated that NF-κB was activated in GES-1 cells treated with bile acids.
Figure 3.
NF-κB is involved in the regulation of bile acid-induced FXR/CDX2/MUC2 signalling pathway activation. (A) Dual-luciferase reporter assay was conducted to determine the effect of bile acids on NF-κB activity. (B) Western blotting was conducted to detect the effects of bile acids on p50/p65 protein expression. (C) Comparison of p50/p65 protein levels in each group of GES-1 cells. (D) Dual-luciferase reporter assay was performed to detect the effect of the FXR agonist GW4064 or the antagonist Z-gug on bile acid-enhanced NF-κB activity. (E) Western blotting was conducted to determine the effect of the FXR agonist GW4064 or the antagonist Z-gug on bile acid-induced alterations in p50 and p65 protein levels. (F) Comparison of p50 and p65 protein levels in each group of GES-1 cells. (G) Dual-luciferase reporter assay was conducted to determine the effect of the NF-κB inhibitor PDTC on bile acid-enhanced CDX2 promoter activity. (H) Western blotting was performed to determine the effect of the NF-κB inhibitor PDTC on bile acid-induced alterations in CDX2 and MUC2 protein expression levels. (I) Comparison of CDX2 and MUC2 protein levels in each group of GES-1 cells. (J) Dual-luciferase reporter assay was conducted to detect the luciferase activities in GES-1 cells transfected with CDX2 promoter reporter construct or mutated construct. (K and L) Quantitative chromatin immunoprecipitation assay was performed to investigate the effect of the FXR agonist GW4064 or the antagonist Z-gug on the binding of NF-κB (K) p50 or (L) p65 protein to the CDX2 promoter. Data are presented as the means ± standard deviation from three independent experiments. *P<0.05, **P<0.01. CDCA, chenodeoxycholic acid; CDX2, caudal-related homeobox transcription factor 2; DCA, deoxycholic acid; DMSO, dimethyl sulfoxide; MT, mutated type; MUC2, mucin 2; NF-κB, nuclear factor-κB; n.s., not significant; PDTC, pyrrolidine dithiocarbamate; WT, wild-type; Z-gug, Z-guggulsterone.
The interaction between FXR and NF-κB has been reported in the hepatic inflammatory response (30). This study aimed to investigate whether FXR is involved in regulating bile acid-induced NF-κB activity. Compared with in the control group, treatment with GW4064 enhanced bile acid-induced NF-κB activity, whereas Z-guggulsterone attenuated this activity (Fig. 3D), indicating that NF-κB activity was regulated by FXR following treatment of GES-1 cells with bile acids. However, treatment with GW4064 or Z-guggulsterone did not affect nuclear p50 and p65 protein expression (Fig. 3E and F).
A previous study demonstrated that the CDX2 promoter contains an NF-κB binding site and can be bound by p50/p65 (31). Recently, Chen et al (32) reported that CDX2 can be activated by the NF-κB signalling pathway in gastric IM. Therefore, the present study assessed whether NF-κB was involved in regulating bile acid-induced CDX2 expression in GES-1 cells. Treatment with PDTC, an inhibitor of NF-κB activation, strongly attenuated bile acid-induced CDX2 promoter-driven luciferase activity (Fig. 3G). Furthermore, PDTC attenuated bile acid-induced protein expression of CDX2 and MUC2 (Fig. 3H and I). These results suggested that bile acid-induced transactivation of CDX2 may be regulated by NF-κB.
In order to determine whether NF-κB transcriptionally activated CDX2 expression by binding to the special motif in the CDX2 promoter, mutagenesis of the predicted NF-κB binding site was performed in the CDX2 promoter using a site-directed mutagenesis kit. Transfection with the CDX2 mutant construct led to a decrease in luciferase activity in GES-1 cells following treatment with bile acids (Fig. 3J). Furthermore, a qChIP assay was conducted to investigate the effect of bile acids on the binding of NF-κB to the CDX2 promoter. The present study indicated that bile acid treatment enhanced the binding of p50, but not p65, to the CDX2 promoter (Fig. 3K and L). The present study also explored whether FXR participated in the regulation of the binding of NF-κB to the CDX2 promoter. Treatment with GW4064 or Z-guggulsterone enhanced or attenuated bile acid-induced binding of p50 to the CDX2 promoter, respectively. However, the binding activity of p65 to the CDX2 promoter exhibited no obvious alterations in response to treatment with GW4064 or Z-guggulsterone. These results demonstrated that bile acid-induced transactivation of CDX2 may be mediated by enhanced p50 binding to the CDX2 promoter and that this effect is regulated by FXR. Taken together, these data indicated that NF-κB may be involved in regulation of the bile acid-induced FXR/CDX2/MUC2 signalling pathway.
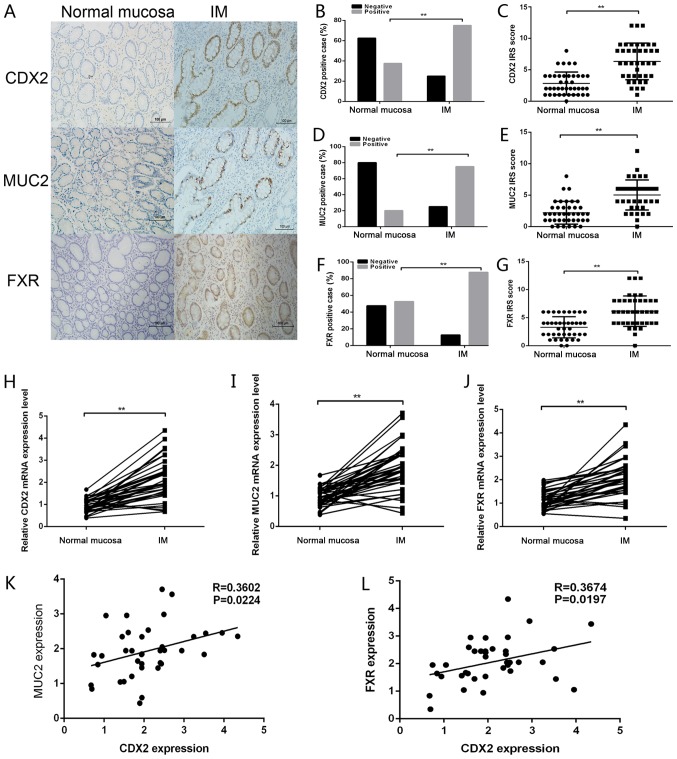
High expression levels of CDX2 are correlated with FXR and MUC2 in gastric IM tissues
To further elucidate the expression pattern of MUC2, CDX2 and FXR in the formation of gastric IM, immunohistochemistry was conducted using paraffin-embedded gastric IM and paired normal gastric mucosa tissues (Fig. 4A–G). Representative CDX2, MUC2 and FXR staining images in normal mucosa and IM tissues are shown in Fig. 4A. Positive CDX2, MUC2 and FXR staining was increased in IM tissues compared with in normal gastric mucosa to (Fig. 4B, D and F). Analysis of the IHC scores also revealed that the IRS of CDX2, MUC2 and FXR staining was higher in IM tissues than in normal tissues (Fig. 4C, E and G). RT-qPCR was further conducted to detect the mRNA expression levels of CDX2, FXR and MUC2 in gastric IM tissues and paired normal gastric tissues. As shown in Fig. 4H–J, the mRNA expression levels of CDX2, FXR and MUC2 were significantly upregulated in IM tissues compared with in normal gastric mucosa. Furthermore, CDX2 mRNA levels were positively correlated with FXR and MUC2 mRNA levels in 40 gastric IM tissues (Fig. 4K and L). Taken together, these data suggested that the expression levels of CDX2, FXR and MUC2 were upregulated in IM tissues relative to normal tissues, and were positively correlated with one another.
Figure 4.
CDX2 expression is correlated with FXR and MUC2 expression in gastric IM tissues. (A) Immunohistochemical staining of CDX2, MUC2 and FXR in gastric mucosal tissues with or without IM. Scale bar, 100 µm. (B) CDX2, (D) MUC2 and (F) FXR staining was classified into negative and positive, and the percentage of tissues in each group was shown. IRS of (C) CDX2, (E) MUC2 and (G) FXR staining in normal mucosa and IM tissues. Reverse transcription-quantitative polymerase chain reaction was performed to determine the mRNA expression levels of (H) CDX2, (I) MUC2 and (J) FXR in mucosal tissues with or without IM. Correlations between the mRNA expression levels of CDX2 and (K) MUC2 (r=0.3602; P<0.05) and (L) FXR (r=0.3674; P<0.05) in gastric IM tissues. Data are presented as the means ± standard deviation from three independent experiments. **P<0.01. CDX2, caudal-related homeobox transcription factor 2; FXR, farnesoid X receptor; IM, intestinal metaplasia; IRS, immunoreactivity score; MUC2, mucin 2.
Bile acids induce molecular alterations linked to premalignant lesions in murine gastric mucosa in vivo
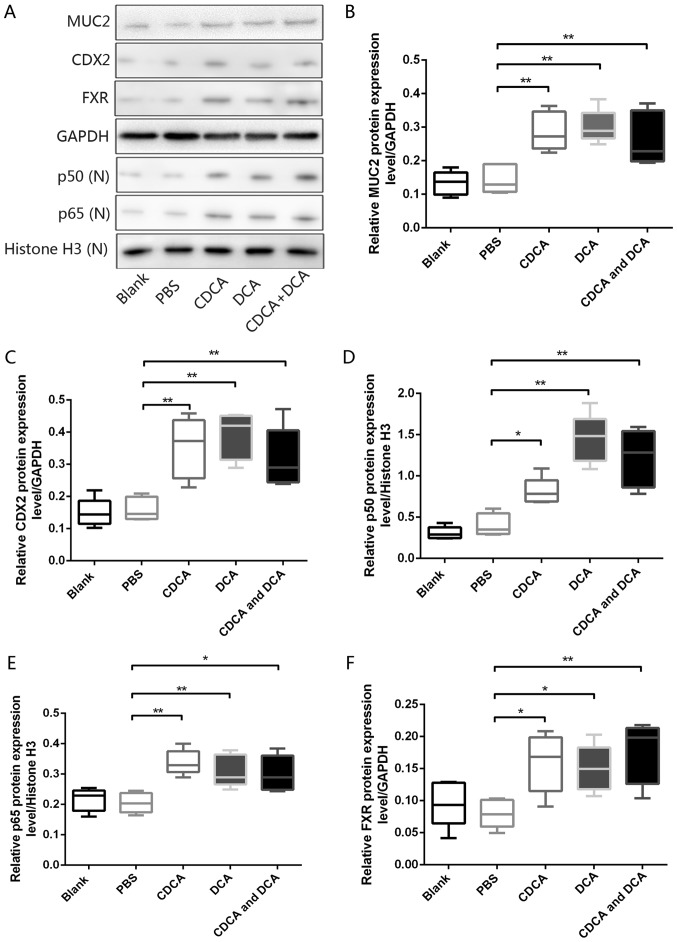
To further investigate the chronic effects of bile acids on gastric IM formation in vivo, an animal model of DGR was generated by administering bile acids to the stomach of mice (33). Western blot analyses revealed that, compared with in mice exposed to PBS alone, the murine gastric mucosa exposed to DCA, CDCA, or a mixture of DCA and CDCA, exhibited significant increases in the expression levels of FXR, p50, p65, CDX2 and MUC2 (Fig. 5A–F). However, H&E-stained murine mucosa revealed no obvious IM-like alterations in mice treated with DCA, CDCA or a mixture of both (data not shown). These results further confirmed the pivotal role of bile acids in gastric IM formation via the induction of molecular alterations linked to premalignant lesions.
Figure 5.
Bile acids induce molecular alterations linked to premalignant lesions in murine gastric mucosa in vivo. (A) Western blot analysis of MUC2, CDX2, p50, p65 and FXR protein expression in murine gastric mucosa samples from each group. Comparison of (B) MUC2, (C) CDX2, (D) p50, (E) p65 and (F) FXR protein expression levels in each group. *P<0.05, **P<0.01. CDCA, chenodeoxycholic acid; CDX2, caudal-related homeobox transcription factor 2; DCA, deoxycholic acid; FXR, farnesoid X receptor; MUC2, mucin 2; N, nuclear.
Discussion
GC is one of the leading causes of cancer-associated morbidity and mortality worldwide; however, the exact molecular mechanisms that underlie GC tumourigenesis remain elusive. IM is considered to be a precancerous lesion of the gastric mucosa. IM places patients at a higher risk of developing gastric adenocarcinoma than patients without IM (34). In addition, mounting epidemiological evidence has demonstrated that exposure to bile acids can induce IM in the stomach (19). Tatsugami et al reported that grades of atrophy and IM are significantly positively correlated with bile acid concentrations in patients with Helicobacter pylori infections (18). According to a multi-centric and large-scale cross-sectional study in Japan (35), the risk of developing IM is significantly higher in patients with high bile acid concentrations, regardless of H. pylori infection status. The critical role of bile acids in IM formation in the stomach is widely accepted; however, the molecular mechanisms by which bile acids promote IM formation in the human stomach remain to be characterized.
Increasing evidence has indicated that CDX2 has a crucial role in gastric IM formation and gastric carcinogenesis. Silberg et al (36) observed that overexpression of CDX2 in transgenic mice leads to IM formation in the stomach, alongside the presence of intestinal-type goblet cells and the expression of intestine-specific genes. This result indicated that activated CDX2 is involved in the initiation of IM formation, which can lead to intestinal neoplasia of the gastric mucosa. As an intestinal-specific transcription factor, CDX2 directly regulates several genes involved in proliferation, differentiation, and intestinal cell fate specification, including sucrose-isomaltase (37), MUC2 (5), TFF3 (6) and guanylyl cyclase C (38). This study revealed that treatment with bile acids increased the mRNA and protein expression levels of CDX2 and MUC2 in GES-1 cells. Xu et al reported that RGM-1 normal rat gastric epithelial cells exposed to bile acids exhibit upregulated CDX2 and MUC2 expression (21), which is consistent with the present findings. In addition, the transcriptional activity of CDX2 and MUC2 was increased in GES-1 cells treated with bile acids in a dose-dependent manner, thus indicating that treatment with bile acids increased CDX2 and MUC2 expression at the transcriptional level. Furthermore, this study revealed that CDX2 regulated bile acid-induced MUC2 transcription in GES-1 cells by binding to the MUC2 promoter, which was consistent with the results of a previous study (5). However, several studies have indicated that bile acids stimulate MUC2 transcription via activation of NF-κB, but not CDX2, in oesophageal adenocarcinoma (39) and colorectal cancer (40), thus suggesting that the pathogenesis of different tumour types may differ. In addition, immunohistochemistry demonstrated that CDX2 and MUC2 expression levels were significantly upregulated in IM tissue compared with in normal gastric mucosa tissue, and CDX2 expression was positively correlated with that of MUC2 in IM tissues. This result supported the present in vitro findings. Taken together, these results suggested that treatment of GES-1 cells with bile acids may lead to the upregulation of CDX2 and MUC2 expression at the mRNA and protein levels in a dose-dependent manner.
FXR is a potent nuclear receptor of bile acid, which exerts important effects on the homeostasis of bile acids, cholesterol and lipids (11,41,42). Aberrant expression of FXR has been associated with human colorectal cancer (43), prostate cancer (44) and breast cancer (45). FXR is slightly detectable in normal human stomach tissue. In a previous study, high expression levels of FXR were reported to be associated with gastric IM formation (15). The present study revealed that treatment of GES-1 cells with bile acid induced overexpression of FXR. The immunohistochemistry staining results indicated that FXR expression was significantly increased in IM tissues compared with in normal mucosa. Consistent with this study, De Gottardi et al (46) reported that Barrett's oesophagus, a precancerous lesion of the oesophageal mucosa, also exhibits high expression levels of FXR, implying that this protein may be involved in the pathogenesis of Barrett's oesophagus. The role of FXR in bile acid-induced CDX2 and MUC2 expression in GES-1 cells was also investigated. The present results indicated that FXR participated in bile acid-induced transactivation of CDX2 in GES-1 cells, a result that was consistent with the work of Li et al (47). In oesophageal adenocarcinoma, the FXR antagonist Z-guggulsterone has been reported to suppress bile acid-induced constitutive CDX2 expression, implying that FXR may regulate bile acid-induced CDX2 expression (48). In the present study, FXR expression was positively correlated with CDX2 expression in gastric IM tissues. Taken together, these results indicated that FXR may be involved in the regulation of bile acid-induced CDX2 and MUC2 expression in gastric IM formation. Lian et al reported that proper activation of FXR is a protective mechanism by which gastric mucosa resists inflammation-mediated damage (49). In addition, colon cells with decreased FXR expression fail to prevent intracellular bile acid accumulation to cytotoxic levels, thus predisposing these cells to tumourigenesis (50). It is possible that the upregulation of FXR in response to bile acids is a protective mechanism by which the gastric mucosa resists bile acid-induced damage. However, further studies are required to investigate the role of FXR in gastric IM formation.
NF-κB is involved in several important biological phenomena, including proliferation, inflammation, apoptosis and differentiation (51). Bile acids can induce the activation of NF-κB in oesophageal cancer (52) and colon cancer (39), triggering a wide array of pro-inflammatory cascades and disturbing cellular homeostasis. The present study demonstrated that exposure of GES-1 cells to bile acids increased NF-κB activity, as well as p50 and p65 protein expression. The potential role of FXR in bile acid-induced NF-κB activity was subsequently investigated. Our preliminary data demonstrated that treatment with GW4064 or Z-guggulsterone enhanced or attenuated bile acid-induced NF-κB activity, respectively. This result suggested that FXR may be a positive modulator of bile acid-induced NF-κB activity in GES-1 cells. However, bile acid-induced increases in nuclear p50 and p65 protein levels were not significantly affected by treatment with GW4064 or Z-guggulsterone, indicating that FXR may affect the activity of NF-κB but not its nuclear expression. Consistent with these results, Lee et al (53) reported that downregulation of FXR in pancreatic carcinoma cells decreases NF-κB activity and consequently inhibits its target genes. However, in inflammatory bowel disease, overexpression of NF-κB results in suppression of FXR activity, indicating that NF-κB may participate in the regulation of FXR activity (54). Wang et al (30) also described a negative crosstalk between the FXR and NF-κB signalling pathways in the hepatic inflammatory response. Therefore, the exact molecular mechanism underlying the effects of FXR and NF-κB on gastric IM formation remains to be further explored.
This study aimed to investigate whether NF-κB was involved in the regulation of bile acid-induced CDX2 expression. The results indicated that PDTC, an inhibitor of NF-κB activation, attenuated CDX2 promoter-driven luciferase activity, as well as CDX2 and MUC2 protein expression, following bile acid treatment, thus preliminarily verifying our hypothesis. A previous study demonstrated that bile acids activate CDX2 transcription via NF-κB and upregulate CDX2 expression in oesophageal keratinocytes (55). Different NF-κB subunits exert different functions on CDX2 transcriptional activity. Debruyne et al (38) reported that DCA induces p50 nuclear translocation and enhances the binding activity of p50 to the CDX2 promoter, whereas p65 remains in the cytoplasm in human oesophageal cells. Kim et al (31) also reported that the transcriptional activity of CDX2 depends on the balance between the p50 and p65 subunits. The present study observed that mutagenesis of the predicted NF-κB binding site in the CDX2 promoter abolished enhanced luciferase activity in GES-1 cells following bile acid treatment. A qChIP assay indicated that bile acid treatment led to increased binding of p50, but not p65, to the CDX2 promoter. Furthermore, treatment with GW4064 or Z-guggulsterone enhanced or attenuated the binding activity of p50 to the CDX2 promoter, respectively. Yamada et al demonstrated that CDCA enhances p50 protein binding to the CDX2 promoter in oesophageal adenocarcinoma. However, treatment with Z-guggulsterone does not reduce CDCA-induced binding activity of p65 to the CDX2 promoter (48). Taken together, these results indicated that bile acid-induced transcriptional activation of CDX2 in GES-1 cells may be increased via the enhancement of p50 binding to the CDX2 promoter; this effect may be regulated by FXR.
The results of the DGR animal model experiment demonstrated an increase in the protein expression levels of FXR, p50, p65, CDX2 and MUC2 in gastric mucosa exposed to DCA, CDCA, or a mixture of both, providing further evidence to verify the present findings. However, H&E-stained murine mucosa revealed no obvious IM-like alterations in mucosa treated with DCA, CDCA or a mixture of both. There are three potential reasons for this phenomenon: i) The time period over which the animals were treated with bile acids was not sufficient to stimulate gastric IM formation. ii) Gastric IM formation is a complex process that depends on multifactorial interactions, such as H. pylori infection, bile acids and acid. iii) The animal model of DGR used in this study did not wholly mimic the complex dynamic environment of DGR in vivo, including the time interval of DGR and the fluctuating concentration of bile acids. In our future in vivo studies, we aim to mimic the actual DGR environment and thereby clarify the molecular mechanisms underlying IM formation in the human stomach.
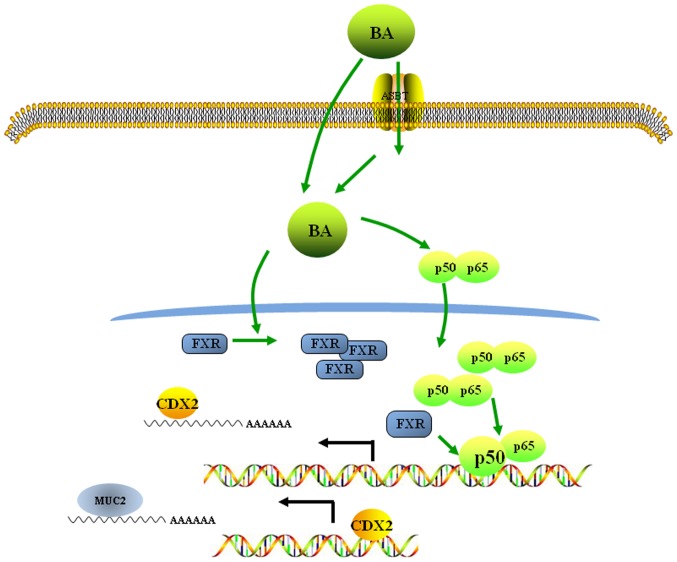
In conclusion, the results of the present study suggested a molecular mechanism underlying the bile acid-induced acquisition of an intestinal phenotype in normal gastric epithelial cells. These data revealed that bile acids may activate the FXR/NF-κB signalling pathway, inducing the ectopic expression of CDX2, an intestinal-specific transcription factor, and MUC2, an intestinal metaplasia marker, in normal gastric epithelial cells (Fig. 6). These observations of IM formation in the human stomach may contribute to our understanding of the pathogenic mechanisms underlying neoplastic transformation in the stomach.
Figure 6.
BAs promote gastric intestinal metaplasia by upregulating CDX2 and MUC2 expression via the FXR/nuclear factor-κB signalling pathway. BAs enter the cell by active transport or by diffusion and induce the upregulation and nuclear location of p50/p65 expression. Nuclear p50 binds to the CDX2 promoter and promotes CDX2 transactivation. The BA-induced increase in FXR expression enhances the binding of p50 to the CDX2 promoter. The CDX2 protein then binds to the MUC2 promoter and increases MUC2 transactivation. ASBT, apical sodium-dependent bile acid transporter; BA, bile acid; CDX2, caudal-related homeobox transcription factor 2; FXR, farnesoid X receptor; MUC2, mucin 2.
Acknowledgments
Not applicable.
Funding
The present study was supported by The National Natural Science Foundation of China (grant nos. 81101874 and 81172362), The Science and Technology Project of Shaanxi Province (grant nos. 2016SF-015 and 2016SF-157), and The Coordinative and Innovative Plan Projects of the Science and Technology Program in Shaanxi Province (grant no. 2013KTCQ03-08).
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
JHY and JBZ performed the experiments, acquired the data and drafted the manuscript. KY, JQ and CBW collected human gastric IM tissues, and assisted with qPCR and western blotting. YHW and KW analysed and interpreted data. XJS and JHY substantially contributed to the study conception and design. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Written informed consent was obtained from all patients, and the study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University. All animal research was carried out following approval by the Institutional Animal Care and Use Committee of The First Affiliated Hospital of Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Costamagna G, Cesaro P. Early gastric cancer: Detection and endoscopic treatment. Ann Ital Chir. 2012;83:183–191. [PubMed] [Google Scholar]
- 3.Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis. 2012;13:2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Compare D, Rocco A, Nardone G. Screening for and surveillance of gastric cancer. World J Gastroenterol. 2014;20:13681–13691. doi: 10.3748/wjg.v20.i38.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto H, Bai YQ, Yuasa Y. Homeodomain protein CDX2 regulates goblet-specific MUC2 gene expression. Biochem Biophys Res Commun. 2003;300:813–818. doi: 10.1016/S0006-291X(02)02935-2. [DOI] [PubMed] [Google Scholar]
- 6.Shimada T, Koike T, Yamagata M, Yoneda M, Hiraishi H. Regulation of TFF3 expression by homeodomain protein CDX2. Regul Pept. 2007;140:81–87. doi: 10.1016/j.regpep.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Teh M, Ito K, Shah N, Ito Y, Yeoh KG. CDX2 expression is progressively decreased in human gastric intestinal metaplasia, dysplasia and cancer. Modern pathology: An official journal of the United States and Canadian Academy of Pathology Inc. 2007;20:1286–1297. doi: 10.1038/modpathol.3800968. [DOI] [PubMed] [Google Scholar]
- 8.Eda A, Osawa H, Yanaka I, Satoh K, Mutoh H, Kihira K, Sugano K. Expression of homeobox gene CDX2 precedes that of CDX1 during the progression of intestinal metaplasia. J Gastroenterol. 2002;37:94–100. doi: 10.1007/s005350200002. [DOI] [PubMed] [Google Scholar]
- 9.Mutoh H, Hakamata Y, Sato K, Eda A, Yanaka I, Honda S, Osawa H, Kaneko Y, Sugano K. Conversion of gastric mucosa to intestinal metaplasia in Cdx2-expressing transgenic mice. Biochem Biophys Res Commun. 2002;294:470–479. doi: 10.1016/S0006-291X(02)00480-1. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 11.Makishima M. Nuclear receptors as targets for drug development: Regulation of cholesterol and bile acid metabolism by nuclear receptors. J Pharmacol Sci. 2005;97:177–183. doi: 10.1254/jphs.FMJ04008X4. [DOI] [PubMed] [Google Scholar]
- 12.Redinger RN. The role of the enterohepatic circulation of bile salts and nuclear hormone receptors in the regulation of cholesterol homeostasis: Bile salts as ligands for nuclear hormone receptors. Can J Gastroenterol. 2003;17:265–271. doi: 10.1155/2003/190784. [DOI] [PubMed] [Google Scholar]
- 13.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 14.Maran RR, Thomas A, Roth M, Sheng Z, Esterly N, Pinson D, Gao X, Zhang Y, Ganapathy V, Gonzalez FJ, et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J Pharmacol Exp Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ku HJ, Kim HY, Kim HH, Park HJ, Cheong JH. Bile acid increases expression of the histamine-producing enzyme, histidine decarboxylase, in gastric cells. World J Gastroenterol. 2014;20:175–182. doi: 10.3748/wjg.v20.i1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobala GM, O'Connor HJ, Dewar EP, King RF, Axon AT, Dixon MF. Bile reflux and intestinal metaplasia in gastric mucosa. J Clin Pathol. 1993;46:235–240. doi: 10.1136/jcp.46.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon MF, Mapstone NP, Neville PM, Moayyedi P, Axon AT. Bile reflux gastritis and intestinal metaplasia at the cardia. Gut. 2002;51:351–355. doi: 10.1136/gut.51.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatsugami M, Ito M, Tanaka S, Yoshihara M, Matsui H, Haruma K, Chayama K. Bile acid promotes intestinal metaplasia and gastric carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2012;21:2101–2107. doi: 10.1158/1055-9965.EPI-12-0730. [DOI] [PubMed] [Google Scholar]
- 19.Jiang JX, Liu Q, Zhao B, Zhang HH, Sang HM, Djaleel SM, Zhang GX, Xu SF. Risk factors for intestinal metaplasia in a southeastern Chinese population: An analysis of 28,745 cases. J Cancer Res Clin Oncol. 2017;143:409–418. doi: 10.1007/s00432-016-2299-9. [DOI] [PubMed] [Google Scholar]
- 20.Tamagawa Y, Ishimura N, Uno G, Aimi M, Oshima N, Yuki T, Sato S, Ishihara S, Kinoshita Y. Bile acids induce Delta-like 1 expression via Cdx2-dependent pathway in the development of Barrett's esophagus. Lab Invest. 2016;96:325–337. doi: 10.1038/labinvest.2015.137. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Watanabe T, Tanigawa T, Machida H, Okazaki H, Yamagami H, Watanabe K, Tominaga K, Fujiwara Y, Oshitani N, et al. Bile acids induce cdx2 expression through the farnesoid x receptor in gastric epithelial cells. J Clin Biochem Nutr. 2010;46:81–86. doi: 10.3164/jcbn.09-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Qin R, Wang NN, Chu J, Wang X. Expression and significance of homeodomain protein Cdx2 in gastric carcinoma and precancerous lesions. World J Gastroenterol. 2012;18:3296–3302. doi: 10.3748/wjg.v18.i25.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, Zheng PS, Yang WT. EZH2-mediated repression of GSK-3β and TP53 promotes Wnt/β-catenin signaling-dependent cell expansion in cervical carcinoma. Oncotarget. 2016;7:36115–36129. doi: 10.18632/oncotarget.8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kauer WK, Peters JH, DeMeester TR, Feussner H, Ireland AP, Stein HJ, Siewert RJ. Composition and concentration of bile acid reflux into the esophagus of patients with gastroesophageal reflux disease. Surgery. 1997;122:874–881. doi: 10.1016/S0039-6060(97)90327-5. [DOI] [PubMed] [Google Scholar]
- 26.Liu T, Zhang X, So CK, Wang S, Wang P, Yan L, Myers R, Chen Z, Patterson AP, Yang CS, et al. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis. 2007;28:488–496. doi: 10.1093/carcin/bgl176. [DOI] [PubMed] [Google Scholar]
- 27.Huo X, Zhang HY, Zhang XI, Lynch JP, Strauch ED, Wang JY, Melton SD, Genta RM, Wang DH, Spechler SJ, et al. Acid and bile salt-induced CDX2 expression differs in esophageal squamous cells from patients with and without Barrett's esophagus. Gastroenterology. 2010;139:194–203.e1. doi: 10.1053/j.gastro.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merga YJ, O'Hara A, Burkitt MD, Duckworth CA, Probert CS, Campbell BJ, Pritchard DM. Importance of the alternative NF-κB activation pathway in inflammation-associated gastrointestinal carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G1081–G1090. doi: 10.1152/ajpgi.00026.2016. [DOI] [PubMed] [Google Scholar]
- 29.Gambhir S, Vyas D, Hollis M, Aekka A, Vyas A. Nuclear factor kappa B role in inflammation associated gastrointestinal malignancies. World J Gastroenterol. 2015;21:3174–3183. doi: 10.3748/wjg.v21.i11.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Domon-Dell C, Wang Q, Chung DH, Di Cristofano A, Pandolfi PP, Freund JN, Evers BM. PTEN and TNF-alpha regulation of the intestinal-specific Cdx-2 homeobox gene through a PI3K, PKB/Akt, and NF-appaB-dependent pathway. Gastroenterology. 2002;123:1163–1178. doi: 10.1053/gast.2002.36043. [DOI] [PubMed] [Google Scholar]
- 32.Chen BJ, Zeng S, Xie R, Hu CJ, Wang SM, Wu YY, Xiao YF, Yang SM. hTERT promotes gastric intestinal metaplasia by upregulating CDX2 via NF-κB signaling pathway. Oncotarget. 2017;8:26969–26978. doi: 10.18632/oncotarget.15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vageli DP, Prasad ML, Sasaki CT. Gastro-duodenal fluid induced nuclear factor-kappaB activation and early pre-malignant alterations in murine hypopharyngeal mucosa. Oncotarget. 2016;7:5892–5908. doi: 10.18632/oncotarget.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spence AD, Cardwell CR, McMenamin UC, Hicks BM, Johnston BT, Murray LJ, Coleman HG. Adenocarcinoma risk in gastric atrophy and intestinal metaplasia: A systematic review. BMC Gastroenterol. 2017;17:157. doi: 10.1186/s12876-017-0708-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuhisa T, Arakawa T, Watanabe T, Tokutomi T, Sakurai K, Okamura S, Chono S, Kamada T, Sugiyama A, Fujimura Y, et al. Relation between bile acid reflux into the stomach and the risk of atrophic gastritis and intestinal metaplasia: a multicenter study of 2283 cases. Dig Endosc. 2013;25:519–525. doi: 10.1111/den.12030. [DOI] [PubMed] [Google Scholar]
- 36.Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122:689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 37.Boudreau F, Rings EH, van Wering HM, Kim RK, Swain GP, Krasinski SD, Moffett J, Grand RJ, Suh ER, Traber PG. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 38.Debruyne PR, Witek M, Gong L, Birbe R, Chervoneva I, Jin T, Domon-Cell C, Palazzo JP, Freund JN, Li P, et al. Bile acids induce ectopic expression of intestinal guanylyl cyclase C Through nuclear factor-kappaB and Cdx2 in human esophageal cells. Gastroenterology. 2006;130:1191–1206. doi: 10.1053/j.gastro.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Gong J, Geng J, Song Y. Deoxycholic acid induces the overexpression of intestinal mucin, MUC2, via NF-kB signaling pathway in human esophageal adenocarcinoma cells. BMC Cancer. 2008;8:333. doi: 10.1186/1471-2407-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HY, Crawley S, Hokari R, Kwon S, Kim YS. Bile acid regulates MUC2 transcription in colon cancer cells via positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and p38/MSK1/CREB pathways and negative JNK/c-Jun/AP-1 pathway. Int J Oncol. 2010;36:941–953. doi: 10.3892/ijo_00000573. [DOI] [PubMed] [Google Scholar]
- 41.Yuan ZQ, Li KW. Role of farnesoid X receptor in cholestasis. J Dig Dis. 2016;17:501–509. doi: 10.1111/1751-2980.12378. [DOI] [PubMed] [Google Scholar]
- 42.Dong B, Young M, Liu X, Singh AB, Liu J. Regulation of lipid metabolism by obeticholic acid in hyperlipidemic hamsters. J Lipid Res. 2017;58:350–363. doi: 10.1194/jlr.M070888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lax S, Schauer G, Prein K, Kapitan M, Silbert D, Berghold A, Berger A, Trauner M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer. 2012;130:2232–2239. doi: 10.1002/ijc.26293. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Tong SJ, Wang X, Qu LX. Farnesoid X receptor inhibits LNcaP cell proliferation via the upregulation of PTEN. Exp Ther Med. 2014;8:1209–1212. doi: 10.3892/etm.2014.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giordano C, Barone I, Vircillo V, Panza S, Malivindi R, Gelsomino L, Pellegrino M, Rago V, Mauro L, Lanzino M, et al. Activated FXR Inhibits Leptin Signaling and Counteracts Tumor-promoting Activities of Cancer-Associated Fibroblasts in Breast Malignancy. Sci Rep. 2016;6:21782. doi: 10.1038/srep21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Gottardi A Dumonceau JM, Bruttin F, Vonlaufen A, Morard I, Spahr L, Rubbia-Brandt L, Frossard JL, Dinjens WN, Rabinovitch PS. et al. Expression of the bile acid receptor FXR in Barrett's esophagus and enhancement of apoptosis by guggulsterone in vitro. Mol Cancer. 2006;5:48. doi: 10.1186/1476-4598-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Chen X, Zhou L, Wang BM. Farnesoid X receptor signal is involved in deoxycholic acid-induced intestinal metaplasia of normal human gastric epithelial cells. Oncol Rep. 2015;34:2674–2682. doi: 10.3892/or.2015.4207. [DOI] [PubMed] [Google Scholar]
- 48.Yamada T, Osawa S, Hamaya Y, Furuta T, Hishida A, Kajimura M, Ikuma M. Guggulsterone suppresses bile acid-induced and constitutive caudal-related homeobox 2 expression in gut-derived adenocarcinoma cells. Anticancer Res. 2010;30:1953–1960. [PubMed] [Google Scholar]
- 49.Lian F, Xing X, Yuan G, Schäfer C, Rauser S, Walch A, Röcken C, Ebeling M, Wright MB, Schmid RM, et al. Farnesoid X receptor protects human and murine gastric epithelial cells against inflammation-induced damage. Biochem J. 2011;438:315–323. doi: 10.1042/BJ20102096. [DOI] [PubMed] [Google Scholar]
- 50.Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall HU, et al. Coordinated induction of bile acid detoxification and alternative elimination in mice: Role of FXR-regulated organic solute transporter-alpha/beta in the adaptive response to bile acids. Am J Physiol Gastrointest Liver Physiol. 2006;290:G923–G932. doi: 10.1152/ajpgi.00490.2005. [DOI] [PubMed] [Google Scholar]
- 51.Sarkar FH, Li Y, Wang Z, Kong D. NF-kappaB signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol. 2008;27:293–319. doi: 10.1080/08830180802276179. [DOI] [PubMed] [Google Scholar]
- 52.Abdel-Latif MM, Inoue H, Kelleher D, Reynolds JV. Factors regulating nuclear factor-kappa B activation in esophageal cancer cells: Role of bile acids and acid. J Cancer Res Ther. 2016;12:364–373. doi: 10.4103/0973-1482.174525. [DOI] [PubMed] [Google Scholar]
- 53.Lee JY, Lee KT, Lee JK, Lee KH, Jang KT, Heo JS, Choi SH, Kim Y, Rhee JC. Farnesoid X receptor, overexpressed in pancreatic cancer with lymph node metastasis promotes cell migration and invasion. Br J Cancer. 2011;104:1027–1037. doi: 10.1038/bjc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gadaleta RM, Oldenburg B, Willemsen EC, Spit M, Murzilli S, Salvatore L, Klomp LW, Siersema PD, van Erpecum KJ, van Mil SW. Activation of bile salt nuclear receptor FXR is repressed by pro-inflammatory cytokines activating NF-κB signaling in the intestine. Biochim Biophys Acta. 2011;1812:851–858. doi: 10.1016/j.bbadis.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 55.Kazumori H, Ishihara S, Rumi MA, Kadowaki Y, Kinoshita Y. Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett's epithelium. Gut. 2006;55:16–25. doi: 10.1136/gut.2005.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.