Abstract
Non-alcoholic fatty liver disease (NAFLD) is the consequence of insulin resistance, fatty acid accumulation, oxidative stress and lipotoxicity. The present study aimed to elucidate the effect of Quaking 5 (QKI 5) as mediated by Sirtuin 1 (SIRT1) on triglyceride (TG) synthesis in the liver of an NAFLD mouse model. A high-fat diet-induced NAFLD model was established in mice, and mouse hepatocytes were isolated to characterize the effects of QKI 5 mediated by SIRT1 on TG synthesis in the liver. Body weight and liver wet weight were recorded. In addition, serum levels of total cholesterol, TG, alanine aminotransferase and aspartate aminotransferase were assessed using an automatic biochemistry analyzer. Hematoxylin and eosin staining was performed to observe the histological morphological alterations of the liver tissues. The concentration of SIRT1 in the serum was also detected. The NAFLD activity score (NAS) was used to evaluate disease severity. The synthesis of TGs in cells or tissues was determined, and the protein levels of SIRT1, QKI 5, peroxisome proliferator-activated receptor (PPAR)α and Forkhead box protein O1 (FoxO1) were examined. The expression levels of SIRT1 or QKI 5, and the acetylation level of QKI 5 were decreased in the mouse model of NAFLD. QKI 5 was deacetylated by SIRT1, which contributed in suppressing the progression of NAFLD in the mice, and inhibiting TG synthesis in vivo and in vitro via the PPARα/FoxO1 signaling pathway. Taken together, the results of the present study demonstrated that SIRT1 deacetylated QKI 5, an RNA-binding protein significantly affecting the synthesis of TG in the liver of the NAFLD mouse model. Furthermore, it activated transcription factor FOXO1 through post-transcriptional regulation of the expression of PPARα and further inhibited the synthesis of TGs, thereby restraining the progression of NAFLD.
Keywords: non-alcoholic fatty liver disease, Quaking 5, Sirtuin 1, Forkhead box protein O1, peroxisome proliferator-activated receptor α, synthesis of triglyceride
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most widespread metabolic syndrome characterized by aberrant lipid accumulation in the hepatocyte cytoplasm, affecting ~25% of adults worldwide (1). NAFLD refers to a variety of liver diseases ranging from non-alcoholic simple fatty liver, to liver fibrosis/cirrhosis, non-alcoholic steatohepatitis (NASH) and NASH-associated hepatocellular carcinoma (NASH-HCC) (2,3). NASH is a form of NAFLD characterized by steatosis, hepatocellular necrosis and fibrosis, and is associated with the development of obesity, metabolic syndrome and diabetes (4,5). There is increasing evidence that NASH can eventually lead to irreversible liver injury, or even the development and progression of HCC (6). Therefore, basic and clinical studies in the coming decades are focusing on the pathogenesis and latent therapeutic targets of NAFLD.
Traditionally, the development of NAFLD has been associated with genetics, systolic blood pressure, serum cholesterol, fasting glucose, sex, age and waist circumference (7). It has been gradually realized that sirtuins and associated metabolic processes are also involved in the development of NAFLD (8). Sirtuins (SIRT1-7) have a spectrum of functions through regulating post-translational protein modification. SIRT1 is closely associated with cellular metabolism through deacetylating cellular proteins at the post-translational level, and serves an important role in a variety of metabolic diseases, including NAFLD (9).
Quaking (QKI) is a member of the STAR family of RNA-binding proteins with diverse functions in mRNA stability (10,11) and translation (12,13), microRNA processing (14,15) and alternative splicing (13,16–20). It was demonstrated that SIRT1 increased the acetylation level of QKI 5 through inhibiting SIRT1 activity. By contrast, increased SIRT1 activity resulted in QKI 5 deacetylation. The acetylation level of QKI 5 was increased in liver cells in which the SIRT1 gene was silenced and in the liver tissues of SIRT1-knockdown mice (13–15,18). These results implicate alternative splicing as a key regulatory factor in the lipid metabolism of hepatocytes.
In the present study, the effect of QKI 5 mediated by SIRT1 on triglyceride synthesis of the liver in an NAFLD model was investigated. The NAFLD model mice, induced by a high-fat diet (HFD), was successfully established, and mouse hepatocytes were isolated to characterize the effects of QKI 5 mediated by SIRT1 on the synthesis of triglycerides in the liver and the activation of the peroxisome proliferator-activated receptor (PPAR)α, Forkhead box protein O1 (FoxO1) signaling pathway.
Materials and methods
Animal experiment
C57BL/6 mice (male, 8–10-weeks old) obtained from the Animal Experimental Center at Tsinghua University (Beijing, China) was housed with standard cages at constant room temperature (22±2°C) and relative humidity of 45±15%, with free access to food and water on a 12/12 h light/dark cycle. Then, these mouse were fed with standard food as controls or the HFD (fat 60%, carbohydrate 20.6%, protein 19.4%; Research Diets, Inc., New Brunswick, NJ, USA) for 8 weeks. All mice were then sacrificed. The liver wet weight and body weight were measured, and alanine aminotransferase (ALT), and aspartate aminotransferase (AST) were detected using an automatic biochemical analyzer. The histopathological changes of the liver were evaluated by hematoxylin and eosin (H&E) staining. Furthermore, the level of SIRT1 in the serum was determined using the ELISA method.
To investigate the functional roles of QKI 5 in NAFLD, 24 C57BL/6 mice were randomized into four groups as follows (n=6 per group): Control group, mice fed with a HFD (model group), mice fed with a HFD and treated with 100 mg/kg/day per os of SRT1720 (SRT1720 group) or vehicle (1% DMSO in 20% cyclodextrin; vehicle group) (21). Following 8 weeks of continuous administration, the mice were sacrificed. Blood samples were collected for the determination of serum SIRT1, ALT, AST, total cholesterol (TC) and triglyceride (TG) levels. Additionally, liver tissues were subjected to pathological H&E staining, determination of NAFLD activity score (NAS), and an assay of the synthesis of TG. All animal experiments were approved by the Animal Ethics Committee of Tsinghua University.
Metabolic phenotyping
The mouse body composition, including fat mass, was assessed with non-invasive quantitative magnetic resonance using an EchoMRI700 instrument. Values are expressed as a percentage of body weight (BW). All experiments were performed at Tsinghua University. Homogenization and protein extraction from the liver tissue-homogenization and extraction of individual liver sections were performed in NP-40 lysis buffer.
Liver histology and analysis
To observe the liver histo-pathological changes, standard H&E staining was performed. Briefly, following dehydration with an ethanol gradient, the liver tissues fixed using formalin (4%) were embedded in paraffin, then cut into 5-µm sections. The sections were then stained with H&E or Oil Red O staining, and immunostaining with anti-SIRT1 or anti-QKI 5 was performed. The slides were mounted with aqueous mountant and viewed with a fluorescent microscope (IX71; Olympus Corporation, Tokyo, Japan). Hepatic TG and TC or TG in cells were measured using the infinity TG and TC reagent kits (cat. nos. TR13421 and TR-22421; Thermo Fisher Scientific, Inc., Waltham, MA, USA), respectively.
Circulating TG, TC, ALT and AST content
The serum levels of TG, TC, ALT and AST were assessed with an automatic biochemical analyzer (Olympus Corporation).
Serum SIRT1 measurement
Based on the manufacturer's protocol, the serum SIRT1 concentration was measured by ELISA using an SIRT1 mouse ELISA kit (Abbexa, Cambridge, UK).
Pathological examination and assessment of disease severity
For liver biopsies of NAFLD, the novel NAS system is suggested for the grading of steatosis, hepatocellular ballooning and inflammatory activity, referring to a guide on the diagnosis and treatment for NAFLD (22).
Mouse hepatocyte isolation and treatment
Mouse hepatocytes were dissociated from the tissues of the C57BL/6 mice using an in situ recirculating collagenase perfusion method (23). The liver tissues were transferred into Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) on ice and filtered through a multi-layer gauze to obtain a hepatocyte suspension. Following washing with PBS, the supernatant was discarded. The primary hepatocytes were maintained in DMEM with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) for cell adherence with 5% CO2 at 37°C for 4–6 h.
The obtained cells were rinsed with fresh medium to remove the dead cells and cell fragments, and then washed with PBS and re-suspended in serum-free medium containing 50 ng/ml SRT1720 or 10 µM niacinamide for 48 h at 37°C, respectively. The cells were collected for synthesis of TG at each time point using a TG assay kit for quantification (cat. no. ab65336; Abcam, Cambridge, MA, USA).
To initially investigate the effect of SRT1720 treatment on the FOXO1 and PPARα signaling pathway, the mouse hepatocytes (5×105 cells/well) were cultured in a 12-well plate at 37°C, and treated with either DMSO (control), SRT1720 (50 ng/ml), FOXO1 inhibitor AS1842856 (100 nM; cat. no. 344355; Merck KGaA, Darmstadt, Germany) or SRT1720 (50 ng/ml) + FOXO1 inhibitor AS1842856 (10 nM). Following 48 h of treatment, the protein expression levels of FOXO1 and PPARα were determined by western blotting. The synthesis of TGs in the mouse hepatocytes was detected.
Adenovirus construction and transfection
Recombinant adenoviruses (Ads) containing SIRT1 were produced with an AdEasy (Ad) Vector system (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). The concentration of 10 plaque-forming units/hepatocyte was used to infect cells. AdGFP was used as the infection control.
Cell transfection
Whether SIRT1 inhibited the activation of FOXO1 was then determined, in addition to the PPARα signaling pathway, in mouse hepatocytes. The mouse hepatocytes were transfected using Invitrogen™ Lipofectamine® 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) with either scrambled small interfering (si)RNA or siRNA of SIRT1, followed by treatment with or without SRT1720 (50 ng/ml). Western blotting was used to quantify the protein levels of SIRT1, and the synthesis of TGs was measured. The sequences of scrambled siRNA and siRNA of SIRT1 were as follows: Sense, 5′-ACUUUGCUGUAACCCUGUAdTdT-3′ and antisense, 5′-UACAGGGUUACAGCAAAGUdTdT-3′ (scrambled siRNA); sense, 5′-CCUACGCCACCAAUUUCGU-3′ and antisense, 5′-ACGAAAUUGGUGGCGUAGG-3′ (siRNA of SIRT1).
Western blotting
A lysis buffer (150 mM NaCl, 0.1% SDS, 0.02% NaN3, 1% NP-40, and 50 mM pH 8.0 Tris) containing a cocktail of inhibitors for proteases and phenylmethylsulfonyl fluoride (1 mM) was used to lyse cells. Western blotting was performed using the Bio-Rad Protean II minigel system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The concentrations of total proteins were quantified by bicinchoninic acid assay. A 50 µg protein sample was injected into every gel (12%) well, and then transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Bedford, MA, USA) following electrophoresis. Subsequently, 1× Tween-20 TBS (5% dry skimmed milk) was used to block the non-specific binding. The membranes were then sequentially incubated with primary at 4°C overnight, and secondary antibodies for 1 h at room temperature. The membranes were visualized with ECL chemiluminescence reagent using the Hyperfilm ECL kit, and exposed using x-ray film. The primary antibodies included anti-SIRT1 (D739; cat. no. CST 2493; Cell Signaling Technology, Inc., Danvers, MA, USA), QKI 5 (cat. no. BL 1041; Bethyl Laboratories, Inc., Montgomery, TX, USA), FOXO1 (L27; cat. no. CST 9454; Cell Signaling Technology, Inc.), PPARα (467D1a; cat. no. sc-130640; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) with dilution 1:1,000, and β-actin (13E5; cat. no. CST 4970S; 1:4,000; Cell Signaling Technology, Inc.). The secondary antibody was horseradish peroxidase-conjugated secondary antibody (cat. no. CST 4410S; Cell Signaling Technology, Inc.).
Determination of total TG in cells
Mouse primary hepatocytes were seeded into a 12-well cell culture plate, following treatment with SRT1720 for 48 h as described above. The supernatant was discarded, then 3×105 cells were collected and lysed. The cell lysate was transferred into a 1.5-ml centrifuge tube and a sample was obtained to be analyzed. The standard protein concentration gradient was performed according to the kit protocol. A total of 50 µl of gradient standard and 10 µl of the sample were added into the 96-well microtiter plate. Following the addition of lipase into the main reaction system, the absorbance at 570 nm was measured with a microplate reader, and the absorbance of the sample was calculated according to the standard curve. The concentration of TG was normalized by the quantity of protein per unit (µg).
Analysis of acetylation modification of QKI 5 induced by SIRT1
The local tool of KA-predictor, which can be freely downloaded (http://sourceforge.net/p/ka-predictor), was used to predict the acetylation modification site of QKI 5.
Immunoprecipitation (IP) with acetylated antibody
The cells/tissues were fully lysed with IP lysate and then centrifuged at 6,720 × g for 10 min at 4°C to remove the precipitate. The protein (1 mg) solution was added to 20 µl agarose cross-linked acetylated lysine antibody (acetyl lysine antibody, agarose) (cat. no. CST 9441S; Cell Signaling Technology, Inc.; dilution, 1:800). Following mixing and incubation overnight at 4°C, the supernatant was discarded. The sediment was washed five times with ice pre-cooled PBS, following which 50 µl of elution buffer was used to elute the proteins which were mixed gently. Following centrifugation at 336 × g for 1 min at 4°C, the IP product was assessed using the aforementioned western blotting protocol. The primary antibody of QKI 5 (cat. no. BL 1041; Bethyl Laboratories, Inc.; dilution, 1:500) was used to detect QKI 5 content in the IP products.
Statistical analysis
The differences between two groups were statistical analyzed using Student's t-test. Normally distributed continuous variables are presented as the mean ± standard deviation. Abnormally distributed data among groups was analyzed with the Kruskal-Wallis one-way analysis of variance method. SPSS (version 18.0) software (SPSS, Inc., Chicago, IL, USA) was used for all statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Metabolic disorders of HFD mice are reversed by SRT1720
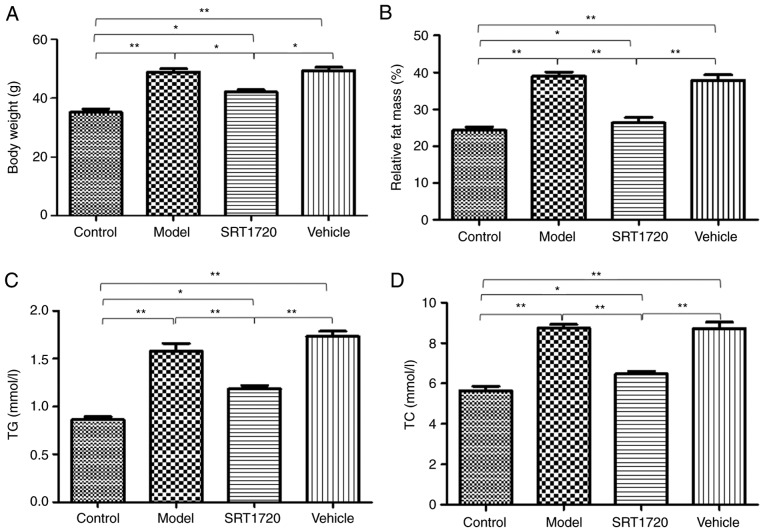
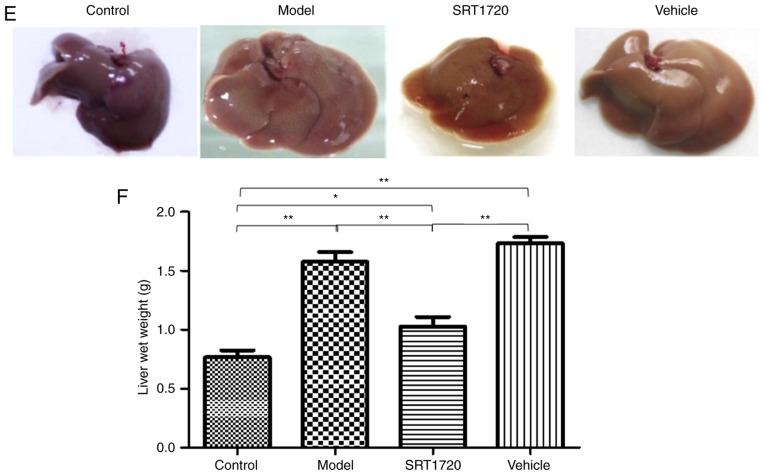
The mice fed the HFD became obese at 8 months of age. The BW (Fig. 1A) and the relative fat mass ratio (Fg. 1B) of the mice in the model group and vehicle group increased significantly, but were only enhanced ~20% in the mice treated with SRT1720. Furthermore, the model and vehicle mice developed hyperlipidemia, as measured by total plasma TG (Fig. 1C) and TC levels (Fg. 1D). Furthermore, these changes were reversed by SRT1720. Consistent with an increase in plasma lipids, the HFD model mice exhibited significantly enlarged fatty livers at 8 months of age (Fig. 1E and 1F), which was reduced by SRT1720.
Figure 1.
Metabolic disorders of HFD mice are reversed by SRT1720. The mice fed with the HFD became obese by 8 months of age compared with the control mice. (A) Body weight and (B) relative fat mass ratio of mice in the model group and vehicle group increased significantly, but only by ~20% in the mice treated with SRT1720. The model and vehicle mice developed hyperlipidemia as measured by (C) total plasma TG and (D) TC levels. Hyperlipidemia was reversed by SRT1720. (E) Images of the livers. (F) Consistent with an increase in plasma lipids, the HFD model mice exhibited significantly enlarged fatty livers at 8 months of age, which was reduced by SRT1720. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01. HDF, high-fat diet; TG, triglycerides; TC, total cholesterol.
HFD mice develop NAFLD, which is reversed by SRT1720
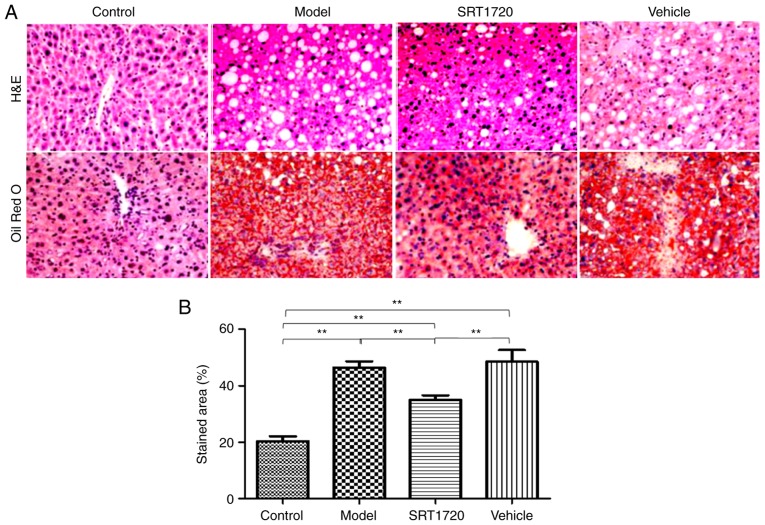
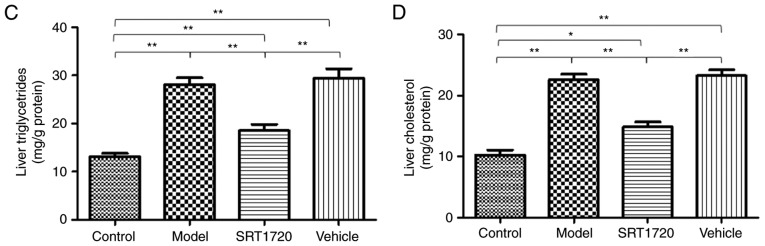
The hepatic lipid content of the HFD model mice at 8 months of age was significantly increased compared with their litter-mate controls, as separately measured using H&E and Oil Red O staining of liver sections (Fig. 2A), and quantification of Oil Red O staining (Fig. 2B). These results indicated the typical pathology of liver steatosis. Furthermore, the TG (Fig. 2C) and TC (Fig. 2D) contents of the extracted liver tissues from the HFD model were significantly increased compared with those of the controls, but were reduced by SRT1720. This demonstrated that the developmental process of liver steatosis was reversed by SRT1720 (Fig. 2).
Figure 2.
Pathological and lipid metabolic changes of NAFLD mice are reversed by SRT1720. (A) Hepatic lipid content of the HFD model mice at 8 months of age was significantly increased compared with littermate controls, as separately measured using H&E and Oil Red O staining and (B) quantification of Oil Red O staining of liver sections (magnification, ×400). These data indicate typical pathology of liver steatosis. The content of extracted (C) liver triglycerides and (D) cholesterol from the HFD model were significantly increased compared with the control, but were reduced by SRT1720. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01. HDF, high-fat diet; NAFLD, non-alcoholic fatty liver disease; H&E, hematoxylin and eosin.
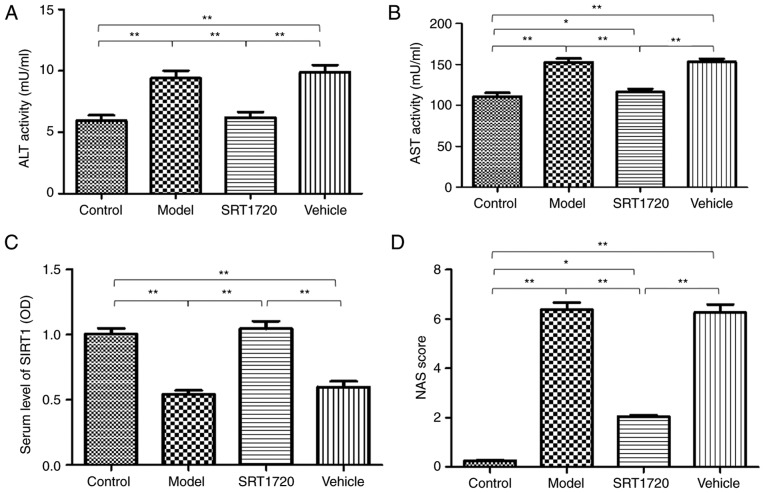
In addition, mildly increased plasma activities of ALT (Fig. 3A) and AST (Fig. 3B) were identified in the model mice. However, the mice treated with SRT1720 demonstrated no signs of liver damage or inflammatory changes in plasma ALT or AST activity. The serum level of SIRT1 in the model mice was significantly decreased compared with that in the control group, however, this was reversed by SRT1720 (Fig. 3C). Furthermore, the NAS was elevated in the model mice; the results revealed significant differences in the model and vehicle groups compared with the control group mice. The NAS of the group treated with SRT1720 was reduced compared with that in the model and vehicle groups (Fig. 3D), however, the NAS score of the SRT1720 group was increased compared with that in the control mice. These results suggested that the administration of SRT1720 improved the pathophysiological process of NAFLD in mice.
Figure 3.
Serum changes and increased NAS score of non-alcoholic fatty liver disease mice are reversed by SRT1720. Mildly increased plasma activities of (A) ALT and (B) AST were determined in model mice. However, mice treated with SRT1720 exhibited no signs of liver damage or inflammatory changes of plasma ALT and AST activity. (C) Serum level of SIRT1 in model mice was significantly lower than that in the control group, but was reversed by SRT1720. (D) NAS was elevated significantly in the model and vehicle groups compared with the control mice. NAS was improved in the SRT1720 group compared with that in the model and vehicle groups, but remained higher than that of the control mice. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01. ALT, alanine aminotransferase; AST, aspartate aminotransferase; SIRT1, Sirtuin 1; NAS, non-alcoholic fatty liver disease activity score.
Hepatic SIRT1 regulates the synthesis of TGs in NAFLD mice via the PPARα/FoxO1 signaling pathway in vitro
An inhibitor of FOXO1 was also used in the present study. The results, as shown in Fig. 4, demonstrated that the inhibitor of FOXO1 (AS1842856) suppressed FOXO1 and PPARα, however, this was reversed by SRT1720. AS1842856 did not inhibit the expression levels of SIRT1 or QKI 5. This demonstrated that hepatic SIRT1 regulated the expression of QKI 5 via the PPARα/FoxO1 signaling pathway.
Figure 4.
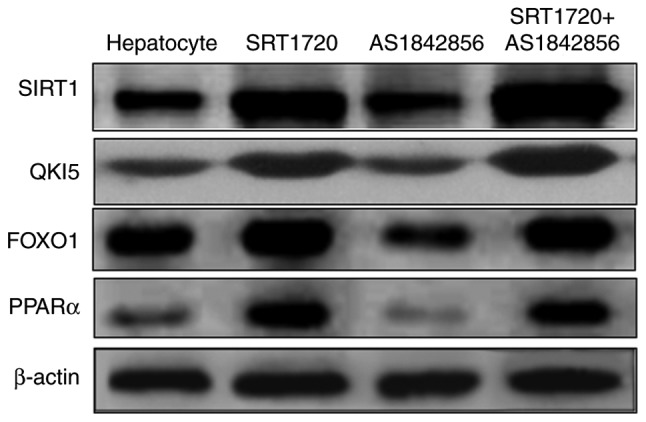
SIRT1 mediates the synthesis of triglycerides in NAFLD mice, which is associated with QKI 5 and the PPARα/FoxO1 signaling pathway in vitro. The inhibitor of FOXO1 (AS1842856) was used in the present study. The results demonstrated that the inhibitor of FOXO1 suppressed FOXO1 and PPARα, and this was reversed by SRT1720. AS1842856 did not inhibit the expression of SIRT1 or QKI 5. Hepatic SIRT1 regulated the expression of QKI 5 via the PPARα/FOXO1 signaling pathway. NAFLD, non-alcoholic fatty liver disease; PPARα, peroxisome proliferator-activated receptor α; FOXO1, Forkhead box protein O1; QKI 5, Quaking 5; SIRT1, Sirtuin 1.
SIRT1 regulates the acetylation level of QKI 5
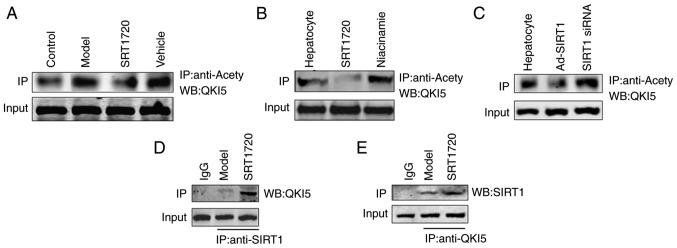
In the liver tissues of the model mice, the acetylation level of QKI 5 increased, and this was reversed by SRT1720 (Fig. 5A). An increase in the acetylation level of QKI 5 was induced by the inhibitor of SIRT1 (niacinamide), whereas the acetylation of QKI 5 decreased in hepatocytes treated with SRT1720 (Fig. 5B). Furthermore, the increased acetylation level of QKI 5 was induced by siRNA of SIRT1, whereas the acetylation of QKI 5 decreased in hepatocytes treated with Ad-SIRT1 (Fig. 5C). The results of the protein interaction analysis confirmed the interaction between SIRT1 and QKI 5, and its level in the SRT1720 group was higher than in the model mice (Fig. 5D and E).
Figure 5.
SIRT1 regulates the acetylation level of QKI 5. (A) In the liver tissues of model mice, the acetylation level of QKI 5 increased, but this was reversed by SRT1720. (B) Increased acetylation of QKI 5 was induced by the inhibition of SIRT1 (niacinamide), but was decreased in hepatocytes treated with SRT1720. (C) Increasing acetylation of QKI 5 was induced by siRNA of SIRT1, but was decreased in hepatocytes treated with Ad-SIRT1. The results of protein interaction confirmed the interaction between (D) SIRT1 and (E) QKI 5, and its level in the SRT1720 group was increased compared with model mice. QKI 5, Quaking 5; SIRT1, Sirtuin 1; siRNA, small interfering RNA; Ad, adenovirus; IP, immunoprecipitation; WB, western blotting.
Hepatic SIRT1 regulates the synthesis of TGs in NAFLD mice via the PPARα/FoxO1 signaling pathway in vivo
To evaluate the ability of hepatic SIRT1 to maintain lipid homeostasis, the agonist (SRT1720) and inhibitor (niacinamide) of SIRT1 was used in the present study. Furthermore, Ad-mediated gene repletion of SIRT1 was used in mice primary hepatocytes, and siRNA of SIRT1 was used to downregulate the expression of SIRT1.
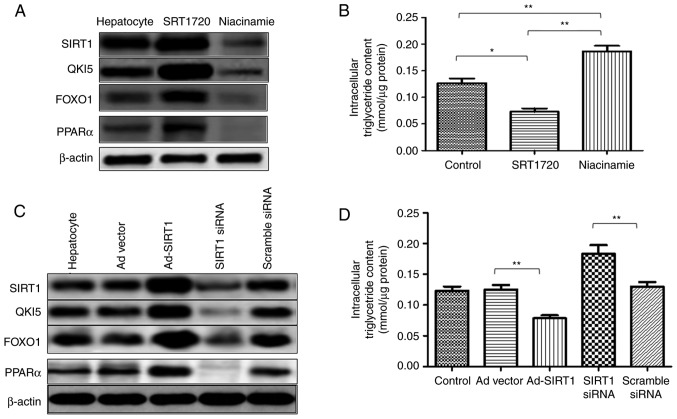
In the primary hepatocytes, SRT1720 enhanced the expression of SIRT1, and the expression levels of QKI 5, FOXO1 and PPARα were increased. However, the expression level of SIRT1 was reduced by niacinamide, which also induced the downregulation of QKI 5, FOXO1 and PPARα (Fig. 6A). In addition, a decrease in intracellular TG content was induced by SRT1720, whereas niacinamide increased the content of TG in the primary hepatocytes (Fig. 6B). Additionally, in the primary hepatocytes, Ad-SIRT1 enhanced the expression of SIRT1 QKI 5, FOXO1 and PPARα, and this was inhibited by SIRT1 siRNA (Fig. 6C). SRT1720 reduced the TG content of the primary hepatocytes, which was promoted by SIRT1 siRNA (Fig. 6D). Therefore, SIRT1 mediated the synthesis of TGs in NAFLD mice, and this was associated with the QKI 5 and the PPARα/FoxO1 signaling pathway.
Figure 6.
Hepatic SIRT1 regulates the synthesis of triglycerides in non-alcoholic fatty liver disease mice via the PPARα/FoxO1 signaling pathway in vivo. In primary hepatocytes, SRT1720 enhanced the expression of SIRT1, QKI 5, FOXO1 and PPARα. (A) Expression of SIRT1 was reduced by niacinamide, which also induced the downregulation of QKI 5, FOXO1 and PPARα. (B) Decreased intracellular triglyceride content was caused by SRT1720, whereas niacinamide enhanced the triglyceride content in primary hepatocytes. (C) In primary hepatocytes, Ad-SIRT1 enhanced the expression of SIRT1, QKI 5, FOXO1 and PPARα, which was inhibited by siRNA of SIRT1. (D) SRT1720 reduced the triglyceride content in primary hepatocytes, which was promoted by SIRT1 siRNA. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01. PPARα, peroxisome proliferator-activated receptor α; FOXO1, Forkhead box protein O1; QKI 5, Quaking 5; SIRT1, Sirtuin 1; siRNA, small interfering RNA; Ad, adenovirus.
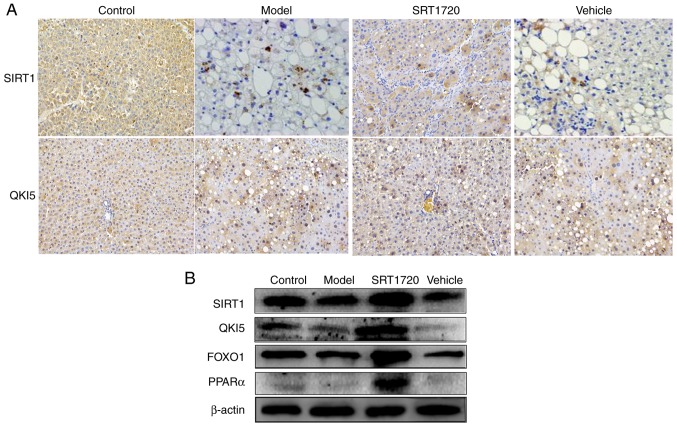
The results of the immunohistochemistry assay indicated that, the expression levels of SIRT1 and QKI 5 were downregulated in the NAFLD model mice, but were promoted by SRT1720 (Fig. 7A). In addition, the western blot assay demonstrated that the expression levels of SIRT1, QKI 5, FOXO1 and PPARα were decreased in the model mice, and this was also reversed by SRT1720 (Fig. 7B).
Figure 7.
Expression levels of SIRT1, QKI 5, FOXO1 and PPARα in mice using immunohistochemistry and western blot assays. (A) Results of immunohisto-chemistry (magnification, ×400) confirmed that, in non-alcoholic fatty liver disease model mice, the expression levels of SIRT1 and QKI 5 were downregulated, but were promoted by SRT1720. (B) Western blotting demonstrated that the expression levels of SIRT1 and QKI 5 were decreased in the model mice, and this was reversed by SRT1720. PPARα, peroxisome proliferator-activated receptor α; FOXO1, Forkhead box protein O1; QKI 5, Quaking 5; SIRT1, Sirtuin 1.
Discussion
Hepatic lipid accumulation is a typical characteristic of NAFLD. There is an ongoing research effort to investigate the mechanism of hepatic steatosis in NAFLD to identify potential novel therapeutic targets (24). Furthermore, de novo lipogenesis has an important effect on the pathogenesis of NAFLD (25). Therefore, in the present study, the process and mechanism of TG synthesis in NAFLD were investigated. NAFLD model mice were produced, with the mice fed an HFD becoming obese at 8 months of age. The results indicated that the BW and the relative fat mass ratio of mice in the model group and vehicle group increased. Furthermore, the model and vehicle mice developed hyperlipidemia, as measured by total plasma TG and TC levels. Consistent with an increase in plasma lipids, the HFD model mice exhibited enlarged fatty livers at 8 months of age. The hepatic lipid content of the HFD model mice at 8 months of age was increased compared with that in their littermate controls, as separately measured using H&E and Oil Red O staining of liver sections, and quantification of the Oil Red O staining. These data indicate a typical pathology of liver steatosis. Furthermore, the TG and TC content in the extracted livers from the NAFLD model mice were increased compared with the non-NAFLD mice. In addition, mildly increased plasma activities of ALT and AST were identified in the model mice. The serum level of SIRT1 in the model mice was reduced compared with that in the control group. Furthermore, the NAS was elevated in the mice; the results exhibited a difference in the model and vehicle group mice compared with the control group mice. These results indicated that the NAFLD model was successfully established.
Carbohydrate and lipid metabolism is regulated by SIRT1, which is associated with the development of NAFLD (26–31). In vivo and in vitro studies of NAFLD have confirmed that SIRT1 is a metabolic sensor, and may improve NAFLD. Through the upregulation of gluconeogenesis-associated genes and de novo lipogenesis, the levels of intracellular lipid and glucose increase, induced by suppressing the expression of SIRT1 (31). Furthermore, the downregulation of SIRT1 in mice treated with small hairpin RNA caused hepatic steatosis (32,33). Therefore, the synthesis of TG in NAFLD associated with SIRT1 was investigated in the present study.
The results of the immunohistochemistry in the present study indicated the decreased expression level of SIRT1in the NAFLD model mice. Furthermore, the western blot assay demonstrated that the expression level of SIRT1 was decreased in model mice. Therefore, to evaluate the ability of hepatic SIRT1 to maintain lipid homeostasis, the agonist (SRT1720) and inhibitor (niacinamide) of SIRT1 were used. In addition, adenovirus-mediated gene repletion of SIRT1 was used in primary hepatocytes of mice, and siRNA of SIRT1 was used to downregulate the expression of SIRT1. The increased BW, relative fat mass ratio, hyperlipidemia levels (total plasma TG and TC), enlarged fatty liver, the hepatic lipid content of the NAFLD model mice were reversed by SRT1720. Additionally, SRT1720 reduced the typical pathology of liver steatosis, increased damage or inflammatory changes of plasma ALT or AST activity, and NAS score. These results demonstrated that the developmental processes of the NAFLD model were reversed by SRT1720.
RNA-binding protein-mediated post-transcriptional regulation is rarely investigated in liver metabolism. STAR family member QKI is an RNA-binding protein that produces multiple isoforms in the human body, and demonstrates a variety of expression patterns in the cells of various types of tissues (10). Among them, QKI 5 is mainly located in the nucleus but can also shuttle to the cytoplasm (13,14), whereas QKI 6 and QKI 7 are mainly distributed in the cytoplasm. By binding to the specific recognition element of the 3′ untranslated region of mRNA (11,12), QKI is involved in regulating the cytoplasmic/nuclear localization, stability and translation efficiency of mRNA (15,16). It has been demonstrated that QKI is expressed in the liver, and the most commonly expressed isomer is QKI 5, although its function in the liver has not been reported.
SIRT1 is mediated by post-translational regulation, and a number of transcription factors, including FOXO1, sterol regulatory element-binding protein 1, carbohydrate-responsive element-binding protein, PPARα and PPARγ coactivator 1α are regulated at the transcriptional level (21). QKI is an RNA-binding protein that is regulated at the post-transcriptional level of RNA. Specific to RNA sequence binding, QKI is widely involved in variable splicing, subcellular localization, stability maintenance and the translational regulation of RNA.
In the present study, primary hepatocytes were extracted to examine the mechanism associated with SIRT1. It was demonstrated that SRT1720 and Ad-SIRT1 enhanced the expression of SIRT1, QKI 5, FOXO1 and PPARα. However, the reduced expression level of SIRT1 induced by niacinamide and siRNA of SIRT1, resulted in the downregulation of QKI 5, FOXO1 and PPARα. In addition, a decrease in intercellular TG content was caused by SRT1720 and Ad-SIRT1, whereas niacinamide and siRNA of SIRT1 enhanced the TG content of primary hepatocytes.
The results of the immunohistochemistry in the present study indicated that the expression levels of SIRT1 and QKI 5 were downregulated in the NAFLD model mice, but were promoted by SRT1720. Furthermore, the results of the western blot assay demonstrated that the expression levels of SIRT1, QKI 5, FOXO1 and PPARα were decreased in the model mice, and this was reversed by SRT1720; therefore, the inhibitor of FOXO1 (AS1842856) was used. The data indicated that AS1842856 suppressed FOXO1 and PPARα, and that this was reversed by SRT1720. AS1842856 did not inhibit the expression levels of SIRT1 or QKI 5. The results demonstrated that hepatic SIRT1 regulated the expression of QKI 5 via the PPARα/FoxO1 signaling pathway. In addition, SIRT1 mediated the synthesis of TGs in NAFLD mice, which was associated with QKI 5 and the PPARα/FoxO1 signaling pathway.
The role of SIRT1 in regulating the acetylation level of QKI 5 was also investigated. In the liver tissues of the model mice, the acetylation level of QKI 5 increased, but this was reversed by SRT1720. In addition, the increasing acetylation level of QKI 5 was induced by the inhibitor of SIRT1 (niacinamide), whereas the acetylation of QKI 5 was decreased in the hepatocytes treated with SRT1720. An increase in the acetylation level of QKI 5 was also induced by SIRT1 siRNA. In addition, the acetylation of QKI 5 was decreased in hepatocytes treated with Ad-SIRT1. The results of this protein interaction confirmed that the interaction between SIRT1 and QKI 5, and its level in the SRT1720 group were increased compared with the model mice.
In conclusion, it was shown that SIRT1 deacetylates QKI 5, which was the RNA-binding protein affecting the synthesis of TGs in the liver of NAFLD mouse model. Furthermore, it activated the transcription factor FOXO1 through post-transcriptional regulation of the expression of PPARα, and further inhibited the synthesis of TG, thereby restricting the progression of NAFLD.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 30600524 and 81341067), the National Natural Science Foundation of Guangdong Province, China (grant no. 2017A030313510), the Introduction of Talent Fund of Guangdong Second Provincial General Hospital (grant no. YY2016-006), the Capital Clinical Featured Applied Research and Results Promotion projects (grant no. Z161100000516141) and Zhejiang Provincial Medical Health Science Technology Project (grant no. 2015KYB382). The study sponsors had no involvement in the study.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
WZ and YS conducted the experiments. WL and JD were involved in the experiments. JC designed the experiments and wrote the manuscript.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Ethics Committee of Tsinghua University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chang Y, Cho YK, Kim Y, Sung E, Ahn J, Jung HS, Yun KE, Shin H, Ryu S. Non-heavy drinking and worsening of non-invasive fibrosis markers in nonalcoholic fatty liver disease: A cohort study. Hepatology. 2018 Jul 17; doi: 10.1002/hep.30170. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Yang W, Chen Z, Chen J, Meng Y, Feng B, Sun L, Dou L, Li J, Cui Q, Yang J. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes. 2018;67:581–593. doi: 10.2337/db17-0799. [DOI] [PubMed] [Google Scholar]
- 3.Jeong HS, Kim KH, Lee IS, Park JY, Kim Y, Kim KS, Jang HJ. Ginkgolide A ameliorates non-alcoholic fatty liver diseases on high fat diet mice. Biomed Pharmacother. 2017;88:625–634. doi: 10.1016/j.biopha.2017.01.114. [DOI] [PubMed] [Google Scholar]
- 4.Tarantino G, Saldalamacchia G, Conca P, Arena A. Non-alcoholic fatty liver disease: Further expression of the metabolic syndrome. J Gastroenterol Hepatol. 2007;22:293–303. doi: 10.1111/j.1440-1746.2007.04824.x. [DOI] [PubMed] [Google Scholar]
- 5.Affo S, Yu LX, Schwabe RF. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estes C, Anstee QM, Arias-Loste MT, Bantel H, Bellentani S, Caballeria J, Colombo M, Craxi A, Crespo J, Day CP, et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J Hepatol. 2018;69:896–904. doi: 10.1016/j.jhep.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 7.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Nassir F, Ibdah JA. Sirtuins and nonalcoholic fatty liver disease. World J Gastroenterol. 2016;22:10084–10092. doi: 10.3748/wjg.v22.i46.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillman KA, Phillips CA, Roslan S, Toubia J, Dredge BK, Bert AG, Lumb R, Neumann DP, Li X, Conn SJ, et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018;37:e99016. doi: 10.15252/embj.201899016. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Ku L, Chen Y, Xia M, LoPresti P, Feng Y. QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol Biol Cell. 2006;17:4179–4186. doi: 10.1091/mbc.e06-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saccomanno L, Loushin C, Jan E, Punkay E, Artzt K, Goodwin EB. The STAR protein QKI-6 is a translational repressor. Proc Natl Acad Sci USA. 1999;96:12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bruin RG, Shiue L, Prins J, de Boer HC, Singh A, Fagg WS, van Gils JM, Duijs JM, Katzman S, Kraaijeveld AO, et al. Quaking promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nat Commun. 2016;7:10846. doi: 10.1038/ncomms10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Vogel G, Yu Z, Richard S. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol Cell Biol. 2013;33:1233–1243. doi: 10.1128/MCB.01604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Song W, Zhao H, Ma Y, Li Y, Zhai D, Pi J, Si Y, Xu J, Dong L, et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017;27:416–439. doi: 10.1038/cr.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall MP, Nagel RJ, Fagg WS, Shiue L, Cline MS, Perriman RJ, Donohue JP, Ares M., Jr Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 2013;19:627–638. doi: 10.1261/rna.038422.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Veer EP, de Bruin RG, Kraaijeveld AO, de Vries MR, Bot I, Pera T, Segers FM, Trompet S, van Gils JM, Roeten MK, et al. Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ Res. 2013;113:1065–1075. doi: 10.1161/CIRCRESAHA.113.301302. [DOI] [PubMed] [Google Scholar]
- 18.Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, Hui J. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagg WS, Liu N, Fair JH, Shiue L, Katzman S, Donohue JP, Ares M., Jr Autogenous cross-regulation of Quaking mRNA processing and translation balances Quaking functions in splicing and translation. Genes Dev. 2017;31:1894–1909. doi: 10.1101/gad.302059.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayakawa-Yano Y, Suyama S, Nogami M, Yugami M, Koya I, Furukawa T, Zhou L, Abe M, Sakimura K, Takebayashi H, et al. An RNA-binding protein, Qki5, regulates embryonic neural stem cells through pre-mRNA processing in cell adhesion signaling. Genes Dev. 2017;31:1910–1925. doi: 10.1101/gad.300822.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnemann J, Kahl M, Siranjeevi PM, Blumrich A, Blümel L, Becker S, Wittig S, Winkler R, Krämer OH, Beck JF. Reverse chemomodulatory effects of the SIRT1 activators resveratrol and SRT1720 in Ewing's sarcoma cells: Resveratrol suppresses and SRT1720 enhances etoposide- and vincristine-induced anticancer activity. J Cancer Res Clin Oncol. 2016;142:17–26. doi: 10.1007/s00432-015-1994-2. [DOI] [PubMed] [Google Scholar]
- 22.Bechmann LP, Kocabayoglu P, Sowa JP, Sydor S, Best J, Schlattjan M, Beilfuss A, Schmitt J, Hannivoort RA, Kilicarslan A, et al. Free fatty acids repress small heterodimer partner (SHP) activation and adiponectin counteracts bile acid-induced liver injury in superobese patients with nonalcoholic steatohepatitis. Hepatology. 2013;57:1394–1406. doi: 10.1002/hep.26225. [DOI] [PubMed] [Google Scholar]
- 23.Biel TG, Lee S, Flores-Toro JA, Dean JW, Go KL, Lee MH, Law BK, Law ME, Dunn WA, Jr, Zendejas I, et al. Sirtuin 1 suppresses mitochondrial dysfunction of ischemic mouse livers in a mitofusin 2-dependent manner. Cell Death Differ. 2016;23:279–290. doi: 10.1038/cdd.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48:434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, Liu YH, Fu YC, Liu XM, Zhou XH. Direct evidence of sirtuin downregulation in the liver of non-alcoholic fatty liver disease patients. Ann Clin Lab Sci. 2014;44:410–418. [PubMed] [Google Scholar]
- 27.Bruce KD, Szczepankiewicz D, Sihota KK, Ravindraanandan M, Thomas H, Lillycrop KA, Burdge GC, Hanson MA, Byrne CD, Cagampang FR. Altered cellular redox status, sirtuin abundance and clock gene expression in a mouse model of developmentally primed NASH. Biochim Biophys Acta. 2016;1861:584–593. doi: 10.1016/j.bbalip.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng C, Zhang Y, Gao Y, Tao W, Zhang H, Liu X, Fang F, Chang Y. Mst1 regulates hepatic lipid metabolism by inhibiting Sirt1 ubiquitination in mice. Biochem Biophys Res Commun. 2016;471:444–449. doi: 10.1016/j.bbrc.2016.02.059. [DOI] [PubMed] [Google Scholar]
- 29.Colak Y, Yesil A, Mutlu HH, Caklili OT, Ulasoglu C, Senates E, Takir M, Kostek O, Yilmaz Y, Yilmaz Enc F, et al. A potential treatment of non-alcoholic fatty liver disease with SIRT1 activators. J Gastrointestin Liver Dis. 2014;23:311–319. doi: 10.15403/jgld.2014.1121.233.yck. [DOI] [PubMed] [Google Scholar]
- 30.Colak Y, Ozturk O, Senates E, Tuncer I, Yorulmaz E, Adali G, Doganay L, Enc FY. SIRT1 as a potential therapeutic target for treatment of nonalcoholic fatty liver disease. Med Sci Monit. 2011;17:HY5–HY9. doi: 10.12659/MSM.881749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobita T, Guzman-Lepe J, Takeishi K, Nakao T, Wang Y, Meng F, Deng CX, Collin de l'Hortet A, Soto-Gutierrez A. SIRT1 disruption in human fetal hepatocytes leads to increased accumulation of glucose and lipids. PLoS One. 2016;11:e0149344. doi: 10.1371/journal.pone.0149344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim KE, Kim H, Heo RW, Shin HJ, Yi CO, Lee DH, Kim HJ, Kang SS, Cho GJ, Choi WS. Myeloid-specific SIRT1 deletion aggravates hepatic inflammation and steatosis in high-fat diet-fed mice. Korean J Physiol Pharmacol. 2015;19:451–460. doi: 10.4196/kjpp.2015.19.5.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.