Abstract
Porphyromonas gingivalis (P. gingivalis) is a periodontal pathogen that may accumulate with other organisms in subgingival plaque biofilms and is associated with periodontal disease. P. gingivalis fimbriae (FimA) is a filamentous structure on the surface of bacteria that is closely associated with bacterial adhesion to and colonization of host tissues, and serves an essential role in biofilm formation. The present study aimed to construct P. gingivalis FimA prokaryotic expression plasmids, purify a FimA fusion protein and explore the effect of a recombinant FimA protein on the inflammatory response in human peripheral blood mononuclear cells (PBMCs). P. gingivalis FimA prokaryotic expression plasmids were constructed by gene cloning and recombination technology. SDS-PAGE was used to evaluate the purified recombinant FimA protein. The cell proliferation rate and inflammatory cytokine expression of PBMCs treated with the FimA fusion protein with or without transfection with toll-like receptor 4 (TLR4) small interfering (si)RNA were detected by CCK-8 assays and ELISAs, respectively. The expression levels of TLR4, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and myeloid differentiation primary response 88 (MyD88) in PBMCs were detected by western blot analysis and reverse transcription quantitative polymerase chain reaction. A FimA fusion protein with high purity was obtained. FimA fusion protein treatment significantly increased PBMC proliferation and promoted the release of tumor necrosis factor-α (TNF-α), interleukin (IL)-6, matrix metalloproteinase (MMP)-8 and MMP-9 in PBMCs. TLR4 interference reversed the effects of the FimA fusion protein on PBMC proliferation and inflammatory cytokine release. The expression levels of TLR4, NF-κB and MyD88 in PBMCs were significantly increased following treatment with the FimA fusion protein, while the expression levels of these genes at the mRNA and protein levels decreased significantly in PBMCs following FimA fusion protein treatment and TLR4 interference. The FimA fusion protein increased PBMC proliferation and promoted the release of the inflammatory cytokines TNF-α, IL-6, MMP-8 and MMP-9 via the TLR4/NF-κB signaling pathway. FimA may serve as a promising therapeutic strategy for periodontal disease.
Keywords: Porphyromonas gingivalis, P. gingivalis fimbriae, cell proliferation, inflammatory cytokines, toll-like receptor 4/nuclear factor kappa-light-chain-enhancer of activated B cells signaling pathway
Introduction
Periodontal disease is a chronic inflammatory disease in which periodontal tissue is infected with pathogenic microorganisms (1). The incidence of chronic periodontal disease, which is the leading cause of tooth loss in adults, is as high as 85% (2-4). Although clinical treatment technology is becoming increasingly sophisticated, controlling the occurrence of periodontal disease and understanding its pathogenesis remain challenging. Bacteria serve a particularly important role in the search for effective therapeutic targets for the treatment of periodontal disease (5).
Porphyromonas gingivalis is a well-known periodontal pathogen that accumulates in subgingival plaque biofilms and invades periodontal tissue (6-8). In addition, P. gingivalis may release a variety of virulence factors, including cysteine prote-ases, hemagglutinins, lipopolysaccharides and P. gingivalis fimbriae (FimA). FimA is important for the invasion of tissue structures, and P. gingivalis FimA, a filamentous structure identified on the surfaces of these bacteria, is closely associated with bacterial adhesion to and colonization of host tissues, and serves an essential role in biofilm formation (9,10). The major gene coding for the P. gingivalis pilin locus is the FimA gene, which serves a decisive role in the pathogenicity of P. gingivalis (11,12). The invasion of periodontal tissue by inflammatory cells and the inflammatory response are major factors in the development of periodontal disease (13). However, the detailed mechanism by which P. gingivalis FimA is closely associated with the inflammatory response in periodontal disease development has not been completely elucidated.
Mononuclear/macrophage cells, as a crucial type of peripheral blood mononuclear cells (PBMCs), serve an important role in the development of periodontal disease (14). When mononuclear/macrophage cells are stimulated, the immune response may be mediated via the secretion of cytokines, but excessive amounts of cytokines may destroy the periodontal tissue and aggravate the progression of periodontal disease (15). The cytokine tumor necrosis factor (TNF)-α is a low-molecular-weight protein with a wide range of biological activities. This cytokine is secreted in large quantities during the early stages of inflammation and is a trigger factor for inflammation (16). The cytokine interleukin (IL)-6 is a proinflammatory factor that regulates the production of proteins in the acute phase, induces and aggravates the inflammatory response, weakens periodontal tissue repair capacity and accelerates the maturation of osteoclasts, resulting in destruction of the alveolar bone (17). MMP-8 is a type of interstitial collagenase, and MMP-9 is a type IV collagenase. Matrix metalloproteinase (MMP)-8 and MMP-9, secreted by neutrophils, may break down collagen, gelatin and elastin, which serve an important role in the process of periodontal tissue destruction (18). A previous study has indicated that FimA may regulate the inflammatory reaction mediated by PBMCs (19).
The present study aimed to construct P. gingivalis FimA prokaryotic expression plasmids by gene cloning and recombination technology, in order to purify FimA fusion proteins and investigate the effects of these proteins on inflammatory cytokines in PBMCs, to study the pathogenic mechanism of P. gingivalis and further the development of clinical treatments for periodontal disease.
Materials and methods
Subject recruitment, blood sampling and culturing of PBMCs
A total of 12 healthy volunteers with good periodontal health (37.5±6.3 years) with no known periodontal pocket formation or attachment loss, no alveolar bone resorption and no systemic disease were selected from the Second Affiliated Hospital of Jinzhou Medical University from September 2017 to December 2017. Additional exclusion criteria were presence of gingivitis, periodontal disease, orthodontic treatment or a history of long-term use of antibiotics or other drugs. All subjects participating in the experiment were subjected to periodontal examination by the same dentist. The subjects clearly understood the purpose of the experiment, agreed to participate and provided written informed consent. The collection of specimens was approved by the Ethics Committee of the Second Affiliated Hospital of Jinzhou Medical University (Jinzhou, China). A total of 10 ml venous blood was collected from each volunteer and stored in EDTA anticoagulant tubes (BD Biosciences, San Jose, CA, USA). PBMCs were isolated by the Ficoll-Paque density gradient centrifugation method (650 × g; 18°C for 20 min) and cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). All cells were grown in an incubator containing 5% CO2 at 37°C in 100% humidity.
P. gingivalis culture and genome extraction
A frozen (−80°C) liquid culture of the P. gingivalis 33277 strain [American Type Culture Collection (ATCC), Manassas, VA, USA] was thawed at room temperature and inoculated on a petri dish containing brain heart infusion (BHI) agar with 1 mg/l vitamin K, 1 g/l yeast extract and 5 mg/l hemin (China Medical University, Shenyang, China). The cells were cultured under anaerobic conditions at 37°C for 4 days. The surface colonies were scraped off with sterile loops on a sterile, ultraclean bench and inoculated in BHI liquid culture medium. Cells were grown for ~24 h, until the bacterial growth reached the logarithmic phase. Then, the supernatant was discarded following centrifu-gation at 4,000 x g for 15 min at 4°C to obtain the bacteria, and the P. gingivalis genomic DNA was extracted. Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA), following the manufacturer's instructions.
Semi-quantitative polymerase chain reaction (PCR) ampli- fication of target genes
Based on the P. gingivalis ATCC 33277 FimA sequence available in the National Center for Biotechnology Information GenBank database (http://www.ncbi.nlm.nih.gov/protein/), primers (forward, 5′-ATT AGG ATC CAT GGT GGT ATT GAA GAC CAG C-3′; reverse, 5′-ATA TCT CGA GCC AAG TAG CAT TCT GAC CAA CGA G-3′) were designed using Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) to remove the stop codon from the original sequence and add a start codon. The primers were synthesized by Nanjing GenScript Biotechnology Corporation (Nanjing, China). The FimA gene was amplified by LA Taq enzyme (Takara Biotechnology Co., Ltd., Dalian, China). The cycling conditions included 94°C for 5 min, followed by 25 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 2 min, with a final extension at 72°C for 10 min. The PCR products were identified by 1% agarose gel electrophoresis in Tris-acetate buffer (40 mM Tris acetate, 1 mM EDTA; pH, 8.0). The products were visualized with ethidium bromide by UV transillumination.
Plasmid construction
The purified PCR product was ligated to the PMD19-T vector at 16°C overnight. The next day, 10 μl ligation reaction mixture was added to 200 μl BL21 (DE3) competent cells. The cells were mixed thoroughly, placed in an ice bath for 30 min, heat-shocked at 42°C for 1 min and the placed in an ice bath for an additional 2 min. Then, 400 μl antibiotic-free lysogeny broth (LB) liquid medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) was added at room temperature, and the cells were shaken gently at 3°C for 60 min. Then, the mixture was added to an ampicillin-containing LB solid medium plate, and the plate was incubated at 37°C overnight. The next day, a single white colony was selected from the petri dish and inoculated in 5 ml LB liquid medium containing 100 mg/ml ampicillin, and the culture was shaken at 37°C until the logarithmic phase [optical density (OD) 0.6-0.8]. The cells were centrifuged at 1,000 x g for 3 min at 4°C, and the supernatant was discarded. A small amount of plasmid DNA was extracted, and the plasmid concentration was measured by UV absorption at 260 nm. The recombinant plasmid was digested with XhoI/BamH I at 37°C and visualized by 1% agarose gel electrophoresis and stained with ethidium bromide. An appropriate amount (200 μl) of plasmid DNA was sent to the Beijing Boao Medical Laboratory (Beijing, China) for sequencing. The recombinant plasmid was named PMD19-T-FimA. The verified PMD19-T-FimA plasmid and the empty PGEX-6P-1 plasmid were digested with XhoI/BamH I, and the target fragments were recovered. The recovered products were ligated in a fragment:vector ratio of 7:3. The ligated product was uniformly mixed with Escherichia coli BL21 (DE3) competent cells for transformation. The cells were subjected to plasmid miniprep according to the aforementioned method, and the extracted DNA was analyzed by 1% agarose gel electrophoresis and stained with ethidium bromide. The recombinant plasmid was named PGEX-6P-FimA.
Expression and purification of the FimA fusion protein
A single white colony of cells harboring PGEX-6P-FimA was selected and inoculated in 30 ml LB medium containing ampicillin (100 mg/ml), and the culture was shaken at a constant speed of 180 r/min at 37°C. The OD of the broth was monitored with a wavelength of 600 nm, and when the OD value reached between 0.6-0.8, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at different concentrations (0, 0.1, 0.3, 0.5, 0.7 and 1 mmol/l) to induce expression for different time intervals (4, 6, 8, 12 and 16 h). A total of 2 ml culture from each induction temperature (16, 20, 25, 30 and 37°C) was collected and centrifuged at 6,000 × g for 4 min at 4°C, and the supernatant was discarded. The pellet was resuspended in 200 μl PBS and sonicated in an ice bath (3 pulses; 5 sec; 100 W) until the broth was clear. The cells were then centrifuged at 6,000 × g for 10 min at 4°C. The supernatant and precipitate were collected and stored at −20°C. The optimal conditions for induction were determined by 12% SDS-PAGE and were stained with 0.1% Coomassie brilliant blue R250 (in 50% ethanol and 10% acetic acid) for 2 h followed by visualization of the protein bands after destaining using 3% NaCl. Based on a comparison of the aforementioned experimental results, 600 ml culture at OD(600) of 0.74 was induced for 12 h at 16°C with an IPTG concentration of 1 mmol/l. Following centrifu-gation at 6,000 × g for 10 min at 4°C, the supernatant was discarded and the cells were resuspended in 20 ml PBS. A total of 200 μl phenylmethylsulfonyl fluoride (1:100 dilution) was added to the precipitate, which was then ultrasonicated in an ice bath (3 pulses; 5 sec; 100 W) for 5-8 cycles, until the bacterial solution became clear. The solution was then centrifuged at 4°C at 6,000 × g for 40 min. The supernatant was discarded, and the precipitate was fully dissolved with 20 ml GuMCAC-0 wash buffer (20 mmol/l Na3PO3·12H2O, 0.5 mmol/l NaCl, 6 mol/l guanidine hydrochloride; pH 7.9) and kept overnight at 4°C. The following day, following centrifugation at 4°C at 6,000 × g for 30 min, the oily supernatant was collected, poured into an ultrafiltration centrifuge tube and centrifuged at 4°C at 6,000 × g for 10 min again, gradually replacing the buffer with an imidazole-free MCAC-0 buffer (20 mmol/l Na3PO3·12H2O, 0.5 mmol/l NaCl; pH 7.9). Subsequent to the liquid becoming clear, the clear solution containing the target protein was added to a glutathione S-transferase (GST) gel chromatography column. The solution was passed through the column at a slow flow rate of 0.3 ml/min, which was followed by addition of an extraction/equilibration buffer (140 mmol/l NaCl, 10 mmol/l Na2HPO4 and 1.8 mmol/l K2HPO4; pH 7.5). The gel with the adsorbed protein was washed thoroughly until no protein was present in the eluate. Finally, elution was performed with 5-10 gel volumes of elution buffer (50 mmol/l Tris-HCl, 33 mmol/l glutathione; pH 8.0). The flow rate was controlled, and the eluates were collected and stained with 0.1% Coomassie brilliant blue R250 (in 50% ethanol and 10% acetic acid) for 2 h followed by visualization of the protein bands after destaining using 3% NaCl.
RNA interference assay
Small interfering (si)RNA targeting toll-like receptor 4 (si-TLR4) and negative siRNA with a random sequence were synthesized by Shanghai GenePharma Co., Ltd., (Shanghai, China). The target sequences for TLR4 were: Sense, 5′-GGG CUU AGA ACA ACU AGA ATT-3′; and antisense, 5′-UUC UAG UUG UUC UAA GCC CTT-3′, and the sequences for the negative siRNA were: Sense, 5′-UUC UCC GAA CGU GUC ACG UTT-3′; and antisense, 5′-ACG UGA CAC GUU CGG AGA ATT-3′. The concentration used was 20 nM. All plasmids and oligonucleotides were transfected using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. After 48 h, the cells were collected and subjected to subsequent experimentation.
Cell proliferation assay
PBMCs (1×103 cells/well) were seeded into 96-well plates and treated with FimA fusion protein (2, 4 and 6 μg/ml) with or without transfection with siTLR4 at 37°C and 5% CO2 for 12, 24 and 48 h. Cell proliferation was detected using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). A 10 μl aliquot of CCK-8 solution was added to the treated PBMCs and incubated at 37°C for an additional 4 h. The cells were washed twice with PBS and then incubated at 37°C for 1-4 h. The absorbance value at 450 nm of each well was measured by a 96-well plate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
ELISA
PBMCs were evenly seeded in 6 wells at a density of 1×106 cells/ml and treated with FimA fusion protein (2, 4 and 6 μg/ml) with or without transfection with siTLR4 at 37°C and 5% CO2 for 12, 24 and 48 h. The cell culture media of all groups were collected and centrifuged at 8,000 × g for 20 min at 4°C to remove debris. The supernatant (400 μl) of the PBMCs was collected to measure the concentrations of TNF-α, IL-6, MMP-8 and MMP-9 with an ELISA-based capture assay, using commercial TNF-α (cat. no. PT518) and IL-6 (cat. no. PI330) ELISA kits (Beyotime Institute of Biotechnology, Haimen, China), and MMP-8 (cat. no. DMP800B) and MMP-9 (cat. no. DMP900) ELISA kits (R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturers' instructions. The absorption was measured at 450 nm. The concentration of inflammatory cytokines was determined by comparing the relative absorbance of the samples with the standards.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted from PBMCs treated with the FimA fusion protein with or without transfection with siTLR4 using TRIzol® reagent (Thermo Fisher Scientific, Inc.). The extracted RNA was then transcribed into double-strand cDNAs using a commercial Reverse Transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was amplified using the following primers: TLR4 forward, 5′-CCG CTT TCA CTT CCT CTC AC-3′; TLR4 reverse, 5′-CAT CCT GGC ATC ATC CTC AC-3′; nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) forward, 5′-CTT GCT TAG TTG GTC CTC-3′; NF-κB reverse, 5′-ACC CGA AGA GAA ACG A-3′; myeloid differentiation primary response 88 (MyD88) forward, 5′-AGA TGG ACC TCG GGA G-3′; MyD88 reverse, 5′-ATC AAT CAC GCA CGA TTT-3′; β-actin forward, 5′-TCC CTG TAT GCC TCT G-3′; β-actin reverse, 5′-ATG TCA CGC ACG ATT-3′. The reaction system contained cDNA template (1 μl), primers (1 μl), 2X SYBR Green Mix (10 μl; Shanghai GeneCore BioTechnologies Co., Ltd., Shanghai, China) and RNase-free water (8 μl), with a total volume of 20 μl. PCR was performed using the ABI 7500 system platform (Applied Biosystems; Thermo Fischer Scientific, Inc.). The 2−ΔΔCq method was used to calculate the relative gene expression of TLR4, NF-κB and MyD88 normalized to β-actin (20). PCR amplification was repeated in triplicate for each gene.
Western blot analysis
Total protein was extracted from PBMCs treated with the FimA fusion protein with or without transfection with siTLR4 by a radioimmunopre cipitation assay (Beyotime Institute of Biotechnology). The protein concentration was evaluated by a BCA protein assay kit (Beyotime Institute of Biotechnology). Total lysates (50 mg) were resolved by 10% SDS-PAGE (Beyotime Institute of Biotechnology), followed by blocking for 1 h at room temperature in blocking buffer (cat. no. P0023B; Beyotime Institute of Biotechnology). The membranes were incubated with primary antibodies (rabbit anti-TLR4, dilution 1:500, cat. no. ab13556; rabbit anti-NF-κB, dilution 1:500, cat. no. ab207297; rabbit anti-MyD88, dilution 1:500, cat. no. ab2064; rabbit anti-β-actin, dilution 1:1,000, cat. no. ab8227; Abcam, Cambridge, UK). Following rinsing with TBST, goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (dilution 1:5,000; cat. no. ab6721; Abcam, Cambridge, UK) was added, and the membranes were incubated at room temperature for an additional 2 h. Image J software version 1.48 (National Institutes of Health, Bethesda, MD, USA) was used to analyze relative protein band density.
Statistical analysis
SPSS 17.0 statistical software (SPPS Software, Inc., Chicago, IL, USA) was used for data analysis. The data are expressed as the mean ± standard deviation. All experiments were repeated at least three times. Comparisons between two groups were analyzed using an independent samples t-test, whereas comparisons among multiple groups were analyzed using one-way analysis of variance followed by the Tukey's honest signifi-cant difference test. P<0.05 was considered to indicate a statistically significant difference.
Results
PCR amplification of the FimA gene, recombinant plasmid construction and sequence identification
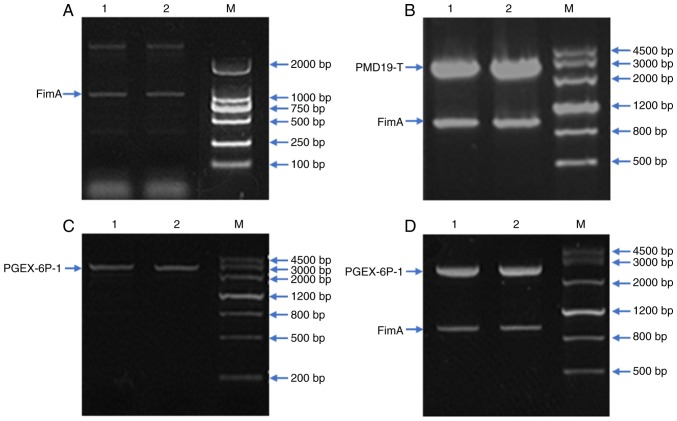
The concentration of the DNA extracted from P. gingivalis ATCC 33277 was determined to be 0.125 μg/μl, and the A260/A280 nm ratio was 1.85, indicating that the DNA purity was good and that there was no RNase contamination (Fig. 1A). The PMD19-T-FimA and PGEX-6P-1 plasmids were subjected to XhoI/BamH I double digestion, the purified target fragment was recovered and ligated, and double digestion was performed again. Electrophoresis indicated a fragment of the expected size, demonstrating that the desired plasmid had been successfully constructed; the plasmid was named PGEX-6P-FimA (Fig. 1B-D).
Figure 1.
PCR amplification of the FimA gene, recombinant plasmid construction and sequence identification. (A) The LA Taq enzyme was used to amplify the FimA gene. Using the extracted DNA as a template, the target gene was amplified by PCR. Electrophoretic detection demonstrated a distinct band with a molecular weight of 1,000 bp, which was consistent with the size of FimA (1,044 bp). Lanes 1 and 2 are FimA target segments. (B) XhoI/BamH I double-digested PMD19-T-FimA plasmid. Lanes 1 and 2 are the PMD19-T-FimA plasmid. (C) XhoI/BamH I double-digested PGEX-6P-1 plasmid. Lanes 1 and 2 are the PGEX-6P-1 plasmid. (D) XhoI/BamH I double-digested PGEX-6P-FimA plasmid. Lanes 1 and 2 are PGEX-6P-FimA plasmid. PCR, polymerase chain reaction; FimA, Porphyromonas gingivalis fimbriae; M, marker.
Expression, purification and identification of the FimA fusion protein
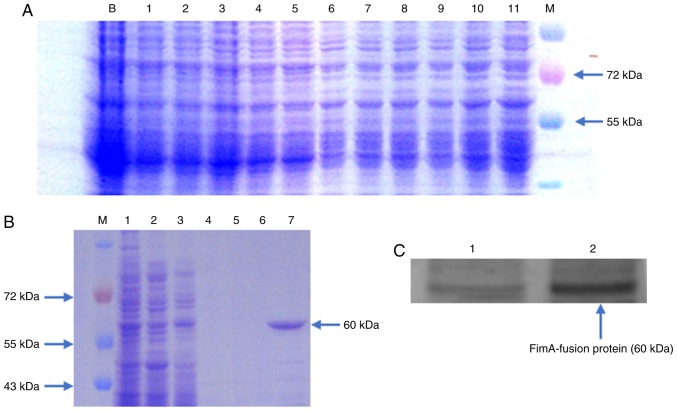
The recombinant plasmid PGEX-6P-FimA was used for expression in E. coli. Following expression under different conditions, the cells were sonicated, and the supernatant and precipitate were collected. SDS-PAGE and Coomassie brilliant blue staining were used to detect whether the protein was expressed under various conditions and to identify the form in which the protein was present. The results indicated that at 16°C, the IPTG concentration was 1 mmol/l; the maximum protein expression level was observed at 12 h after induction; and the protein existed primarily in the form of inclusion bodies (Fig. 2A). A large amount of FimA fusion protein was induced under the optimal conditions. The fusion protein was collected and eluted with a gradient at 4°C in the presence of denaturing agent, and then, gradient renaturation of the inclusion bodies was conducted. The proteins were washed, and the target protein was eluted. Coomassie blue staining and western blot analysis detection were performed. FimA fusion protein with high purity was obtained, and there was a distinct band at ~60 kDa, which was consistent with the expected size of the fusion protein (Fig. 2B-C).
Figure 2.
Expression, purification and identification of the FimA fusion protein. (A) Optimum induction conditions were induction at 16°C for 12 h. Lanes 1, 2, 3, 4 and 5 represent the supernatant following ultrasonication. Lanes 6, 7, 8, 9, 10 and 11 represent the precipitate following ultrasonication; the isopropyl β-D-1-thiogalactopyranoside concentrations were 0, 0.1, 0.3, 0.5, 0.7 and 1 mmol/l, respectively. (B) Eluted protein. Lanes 1 and 2 represent the precipitate and supernatant of the bacterial solution following ultrasonication; lanes 3, 4, 5 and 6 represent the impurities from the four washes; and lane 7 is the eluted FimA fusion protein. (C) Western blot analysis of the FimA fusion protein. Lane 1 contains pre-ultrasonication bacterial solution, and lane 2 represents the purified FimA fusion protein. M, Marker; FimA, Porphyromonas gingivalis fimbriae.
FimA fusion protein treatment increases PBMC proliferation
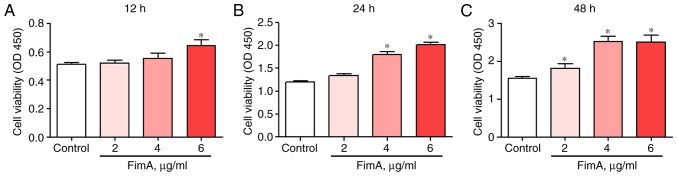
PBMCs were treated with 2, 4 or 6 μg/ml FimA fusion protein for 12, 24 or 48 h. Cell proliferation was detected by CCK-8 at the 3 time points. The results revealed increased proliferation of PBMCs treated with 6 μg/ml FimA fusion protein at 12 h (Fig. 3A) and 4 and 6 μg/ml FimA fusion protein at 24 h (Fig. 3B). All concentrations of the FimA fusion protein resulted in a significant increase in PBMC proliferation at 48 h (Fig. 3C). These data indicated that the FimA fusion protein may promote PBMC proliferation and implicated FimA in the development of the inflammatory response in periodontal disease.
Figure 3.
FimA fusion protein treatment increases PBMC proliferation. Cell proliferation was detected by Cell Counting Kit-8 following treatment of PBMCs with 2, 4 and 6 μg/ml FimA fusion protein for (A) 12, (B) 24 and (C) 48 h. Data are expressed as the means ± standard deviation based on at least three independent experiments; *P<0.05 vs. the control group (0 μg/ml FimA fusion protein). FimA, Porphyromonas gingivalis fimbriae; PBMC, peripheral blood mononuclear cells.
TLR4 interference reverses the effects of the FimA fusion protein on PBMC proliferation
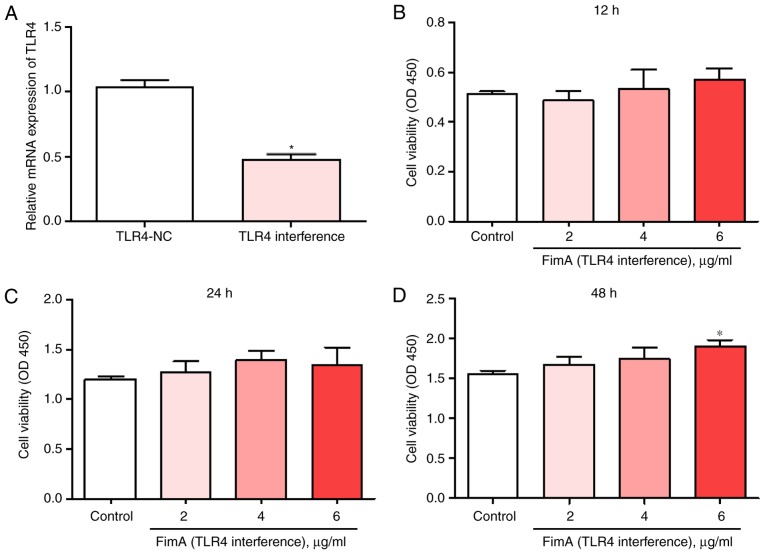
To explore the effects of TLR4 on PBMC proliferation, cells were treated with 2, 4 or 6 μg/ml FimA fusion protein for 12, 24 or 48 h with or without transfection with siTLR4. The mRNA expression of TLR4 was significantly decreased in PBMCs following transfection with siTLR4 (Fig. 4A). The CCK-8 results indicated that treatment with the FimA fusion protein and transfection with siTLR4 did not significantly alter PBMC proliferation at 12 or 24 h compared with that of the control group (Fig. 4B and C). Cell proliferation was significantly increased only in PBMCs treated with 6 μg/ml FimA fusion protein and transfected with siTLR4 at 48 h (Fig. 4D). These results suggested that TLR4 interference inhibited the induction of PBMC proliferation by the FimA fusion protein.
Figure 4.
TLR4 interference reverses the effects of the FimA fusion protein on PBMC proliferation. (A) The expression levels of TLR4 mRNA were detected by reverse transcription quantitative polymerase chain reaction following treatment of PBMCs with or without siTLR4 transfection. Cell proliferation was detected by Cell Counting Kit-8 following treatment of PBMCs with 2, 4 and 6 μg/ml FimA fusion protein for (B) 12, (C) 24 and (D) 48 h and transfection with siTLR4. Data are expressed as the means ± standard deviation based on at least three independent experiments; *P<0.05 vs. the control group (negative siRNA-transfected). TLR4, toll-like receptor 4; FimA, Porphyromonas gingivalis fimbriae; PBMC, peripheral blood mononuclear cells; si, small interfering.
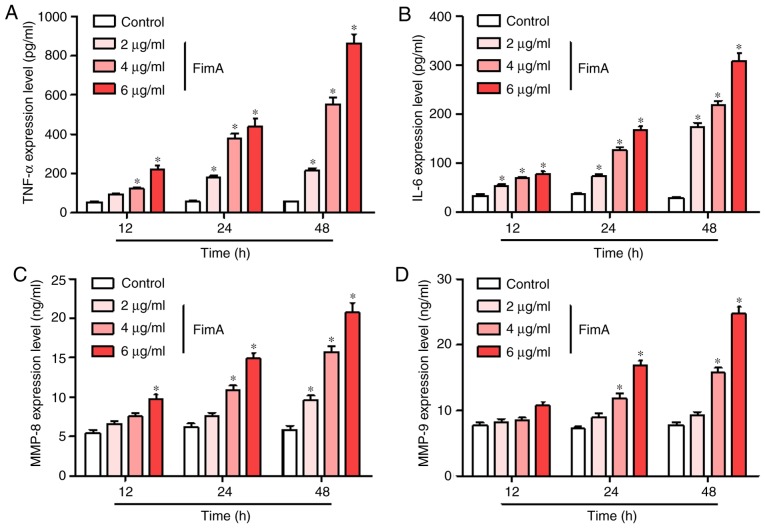
FimA fusion protein treatment promotes the release of inflammatory cytokines by PBMCs
To investigate whether the FimA fusion protein promotes the release of inflammatory cytokines in PBMCs, ELISA was used to detect the expression levels of TNF-α, IL-6, MMP-8 and MMP-9 following treatment of the cells with 2, 4 and 6 μg/ml FimA fusion protein for 12, 24 and 48 h. FimA fusion protein treatment (4 and 6 μg/ml) significantly increased the release of TNF-α in PBMCs at 12, 24 and 48 h, and 2 μg/ml FimA fusion protein treatment significantly increased the release of TNF-α in PBMCs at 24 and 48 h (Fig. 5A). The concentrations of IL-6 in the supernatant were significantly increased at each time point in PBMCs treated with all different dosages of FimA fusion protein (Fig. 5B). The level of MMP-8 release was significantly increased by treatment with 6 μg/ml FimA fusion protein at 12 h, 4 and 6 μg/ml FimA fusion protein at 24 h, and 2, 4 and 6 μg/ml FimA fusion protein at 48 h (Fig. 5C). In addition, the concentrations of MMP-9 were significantly increased at 24 h and 48 h in PBMCs treated with 4 and 6 μg/ml FimA fusion protein (Fig. 5D). These results indicated that the treatment of PBMCs with the FimA fusion protein promoted the release of inflammatory cytokines.
Figure 5.
FimA fusion protein treatment promotes the release of inflammatory cytokines in PBMCs. The expression levels of (A) TNF-α, (B) IL-6, (C) MMP-8 and (D) MMP-9 were detected by ELISA following treatment of PBMCs with 2, 4 and 6 μg/ml FimA fusion protein for 12, 24 and 48 h. Data are expressed as the means ± standard deviation based on at least three independent experiments; *P<0.05 vs. the control group (0 μg/ml FimA fusion protein). FimA, Porphyromonas gingivalis fimbriae; PBMC, peripheral blood mononuclear cells; TNF-α, tumor necrosis factor α; IL, interleukin; MMP, matrix metalloproteinase.
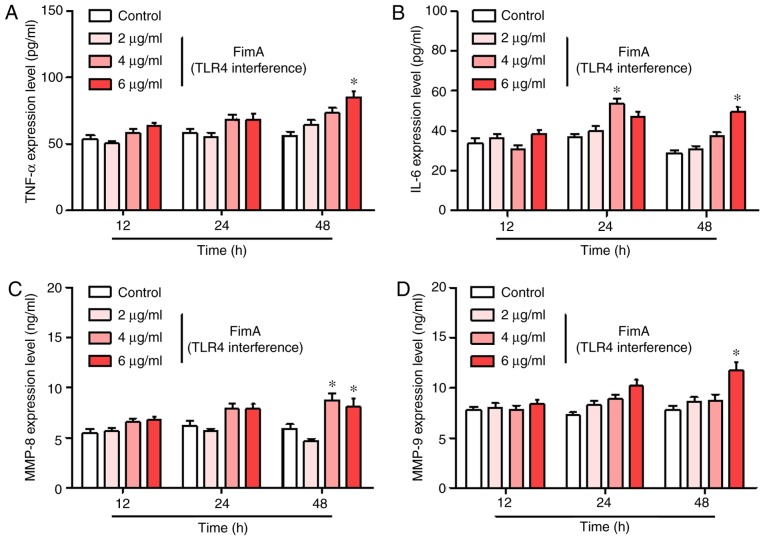
TLR4 interference inhibits the release of inflammatory cytokines induced by the FimA fusion protein in PBMCs
To additionally investigate the role of TLR4 in the release of inflammatory cytokines induced by the FimA fusion protein in PBMCs, PBMCs were treated with the FimA fusion protein with and without transfection with siTLR4 for 12, 24 and 48 h. The ELISA results demonstrated that TLR4 interference significantly decreased the levels of TNF-α, IL-6, MMP-8 and MMP-9 induced by the different dosages of the FimA fusion protein at each time point compared with those induced by the FimA fusion protein treatment alone (Fig. 6A-D). However, FimA fusion protein treatment (6 μg/ml) and transfection with siTLR4 significantly increased the release of TNF-α, IL-6, MMP-8 and MMP-9 in PBMCs at each time point compared with that in the control group. These results suggested that TLR4 serves an important role in the inflammatory response of PBMCs.
Figure 6.
TLR4 interference inhibits the release of inflammatory cytokines induced by the FimA fusion protein in PBMCs. The expression levels of (A) TNF-α, (B) IL-6, (C) MMP-8 and (D) MMP-9 were detected by ELISA following treatment of PBMCs with 2, 4 and 6 μg/ml FimA fusion protein for 12, 24 and 48 h and transfected with siTLR4. Data are expressed as the means ± standard deviation based on at least three independent experiments. *P<0.05 vs. the control group (negative siRNA-transfected). TLR4, toll-like receptor 4; FimA, Porphyromonas gingivalis fimbriae; PBMC, peripheral blood mononuclear cells; TNF-α, tumor necrosis factor α; IL, interleukin; MMP, matrix metalloproteinase; si, small interfering.
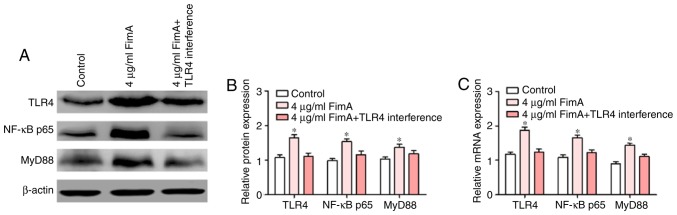
FimA fusion protein induced the inflammatory response via the TLR4/NF-κB signaling pathway in PBMCs
The TLR4/NF-κB signaling pathway serves an important role in the regulation of the inflammatory response (21). To additionally confirm the effects of TLR4 on the induction of PBMC proliferation and inflammatory cytokine release by the FimA fusion protein, TLR4 downstream signaling molecules were detected by western blot analysis and RT-qPCR following treatment of PBMCs with 4 μg/ml FimA fusion protein with or without transfection with siTLR4 for 24 h. Western blot analysis indicated that the expression levels of TLR4, NF-κB and MyD88 proteins in PBMCs treated with 4 μg/ml FimA fusion protein were significantly increased compared with those in the control group at 24 h, and the expression levels of these proteins were significantly decreased in the cells subsequent to treatment with the FimA fusion protein combined with transfection with siTLR4 (Fig. 7A and B). In addition, RT-qPCR data suggested that TLR4, NF-κB and MyD88 mRNA expression levels were increased by FimA fusion protein (4 μg/ml) at 24 h, and the increases in TLR4, NF-κB and MyD88 were reversed by TLR4 interference (Fig. 7C). Taken together, these results indicated that the FimA fusion protein promoted the inflammatory response in a manner associated with the regulation of the TLR4/NF-κB signaling pathway in PBMCs.
Figure 7.
FimA fusion protein induced an inflammatory response via the TLR4/NF-κB signaling pathway in PBMCs. (A) The expression levels of TLR4, NF-κB and MyD88 protein were detected by western blot analysis following treatment of PBMCs with 4 μg/ml FimA fusion protein with or without transfection with siTLR4 for 24 h. (B) Densitometric analysis of western blot data. (C) The expression levels of TLR4, NF-κB and MyD88 mRNA were detected by reverse transcription quantitative polymerase chain reaction following treatment of PBMCs with 4 μg/ml FimA fusion protein with or without transfection with siTLR4 for 24 h. Data are expressed as the means ± standard deviation based on at least three independent experiments. *P<0.05 vs. the control group (negative siRNA-transfected). TLR4, toll-like receptor 4; NF-κB; nuclear factor kappa-light-chain-enhancer of activated B cells; FimA, Porphyromonas gingivalis fimbriae; PBMC, peripheral blood mononuclear cells; MyD88, myeloid differentiation primary response 88; NF-κB p65, NF-κB p65 subunit; si, small interfering.
Discussion
Periodontal disease is the most common plaque-induced inflammatory disease. Actinobacillus actinomycetemcomitans, P. gingivalis and Prevotella intermedia are the most common anaerobic gram-negative bacteria in the context of periodontal disease. These bacteria may lead to the formation of periodontal pockets, causing the destruction of connective tissue and alveolar bone resorption (22). The development of vaccines against periodontal disease caused by bacteria has been one of the auxiliary approaches for the treatment of periodontal disease. Sharma et al (23) described the use of a DNA vaccine against the P. gingivalis FimA protein for the prevention of periodontal disease. However, the underlying molecular mechanisms of FimA in periodontal disease progression are far from clear.
In the present study, the FimA target gene was cloned, and prokaryotic expression plasmids were constructed to purify FimA fusion proteins. The fusion protein was primarily expressed in the form of inclusion bodies. At 16°C, the IPTG concentration was 1 mmol/l, and the maximum protein expression level was observed at 12 h. Notably, GST-tagged proteins in inclusion bodies require denaturation and gradient rena-turation to bind well with glutathione. Following Coomassie blue staining and western blot analysis, the recombinant protein expressed from the prokaryotic expression plasmid PGEX-6P-FimA exhibited a specific band at approximately 60 kDa, which was consistent with the expected protein size.
Inflammation is a biological reaction of tissues to infection with pathogens and cell damage (24). P. gingivalis exhibits relatively mild inflammatory cell infiltration and a weak antigen-specific immune response that induces high proliferation of previously restricted subgingival plaques in the early stages (25). Interference with the cytokine network makes the host vulnerable to the virulence factors of P. gingivalis, including FimA, lipopolysaccharide and various genotoxins, among which FimA is an important virulence factor that stimulates the release of a variety of inflammatory mediators from gingival tissues (26-28). The innate immune system of the host responds to decrease inflammation by activating mechanisms including the mononuclear/macrophage system. Increased levels of inflammatory factors and release of matrix metalloproteinases may cause irreversible decomposition of collagen fibers in the periodontium, resulting in periodontal bone loss and organizational damage (29,30). Ogawa et al (31) demonstrated that the detection rate of P. gingivalis FimA-specific antibody-expressing cells increased with the development of periodontal disease. In the late stage of infection, FimA released by the bacteria may penetrate gingival tissue and enter blood circulation, which may contribute to the systemic inflammatory response, and studies have confirmed that P. gingivalis FimA stimulates endothelial cells to produce IL-8 and monocyte chemoattractant protein 1 in humans and that these cytokines are closely associated with atherosclerosis (32,33).
The present study identified that FimA fusion protein treatment significantly increased PBMC proliferation and promoted the release of TNF-α, IL-6, MMP-8 and MMP-9 by PBMCs. Nagano et al (34) used FimA at a concentration of 1 mg/ml to stimulate mouse macrophages, and the macrophage line exhibited strong activation. Cochran (35) demonstrated that the expression levels of TNF-α and other cytokines in the periodontal tissues of patients with periodontal disease were significantly increased with those in the periodontal tissues of patients in the control group. Following general and periodontal cleaning and treatment of periodontal disease, the expression levels of TNF-α and other cytokines were significantly decreased in the treated group compared with the group with untreated periodontal disease. Ikezawa et al (36) identified that there was a significant difference in the levels of TNF-α in gingival crevicular fluid and serum between patients with chronic periodontal disease and healthy subjects, and that this difference increased significantly with the deepening of periodontal pockets. However, the mechanism via which mononuclear/macrophage cells or other innate immune cells recognize and bind FimA, leading to intracellular signaling and proinflammatory gene activation, is not known.
The present study examining the time- and dose-effect associations of the FimA fusion protein with TNF-α, IL-6, MMP-8 and MMP-9 in healthy subjects is based on a previous hypothesis (37). The experimental results indicated that the levels of TNF-α, IL-6, MMP-8 and MMP-9 secreted by PBMCs increased significantly with increasing concentrations of the fusion protein and prolonged treatment time intervals. This result suggests that the FimA fusion protein may stimulate PBMCs in vivo, additionally increasing inflammatory cytokine expression and participating in the pathogenesis of periodontal disease. Graves et al (38) purified a FimA fusion protein, injected the protein into mouse scalps, and performed an RNase protection assay. The results confirmed that the purified FimA protein may markedly induce the expression of the cytokines TNF-α and MIP-2 (38).
Studies have indicated that P. gingivalis FimA may activate Toll-4, which appears to be an important signaling pathway for host recognition and bodily response to P. gingivalis (39,40). In the present study, the FimA fusion protein induced the expression of TLR4 downstream signaling molecules. To additionally confirm the effects of TLR4 on the induction of PBMC proliferation and inflammatory cytokine release by the FimA fusion protein, cells were treated with the FimA fusion protein simultaneously with transfection with siTLR4. The results revealed that TLR4 interference reversed the effects of the FimA fusion protein on PBMC proliferation and inflammatory cytokine release, and the expression levels of TLR4, NF-κB and MyD88 in PBMCs decreased significantly following FimA fusion protein treatment and transfection with siTLR4. The results suggested that the FimA fusion protein induced an inflammatory response via the TLR4/NF-κB signaling pathway by regulating the TLR4/NF-κB signaling pathway in PBMCs.
In summary, as the major virulence factor of P. gingivalis, FimA serves an important role in mediating the inflammatory response. The PGEX-6P-FimA prokaryotic expression plasmid was successfully constructed based on gene cloning techniques, and the specific FimA fusion protein was purified by GST affinity chromatography. Preliminary experiments indicated that the FimA fusion protein may activate PBMCs and subsequently upregulate TNF-α, IL-6, MMP-8 and MMP-9 expression levels via the TLR4/NF-κB signaling pathway. FimA may serve as a promising therapeutic strategy for periodontal disease.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Local Services Project of the Department of Education in Liaoning Province (grant nos. JYTFUDF201756 and JYTFUDF201745), the Clinical Capacity-Building Project of Liaoning Province (grant no. LNCCC-D30-2015) and the College Students' Innovation and Entrepreneurship Training Program of Liaoning Province (grant no. 20181060013).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JCa, JCh, XL and YL contributed to the conception and design of the experiment. JCa, JCh, HG, YP and YL performed all experiments and verified the analytical data. YZ, WZ and XL contributed to the statistical analysis and helped interpret the results. XL supervised the experiments in discussion with YL. JCa, XL and YL wrote the manuscript. All authors discussed the final results and approved the final manuscript.
Ethics approval and consent to participate
The collection of specimens was approved by the Ethics Committee of the Second Affiliated Hospital of Jinzhou Medical University (Jinzhou, China). The subjects clearly understood the purpose of the experiment, agreed to participate and provided written informed consent.
Patient consent for publication
The subjects provided written informed consent.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pennisi E. A mouthful of microbes. Science. 2005;307:1899–1901. doi: 10.1126/science.307.5717.1899. [DOI] [PubMed] [Google Scholar]
- 2.Schutzhold S, Kocher T, Biffar R, Hoffmann T, Schmidt CO, Micheelis W, Jordan R, Holtfreter B. Changes in prevalence of periodontitis in two German population-based studies. J Clin Periodontol. 2015;42:121–130. doi: 10.1111/jcpe.12352. [DOI] [PubMed] [Google Scholar]
- 3.Hasiuk P, Hasiuk N, Kindiy D, Ivanchyshyn V, Kalashnikov D, Zubchenko S. Characteristics of cellular composition of periodontal pockets. Interv Med Appl Sci. 2016;8:172–177. doi: 10.1556/1646.8.2016.4.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aimetti M, Perotto S, Castiglione A, Mariani GM, Ferrarotti F, Romano F. Prevalence of periodontitis in an adult population from an urban area in North Italy: Findings from a cross-sectional population-based epidemiological survey. J Clin Periodontol. 2015;42:622–631. doi: 10.1111/jcpe.12420. [DOI] [PubMed] [Google Scholar]
- 5.Souza JA, Medeiros MC, Rocha FR, de Aquino SG, Ávila-Campos MJ, Spolidorio LC, Zamboni DS, Graves DT. Role of NOD2 and RIP2 in host-microbe interactions with Gram-negative bacteria: Insights from the periodontal disease model. Innate Immun. 2016;22:598–611. doi: 10.1177/1753425916666652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsuda K, Furuta N, Inaba H, Kawai S, Hanada K, Yoshimori T, Amano A. Functional analysis of alpha5beta1 integrin and lipid rafts in invasion of epithelial cells by Porphyromonas gingivalis using fluorescent beads coated with bacterial membrane vesicles. Cell Struct Funct. 2008;33:123–132. doi: 10.1247/csf.08012. [DOI] [PubMed] [Google Scholar]
- 8.Bozkurt SB, Hakki SS, Hakki EE, Durak Y, Kantarci A. Porphyromonas gingivalis lipopolysaccharide induces a pro-inflammatory human gingival fibroblast phenotype. Inflammation. 2017;40:144–153. doi: 10.1007/s10753-016-0463-7. [DOI] [PubMed] [Google Scholar]
- 9.Zheng C, Wu J, Xie H. Differential expression and adherence of Porphyromonas gingivalis FimA genotypes. Mol Oral Microbiol. 2011;26:388–395. doi: 10.1111/j.2041-1014.2011.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dashper SG, Mitchell HL, Seers CA, Gladman SL, Seemann T, Bulach DM, Chandry PS, Cross KJ, Cleal SM, Reynolds EC. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol. 2017;8:48. doi: 10.3389/fmicb.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo NR, Ji MK, Park SW, Lee K, Bae JC, Yun KD, Park C, Oh GJ, Kim JH, Lim HP. Effect on adhesion of porphyromonas gingivalis by titanium nitride sputter coating or plasma nitriding of titanium. J Nanosci Nanotechnol. 2017;17:2633–2636. doi: 10.1166/jnn.2017.13320. [DOI] [PubMed] [Google Scholar]
- 12.Feng X, Zhang L, Xu L, Meng H, Lu R, Chen Z, Shi D, Wang X. Detection of eight periodontal microorganisms and distribution of Porphyromonas gingivalis fimA genotypes in Chinese patients with aggressive periodontitis. J Periodontol. 2014;85:150–159. doi: 10.1902/jop.2013.120677. [DOI] [PubMed] [Google Scholar]
- 13.Zhou W, Zhang X, Zhu CL, He ZY, Liang JP, Song ZC. Melatonin receptor agonists as the 'perioceutics' agents for periodontal disease through modulation of porphyromonas gingivalis virulence and inflammatory response. PLoS One. 2016;11:e166442. doi: 10.1371/journal.pone.0166442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikado A, Uesaki S, Suido H, Nomura Y, Sumikawa K, Maeda M, Miyauchi M, Takata T, Makino T. Human trial of liposomal lactoferrin supplementation for periodontal disease. Biol Pharm Bull. 2010;33:1758–1762. doi: 10.1248/bpb.33.1758. [DOI] [PubMed] [Google Scholar]
- 15.Basic A, Alizadehgharib S, Dahlén G, Dahlgren U. Hydrogen sulfide exposure induces NLRP3 inflammasome-dependent IL-1β and IL-18 secretion in human mononuclear leukocytes in vitro. Clin Exp Dent Res. 2017;3:115–120. doi: 10.1002/cre2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutando A, Montero J, Gómez-de Diego R, Ferrera MJ, Lopez-Valverde A. Effect of topical application of melatonin on serum levels of C-reactive protein (CRP), interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α) in patients with type 1 or type 2 diabetes and periodontal disease. J Clin Exp Dent. 2015;7:e628–e633. doi: 10.4317/jced.52604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otenio CC, Fonseca I, Martins MF, Ribeiro LC, Assis NM, Ferreira AP, Ribeiro RA. Expression of IL-1β, IL-6, TNF-α, and iNOS in pregnant women with periodontal disease. Genet Mol Res. 2012;11:4468–4478. doi: 10.4238/2012.September.20.3. [DOI] [PubMed] [Google Scholar]
- 18.Marcaccini AM, Novaes AB, Jr, Meschiari CA, Souza SL, Palioto DB, Sorgi CA, Faccioli LH, Tanus-Santos JE, Gerlach RF. Circulating matrix metalloproteinase-8 (MMP-8) and MMP-9 are increased in chronic periodontal disease and decrease after non-surgical periodontal therapy. Clin Chim Acta. 2009;409:117–122. doi: 10.1016/j.cca.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Q, Amar S. Identification of signaling pathways in macrophage exposed to Porphyromonas gingivalis or to its purified cell wall components. J Immunol. 2007;179:7777–7790. doi: 10.4049/jimmunol.179.11.7777. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Tao K, Xion J, Xu S, Jiang Y, Chen Q, He S. TAK-242 attenuates acute cigarette smoke-induced pulmonary inflammation in mouse via the TLR4/NF-κB signaling pathway. Biochem Biophys Res Commun. 2016;472:508–515. doi: 10.1016/j.bbrc.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Sanz M, van Winkelhoff AJ, Herrera D, Dellemijn-Kippuw N, Simón R, Winkel E. Differences in the composition of the subgingival microbiota of two periodontitis populations of different geographical origin. A comparison between Spain and The Netherlands. Eur J Oral Sci. 2000;108:383–392. doi: 10.1034/j.1600-0722.2000.108005383.x. [DOI] [PubMed] [Google Scholar]
- 23.Sharma A, Honma K, Evans RT, Hruby DE, Genco RJ. Oral immunization with recombinant Streptococcus gordonii expressing porphyromonas gingivalis FimA domains. Infect Immun. 2001;69:2928–2934. doi: 10.1128/IAI.69.5.2928-2934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Q, Desta T, Fenton M, Graves DT, Amar S. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect Immun. 2005;73:935–943. doi: 10.1128/IAI.73.2.935-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suwatanapongched P, Surarit R, Srisatjaluk R, Offenbacher S. Translocation of Porphyromonas gingivalis infected mono-cytes and associated cellular responses. Asian Pac J Allergy Immunol. 2010;28:192–199. [PubMed] [Google Scholar]
- 27.Nozoe K, Sanui T, Takeshita M, Fukuda T, Haraguchi A, Aida Y, Nishimura F. Innate immune-stimulatory activity of Porphyromonas gingivalis fimbriae is eliminated by phase separation using Triton X-114. J Immunol Methods. 2017;441:31–38. doi: 10.1016/j.jim.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Yun PL, Decarlo AA, Collyer C, Hunter N. Hydrolysis of interleukin-12 by Porphyromonas gingivalis major cysteine proteinases may affect local gamma interferon accumulation and the Th1 or Th2 T-cell phenotype in periodontitis. Infect Immun. 2001;69:5650–5660. doi: 10.1128/IAI.69.9.5650-5660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhuri S, Pratap S, Paromov V, Li Z, Mantri CK, Xie H. Identification of a diguanylate cyclase and its role in Porphyromonas gingivalis virulence. Infect Immun. 2014;82:2728–2735. doi: 10.1128/IAI.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantri CK, Chen CH, Dong X, Goodwin JS, Pratap S, Paromov V, Xie H. Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis. Microbiologyopen. 2015;4:53–65. doi: 10.1002/mbo3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa T, Asai Y, Hashimoto M, Uchida H. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur J Immunol. 2002;32:2543–2550. doi: 10.1002/1521-4141(200209)32:9<2543::AID-IMMU2543>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Deng B, Fang F, Yang T, Yu Z, Zhang B, Xie X. Ghrelin inhibits AngII-induced expression of TNF-α, IL-8, MCP-1 in human umbilical vein endothelial cells. Int J Clin Exp Med. 2015;8:579–588. [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigues PH, Reyes L, Chadda AS, Bélanger M, Wallet SM, Akin D, Dunn W, Jr, Progulske-Fox A. Porphyromonas gingivalis strain specific interactions with human coronary artery endothelial cells: A comparative study. PLoS One. 2012;7:e52606. doi: 10.1371/journal.pone.0052606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagano K, Hasegawa Y, Abiko Y, Yoshida Y, Murakami Y, Yoshimura F. Porphyromonas gingivalis FimA Fimbriae: Fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA genotypes. PLoS One. 2012;7:e43722. doi: 10.1371/journal.pone.0043722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cochran DL. Inflammation and bone loss in periodontal disease. J Periodontol. 2008;79:1569–1576. doi: 10.1902/jop.2008.080233. [DOI] [PubMed] [Google Scholar]
- 36.Ikezawa I, Tai H, Shimada Y, Komatsu Y, Galicia JC, Yoshie H. Imbalance between soluble tumour necrosis factor receptors type 1 and 2 in chronic periodontitis. J Clin Periodontol. 2005;32:1047–1054. doi: 10.1111/j.1600-051X.2005.00832.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Zhou R, Yi Z, Li Y, Fu Y, Zhang Y, Li P, Li X, Pan Y. Porphyromonas gingivalis induced inflammatory responses and promoted apoptosis in lung epithelial cells infected with H1N1 via the Bcl-2/Bax/Caspase-3 signaling pathway. Mol Med Rep. 2018;18:97–104. doi: 10.3892/mmr.2018.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graves DT, Naguib G, Lu H, Desta T, Amar S. Porphyromonas gingivalis fimbriae are pro-inflammatory but do not play a prominent role in the innate immune response to P. gingivalis. J Endotoxin Res. 2005;11:13–18. doi: 10.1177/09680519050110010501. [DOI] [PubMed] [Google Scholar]
- 39.Hajishengallis G, Sojar HT, Sharma A, Martin M, Denardini E, Genco RJ. TLR4-dependent upregulation of costimulatory molecules in macrophages by p. gingivalis FimA. J Dent Res. 2002;81:A53. [Google Scholar]
- 40.Hajishengallis G, Martin M, Schifferle RE, Genco RJ. Counteracting interactions between lipopolysaccharide molecules with differential activation of toll-like receptors. Infect Immun. 2002;70:6658–6664. doi: 10.1128/IAI.70.12.6658-6664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.