Abstract
Inflammatory response has an important role in the outcome of cerebral ischemia reperfusion injury (CIR). Biliverdin (BV) administration can relieve CIR in rats, but the mechanism remains unknown. The aim of the present study was to explore the expressional network of microRNA (miRNA)-mRNA in CIR rats following BV administration. A rat middle cerebral artery occlusion model with BV treatment was established. After neurobehavior was evaluated by neurological severity scores (NSS), miRNA and mRNA expressional profiles were analyzed by microarray technology from the cerebral cortex subjected to ischemia and BV administration. Then, bioinformatics prediction was used to screen the correlation between miRNA and mRNA, and 20 candidate miRNAs and 33 candidate mRNAs were verified by reverse transcription-quantitative polymerase chain reaction. Furthermore, the regulation relationship between ETS proto-oncogene 1 (Ets1) and miRNA204-5p was examined by luciferase assay. A total of 86 miRNAs were differentially expressed in the BV group compared with the other groups. A total of 10 miRNAs and 26 candidate genes were identified as a core 'microRNA-mRNA' regulatory network that was linked with the functional improvement of BV administration in CIR rats. Lastly, the luciferase assay results confirmed that miRNA204-5p directly targeted Ets1. The present findings suggest that BV administration may regulate multiple miRNAs and mRNAs to improve neurobehavior in CIR rats, by influencing cell proliferation, apoptosis, maintaining ATP homeostasis, and angiogenesis.
Keywords: inflammatory response, cerebral ischemia reperfusion injury, biliverdin, microRNA, mRNA
Introduction
Brain ischemic stroke, a serious disease in the central nervous system (CNS), is becoming a prominent public health risk (1-3). Generally, vascular recanalization to obtain timely reperfusion is the preferred treatment in clinical practice. However, reperfusion can induce further unexpected brain injury, termed cerebral ischemia reperfusion injury (CIR) (4). Previous studies have demonstrated that the inflammatory response has an important role in the outcome of stroke (5,6). Thus, anti-inflammatory therapy may lessen neurological deficits by ischemic stroke and it may serve as a potential therapeutic strategy following ischemic stroke (3).
Biliverdin (BV) is a metabolite of heme catabolism that has a protective role in lung graft injury, hemorrhagic shock and resuscitation-induced lung injury through anti-inflammatory and antioxidant mechanisms (7,8). A previous study has suggested that exogenously administered carbonic oxide (CO) and BV have potent cytoprotective effects on intestinal ischemia reperfusion injury (9). In addition, BV administration can ameliorate CIR in rats, and the mechanism may be related to the downregulation of proinflammatory factors. To date, two pathways are known to be involved in the anti-inflammatory mechanism of BV. By activating the nitric oxide-dependent BV reductase, BV reduces the expression of toll like receptor-4 (TLR-4) in murine macrophages. BV regulates the lipopolysaccharide (LPS)-mediated expression of complement C5a receptor 1 via the mammalian target of rapamycin (mTOR) pathway (10,11). While the anti-inflammatory mechanism of BV has been the focus of previous studies, the molecular network upstream and downstream of BV is largely unknown.
MicroRNAs (miRNAs), small non-coding RNAs of 21-23 bp in length, serve crucial roles in several biological processes. By binding to the 3'-untranslated region (UTR) of target mRNA, miRNAs induce mRNA cleavage or translation inhibition (12). Each miRNA has multiple potential mRNA targets, and therefore, a broad-spectrum gene expression can be affected by an specific miRNA (13,14). Recent studies have demonstrated that miRNA inhibitory activity can be quantified by examining their target mRNA expression levels (15). However, the relation between miRNA and mRNA during CIR pathogenesis following BV administration remains to be determined.
In the present study, it was hypothesized that miRNA and mRNA expression may be regulated by the BV anti-inflammatory mechanism in CIR rats. To explore the potential miRNA-mRNA regulatory mechanism and relevant signaling pathways, a rat middle cerebral artery occlusion (MCAO) model was first established and then BV treatment was performed. Subsequently, the expressional network of miRNAs and mRNAs was examined by microarray, bioinformatics, and integrated genomics analyses. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to validate the reliability of microarray data. Lastly, miRNA204-5p was confirmed to directly target ETS proto-oncogene 1 (Ets1) following BV administration in CIR rats. To the best of our knowledge, this is the first report of the integrated molecular network of miRNA with gene microarray expression following BV treatment in CIR rats. The present data suggested that miRNAs, such as miRNA204-5p, can act as key regulators to target Ets1 in endogenous responses to stroke. Therefore, manipulation of these miRNAs might serve as a novel diagnosis or therapeutic indication for acute stroke patients.
Materials and methods
Animals and experimental groups
Animal care and all experimental protocols involving animals were approved by the Animal Care and Welfare Committee of Kunming Medical University (Kunming, China). A total of 36 adult male Sprague-Dawley (SD) rats (specific pathogen-free; age, 8-12 weeks; weight, 220±10 g; Kunming Medical University Laboratory Animal Research Center, Kunming, China) were used in the study. All rats were housed at room temperature (21-25°C) with 45-50% humidity and a 12-h light/dark cycle, with free access to soft food and tap water. Rats were randomly divided into three groups (Table I): Sham group (S, sham operation); Brain ischemic and vehicle control group (C, CIR + normal saline); and brain ischemic with BV treatment group (BV, CIR+BV).
Table I.
Experimental groups and methods.
| Number of rats
|
|||
|---|---|---|---|
| Group | TTC stain/NSS | Microarray | qPCR |
| S | 3 | 3 | 6 |
| C | 3 | 3 | 6 |
| BV | 3 | 3 | 6 |
TTC, 2,3,5-triphenyltetrazolium chloride; NSS, neurological severity score; qPCR, quantitative polymerase chain reaction; S, sham; C, cerebral ischemia reperfusion injury; BV, Biliverdin.
Animal model of MCAO and BV administration
The right middle cerebral artery was occluded according to the standard operation procedures for MCAO in rats (16). Briefly, animals were anaesthetized by an intraperitoneal (ip) injection of 5 mg/kg xylazine HCl and 40 mg/kg ketamine HCl, and then a midline neck incision was made. The right common carotid artery (RCCA), right external carotid artery, and right internal carotid artery were isolated for inserting the nylon monofilament (diameter 0.24 mm; Johnson & Johnson, New Brunswick, NJ, USA) through a small incision in the RCCA. The monofilament was fixed in position tightly and then the incision was sutured. Rat body temperature was maintained at 36.5±0.5°C using a heating lamp. A laser Doppler system (Peri-Flux System 5000; Perimed, Jarfalla, Sweden) was used to supervise regional cerebral blood flow (rCBF). Rats in which rCBF did not drop <20% of the baseline levels following MCAO were excluded from analysis. Sham group rats underwent the same procedures without inserting a nylon thread. After 2 h of tMCAO, CBF was recovered by removing the nylon thread, and the incision was closed.
BV HCl (Frontier Scientific, Inc., Logan, UT, USA) was dissolved in 0.2 N NaOH and adjusted to final pH 7.4 with HCl. As previously published, BV was diluted in saline and injected (35 mg/kg ip) to rats 15 min prior to reperfusion, then once again 4 h after reperfusion, and twice per day thereafter (17). In the vehicle control group, the same volume of saline alone was injected in the same way. Stroke-onset was assessed by circadian rhythm disturbances in heart rate (HR) and mean arterial blood pressure (MABP).
Neural behavioral test
Rats from the three groups were scored by an evaluator with neurological severity scores (NSS), as previously reported (17), at day 1 and 2 post-reperfusion. The NSS includes four physiological function evaluation scores: Feeling, movement, reflection and balance. Scores 1-6 indicate mild injury, 7-12, moderate injury, and 13-18, severe injury. The neural behavioral test was conducted by an evaluator that was blinded to the treatments.
Evaluation of cerebral infarct volume indicated by 2,3,5-triphenyltetrazolium chloride (TTC) staining
Whole brain tissue was harvested after 2 h of ischemia, followed by 48 h of reperfusion, and the integrity of the brain was maintained upon removal (n=3 for each group). Brain tissue from bregma was cut into 2 mm thick coronal slices. Brain slices were then incubated in 2% TTC solution at 37°C for 30 min in the dark, as previously described (18). After staining, the sections were washed with PBS (3 times, 1 min each) and fixed in 4% paraformaldehyde for 24 h. Color images of these sections were directly obtained with a stereomicroscope by an evaluator that was blinded to the treatments, and the infarct areas of each section were measured with ImageJ 1.4 software (National Institutes of Health, Bethesda, MD, USA). To compensate for the effect of brain edema following cerebral infarction, the corrected infarct volume was calculated as the sum of the infarct areas multiplied by the section thickness (2 mm), and expressed as a % of the contralateral (non-occluded) hemisphere.
RNA isolation
Expression levels of inflammatory factors were the most obvious at 2 h ischemia and 6 h following reperfusion, according to our previous study (17); therefore, the ipsilateral ischemic cortex at 6 h following reperfusion was selected to perform miRNA and mRNA assays. In the C and BV groups, the lesion tissue from the ipsilateral ischemic cortex was obtained from rats at 2 h ischemia and 6 h post-reperfusion, with the corresponding cortex harvested from the S group as well. Three samples of each group were pooled into one S pool, one C pool and one BV pool for microarray analysis, and each pool was on a different microarray chip. Briefly, after rats were anaesthetized by an ip injection of 5 mg/kg xylazine HCl and 40 mg/kg ketamine HCl, the brain cortex of ischemia regions was harvested and fresh-frozen in liquid nitrogen. Total RNA was isolated using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and purified with RNeasy mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. RNA quality and quantity was measured with a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies; Thermo Fisher Scientific, Inc.) and RNA integrity was determined by gel electrophoresis.
miRNA microarray analysis
After total RNA samples were labelled using the miRCURY Hy3/Hy5 Power labeling kit (Exiqon; Qiagen), they were hybridized to a 12-Bay Hybridization System (Nimblegen Systems, Inc., Madison, WI, USA) to detect miRNA expressional profiles. Then, the positive signal was recorded using the Axon GenePix 4000B micro-array scanner (Axon Instruments; Molecular Devices, LLC, Sunnyvale, CA, USA). Subsequently, the scanned images were imported into GenePix Pro 6.0 software (Axon Instruments; Molecular Devices, LLC) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs with intensities ≥30 in all samples were selected for calculating the normalization factor. Expressed data were normalized using the Median normalization. Following normalization, differentially expressed miRNAs between two samples were filtered through fold change. The miRNA array results following normalization and fold changes are listed in Table II. Finally, hierarchical clustering was performed to display distinguishable miRNA expression profiles among samples.
Table II.
miRNA array ratios following normalization and fold change.
| microRNA array ratio after normalization
|
Fold change
|
|||||
|---|---|---|---|---|---|---|
| microRNA | S | C | BV | S vs. C | C vs. BV | Change trend |
| rno-miR-324-5p | 0.09 | 0.62 | 0.04 | 6.59 | 17.59 | Up/down |
| rno-miR-3072 | 0.29 | 1.15 | 0.23 | 3.91 | 5.03 | Up/down |
| rno-miR-124-3p | 26.93 | 126.82 | 37.02 | 4.71 | 3.43 | Up/down |
| rno-miR-375-5p | 0.06 | 0.264 | 0.07 | 4.51 | 3.62 | Up/down |
| rno-miR-3573-3p | 6.33 | 28.02 | 10.82 | 4.43 | 2.59 | Up/down |
| rno-miR-150-5p | 2.11 | 8.58 | 2.35 | 4.07 | 3.65 | Up/down |
| rno-miR-935 | 0.20 | 0.73 | 0.23 | 3.60 | 3.21 | Up/down |
| rno-miR-133a-3p | 0.16 | 0.57 | 0.11 | 3.53 | 4.94 | Up/down |
| rno-miR-539-3p | 1.37 | 4.40 | 0.75 | 3.20 | 5.88 | Up/down |
| rno-miR-370-3p | 0.14 | 0.44 | 0.16 | 3.14 | 2.77 | Up/down |
| rno-miR-181b-5p | 3.17 | 0.03 | 1.51 | 116.9 | 55.69 | Down/up |
| rno-miR-204-5p | 1.55 | 0.02 | 0.82 | 68.77 | 36.55 | Down/up |
| rno-miR-664-1-5p | 0.81 | 0.02 | 0.60 | 32.67 | 24.00 | Down/up |
| rno-miR-136-5p | 19.47 | 0.64 | 14.36 | 30.41 | 22.42 | Down/up |
| rno-miR-126a-5p | 4.24 | 0.11 | 2.24 | 37.64 | 19.86 | Down/up |
| rno-miR-124-5p | 0.75 | 0.02 | 0.30 | 47.74 | 19.25 | Down/up |
| rno-miR-363-5p | 2.09 | 0.06 | 0.78 | 35.69 | 13.28 | Down/up |
| rno-miR-27a-3p | 4.85 | 0.23 | 2.88 | 21.51 | 12.76 | Down/up |
| rno-miR-29b-3p | 24.68 | 1.41 | 15.96 | 17.51 | 11.32 | Down/up |
| rno-miR-376a-5p | 0.84 | 0.02 | 0.34 | 41.32 | 16.92 | Down/up |
S, sham; C, cerebral ischemia reperfusion injury; BV, Biliverdin.
Gene microarray analysis
Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, Inc., Santa Clara, CA, USA). Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY Eukaryotic mRNA Isolation kit; Epicentre; Illumina, Inc., San Diego, CA, USA). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3' bias utilizing a random priming method (Arraystar Flash RNA Labeling kit; Arraystar, Inc. Rockville, MD, USA). The labeled cRNAs were purified with RNeasy Mini kit (Qiagen GmbH). Labeled samples hybridized for 17 h at 65°C in an Agilent hybridization oven. Then, the hybridized arrays were washed, fixed and scanned with the Agilent DNA Microarray Scanner (part no. G2505C). Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed with GeneSpring GX v12.1 software package (Agilent Technologies, Inc.). Differentially expressed mRNAs between the two samples were identified through fold change filtering.
qPCR verification for miRNA microarray data
To validate the microarray data, first 20 differentially expressed miRNAs were selected for qPCR. Total RNA from the brain cortex of ischemia region was extracted with TRIzol (Thermo Fisher Scientific, Inc.). Poly(A) Tailing and reverse transcription were performed with the All-in-one miRNA qRNA miRNA qPCR Start kit (Yijing, Guangzhou, China) in a DNA thermal cycler (T100 TM; Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 85°C for 5 min. Then, qPCR was performed using a SYBR-Green RT-PCR Master Mix kit (Takara Biotechnology Co., Ltd., Dalian, China). The annealing temperature was 53°C. The reaction was performed at 95°C for 2 min and 40 cycles of 95°C for 20 sec, 53°C for 30 sec and 60°C for 40 sec. The primers used (Sangon Biotech Co., Ltd., Shanghai, China) are listed in Table III. The U6 gene was used as the internal control. Relative levels of miRNA were calculated using the formula 2−ΔΔCq (19). At least three independent biological replicates were used for miRNA.
Table III.
Primer sequence cat. nos for microRNAs.
| microRNA | Catalog number |
|---|---|
| rno-miR-29c-3p | RmiRQP0375 |
| rno-miR-181b-5p | RmiRQP0234 |
| rno-miR-204-5p | RmiRQP0306 |
| rno-miR-664-1-5p | RmiRQP3276 |
| rno-miR-136-5p | RmiRQP0173 |
| rno-miR-126a-5p | RmiRQP0098 |
| rno-miR-124-5p | RmiRQP0073 |
| rno-miR-376a-5p | RmiRQP1253 |
| rno-miR-363-5p | RmiRQP0450 |
| rno-miR-27a-3p | RmiRQP0359 |
| rno-miR-29b-3p | RmiRQP0373 |
| rno-miR-324-5p | RmiRQP0412 |
| rno-miR-3072 | RmiRQP2830 |
| rno-miR-124-3p | RmiRQP0074 |
| rno-miR-375-5p | RmiRQP3367 |
| rno-miR-3573-3p | RmiRQP1852 |
| rno-miR-150-5p | RmiRQP0210 |
| rno-miR-935 | RmiRQP0840 |
| rno-miR-133a-3p | RmiRQP0166 |
| rno-miR-539-3p | RmiRQP3241 |
| rno-miR-370-3p | RmiRQP0456 |
| U6 | RmiRQP9003 |
Bioinformatics prediction
Predicted target genes of candidate miRNAs were determined using three bioinformatics prediction tools: TargetScanv6.2 (http://www.targetscan.org/mamm_31/), miRmap (http://mirmap.ezlab.org/), and miRDB (http://www.mirdb.org/miRDB/). The selection criteria were correlation >0.99 or correlation <-0.99, and P-value <0.05. The genes that overlapped in all three databases were selected for further functional analyses. The miRNAs and predicted mRNA target genes were then subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses using David v6.7 (http://david.abcc.ncifcrf.gov/) online. Cytoscape version 3.4.0 was used to visualize the connections between the miRNAs and their predicted gene targets. Additionally, the sequences of the target genes were obtained from the TargetScanv6.2 (http://www.targetscan.org/mamm_31/), miRmap (http://mirmap.ezlab.org/) and miRDB (http://www.mirdb.org/miRDB/) databases, and the complementarity of the miRNAs with the 3'UTR regions of target genes was examined by manual method.
Integration of miRNA and mRNA
First, significant differentially expressed mRNAs were identified by fold change ≥2 of each data set for further data analysis. Further testing involved determination of the overlap between predicted gene targets of candidate miRNAs and mRNAs array. In order to select relatively novel genes, those overlapped mRNAs were searched in PubMed web source (https://www.ncbi.nlm.nih.gov/pubmed/), and candidate genes were verified by qPCR.
qPCR verification for candidate genes
A total of 33 candidate genes were verified using qPCR. Total RNA from the brain cortex of ischemia region was extracted with TRIzol (Thermo Fisher Scientific, Inc.). After RNA samples were reverse transcribed into cDNA using the RevertAidTM First Strand cDNA Synthesis kit (cat. no. K1622; Thermo Fisher Scientific, Inc.), qPCR was performed using a SYBR Green RT-PCR Master Mix kit (Takara Biotechnology Co., Ltd.). The annealing temperature was 53°C. The reaction was performed at 95°C for 2 min and 40 cycles of 95°C for 20 sec, 53°C for 30 sec and 60°C for 40 sec. The primers are listed in Table IV. β-actin was used as the internal control. Relative expression levels of mRNA were calculated using the formula 2−ΔΔCq (19). At least three independent biological replicates were used for mRNA.
Table IV.
Primer sequences and annealing temperatures.
| Gene | Primer | Sequence | Annealing temperature |
|---|---|---|---|
| Sox7 | Forward | CCTCTGCAGTCACCTTAGCC | 51°C |
| Reverse | GTCCATGTCGCCAAGAAGTT | ||
| Fbxo33 | Forward | TCCAGCACATACCTCAGCAG | 50°C |
| Reverse | TGGTGTTGGCAATGGAGTTA | ||
| Plekha8 | Forward | TGCTGAGATCAACCTGCAAC | 50°C |
| Reverse | GCAATCTGCCTTTGTGGATT | ||
| Ets1 | Forward | ACTGTGTGCCCTGGGTAAAG | 53°C |
| Reverse | GAGTTCTTCCGAGCTGATGG | ||
| Gcnt2 | Forward | AGCGAAAGGACGTCTGTCAT | 51°C |
| Reverse | GTAGACGTTCTGGGGCATGT | ||
| Csrnp1 | Forward | GCTTTCAGTGTCCGGAGTTC | 53°C |
| Reverse | GCCATCACAGTGACAACCAC | ||
| En2 | Forward | CCAGGTCTCGAAAACCAAAG | 50°C |
| Reverse | ACCGCCAAAGTGTTCTTGTT | ||
| Rgs1 | Forward | AACTCCTTGCCAACCAGATG | 50°C |
| Reverse | TGTGGGAGTTGGTGTTTTGA | ||
| Litaf | Forward | TCCAGGACCTTACCAAGCAG | 50°C |
| Reverse | AGGAAGGACAGCACATCTGG | ||
| Rnd3 | Forward | CGGACAGATGTCAGCACACT | 51°C |
| Reverse | GTGGCCCTCTGTGATTTGTT | ||
| Zc3h12d | Forward | GAAGGATGGCGTCATTGTCT | 51°C |
| Reverse | TCTCCGGGTGGTAGAATCTG | ||
| P4hb | Forward | CAATTTTGCCACCACTTCCT | 50°C |
| Reverse | CTTCCACCTCATTGGCTGTT | ||
| Mthfd2 | Forward | TCCTTGCGGACATTGTGATA | 50°C |
| Reverse | TCTGGCCTGAGCACTTTCTT | ||
| Mafk | Forward | GACTAATCCCAAGCCCAACA | 51°C |
| Reverse | TCCAGCTCCTCCTTCTGTGT | ||
| B4galt1 | Forward | GGCCTGAAGAGCAACTTGAC | 50°C |
| Reverse | AATGATGGCCACCTTGTGA | ||
| Slc25a25 | Forward | CTTGATGGGCAACTGGACTT | 51°C |
| Reverse | ACTCGTTCCAGTCGATGGTC | ||
| Esm1 | Forward | TGATTTCGGTGACGAGTTTG | 49°C |
| Reverse | TCTCTCACAGCATTGCCATC | ||
| Vps37c | Forward | GCATGAAGATCGAGGAGGAG | 54°C |
| Reverse | TAGGGCAAAGGGTAGGGAGT | ||
| Zmiz1 | Forward | ACTCTGTCGCACAGTGATGG | 51°C |
| Reverse | TGGAGAACTGTTGCTGTTGC | ||
| Rhoq | Forward | TTCGACCACTACGCAGTCAG | 50°C |
| Reverse | AAGGGGACATTTGGTGCATA | ||
| Plp2 | Forward | CTACTCCTCCCTGTCGGTGA | 51°C |
| Reverse | TCGTAGCCAAAGAGCAAGGT | ||
| Zkscan1 | Forward | GGTTCAGGCGCTTCTGTTAC | 53°C |
| Reverse | GCCTCTTCTCCACTGTCAGG | ||
| Dgkg | Forward | TGTCCACCAACGCTGTGTAT | 51°C |
| Reverse | GGTAGGCAGCAAAATGTGGT | ||
| Megf9 | Forward | TACTCCTCCTCCCCCAGAGT | 51°C |
| Reverse | GTGACACCCAGTTTGCATTG | ||
| Zbtb3 | Forward | CAGGTCCAGGGCAGTATCAT | 53°C |
| Reverse | GAGGTCCCCTGACTGTGTGT | ||
| Guca1b | Forward | TGGAACACAAGCTCAAGTGG | 52°C |
| Reverse | AGACAGCTGACCGTCTCCAT | ||
| Pnrc1 | Forward | GAAAATTTCCCCTCCCCATA | 50°C |
| Reverse | TCTTCCCTCGGTTTTCCTTT | ||
| Ankrd12 | Forward | CAGCAGTGGGCACAGAGATA | 51°C |
| Reverse | GGCTTCCACAGACGAAAGAG | ||
| Kcnab3 | Forward | ATTTGCCAATCGTTCAGACC | 51°C |
| Reverse | TCTCCACCTTCTCCCTCTGA | ||
| Map2k6 | Forward | AGTGGACTGTCCGTTTACCG | 50°C |
| Reverse | TGAGCACATTTGAAGGCTTG | ||
| Scn4b | Forward | GCCACCACCATCTACGCTAT | 53°C |
| Reverse | CTCTAGGGTGATGCGGTCAT | ||
| Cmklr1 | Forward | GTGCCCCTACCACACACTCT | 53°C |
| Reverse | TGGTGAAGCTCCTGTGACTG | ||
| β-actin | Forward | TACTCCTGCTTGCTGATCCA | 53°C |
| Reverse | CTGTCCACCTTCCAGCAGA |
Luciferase assay
The miRNA mimics and the 3'UTR plasmids of Ets1, glucosaminyl N-acetyl transferase 2 (GcnT2), Rho family GTPase 3 (Rnd3), prolyl 4-hydroxylase subunit β (P4hb), MAF bZIP transcription factor B (mafb), and mitogen-activated protein kinase kinase 6 (map2K6) were supplied by RiboBio Co., Ltd. (Guangzhou, China). 293T cells (5×104 per well) were seeded in a 96-well plate 24 h prior to transfection. Each well was transfected with 1 ng/µl of the 3'UTR luciferase vector and 50 nM of the miRNA mimic using a FECT transfection kit (RiboBio Co., Ltd.). The assay was performed using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) 48 h after transfection, using Renilla luciferase as the reporter and firefly luciferase as the control. Luminescence was measured with a Synergy 2 microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Statistical analysis
All data were represented as mean ± standard deviation. For multiple group comparisons, one-way analysis of variance with Tukey's post hoc test was applied. All statistical analyses were performed with SPSS 19.0 software (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
BV treatment decreases cerebral infarction volume and may improve functional recovery
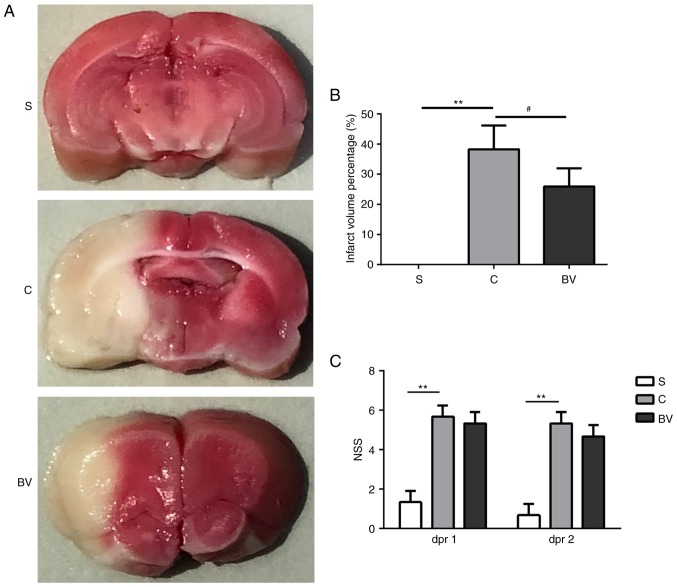
To examine whether BV treatment following MCAO could reduce the functional deficits caused by ischemic brain injury, TTC staining was used to detect the infarct volume at 48 h post-reperfusion. The borders of the TTC stain enclosing the white infarct area were readily distinguishable in contrast to the red color which is the normal tissue (Fig. 1A). The infarct volumes % observed in the C group was significantly larger compared with the S group (38.25±7.87 vs. 00.00±0.00; P<0.01; Fig. 1A and B). The infarct volumes observed in the BV group were obviously decreased compared with the C group (38.25±7.87 vs. 25.93±6.02; P<0.05; Fig. 1A and B).
Figure 1.
TTC stain and NSS test in the experimental groups. (A) Representative images of TTC-stained coronal slices at 48 h post-reperfusion from the three groups. Non-ischemic region is red, and infarct region appears as white color. (B) The infarct volume % of cerebral hemisphere in the three groups. (C) NSS test results in the three groups. Data are presented as mean ± standard deviation (n=3). **P<0.01 vs. group S; #P<0.05 vs. group C. TTC, 2,3,5-triphenyltetrazolium chloride; NSS, neurological severity score; BV, biliverdin; dpr, days post reperfusion.
To determine the effect of BV treatment on the neurological function following MCAO, NSS was used to assess the functional recovery. The NSS test was performed at day 1 and 2 post-reperfusion. Compared with the S group, functional deficits were impaired by ischemic insult in the C group (P<0.01; Fig. 1C). However, a slight recovery of neurological functions was observed in the BV group (Fig. 1C). Therefore, it can be concluded that BV treatment effectively decreased cerebral infarction volume and may improve functional recovery.
In addition, the effects of BV administration on MABP and heart rate (HR) were evaluated in the S, C and BV groups (Table V). Compared with the S group, MABP was decreased in the C group, while BV treatment reversed this decline. Compared with the S group, HR was increased in the C group, and BV treatment significantly alleviated this increase.
Table V.
MABP and HR in the different experimental groups.
| Group | MABP (mmHg)
|
HR (bpm)
|
||
|---|---|---|---|---|
| Pre-operation | 48 h post-operation | Pre-operation | 48 h post-operation | |
| S | 76.6+7.5 | 76.4+4.3 | 329.2+9.1 | 330.2+15.3 |
| C | 75.6+3.9 | 63.2+4.3a | 327.2+13.1 | 351.2+24.1b |
| BV | 75.8+4.5 | 68.8+3.6b, c | 328.8+8.8 | 343.4+10.3b, c |
P<0.05 compared with the S group;
P<0.01 compared with the S group;
P<0.05 compared with the C group. MABP, mean arterial blood pressure; HR, heart rate; S, sham; C, cerebral ischemia reperfusion injury; BV, Biliverdin; bpm, beats per minute.
BV administration alters miRNA expression in the rat MCAO model
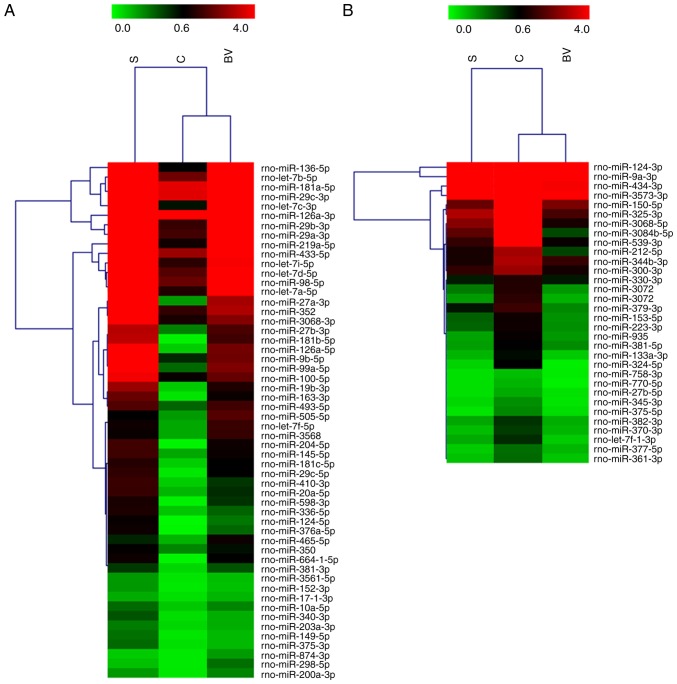
miRNA microarray analysis was used to detect the expression profiles of miRNAs in brain tissues isolated from the three groups (n=3 rats per group), in order to examine whether BV treatment can alter the expression profile of miRNAs following cerebral ischemia. miRNA microarray analysis revealed that 86 miRNAs were differentially expressed (fold change ≥2) in the BV group compared with the other groups (Fig. 2). The dramatic expression change in the three groups indicates that the BV administration altered the miRNA response to stroke.
Figure 2.
Heatmap of miRNA profiling following different treatments. Heatmap of hierarchical clustering showing 86 significantly regulated miRNAs in S, C, and BV groups. (A) A total of 54 miRNAs were downregulated in the C group compared with eth S group, and upregulated in the BV group compared with the C group. (B) The other 32 miRNAs displayed an opposite trend to panel A. The color code in the heat maps is linear with green as the lowest and red as the highest expression. BV, biliverdin.
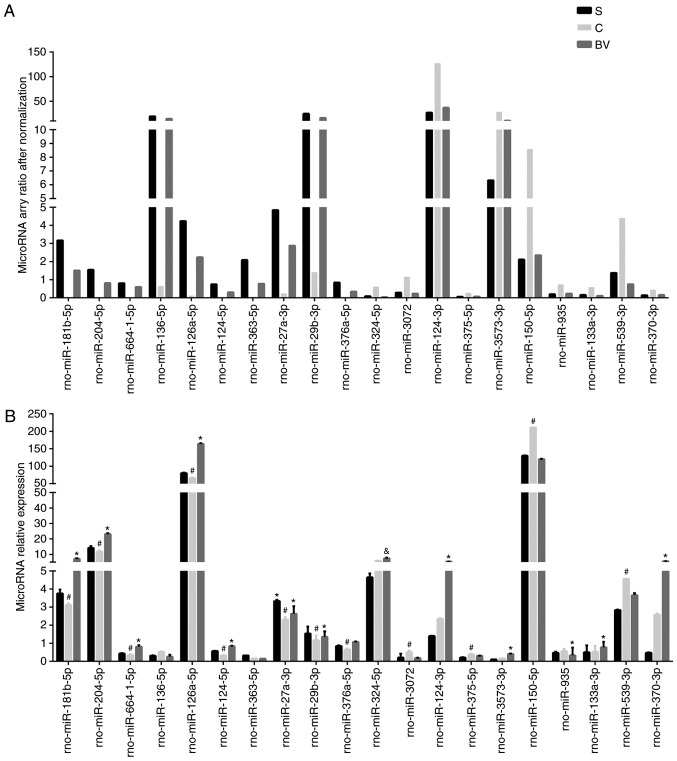
qPCR verification of miRNA array results
A total of 20 miRNAs were selected from the miRNA array with the top 10 fold change of 'Up/Down' or 'Down/Up' groups. These included rno-miR-181b-5p, -204-5p, -664-1-5p, -126a-5p, -124-5p, -376a-5p, -27a-3p, -29b-3p, -136-5p, -363-5p, -324-5p, -3072, -124-3p, -375-5p, -3573-3p, -150-5p, -935, -133a-3p, -539-3p, -370-3p (Fig. 3A). Compared with the S group, miRNAs were upregulated in the C group, but downregulated in the BV group, from the group referred as 'Up/Down', which contains 32 miRNAs. By contrast, the other 54 microRNAs changed trends were on the contrary referred to as 'Down/Up'. Except for rno-miR-136-5p, -363-5p, -324-5p, -124-3p, 3573-3p, 133a-3p, -370-3p, all other miRNAs verified by qPCR exhibited similar alterations as detected by microarray (Fig. 3B).
Figure 3.
qPCR validation of miRNA array. (A) Normalizing ratios of candidate miRNAs detected on the microarrays. (B) qPCR results of selected miRNAs. Results are presented as mean ± standard deviation (n=6). &P<0.05 group BV vs. group S; #P<0.01 group C vs. group S; *P<0.01 group BV vs. group C. qPCR, quantitative polymerase chain reaction; BV, biliverdin.
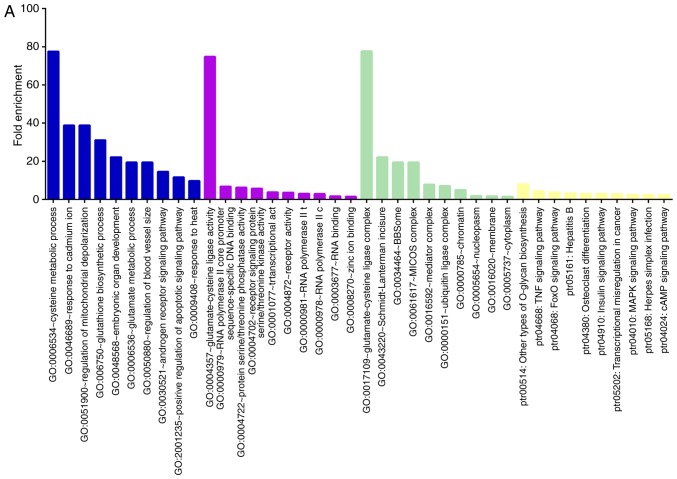
GO and KEGG analysis of miRNAs
To determine the potential influence of the differentially expressed miRNAs following BV treatment in MCAO, the potential target genes of these miRNAs were predicted by Targetscan v6.2, miRmap and miRDB analyses. Some miRNAs had a large number of target genes, including rno-miR-181b-5p, -124-5p, -376a-5p, -664-1-5p, -126a-5p; -27a-3p, -3072, -935, 539-3p, while others had just one target, such as rno-miR-375-5p.
A total of 1,052 predicted target genes were identified ('Up/Down' miRNAs group, 225 genes; 'Down/Up' miRNAs group, 727 genes). Among them, 970 predicted genes had David IDs and were subjected to GO and KEGG analysis in DAVID v6.7. ('Up/Down' miRNAs group, 222 genes; 'Down/Up' miRNAs group, 718 genes).
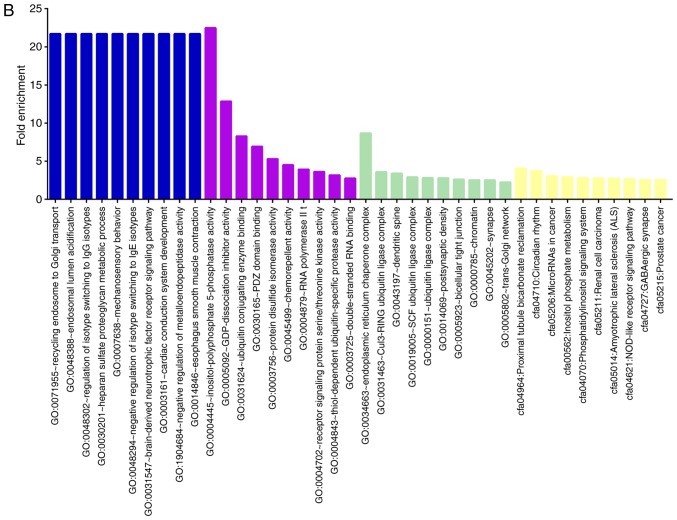
Analyzing the target genes of the 'Up/Down' group revealed that GO processes associated with metabolism were significantly overrepresented in these genes, such as regulation of mitochondrial depolarization, glutamate metabolic process, glutathione biosynthetic process, and protein serine/threonine phosphatase activity. In addition, overrepresented pathways included the TNF, MAPK, cAMP, FoxO and insulin signaling pathways, as well as herpes simplex infection and transcriptional misregulation in cancer (Fig. 4A).
Figure 4.
GO and KEGG analysis of candidate miRNA target genes. (A) The top 10 significantly GO and KEGG terms of 'Up/Down' miRNA group target genes. (B) The top 10 GO and KEGG terms of 'Down/Up' miRNA group target genes. The shared GO terms of the two microRNA target gene groups are associated with receptor signaling protein serine/threonine kinase activity (molecular function), ubiquitin ligase complex, and chromatin (cellular component). Blue, biological process; purple, molecular function; green, cellular component complete; yellow, KEGG. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Analyzing the target genes of the 'Down/Up' group revealed overrepresentation of GO processes associated with ubiquitination, such as ubiquitin conjugating enzyme binding, thiol-dependent ubiquitin-specific protease activity, thiol-dependent ubiquitin-specific protease activity, SCF and Cul3-RING ubiquitin ligase complex, ubiquitin ligase complex, the pathway including proximal tubule bicarbonate reclamation, inositol phosphate metabolism, phosphatidylinositol signaling system, GABAergic synapse, and the NOD-like receptor signaling pathway. The inflammation-related TNF pathway and the neuronal function-related biological functions, including positive regulation of apoptotic signaling pathway, postsynaptic density, synapse, GABAergic synapse and regulation of blood vessel size, may explain how BV exerts its neuroprotective effects in MCAO (Fig. 4B).
Integration analysis of miRNA and mRNA
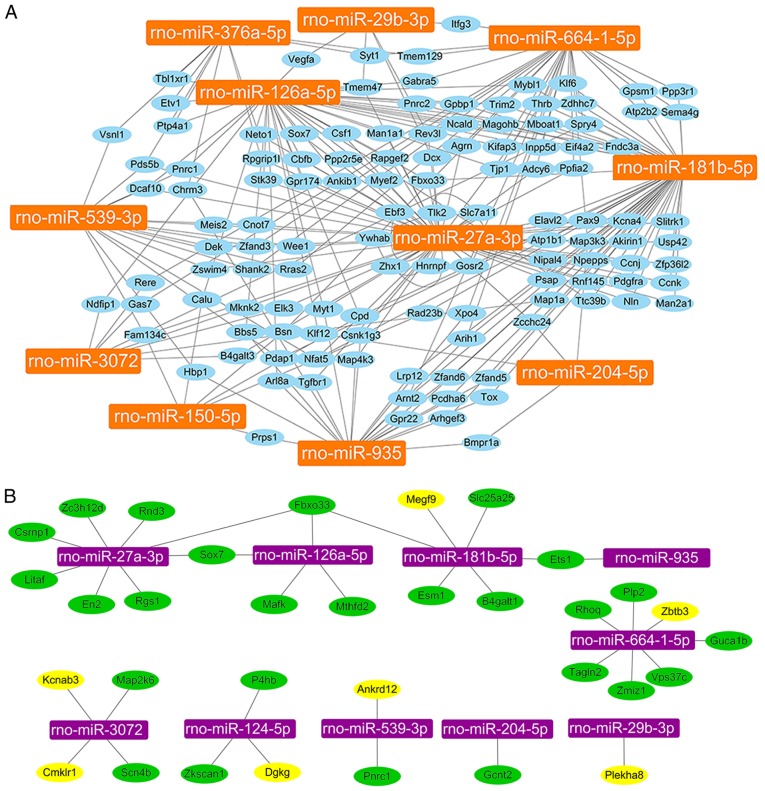
Because each miRNA has multiple potential mRNA targets, one mRNA can regulated by multiple miRNAs. The 13 miRNAs that were confirmed by qPCR were further analyzed and their 1,052 predicted target genes were screened. The results demonstrated that except for rno-miR-124-5p and rno-miR-375-5p, 126 genes had at least two miRNAs co-regulation (Fig. 5A). In order to screen more accurate target genes of miRNAs, the results from the miRNA microarray were compared to the results from the mRNA microarray. The mRNA microarray assay revealed a total of 1,718 genes with fold change ≥2 (group C vs. group S, group BV vs. group C). Of the 1,718 genes, 49 genes overlapped with miRNA target genes (Sox7, Fbxo33, Spry4, Plekha8, Ets1, Cd4, Gcnt2, Dll4, Nfe2l2, Pparg, Csrnp1, En2, Rgs1, Litaf, Rnd3, Zc3h12d, Hbegf, Ier3, P4hb, Mthfd2, Mafk, Nr4a3, Tnf, Il1a, Adamts1, Dusp5, B4galt1, Slc25a25, Cyr61, Esm1, Pgf, Vps37c, Zmiz1, Rhoq, Emp1, Tagln2, Plp2, Zkscan1, Dgkg, Megf9, Zbtb3, Guca1b, Pnrc1, Ankrd12, Kcnab3, Map2k6, Scn4b, Tlr5, Cmklr1). Among the 49 genes, 33 candidate genes were selected for qPCR verification, including Sox7, Fbxo33, Plekha8, Ets1, Gcnt2, Csrnp1, En2, Rgs1, Litaf, Rnd3, Zc3h12d, P4hb, Mthfd2, Mafk, B4galt1, Slc25a25, Esm1, Vps37c, Zmiz1, Rhoq, Tagln2, Plp2, Zkscan1, Dgkg, Megf9, Zbtb3, Guca1b, Pnrc1, Ankrd12, Kcnab3, Map2k6, Scn4b, Cmklr1 (Fig. 5B).
Figure 5.
Relationships between miRNAs and their predicted gene targets. (A) Relationships between 11 miRNAs and their 126 overlapped predicted target genes. (B) Relationships between 10 miRNAs and their 33 candidate genes verified through qPCR testing. Orange, qPCR-verified miRNAs; blue, overlapped predicted target genes of 10 microRNAs verified through qPCR; purple, qPCR-verified miRNAs; green, the candidate genes of the core 'miRNA-mRNA' regulatory network after qPCR testing; yellow, the candidate genes not consistent with the 'microRNA-mRNA' regulatory mechanism after qPCR testing. qPCR, quantitative polymerase chain reaction.
qPCR verification of target mRNAs
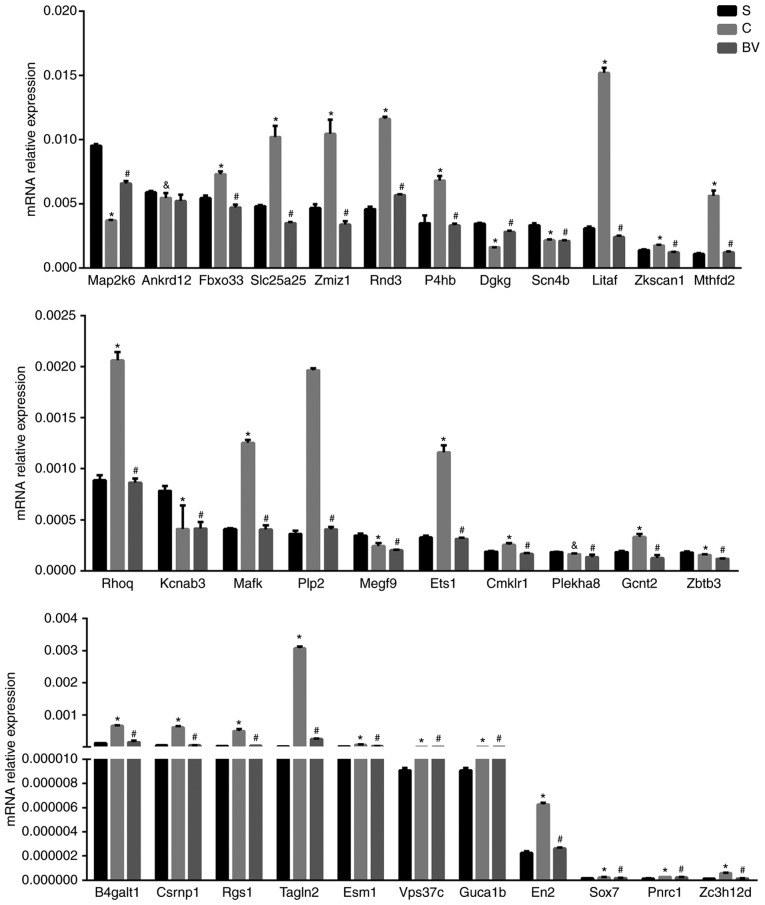
qPCR was performed to identify target mRNAs of miRNAs, which may be related with BV treatment in MCAO. Based on the criteria that the target mRNA should display an inverse expression correlation with the miRNA, 33 mRNAs were tested. These included the genes Sox7, Fbxo33, Plekha8, Ets1, Gcnt2, Csrnp1, En2, Rgs1, Litaf, Rnd3, Zc3h12d, P4hb, Mthfd2, Mafk, B4galt1, Slc25a25, Esm1, Vps37c, Zmiz1, Rhoq, Tagln2, Plp2, Zkscan1, Dgkg, Megf9, Zbtb3, Guca1b, Pnrc1, Ankrd12, Kcnab3, Map2k6, Scn4b, Cmklr1. Excepting Plekha8, Zbtb3, Megf9, Dgkg, Ankrd12, Kcnab3 and Cmklr1. The results demonstrated that out of these 33 genes, 26 were changed in accordance to the 'microRNA-mRNA' regulatory mechanism identified by the microarray analyses (Figs. 5B and 6).
Figure 6.
qPCR validation of target mRNAs. Data are presented as mean ± standard deviation (n=6). &P<0.05 group BV vs. group S; #P<0.01 group C vs. group S; *P<0.01 group BV vs. group C. qPCR, quantitative polymerase chain reaction; BV, biliverdin.
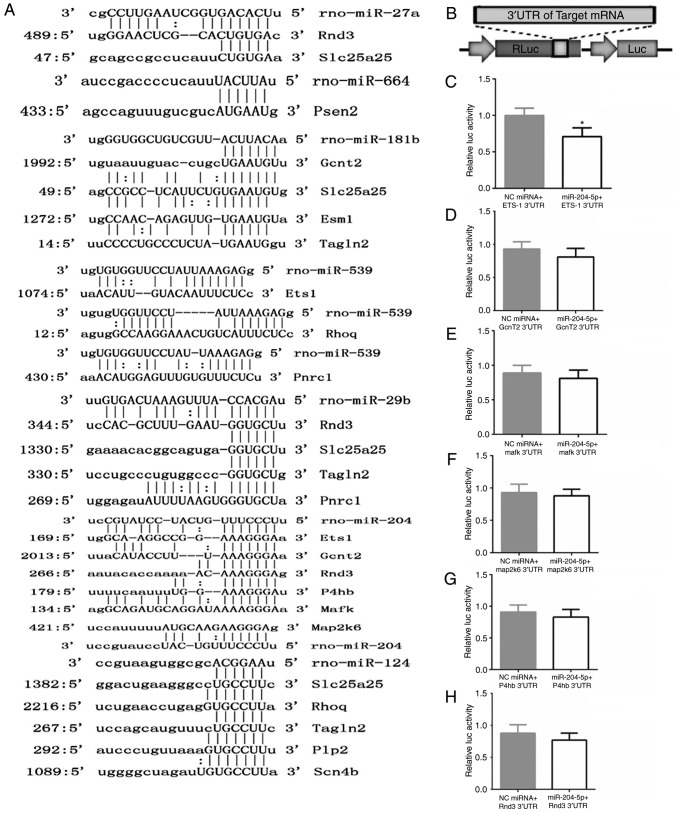
Additionally, using luciferase assay, the regulation relationship of Ets1 and miR-204-5p was examined. Firstly, the conserved binding site of the target gene was compared with the miRNAs, and it was hypothesized that an effect of miR-204-5p on the Ets1 3'UTR was mediated via a single, highly conserved binding site (Fig. 7A). Then, the full-length 3'UTR of the transcript was cloned downstream of a luciferase reporter vector (Fig. 7B). The reporter vector, in combination with miRNA mimics, was co-transfected into 293T cells, and luciferase activity was monitored 48 h later. A robust decrease in luciferase activity was observed in the miR-204-5p group, while the negative control (NC) miRNA had no effect on luciferase activity (Fig. 7C).
Figure 7.
Determination of Ets1 as a direct target of mo-miR-204-5p. (A) The miR-204-5p binding site in the 3'UTR region of Ets1 was highly conserved among several miRNAs. (B) 293T cells were transfected with the luciferase construct containing the 3'UTR of 6 targeted genes of miR-204-5p together with the miR-204-5p mimics or NC miRNA. (C-H) Quantification of luciferase activity. *P<0.05 compared with the NC plasmid. UTR, untranslated region; NC, negative control.
Discussion
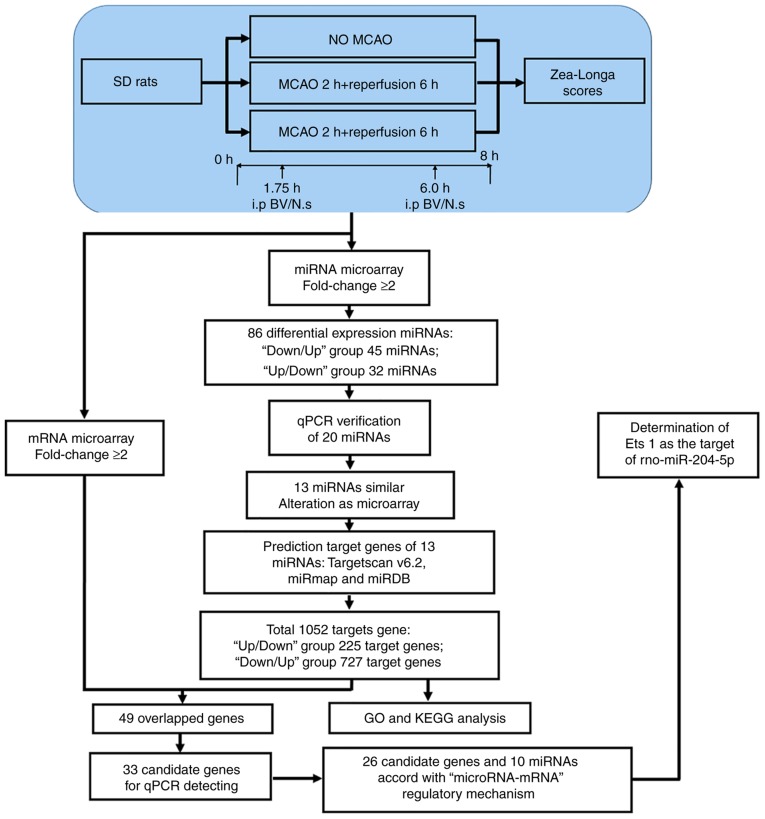
The present study investigated the molecular network of miRNA and mRNA expression to modulate critical processes in CIR rats following BV administration. To clarify the influence of miRNA and mRNA on CIR rats, a model of BV treatment in MCAO rats was established and the expression change of mRNA and miRNA were evaluated by microarray analysis. The microarray data were then verified using qPCR. Furthermore, combined with bioinformatics prediction, BV treatment was demonstrated to alter the expression profile of multiple miRNAs and mRNAs in the cortex of rats. Among them, there were 10 miRNAs and 26 candidate genes that comprised a core 'microRNA-mRNA' regulatory mechanism (Fig. 8).
Figure 8.
Experimental flow diagram. The blue portion explains the animal experimental groups. The white portion of the diagram describes the miRNA-gene integration and analysis. Significantly regulated miRNAs were selected and separated into two groups based on expression pattern. miRNAs that were succesfully validated by qPCR were selected for further analysis. TargetScanv6.2, miRmap, miRDB analyses were used to identify gene targets of candidate miRNAs; only those genes that overlapped in all three analyses were included for GO and KEGG analysis. A total of 49 genes from the mRNA array overlapped with miRNA target genes, and 33 of them were selected for qPCR testing. As a result, 26 candidate genes and 10 miRNAs were identified as a core 'microRNA-mRNA' regulatory network. qPCR, quantitative polymerase chain reaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MCAO, middle cerebral artery occlusion.
First, the present study successfully established the MCAOs model, as confirmed by TTC staining. At 48 h post-reperfusion, BV treatment markedly reduced the infarct size and slightly decreased the NSS score, compared with the vehicle control group. Generally, the changes of morphology and function were followed by changes in gene expression. A previous study has indicated that administration of BV resulted in rapid appearance of bilirubin in the serum and significantly suppressed IRI-induced liver dysfunction in swine (20). Exogenously administered CO and BV have been demonstrated to exhibit potent cytoprotective effects on intestinal IRI (9). Recent evidence has demonstrated that BV has a protective role against lung graft injury, hemorrhagic shock and resuscitation-induced lung injury through anti-inflammatory and antioxidant mechanisms (8,21). The present findings suggested that the molecular network of miRNA-mRNA expression may regulate critical processes to attenuate the release of inflammatory factors in CIR rats following BV administration.
Next, through GO analysis, the function of 26 candidate genes was divided into six categories. The first category was related to cell proliferation, and included the genes En2, Rnd3, Sox7, Mthfd2, β4GalT, MaP2K6, Gcnt2, and Zmiz1. Engrailed 2 (En2) is expressed in all cerebellar cell types, and is critical for regulating the formation of specific fissures and general growth of the cerebellum (22). Downregulation of Rnd3 promotes glioma cell proliferation (23,24). Overexpression of sex determining region Y-box 7 (Sox7) antagonizes cell growth and promotes apoptosis in a number of cancer cell lines (25-27). Furthermore, the overexpression of Mthfd2 promotes tumor cell proliferation (28,29). β4GalT functions in several types of cancer, such as renal cancer, leukemia and neural crest-derived tumors, and is correlated with cancer cell proliferation, metastasis, invasiveness and drug resistance (30-32), Map2k6 can effectively inhibit cell proliferation and induce apoptosis in ovarian cancer HO8910 cells (33). In addition, downregulation of Gcnt2 can promote DNA hyper-methylation (34), and enhance cell migration and invasion in breast cancer and prostate cancer (35,36). Lastly, ectopic expression of zinc finger MIZ-type containing 1 (Zmiz1) induces cutaneous squamous cell malignancies in a mouse model of cancer (37). In the present study, the expression levels of En2, Rnd3, Sox7, Mthfd2, β4GalT, Gcnt2 and Zmiz1 were downregulated, while MaP2K6 was upregulated following BV administration, which suggested that these genes were involved in the process of CIR following BV treatment.
The second category was related to the development and function of the nervous system, and included the genes Csrnp1 and En2. Cystein-serine-rich nuclear protein 1 (Csrnp1) is essential for cephalic neural progenitor proliferation and survival in zebrafish (38). EN2 has key roles in developing mesencephalic dopa-minergic (mDA) neurons and also impacts on the adult mDA neuronal biological function (39). In the present study, the expression change of Csrnp1 and En2 may regulate the functional remodeling in our experiment condition.
The third category related to the anti-inflammatory response, and included the genes Litaf, Zc3h12d and Rgs1. LPS-induced TNF factor is known to activate the transcription of multiple cytokines, such as TNF-α, IL-8 and IL-β, in a variety of cellular processes and many inflammatory diseases (21,40-42). Zc3h12a and Zc3h12d recognize a set of common target mRNAs encoding proteins that serve important roles in the course of the inflammation (43). By reducing the stability of mRNAs encoding proinflammatory factors, Zc3h12d attenuates inflammatory responses (44,45). Regulator of G-protein Signaling (Rgs1) can inhibit chemokine receptor signaling as potential therapeutic targets in leukocyte trafficking and vascular inflammation (46). Endothelial cell-specific molecule-1 (Esm1) is secreted by endothelial cells and upregulated by inflammatory cytokines, such as TNF (47). In the present study, the increase of Litaf, Zc3h12d and Rgs1 may be associated with the anti-inflammation activity in CIR brains following BV treatment.
The fourth category was related to angiogenesis, and included the genes Rnd3, Sox7, Ets1, Esm1 and Esm1. Rnd3 acts as a novel proangiogenic factor involved in cardiac responsive angiogenesis through HIF1α-VEGFA signaling promotion (24). Sox7 regulates angiogenesis and vasculogenesis through mechanisms that are redundant with those of Sox18 (48,49). TF-bearing mEMPs increase angiogenesis operating via paracrine regulation of neighboring endothelial cells, signaling through the β1-integrin pathway Rac1-ERK1/2-Ets1 and triggering CCL2 production to form new and competent mature neovessels (50). Esm1 promotes the angiogenesis and tumor cell growth of prolactinomas (51). Together, the expression of those molecules may regulate the angiogenesis process that is beneficial to the vessel development in ischemia brains following BV administration.
The fifth category was related to cell apoptosis, and included the genes Mafk, Ets1 and Plp2. Protein K (avian) (Mafk) enhances oxidative stress-induced apoptosis (52). Knockdown of erythroblastosis virus E26 oncogene homolog 1 (Ets1) promotes apoptosis in myocardial ischemic reperfusion injury (53). Downregulation of Plp2 increases endoplasmic reticulum-stress-induced neuronal apoptosis and risk for adverse neurological outcomes after hypoxia ischemia injury (54). Inhibiting Tagln2 expression could mediate the hypoxia-induced apoptosis in cardiac myocytes (55). Therefore, these gene expressions may influence function by regulating cell apoptosis in the present model.
Finally, the sixth category includes genes such as Rhoq, slc25a25, vps37c, P4hb, Scn4b and Zkscan1. Overexpression of member Q (Rhoq) may increase invasion potential in colorectal cancer (56). Solute carrier family 25 (slc25a25) is localized in the mitochondrial inner membrane and maintains ATP homeostasis and endurance performance (57). Derepression of the slc25a25 can boost mitochondrial respiration and promote the product of ATP (58). Hyperoxia reduces the survival benefit of U87 and U251 cells with prolyl 4-hydroxylase β polypeptide (P4hb) over-expression through the unfolded protein response (UPR) (59). When cultured mouse cerebral cortical neurons were exposed to hypoxia for 24 h, P4hb was upregulated, and it may participate protein-ubiquitination pathways in the neuronal response to hypoxia (59). Vacuolar protein sorting 37 homolog C (vps37c) is a component of endosomal sorting complex required for transport-I important for viral budding (60). Guca1b is upregulated in dog heart failure (61). Sodium channel voltage-gated type IV β (Scn4b) function may relate with Na+ channels (62). In adenocarcinomas of the gastroesophageal junction, Zkscan1 mRNA is overexpressed (63). The present study is the first to report that the aforementioned genes are involved in the neuroprotection activity of BV in treated CIR rats.
Furthermore, the present study screened a series of miRNAs and several crucial findings were observed in CIR rats following BV administration. By analysis of bionetwork, these miRNAS were predicted to function with their target mRNAs. Compared with the S group, miR-27a, miR-126a-3p, miR-181b-5p, miR-664, miR-124-5p and miR-204-5p were downregulated in the C group and upregulated in the BV group, while miR-935 was upregulated in the C group and downregulated in the BV group. A previous study indicated that miR-27 might unravel unknown pathways in virus-associated CNS dysfunction (64). The present results revealed that BV administration on CIR rats induced downregulation of En2, Rnd3 and Sox7 (the target genes of miR-27a) which may promote cell proliferation; downregulation of Litaf, Zc3h12d, Rgs1 (the target genes of miR-27a) may promote anti-inflammatory response; downregulation of Rnd3 and Sox7 (the target genes of miR-27a) may inhibit angiogenesis; and downregulation of Csrnp1and En2 (the target genes of miR-27a) are poorly expressed to CNS. In sepsis, the downregulation of miR-126a-3p in endothelial cells resulted in increased apoptosis, and decreased proliferation and migration (65). The present results suggested that BV administration in CIR rats induced downregulation of Mafk (the target gene of miR-126a-3p) and may promote cell apoptosis; while downregulation of Mthfd2 (the target gene of miR-126a-3p) may inhibit cell proliferation; and downregulation of Sox7 (the target gene of miR-126a-3p) may promote cell proliferation. miR-181b-5p participates in eosinophilic airway inflammation by regulating IL-13, IL-1β and CCL11 expression via targeting SPP1 (66). The present results indicated that BV administration in CIR rats induced the change of miR-181b-5p, supporting its role in the current model. miR-664 has been known to negatively regulate proteolipid protein 2 (Plp2), promote cell proliferation and invasion in T-cell acute lymphoblastic leukaemia and melanoma (67,68). Combining knowledge from the PubMed database with the current results suggests that BV administration in CIR rats induced downregulation of Zmiz1 (the target gene of miR-664) to inhibit cell proliferation. miR-124-5p functions are as a tumor suppressor and serve as a molecular marker for glioma diagnosis (69). In addition, BV administration in CIR rats induced downregulation of P4hb (the target gene of miR-124-5p), which may participate in protein-ubiquitination pathways in the neuronal response to hypoxia; moreover, downregulation of Zkscan1 (the target gene of miR-124-5p) mRNA may relate to the gastroesophageal junction. Overexpression of miR-935 promotes cell proliferation of gastric cancer and lung cancer (70,71), where the target gene Ets1 is not consistant with the miRNA-mRNA regulatory mechanism following qPCR testing.
In addition, for miR-204-5p, several studies have demonstrated that miR-204 is significantly increased at 24 h post-oxygen-glucose deprivation (OGD) by using RT-qPCR to determine the expression of 16 miRNAs of interest at 1 and 24-h post-OGD (72). Overexpression of miR-204-5p inhibits cell migration and proliferation in glioma cancer (73). Through the luciferase reporter assay, the present results confirmed the regulation of miR-204-5p on the target gene Ets1, and Ets1 was downregulated in CIR rats following BV administration. It has been reported that Ets1 mRNA is increased following MCAO-induced stroke (74). Ets1 was required for activation of RAS-regulated cell migration genes, but also identified a surprising role for Ets1 in the repression of genes such as DUSP4, DUSP6 and SPRY4 that provide negative feedback to the RAS/ERK pathway (75). The present results suggested that miR-204-5p directly regulates Ets-1, which may indicate a potential mechanism by which BV improves neural behavior in CIR rats.
The present study provides a global view of the influence of miRNA/mRNA expression on the genomic response in CIR rats following BV treatment. miR-27a, miR-181b-5p, Litaf, Zc3h12d and Rgs1 have been demonstrated to participate in inflammatory response. BV treatment upregulated miR-27a, miR-181b-5p and miR-204 5p expression and induced their target genes downregulation to influence the anti-inflammatory effect of BV in CIR. Therefore, BV may influence crucial biological functions, such as cell proliferation, apoptosis, maintaining ATP homeostasis and angiogenesis, by miRNAs regulating target genes, such as miRNA 204-5p directly targeting Ets1.
Acknowledgments
Not applicable.
Abbreviations
- CIR
cerebral ischemia reperfusion injury
- BV
biliverdin
- CO
carbonic oxide
- miRNA
microRNA
- MCAO
middle cerebral artery occlusion
- SPF
specific pathogen free
- ip
intraperitoneally
- RCCA
right common carotid artery
- CBF
cerebral blood flow
- TTC
2,3,5-triphenyltetrazolium chloride
Funding
This research was supported by the Natural Science Foundation of China (grant no. 81760248), the Foundation of Yunnan Provincial Science and Technology Department (grant no. 2017FE468-034), the Foundation of Yunnan Health Department (grant no. 2016NS043), Weifeng Yu Expert Workstation (grant no. 2017IC067) and Key Applied and Basic Research Program in Yunnan Province (grant no. 2018FA042).
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
ZYZ and JLS conceived and designed the project. ZYZ, CC and JJL analyzed and interpreted the data. YJ, JL and ZM provided technical support. ZYZ, THW and JLS wrote and revised the manuscript. THW and JLS supervised the study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All experimental protocols involving animals were approved by the Animal Care and Welfare Committee of Kunming Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sarady-Andrews JK, Liu F, Gallo D, Nakao A, Overhaus M, Ollinger R, Choi AM, Otterbein LE. Biliverdin administration protects against endotoxin-induced acute lung injury in rats. Am J Physiol Lung Cell Mol Physiol. 2005;289:L1131–L1137. doi: 10.1152/ajplung.00458.2004. [DOI] [PubMed] [Google Scholar]
- 2.Zhou XQ, Zeng XN, Kong H, Sun XL. Neuroprotective effects of berberine on stroke models in vitro and in vivo. Neurosci Lett. 2008;447:31–36. doi: 10.1016/j.neulet.2008.09.064. [DOI] [PubMed] [Google Scholar]
- 3.Chen GY, Nuñez G. Sterile inflammation: Sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng H, Zuo X, Zhang J, Liu X, Liu L, Xu Q, Wu Z, Ji A. Alphalipoic acid protects against cerebral ischemia/reperfusion-induced injury in rats. Mol Med Rep. 2015;11:3659–3665. doi: 10.3892/mmr.2015.3170. [DOI] [PubMed] [Google Scholar]
- 5.Yamasaki Y, Matsuura N, Shozuhara H, Onodera H, Itoyama Y, Kogure K. Interleukin-1 as a pathogenetic mediator of ischemic brain damage in rats. Stroke. 1995;26:676–681. doi: 10.1161/01.STR.26.4.676. [DOI] [PubMed] [Google Scholar]
- 6.Lavine SD, Hofman FM, Zlokovic BV. Circulating antibody against tumor necrosis factor-alpha protects rat brain from reperfusion injury. J Cereb Blood Flow Metab. 1998;18:52–58. doi: 10.1097/00004647-199801000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Fondevila C, Shen XD, Tsuchiyashi S, Yamashita K, Csizmadia E, Lassman C, Busuttil RW, Kupiec-Weglinski JW, Bach FH. Biliverdin therapy protects rat livers from ischemia and reperfusion injury. Hepatology. 2004;40:1333–1341. doi: 10.1002/hep.20480. [DOI] [PubMed] [Google Scholar]
- 8.Kosaka J, Morimatsu H, Takahashi T, Shimizu H, Kawanishi S, Omori E, Endo Y, Tamaki N, Morita M, Morita K. Effects of biliverdin administration on acute lung injury induced by hemorrhagic shock and resuscitation in rats. PLoS One. 2013;8:e63606. doi: 10.1371/journal.pone.0063606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakao A, Kaczorowski DJ, Sugimoto R, Billiar TR, McCurry KR. Application of heme oxygenase-1, carbon monoxide and biliverdin for the prevention of intestinal ischemia/reperfusion injury. J Clin Biochem Nutr. 2008;42:78–88. doi: 10.3164/jcbn.2008013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wegiel B, Gallo D, Csizmadia E, Roger T, Kaczmarek E, Harris C, Zuckerbraun BS, Otterbein LE. Biliverdin inhibits Toll-like receptor-4 (TLR4) expression through nitric oxide-dependent nuclear translocation of biliverdin reductase. Proc Natl Acad Sci USA. 2011;108:18849–18854. doi: 10.1073/pnas.1108571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holst B, Raby AC, Hall JE, Labéta MO. Complement takes its Toll: An inflammatory crosstalk between Toll-like receptors and the receptors for the complement anaphylatoxin C5a. Anaesthesia. 2012;67:60–64. doi: 10.1111/j.1365-2044.2011.07011.x. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Yu J, Liu F, Yin P, Zhu X, Cheng G, Wang N, Lu A, Luan W, Zhang N, Li J, et al. 2.2 Integrating miRNA and mRNA expression profiles in response to heat stress-induced injury in rat small intestine. Funct Integr Genomics. 2011;11:203–213. doi: 10.1007/s10142-010-0198-8. [DOI] [PubMed] [Google Scholar]
- 14.Nassar FJ, El Eit R, Nasr R. An integrative analysis of microRNA and mRNA profiling in CML stem cells. Methods Mol Biol. 2016;1465:219–241. doi: 10.1007/978-1-4939-4011-0_18. [DOI] [PubMed] [Google Scholar]
- 15.Chen JH, He HC, Jiang FN, Militar J, Ran PY, Qin GQ, Cai C, Chen XB, Zhao J, Mo ZY, et al. Analysis of the specific pathways and networks of prostate cancer for gene expression profiles in the Chinese population. Med Oncol. 2012;29:1972–1984. doi: 10.1007/s12032-011-0088-5. [DOI] [PubMed] [Google Scholar]
- 16.Chiang T, Messing RO, Chou WH. Mouse model of middle cerebral artery occlusion. J Vis Exp. 2011;2761 doi: 10.3791/2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JJ, Zou ZY, Liu J, Xiong LL, Jiang HY, Wang TH, Shao JL. Biliverdin administration ameliorates cerebral ischemia reperfusion injury in rats and is associated with proinflammatory factor downregulation. Exp Ther Med. 2017;14:671–679. doi: 10.3892/etm.2017.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kam KY, Yu SJ, Jeong N, Hong JH, Jalin AM, Lee S, Choi YW, Lee CK, Kang SG. p-Hydroxybenzyl alcohol prevents brain injury and behavioral impairment by activating Nrf2, PDI, and neurotrophic factor genes in a rat model of brain ischemia. Mol Cells. 2011;31:209–215. doi: 10.1007/s10059-011-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Andria B, Bracco A, Attanasio C, Castaldo S, Cerrito MG, Cozzolino S, Di Napoli D, Giovannoni R, Mancini A, Musumeci A, et al. Biliverdin protects against liver ischemia reperfusion injury in swine. PLoS One. 2013;8:e69972. doi: 10.1371/journal.pone.0069972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Yang Z, Tsuji T, Gong J, Xie J, Chen C, Li W, Amar S, Luo Z. LITAF and TNFSF15, two downstream targets of AMPK, exert inhibitory effects on tumor growth. Oncogene. 2011;30:1892–1900. doi: 10.1038/onc.2010.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orvis GD, Hartzell AL, Smith JB, Barraza LH, Wilson SL, Szulc KU, Turnbull DH, Joyner AL. The engrailed homeobox genes are required in multiple cell lineages to coordinate sequential formation of fissures and growth of the cerebellum. Dev Biol. 2012;367:25–39. doi: 10.1016/j.ydbio.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu BH, Lin X, Yang X, Dong H, Yue X, Andrade KC, Guo Z, Yang J, Wu L, Zhu X, et al. Downregulation of RND3/RhoE in glioblastoma patients promotes tumorigenesis through augmentation of notch transcriptional complex activity. Cancer Med. 2015;4:1404–1416. doi: 10.1002/cam4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue XJ, Lin X, Yang T, Yang X, Yi X, Jiang X, Li X, Li T, Guo J, Dai Y, et al. Rnd3/RhoE modulates hypoxia-inducible factor 1α/vascular endothelial growth factor signaling by stabilizing hypoxia-inducible factor 1α and regulates responsive cardiac angiogenesis. Hypertension. 2016;67:597–605. doi: 10.1161/HYPERTENSIONAHA.115.06412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Zhong D, Lau S, Liu X, Dong XY, Sun X, Yang VW, Vertino PM, Moreno CS, Varma V, et al. Sox7 is an independent checkpoint for beta-catenin function in prostate and colon epithelial cells. Mol Cancer Res. 2008;6:1421–1430. doi: 10.1158/1541-7786.MCR-07-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Huang SY, Dong W, Li L, Feng Y, Pan L, Han Z, Wang X, Ren G, Su D, et al. SOX7, down-regulated in colorectal cancer, induces apoptosis and inhibits proliferation of colorectal cancer cells. Cancer Lett. 2009;277:29–37. doi: 10.1016/j.canlet.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Qin L, Min Z, Zhao Y, Zhu L, Zhu J, Yu S. SOX7 interferes with β-catenin activity to promote neuronal apoptosis. Eur J Neurosci. 2015;41:1430–1437. doi: 10.1111/ejn.12910. [DOI] [PubMed] [Google Scholar]
- 28.Gustafsson Sheppard N, Jarl L, Mahadessian D, Strittmatter L, Schmidt A, Madhusudan N, Tegnér J, Lundberg EK, Asplund A, Jain M, Nilsson R. The folate-coupled enzyme MTHFD2 is a nuclear protein and promotes cell proliferation. Sci Rep. 2015;5:15029. doi: 10.1038/srep15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tedeschi PM, Vazquez A, Kerrigan JE, Bertino JR. Mitochondrial methylenetetrahydrofolate dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Mol Cancer Res. 2015;13:1361–1366. doi: 10.1158/1541-7786.MCR-15-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.la Torre A, Muscarella LA, Parrella P, Balsamo T, Bisceglia M, Valori VM, la Torre A, Barbano R, Perrella E, Poeta ML, et al. Aberrant genes promoter methylation in neural crest-derived tumors. Int J Biol Marker. 2012;27:E389–E394. doi: 10.5301/JBM.2012.9766. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H, Zhang Z, Liu C, Jin C, Zhang J, Miao X, Jia L. B4GALT1 gene knockdown inhibits the hedgehog pathway and reverses multidrug resistance in the human leukemia K562/adriamycin-resistant cell line. IUBMB Life. 2012;64:889–900. doi: 10.1002/iub.1080. [DOI] [PubMed] [Google Scholar]
- 32.Xie H, Zhu Y, An H, Wang H, Zhu Y, Fu H, Wang Z, Fu Q, Xu J, Ye D. Increased B4GALT1 expression associates with adverse outcome in patients with non-metastatic clear cell renal cell carcinoma. Oncotarget. 2016;7:32723–32730. doi: 10.18632/oncotarget.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Kang JL, Liao H, Wang XX, Nie ML, Shuai R, Deng C. Mitogen-activated protein kinase kinase 6-fusion protein (MAP2K6-FP) potentiates the anti-tumor effects of paclitaxel in ovarian cancer. Anticancer Agent Med Chem. 2015;15:1308–1316. doi: 10.2174/1871520615666150629100405. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura K, Yamashita K, Sawaki H, Waraya M, Katoh H, Nakayama N, Kawamata H, Nishimiya H, Ema A, Narimatsu H, Watanabe M. Aberrant methylation of GCNT2 Is tightly related to lymph node metastasis of primary CRC. Anticancer Res. 2015;35:1411–1421. [PubMed] [Google Scholar]
- 35.Zhang H, Meng F, Wu S, Kreike B, Sethi S, Chen W, Miller FR, Wu G. Engagement of I-branching {beta}-1, 6-N-acetylglucosaminyltransferase 2 in breast cancer metastasis and TGF-{beta} signaling. Cancer Res. 2011;71:4846–4856. doi: 10.1158/0008-5472.CAN-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikami J, Tobisawa Y, Yoneyama T, Hatakeyama S, Mori K, Hashimoto Y, Koie T, Ohyama C, Fukuda M. I-branching N-acetylglucosaminyltransferase regulates prostate cancer invasiveness by enhancing α5β1 integrin signaling. Cancer Sci. 2016;107:359–368. doi: 10.1111/cas.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers LM, Riordan JD, Swick BL, Meyerholz DK, Dupuy AJ. Ectopic expression of Zmiz1 induces cutaneous squamous cell malignancies in a mouse model of cancer. J Invest Dermatol. 2013;133:1863–1869. doi: 10.1038/jid.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feijóo CG, Sarrazin AF, Allende ML, Glavic A. Cystein-serine-rich nuclear protein 1, Axud1/Csrnp1, is essential for cephalic neural progenitor proliferation and survival in zebrafish. Dev Dyn. 2009;238:2034–2043. doi: 10.1002/dvdy.22006. [DOI] [PubMed] [Google Scholar]
- 39.Rekaik H, Blaudin de Thé FX, Prochiantz A, Fuchs J, Joshi RL. Dissecting the role of Engrailed in adult dopaminergic neurons-insights into Parkinson disease pathogenesis. FEBS Lett. 2015;589:3786–3794. doi: 10.1016/j.febslet.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiomi N, Myokai F, Naruishi K, Oyaizu K, Senoo K, Yamaguchi T, Amar S, Takashiba S. Cloning and characterization of lipopolysaccharide-induced tumor necrosis factor alpha factor promoter. Fems Immunol Med Mic. 2006;47:360–368. doi: 10.1111/j.1574-695X.2006.00094.x. [DOI] [PubMed] [Google Scholar]
- 41.Bushell KN, Leeman SE, Gillespie E, Gower AC, Reed KL, Stucchi AF, Becker JM, Amar S. LITAF mediation of increased TNF-α secretion from inflamed colonic lamina propria macrophages. PLoS One. 2011;6:e25849. doi: 10.1371/journal.pone.0025849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang XR, Amar S. Kavain involvement in LPS-induced signaling pathways. J Cell Biochem. 2016;117:2272–2280. doi: 10.1002/jcb.25525. [DOI] [PubMed] [Google Scholar]
- 43.Wawro M, Kochan J, Krzanik S, Jura J, Kasza A. Intact NYN/PIN-like domain is crucial for the degradation of inflammation-related transcripts by ZC3H12D. J Cell Biochem. 2017;118:487–498. doi: 10.1002/jcb.25665. [DOI] [PubMed] [Google Scholar]
- 44.Minagawa K, Wakahashi K, Kawano H, Nishikawa S, Fukui C, Kawano Y, Asada N, Sato M, Sada A, Katayama Y, Matsui T. Posttranscriptional modulation of cytokine production in T cells for the regulation of excessive inflammation by TFL. J Immunol. 2014;192:1512–1524. doi: 10.4049/jimmunol.1301619. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H, Wang WC, Chen JK, Zhou L, Wang M, Wang ZD, Yang B, Xia YM, Lei S, Fu EQ, Jiang T. ZC3H12D attenuated inflammation responses by reducing mRNA stability of proinflammatory genes. Mol Immunol. 2015;67:206–212. doi: 10.1016/j.molimm.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 46.Patel J, McNeill E, Douglas G, Hale AB, de Bono J, Lee R, Iqbal AJ, Regan-Komito D, Stylianou E, Greaves DR, Channon KM. RGS1 regulates myeloid cell accumulation in atherosclerosis and aortic aneurysm rupture through altered chemokine signalling. Nat Commun. 2015;6:6614. doi: 10.1038/ncomms7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grigoriu BD, Depontieu F, Scherpereel A, Gourcerol D, Devos P, Ouatas T, Lafitte JJ, Copin MC, Tonnel AB, Lassalle P. Endocan expression and relationship with survival in human non-small cell lung cancer. Clin Cancer Res. 2006;12:4575–4582. doi: 10.1158/1078-0432.CCR-06-0185. [DOI] [PubMed] [Google Scholar]
- 48.Zhang C, Basta T, Klymkowsky MW. SOX7 and SOX18 are essential for cardiogenesis in Xenopus. Dev Dyn. 2005;234:878–891. doi: 10.1002/dvdy.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herpers R, van de Kamp E, Duckers HJ, Schulte-Merker S. Redundant roles for sox7 and sox18 in arteriovenous specification in zebrafish. Circ Res. 2008;102:12–15. doi: 10.1161/CIRCRESAHA.107.166066. [DOI] [PubMed] [Google Scholar]
- 50.Arderiu G, Peña E, Badimon L. Angiogenic microvascular endothelial cells release microparticles rich in tissue factor that promotes postischemic collateral vessel formation. Arterioscl Throm Vasc Biol. 2015;35:348–357. doi: 10.1161/ATVBAHA.114.303927. [DOI] [PubMed] [Google Scholar]
- 51.Cai L, Leng ZG, Guo YH, Lin SJ, Wu ZR, Su ZP, Lu JL, Wei LF, Zhuge QC, Jin K, Wu ZB. Dopamine agonist resistance-related endocan promotes angiogenesis and cells viability of prolactinomas. Endocrine. 2016;52:641–651. doi: 10.1007/s12020-015-0824-2. [DOI] [PubMed] [Google Scholar]
- 52.Bensellam M, Montgomery MK, Luzuriaga J, Chan JY, Laybutt DR. Inhibitor of differentiation proteins protect against oxidative stress by regulating the antioxidant-mitochondrial response in mouse beta cells. Diabetologia. 2015;58:758–770. doi: 10.1007/s00125-015-3503-1. [DOI] [PubMed] [Google Scholar]
- 53.Bian C, Xu TD, Zhu H, Pan D, Liu Y, Luo Y, Wu P, Li D. Luteolin inhibits ischemia/reperfusion-induced myocardial injury in rats via downregulation of microRNA-208b-3p. PLoS One. 2015;10:e0144877. doi: 10.1371/journal.pone.0144877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang LL, Wang T, Valle D. Reduced PLP2 expression increases ER-stress-induced neuronal apoptosis and risk for adverse neurological outcomes after hypoxia ischemia injury. Hum Mol Genet. 2015;24:7221–7226. doi: 10.1093/hmg/ddv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li AY, Yang Q, Yang K. miR-133a mediates the hypoxia-induced apoptosis by inhibiting TAGLN2 expression in cardiac myocytes. Mol Cell Biochem. 2015;400:173–181. doi: 10.1007/s11010-014-2273-2. [DOI] [PubMed] [Google Scholar]
- 56.Han SW, Kim HP, Shin JY, Jeong EG, Lee WC, Kim KY, Park SY, Lee DW, Won JK, Jeong SY, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med. 2014;211:613–621. doi: 10.1084/jem.20132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sako H, Yada K, Suzuki K. Genome-wide analysis of acute endurance exercise-induced translational regulation in mouse skeletal muscle. PLoS One. 2016;11:e0148311. doi: 10.1371/journal.pone.0148311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karunakaran D, Thrush B, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, Ramphos E, Shangari P, Ouimet M, Pezacki JP, et al. Macrophage mitochondrial energy status regulates cholesterol efflux and is enhanced by anti-miR33 in atherosclerosis. Circ Res. 2015;117:266–278. doi: 10.1161/CIRCRESAHA.117.305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee D, Sun S, Ho AS, Kiang KM, Zhang XQ, Xu FF, Leung GK. Hyperoxia resensitizes chemoresistant glioblastoma cells to temozolomide through unfolded protein response. Anticancer Res. 2014;34:2957–2966. [PubMed] [Google Scholar]
- 60.Eastman SW, Martin-Serrano J, Chung W, Zang T, Bieniasz PD. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J Biol Chem. 2005;280:628–636. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- 61.Lanfear DE, Yang JJ, Mishra S, Sabbah HN. Genome-wide approach to identify novel candidate genes for beta blocker response in heart failure using an experimental model. Discov Med. 2011;11:359–366. [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis AH, Raman IM. Resurgent current of voltage-gated Na(+) channels. J Physiol. 2014;592:4825–4838. doi: 10.1113/jphysiol.2014.277582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeh I, Botton T, Talevich E, Shain AH, Sparatta AJ, de la Fouchardiere A, Mully TW, North JP, Garrido MC, Gagnon A, et al. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun. 2015;6:7174. doi: 10.1038/ncomms8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ebrahimie E, Nurollah Z, Ebrahimi M, Hemmatzadeh F, Ignjatovic J. Unique ability of pandemic influenza to downregulate the genes involved in neuronal disorders. Mol Biol Rep. 2015;42:1377–1390. doi: 10.1007/s11033-015-3916-4. [DOI] [PubMed] [Google Scholar]
- 65.Chu M, Qin S, Wu R, Zhou X, Tang X, Zhang S, Zhao Q, Wang H, Liu Y, Han X, et al. Role of MiR-126a-3p in endothelial injury in endotoxic mice. Criti Care Med. 2016;44:e639–e650. doi: 10.1097/CCM.0000000000001629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huo X, Zhang K, Yi L, Mo Y, Liang Y, Zhao J, Zhang Z, Xu Y, Zhen G. Decreased epithelial and plasma miR-181b-5p expression associates with airway eosinophilic inflammation in asthma. Clin Exp Allergy. 2016;46:1281–1290. doi: 10.1111/cea.12754. [DOI] [PubMed] [Google Scholar]
- 67.Ding Z, Jian S, Peng X, Liu Y, Wang J, Zheng L, Ou C, Wang Y, Zeng W, Zhou M. Loss of MiR-664 expression enhances cutaneous malignant melanoma proliferation by upregulating PLP2. Medicine (Baltimore) 2015;94:e1327. doi: 10.1097/MD.0000000000001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu H, Miao MH, Ji XQ, Xue J, Shao XJ. miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic. leukaemia Biochem Biophys Res Commun. 2015;459:340–345. doi: 10.1016/j.bbrc.2015.02.116. [DOI] [PubMed] [Google Scholar]
- 69.Chen Q, Lu G, Cai Y, Li Y, Xu R, Ke Y, Zhang S. MiR-124-5p inhibits the growth of high-grade gliomas through posttranscriptional regulation of LAMB1. Neuro Oncol. 2014;16:637–651. doi: 10.1093/neuonc/not300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang M, Cui G, Ding M, Yang W, Liu Y, Dai D, Chen L. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed Pharmacother. 2016;79:153–158. doi: 10.1016/j.biopha.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L, Zeng D, Chen Y, Li N, Lv Y, Li Y, Xu X, Xu G. miR-937 contributes to the lung cancer cell proliferation by targeting INPP4B. Life Sci. 2016;155:110–115. doi: 10.1016/j.lfs.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 72.Keasey MP, Scott HL, Bantounas I, Uney JB, Kelly S. MiR-132 is upregulated by ischemic preconditioning of cultured hippocampal neurons and protects them from subsequent OGD toxicity. J Mol Neurosci. 2016;59:404–410. doi: 10.1007/s12031-016-0740-9. [DOI] [PubMed] [Google Scholar]
- 73.Stylli SS, Adamides AA, Koldej RM, Luwor RB, Ritchie DS, Ziogas J, Kaye AH. miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J Neurosurg. 2017;126:1131–1139. doi: 10.3171/2016.1.JNS151454. [DOI] [PubMed] [Google Scholar]
- 74.Pulliam JV, Xu Z, Ford GD, Liu C, Li Y, Stovall KC, Cannon VS, Tewolde T, Moreno CS, Ford BD. Computational identification of conserved transcription factor binding sites upstream of genes induced in rat brain by transient focal ischemic stroke. Brain Res. 2013;1495:76–85. doi: 10.1016/j.brainres.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Plotnik JP, Budka JA, Ferris MW, Hollenhorst PC. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res. 2014;42:11928–11940. doi: 10.1093/nar/gku929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.