Abstract
Immune checkpoint inhibitor (ICI) treatments benefit some patients with metastatic cancers, but predictive biomarkers are needed. Findings in select cancer types suggest that tumor mutational burden (TMB) may predict clinical response to ICI.To examine this association more broadly, we analyzed the clinical and genomic data of 1662 advanced cancer patients treated with ICI, and 5371 non-ICI treated patients, whose tumors underwent targeted next-generation sequencing (MSK-IMPACT). Among all patients, higher somatic TMB (highest 20% in each histology) was associated with better OS (HR 0.52; p=1.6 ×10−6). For most cancer histologies, an association between higher TMB and improved survival was observed. The TMB cutpoints associated with improved survival varied markedly between cancer types. These data indicate that TMB is associated with improved survival in patients receiving ICI across a wide variety of cancer types, but that there may not be one universal definition of high TMB.
In recent years, immune checkpoint inhibitors (ICI) have revolutionized the treatment of patients with advanced stage cancers. These agents include antibodies that target CTLA4 or PD-1/PD-L1.1 Durable benefit, however, is limited to a minority of patients. Recently, several large phase 3 trials have reported negative results both in unselected patients and some selected groups, highlighting the clinical need to identify better predictive biomarkers.2–5 Early reports have suggested that PD-L1 immunohistochemistry, T cell infiltration levels, T-cell receptor clonality, gene expression signatures, and peripheral blood markers may correlate with clinical response.6 Additionally, an association between high mutational load and clinical benefit was observed in small cohorts of melanoma patients treated with CTLA4 blockade,7,8 and non-small cell lung cancer (NSCLC), melanoma, and bladder cancer patients treated with PD-1/PD-L1 inhibitors.9–11 However, it is unclear whether TMB is robustly predictive of clinical benefit across diverse human cancers, or outside of these specific clinical trial populations.
In prior studies, mutation load was determined using whole exome sequencing, which is not widely utilized in routine clinical care. Currently, the majority of precision oncology platforms utilize next-generation sequencing of targeted gene panels. At Memorial Sloan Kettering Cancer Center, as part of clinical care, patients undergo genomic profiling using the FDA authorized Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay.12 This test is performed in a CLIA environment and identifies somatic exonic mutations in a pre-defined subset of 468 cancer-related genes (earlier versions included 341 or 410 genes), using both tumor-derived and matched germline normal DNA.
We examined the association between non-synonymous somatic TMB, as measured by MSK-IMPACT, and overall survival (OS) after treatment with ICI. The cohort included 1662 patients whose tumors were profiled by next generation sequencing and who received at least one dose of ICI therapy, representing a variety of cancer types with sufficient number of patients for analysis (Supp-Fig 1). Patients who received atezolizumab, avelumab, durvalumab, ipilimumab, nivolumab, pembrolizumab, or tremelimumab as monotherapy or in combination were included in the study.The vast majority of patients (1446, 94% of tumors excluding glioma) had stage IV or metastatic disease. A small number of patients had locally recurrent disease (n=10), or were melanoma patients with regionally advanced unresectable disease (stage III, n= 989 (Supp Table 1). In total, 146 received anti-CTLA4, 1447 received anti-PD1 or PD-L1, and 189 received both. A large number of patients had cancers for which ICI is FDA-approved, including 350 NSCLCs, 321 melanomas, 151 renal cell carcinomas (RCC), 214 bladder cancers and 138 head and neck squamous cell cancers (Supp-Table-2). To calculate TMB, the total number of somatic non-synonymous mutations was normalized to the total number of megabases sequenced. OS was measured from the date of first ICI treatment to time of death or last follow-up. The median followup was 19 months (range 0–80, with 830 [50%] patients alive and censored at last followup.
We defined TMB subgroups by percentiles within each histology. We took this approach because the median and range of mutational load has been shown to vary across tumor types13; therefore, a universal cutoff for “high TMB” would be enriched for tumor types with higher mutation load.Across the entire cohort, stratifying tumors by TMB decile within histology revealed that a higher number of mutations was associated with improved OS. This significant association, stratified by histology, was seen across a variety of cutpoints chosen to define the high TMB group (ranging from top 10–50%; Fig-1 A, Supp. Fig 3–4). A clear trend toward decreasing hazard ratio (HR) of death with increasing TMB cutoff was observed across cancer types demonstrating increasing benefit from ICI with higher TMB (Fig-1 B, Supp-Fig-3).14
Figure 1: Effect of Mutational Load on Overall Survival after ICI Treatment.
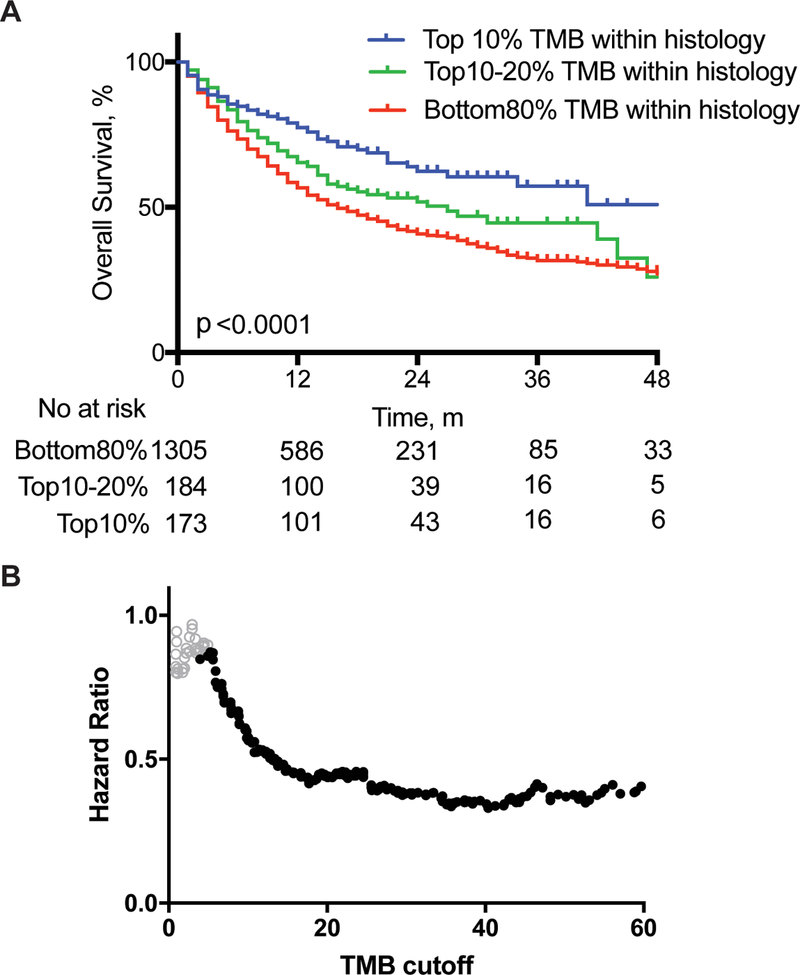
A. Kaplan-Meier curves for patients with tumors falling into the depicted deciles of TMB within each histology. Overall survival is from the first dose of ICI. Two-sided log-rank p value indicated for all patients, with univariate Cox regression hazard ratio of 0.76 (95% CI 0.62–0.94) and 0.52 (95% CI 0.42–0.64) for the 10–20% and Top10% groups, respectively, compared to Bottom80% group. m, monthsB. Cox regression hazard ratios for overall survival on 1662 patients, at depicted cutoffs of TMB across all cancer subtypes. Solid black circles represent hazard ratios with p-values <.05 (two-sided log rank p value).
To confirm that these results were present across multiple cancer types, we performed two additional analyses.First, a multivariable analysis across the entire cohort using Cox proportional-hazards regression demonstrated that the tumor mutation burden was significantly associated with OS both as a continuous variable (HR=0.985, p=3.4×10−7) and with a binary cutoff (top 20% of each histology, HR 0.61 p=1.3×10−7), adjusting for cancer type, age, drug class of ICI, and year of ICI start (Table-1). Furthermore, this association remained significant with removal of melanoma and NSCLC patients from the cohort (Supp Table 2), indicating that this effect was not solely driven by these histologies.
Table 1: Multivariable analysis of factors associated with overall survival.
| HR | 95% CI | P value | |
|---|---|---|---|
| Normalized Mutation Count | |||
| Continuous | 0.985 | 0.979–0.991 | 3.4×10−7 |
| Binary (Top 20%of each histology) | 0.61 | 0.508–0.733 | 1.3×10−7 |
|
Cancer Type |
|||
| Melanoma (reference) | |||
| NSCLC | 2.08 | 1.61–2.68 | 1.9×10−8 |
| Not Melanoma/NSCLC | 1.52 | 1.21–1.92 | 3.7×10−4 |
|
Age |
0.995 |
0.990–1.004 |
0.07 |
|
Drug Class |
|||
| PD-1/PD-L1 (reference) | |||
| CTLA4 | 1.18 | 0.846–1.66 | 0.32 |
| Combo | 0.67 | 0.534–0.844 | 6.6×10−4 |
|
Year of ICI start |
2.3X10−8 |
ICI-Immune Checkpoint Inhibitor
Cox proportional hazards multivariable analysis of overall survival in 1662 patients treated with ICI demonstrating the hazard ratios for individual covariates.
We also performed a stratified analysis within each cancer type, by selecting the higher mutation load quintile (top 20%) in each histology as the TMB-high group. Using this approach, we observed a similar association of longer OS with higher TMB (top 20% within each histology) across multiple cancer types (Fig 2, Sup-Fig-5). Although the effect for some individual cancers did not reach statistical significance, possibly due to smaller sample size, the numerical trend of better OS (HR<1) was observed in nearly all cancer types, with glioma the clearest exception. Taken together, these data indicate that the association between TMB and improved survival after ICI is likely to be present in the majority of cancer histologies.
Figure 2: Effect of Non-synonymous Mutational Load on Overall Survival after ICI Treatment by Cancer Subtype and Drug Class.
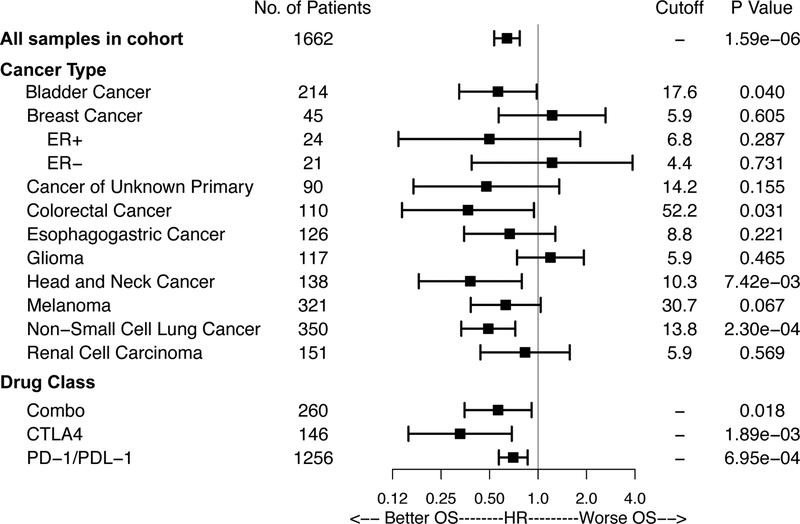
Forest plot for all patients in the identified cohort or individual cancer subtypes.Indicated are the number of patients and hazard ratio comparing overall survival after ICI in patients in the highest 20th percentile TMB within each histology. Horizontal lines represent the 95% confidence interval. The cutoff defining top 20% of normalized mutational burden from MSK-IMPACT for each cancer type is shown, as well as the two-sided log-rank p value for the comparison of high and low mutational burden survival curves. All cancer types in analysis are displayed.
Consistent with varying distributions of TMB across histologies, the TMB cutoff associated with the top 20% of each cancer type varied markedly (Fig 2). Importantly, this suggests that there is not likely to be a universal number defining high TMB that is predictive of clinical benefit to ICI across all cancer types, and that the optimal cutpoint is likely to vary for different cancers.
A similar numerical trend was observed for longer OS with TMB measured as a continuous variable, across many histologies, concordant with the number of patients in the subgroup (Supp-Fig-6). Consistent with the differences in OS, we also observed similar associations between TMB and rates of clinical benefit to ICI, or progression free survival, in patients with cancer types for which response data was available – NSCLC, melanoma, esophagogastric, head and neck, and renal cell cancer (Supp-Fig 7–8). 15,16,17
To investigate the possibility that the observed survival differences among patients with higher TMB tumors could simply be attributable to a general prognostic benefit of high mutational load, unrelated to ICI, we analyzed the outcomes of 5371 patients with metastatic cancers who did not receive ICI, and whose tumors were sequenced with MSK-IMPACT. In these patients, there was no association between higher TMB and improved OS (HR 1.12, p=0.11). This lack of prognostic benefit was also observed within each histology (Supp-Fig-5, 9).
Of note, the TMB cutpoint for the top 20% of colorectal cancer patients was high (52.2/MB), potentially consistent with many MSI-high colorectal tumors receiving ICI treatment. To evaluate the possibility that the ICI-treated cohort of patients might be enriched for patients with higher TMB – if, for example, clinicians were more likely to triage higher TMB patients to ICI therapy – we repeated the survival analyses, instead calculating the top 20% of TMB among all (both ICI and non-ICI treated) patients. The TMB cutpoints in other cancer types were not changed with this calculation, and the associations with survival in each cancer type remained very similar, in both the ICI and non-ICI treated cohorts (Supp Figs 10 and 11).
Distinct from the other cancer types, there was no association between higher TMB and improved survival in patients with glioma; in fact, the trend was toward poorer survival. Although there have been case reports of dramatic responses to ICI in patients with glioblastoma associated with childhood biallelic mismatch repair deficiency18,mismatch repair is very rare in GBM, and higher TMB in many glioma patients may reflect prior exposure to the alkylating agent temozolomide, which can promote the expansion of less immunogenic subclonal mutations.19 Alternatively, anti-tumor immune responses in the CNS may be distinct and less dependent on TMB.
As would be expected in a large multi-cancer analysis of tumors sequenced as part of clinical care, the patients included were heterogeneous, with some having been heavily pre-treated whereas others were treated with a variety of combination therapies. The timing of MSK-IMPACT testing relative to ICI start was also variable. Nevertheless, the finding of a significant association with OS in a heterogeneous cohort underscores the robustness of TMB as a predictive biomarker, suggesting it is likely to be clinically meaningful.
TMB, as measured by targeted NGS panels such as MSK-IMPACT, has been previously shown to have a high correlation with total mutational burden as measured by WES. 20–24 MSK-IMPACT offers the advantage of matched normal germline sequencing for each patient, permitting precise identification of true somatic mutations.
While TMB measured in exome sequencing is highly correlated with measurements in targeted sequencing, it is important to note that numerical cutpoints may differ across platforms. Additionally, we note that TMB cutoffs for individual histologies may not represent the ideal values for use clinically, and are shown primarily to demonstrate that a relationship exists between TMB and survival for each histology. We chose a top 20th percentile cutoff for TMB in order to dichotomize our data, and this does not imply any clinical significance to this threshold.
The variable threshold of TMB across histologies can likely be attributed to distinct tumor microenvironments as well as the numerous other factors shows to independently predict response to ICI including clonality, immune infiltration, immune cell exclusion, HLA genotype and alterations, expression levels of checkpoint molecules, as well as others.19,25–28 Our data overall suggest that TMB is associated with increasing OS in a dose-dependent fashion. The pan-cancer nature of this biomarker likely reflects fundamental mechanisms by which ICI functions. Our data are also consistent with the hypothesis that higher mutation load is associated with a higher number of tumor neoantigens presented on MHC molecules that facilitate immune recognition as foreign and the development of an anti-tumor immune response. 29,30
This finding is in line with the observation that patients with hypermutated tumors as a result of defective mismatch repair have high response rates to pembrolizumab, a finding that had led to the FDA’s tissue/site-agnostic approval of this agent for microsatellite instability-high or mismatch repair deficient tumors31. Further elucidation of appropriate mutational load cutoffs with integration of relevant clinical variables within each cancer type will be necessary, likely in the context of prospective clinical studies, to allow for implementation of TMB as a predictive biomarker.
Our study addresses several fundamentally important questions in immuno-oncology. Mutational load can predict survival across diverse types of human cancers and is relevant in patients treated with either anti-CTLA4 or anti-PD1 therapies. Second, previous studies on the association between mutational load and survival after ICI had examined small cohorts and therefore, the effects of TMB on clinical benefit could not be quantified in a precise manner. This study presents genomic data from the largest cohort of patients treated with ICI to date and demonstrates the continuous association between higher TMB and superior OS. Capturing as little as 3% of the coding exome using targeted panels such as MSK IMPACT appears to provide a sufficient estimation of total tumor mutational load to confer predictive value for patients in whom ICI treatment is being considered.Finally, the mutational number defining TMB high appears to vary across cancer types, and there is unlikely to be a universal number that defines the likelihood of benefit from ICI across all histologies.
Given the potential toxicities of immunotherapy, the highly variable response to ICI, as well as the significant economic cost of these agents, there is an urgent need for biomarkers that can predict for immunotherapy response. Future studies that integrate other genomic or pathologic biomarkers may allow for the development of an even more optimized predictive test to inform clinical decisions on the use of ICI.
Online Methods
Patient Selection
After receiving institutional review board approval from the Memorial Sloan Kettering Cancer Center, institutional pharmacy records were used to identify patients who received at least one dose of immunotherapy (atezolizumab, avelumab, durvalumab, ipilimumab, nivolumab, pembrolizumab, or tremelimumab) and then cross-referenced with patients who had MSK-IMPACT testing done in the context of routine clinical care. Cancer types with greater than 35 patients on initial collection were selected for further analysis in the cohort. The majority of patients who received MSK-IMPACT testing on tumor tissue are enrolled on an institutional IRB-approved research protocol (NCT01775072) with the remaining patients receiving testing as part of routine clinical care; all patients provided informed consent permitting return of results from sequencing analyses and broader characterization of banked specimens for research.Details of tissue processing and next generation sequencing and analysis have been previously described. 11 Importantly, concurrent sequencing of germline DNA from peripheral blood is performed for all samples to identify somatic tumor mutations. Patients enrolled on ongoing clinical trials for which publication of outcomes data was prohibited were removed as well as a small proportion of patients with localized disease treated in the neoadjuvant setting(n=9) or who had localized disease. Other preceding or concurrent non-ICI treatments were not recorded or accounted for in the analysis. The timing of tissue pathology on which MSK-IMPACT was performed relative to ICI administration is also heterogenous with a small portion of patients with testing after ICI administration.
Mutational Load Assessment and Statistical Analysis
The total number of somatic mutations identified was normalized to the exonic coverage of the respective MSK-IMPACT panel in megabases. Mutations in driver oncogenes were not excluded from the analysis. Overall survival analysis on ICI patients was performed from the date of first infusion of any ICI. For patients who received multiple courses of ICI, the first treatment was used for analysis.Patients were censored at the date of last attended appointment at MSK if death was not recorded in the electronic medical record.
For analysis of patients who did not receive ICI, all patients for whom MSK-IMPACT data was available across all histologies were included. Overall survival analysis was performed from the date of first infusional chemotherapy.
Survival analysis was performed using Kaplan Meier with log-rank p values reported. Multivariable analysis was performed using Cox proportional hazard regression with inclusion of variables significant on univariate regression including normalized TMB, cancer type, age, ICI drug class , and year of ICI administration. Year of ICI administration was included in order to avoid any possible differences in patients treated in the early years of MSK-IMPACT testing being available.
For each histology, we subsequently identified cases in the top 20% percentile of TMB and determined the log-rank p-value for difference in OS and the direction of the effect with a HR determined from a coxph model. Additional analyses were performed with the TMB cutoff ranging from 10 to 50%, as well as with the TMB cutoff instead defined among all patients (both ICI-treated and non-ICI-treated).
Response data for individual histologies was obtained from published analyses of clinical outcome in the cohorts of patients with NSCLC or esophagogastric cancer patients.15,16. For patients with head and neck cancer, radiology records were reviewed manually to determine evidence of progression or tumor response. In these tumor types, clinical benefit was defined as any partial/complete response, or evidence of stable disease for ≥6 months. For renal cell carcinoma, time to next treatment was recorded manually for all patients.Statistical analysis was performed in R using the survival package. Graph-Pad Prism was used for basic analysis and generating graphs.Additional information can be found in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgements
We gratefully acknowledge the patients in this study, and their families. We thank Mithat Gonen for statistical advice. We thank members of the Molecular Diagnostics Service in the Department of Pathology and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We acknowledge funding sources including Pershing Square Sohn Cancer Research grant (TAC), the PaineWebber Chair (TAC), Stand Up 2 Cancer (TAC), NIH R01 CA205426, the STARR Cancer Consortium (TAC), NCI R35 CA232097 (TAC), Precision Immunotherapy Kidney Cancer Fund (TAC, RJM), The Frederick Adler Fund, Cycle for Survival, NIH K08 DE024774 and R01 DE027738 (LGTM), and MSKCC through NIH/NCI Cancer Center Support Grant (P30 CA008748).
Footnotes
Data Availability
All data is publicly available at http://www.cbioportal.org/study?id=tmb_mskcc_2018.
Competing Interests Statement
RMS, TAC and LGTM are inventors on a provisional patent application (62/569,053) filed by MSK, relating to the use of TMB in cancer immunotherapy.MDH, NAR and TAC are inventors on a PCT patent application (PCT/US2015/062208) filed by MSK, relating to the use of TMB in lung cancer immunotherapy.MSK and the inventors may receive a share of commercialization revenue from license agreements relating to these patent applications. CHL received research funding from Eisai, BMS, Exelixis, Pfizer, Calithera and consulting fees from Exelixis and Eisai. ANS has received research support from Bristol Myers Squibb, Immunocore, Astra-Zeneca, Xcovery and serves on the advisory board for Bristol Myers Squibb, Immunocore, Castle Biosciences; he also receives royalties from UpToDate. MDH receives research funding from Bristol-Myers Squibb; is paid consultant to Merck, Bristol-Myers Squibb, AztraZeneca, Genentech/Roche, Janssen, Nektar, Syndax, Mirati, and Shattuck Labs. YYJ received research funding from Boehringer Ingelheim, Bayer, Genentech/Roche, Bristol-Myers Squibb, Eli Lilly, Merck and served on advisory boards for Merck Serono, Bristol-Myers Squibb, Eli Lilly, Pfizer, Bayer, Imugene, Merck. SB currently works for Flatiron Health which is a for-profit company. RJM received research support from Pfizer, Genentech/Roche, Bristol Myers Squibb and Eisai and consulting fees from Pfizer, Genentech/Roche, Merck, Incyte, Novartis, Eisai and Exelixis. MHV received commercial research grants from Bristol-Myers Squibb andGenentech/Roche; honoraria from Novartis; travel/accommodation from Eisai, Novartis and Takeda; consultant/advisory board member for- Alexion Pharmaceuticals, Bayer, Calithera Biosciences, Corvus Pharmaceuticals, Exelixis, Eisai,GlaxoSmithKline, Natera; Novartis and Pfizer.
JR receives consulting fees from Merck, AstraZeneca, BMS, EMD-Serono, Roche/Genentech, Sanofi, Seattle Genetics, Agensys, Bayer, Inovio, Lilly, Adicet Bio, Sensei, Chugai, and Inovio. BHB receives consulting fees from Genentech. GJR received research funding from Novartis, Roche/Genentech, Millennium, GlaxoSmithKline, Pfizer, Infinity Pharmaceuticals, Takeda and received travel expense compensation from Merck. ALH receives research funding from Eisai, BMS, Kura Oncology, AstraZeneca, Genentech Roche, Celldex, Pfizer, Lilly, Bayer; consulting fees from BMS, Merck, Novartis, AstraZeneca, Regeneron, Sanofi Aventis, Sun Pharmaceuticals, Eisai, Genentech/Roche, Genzyme, Ayala Pharmaceuticals; and travel fees from Ignyta, and Kura Oncology. CAB receives research funding from Merck, Amgen, Bristol Myers Squibb. JDW was a consultant for Adaptive Biotech, Amgen, Apricity, Array BioPharma, Ascentage Pharma, Beigene, Bristol Myers Squibb, Celgene, Chugai, Elucida, Eli Lilly, F Star, Genentech, Imvaq, Kleo Pharma, MedImmune, Merck, Neon Therapuetics, Ono, Polaris Pharma, Polynoma, Psioxus, Puretech, Recepta, Trienza, Sellas Life Sciences, Serametri,; Surface Oncology, and Syndax; Research support from Bristol Myers Squibb, Medimmune, Merck Pharmaceutical, and Genentech; and Equity in Potenza Therapeutics, Tizona Pharmaceuticals, Adaptive Biotechnologies, Elucida, Imvaq, Beigene, and Trienza.JB is on the Board of Directors for Varian Medical Systems, Bristol-Myers Squibb and Foghorn, and is a past board member of Grail, Aura Biosciences and Infinity Pharmaceuticals.He has performed consulting and/or advisory work for Grail, PMV Pharma, ApoGen, Juno, Roche, Lilly, Novartis and Northern Biologics.He has stock or other ownership interests in PMV Pharma, Grail, Juno, Varian, Foghorn, Aura, Infinity, ApoGen, as well Tango and Venthera, for which is a co-founder.He has previously received Honoraria and/or Travel Expenses from Roche, Novartis, and Lilly.GYK received research funding and consulting fee from AstraZeneca, Bristol-Myers Squibb, and Merck. IKM reports research funding from GE and consulting/speaker fees from Agios Pharmaceuticals, Debiopharm Group, Roche, Merck, Puma Biotechnology, and Deciphera Pharmaceuticals. WDT reports personal fees from Eli Lilly, personal fees from EMD Serono, personal fees from Novartis, personal fees from Eisai, personal fees from Janssen, personal fees from Immune Design, personal fees from Adaptimmune, personal fees from Daiichi Sankyo, personal fees from Blueprint, personal fees from Loxo, personal fees from GlaxoSmithKline, personal fees from Agios Pharmaceuticals,from Plexxikon Pharmaceuticals,outside the submitted work;In addition, WDT has a patent Companion Diagnostic for CDK4 inhibitors - 14/854,329 pending to MSKCC/SKI, and a patent Methods of Treating Metastatic Sarcoma Using Talimogene Laherparepvec (T-Vec) and Pembrolizumab Combination Therapy- 62/671,625 pending to MSKCC/SKI and Scientific Advisory Board - Certis Oncology Solutions, Stock OwnershipÐcientific Advisory Board - Atropos Therapeutics, Stock Ownership. CMR has consulted on oncology drug development with AbbVie, Amgen, Ascentage, BMS, Celgene, Daiichi Sankyo, Genentech/Roche, Harpoon, Loxo, Pharmamar, and Seattle Genetics. AZ, DAB, RS, SMK, GI, DFB, RJW, PR, NL, PHG, TJK, LMD, PR, AAH, HIS, CEA, CAK, JEC, SD, RPD, NHS, ZKS, MMT,HAA,RY, LS have no conflicts of interest to report. VT is a co-founder and consultant for BluRock Therapeutics. NAR received consulting fees from Merck, AstraZeneca, Roche, Bristol-Myers Squibb, Novartis, Pfizer, Lilly, Abbvie, Merck KGaA, Regeneron, Janssen; is a cofounder and shareholder in Gritstone Oncology and serves on the advisory board or Neogenomics, OncoMed, and Bellcum.ML has received ad hoc advisory board compensation from AstraZeneca, Bristol-Myers Squibb, Takeda, and Bayer, and research support from LOXO Oncology and Helsinn Healthcare. MFB received research support from Illumina and consulting fees from Roche. DBS received funding from Pfizer, Loxo Oncology, Illumina, Intezyne, Vivideon Therapuetics.NR receives research support from Bristol-Myers Squibb and Pfizer and speakers fees from Illumina. TAC is a co-founder of Gritstone Oncology and holds equity.TAC holds equity in An2H.TAC acknowledges grant funding from Bristol-Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H, and Eisai. TAC has served as an advisor for Bristol-Myers Squibb, Illumina, Eisai, and An2H. LGTM received consulting fees from Rakuten Aspyrian and speaker fees from Physician Educational Resources.
References
- 1.Callahan MK, Postow MA & Wolchok JD Targeting T Cell Co-receptors for Cancer Therapy. Immunity 44, 1069–1078 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Powles T et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet 391, 748–757 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Carbone DP et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 376, 2415–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen EE et al. LBA45_PRPembrolizumab (pembro) vs standard of care (SOC) for recurrent or metastatic head and neck squamous cell carcinoma (R/M HNSCC): Phase 3 KEYNOTE-040 trial. Annals of Oncology 28, mdx440.040 (2017). [Google Scholar]
- 5.Bendell J et al. LBA-004Efficacy and safety results from IMblaze370, a randomised Phase III study comparing atezolizumab+cobimetinib and atezolizumab monotherapy vs regorafenib in chemotherapy-refractory metastatic colorectal cancer. Annals of Oncology 29, mdy208.003 (2018). [Google Scholar]
- 6.Gibney GT, Weiner LM & Atkins MB Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 17, e542–e551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Allen EM et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg JE et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugo W et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Jordan EJ et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 7, 596–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janjigian YY et al. Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer. Cancer Discov (2017). doi: 10.1158/2159-8290.CD-17-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizvi H et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J. Clin. Oncol 36, 633–641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouffet E et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J. Clin. Oncol 34, 2206–2211 (2016). [DOI] [PubMed] [Google Scholar]
- 19.McGranahan N et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frampton GM et al. Assessment of tumor mutation burden from >60,000 clinical cancer patients using comprehensive genomic profiling in 34, (2016). [Google Scholar]
- 21.Johnson DB et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 4, 959–967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalmers ZR et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roszik J et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med 14, 168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowell D et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 62, eaao4572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balachandran VP et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal NH et al. Epitope landscape in breast and colorectal cancer. Cancer Res 68, 889–892 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Verdegaal EME et al. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 536, 91–95 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Le DT et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


