Abstract
The copper-transporting ATPase ATP7B has an essential role in human physiology, particularly for the liver and brain function. Inactivation of ATP7B is associated with a severe hepatoneurologic disorder, Wilson disease (WD). Hundreds of WD related mutations have been identified in ATP7B to date. The low frequency and the compound-heterozygous nature of causative mutations complicate the analysis of individual mutants and the establishment of genotype–phenotype correlations. To facilitate studies of disease-causing mutations and mechanistic understanding of WD, we have homology-modelled the ATP7B core (residues 643–1377) using the recent structure of the bacterial copper-ATPase LCopA as a template. The model, supported by evolutionary conservation and hydrophobicity analysis, as well as existing and new mutagenesis data, allows molecular interpretations of experimentally characterized clinical mutations. We also illustrate that structure and conservation can be used to grade potential deleterious effects for many WD mutations, which were clinically detected but have not yet been experimentally characterized. Finally, we compare the structural features of ATP7B and LCopA and discuss specific features of the eukaryotic copper pump.
Introduction
The copper transporting ATPases ATP7A and ATP7B play an essential role in normal human physiology.1 By facilitating copper export from cells, these transporters mediate the transfer of dietary copper from the intestine into the blood (ATP7A) and the removal of excess copper from the body via the liver (ATP7B). In addition, within a cell ATP7A and ATP7B transport copper from the cytosol into the lumen of a secretory pathway, where copper is incorporated into various copper-dependent enzymes. Participation in cuproenzymes biosynthesis makes the copper-ATPases essential for the production of neuroendocrine peptides and neurotransmitters, iron balance, vascular tissue formation, pigmentation, protection against radicals during bacterial infection, and many other processes.2–4
Inactivation of either ATP7A or ATP7B is associated with severe multisystem disorders (Menkes disease and Wilson disease, respectively5,6). These diseases have an onset in a childhood and result in a significant neurological impairment and death (Menkes disease6) or require life-long treatment, sometimes involving liver transplantation (Wilson disease, WD5).
In patients, hundreds of sequence variants have been identified for both copper-ATPases; those detected in ATP7B are collected in the WD Mutation Database.7 Most substitutions show low frequency7 and to date only approximately 10% of detected mutations have been experimentally characterized in some detail.8–17 These studies revealed a wide spectrum of consequences on the function, stability and trafficking of copper ATPases. These observations emphasized the importance of detailed analysis of ATP7A and ATP7B variants for better mechanistic understanding of the diseases.
In the last 10 years, significant progress has been made in the analysis of biochemical properties and regulation of Cu-ATPases.18,19 Studies of thermophilic Cu-ATPases from A. fulgidus, T. maritima, and human ATP7A/ATP7B have been particularly informative. It is now well established that these ATPases hydrolyze ATP to transport Cu+ with the formation of a transient acyl-phosphate intermediate.20–23 Analysis of CopA from A. fulgidus suggested that two copper atoms are transported per ATP hydrolyzed;24 similarly to other P-type ATPases, Cu-ATPases were shown to undergo significant ligand dependent conformational transitions.25
The sequence of human Cu-ATPases is divided almost equally between the N-terminal regulatory domain, which differs in the Cu-ATPases from different species, and the C-terminal core structure, shared between all Cu-ATPases. The regulatory N-terminal domain of human ATP7A and ATP7B is composed of 6 metal binding sub-domains (MBDs). Each MBD is folded into a ferredoxin-like structure, as determined by NMR, and contains one copper-binding motif CxxC.26–29 The C-terminal half includes eight transmembrane (TM) helices, as well as the highly conserved A-, N- and P-domains. For the C-terminal portion of human ATP7A/7B, the structural NMR data are available only for the N- and A-domains.13,30
Recently, the first crystallographic structure of the Cu(I)-ATPase LCopA from Legionella pneumophila was solved in the apo-form.31 This pioneering work revealed the organization of a distinct TM domain of P1B-ATPases and pointed to the structural elements that could be important for copper delivery to the intra-membrane sites, as well as copper exit.31 The structure has also provided a platform for modeling the core region of human Cu-ATPases and formulating further hypotheses about their mechanism and specific effects of disease-causing mutations on protein structure and function.
Herein, we present a homology-model of ATP7B, generated using the LCopA structure as a template. The model-structure, along with evolutionary conservation analysis, provides a framework for studies of human mutations linked to WD.
Methods
Model construction
The fold recognition servers FFAS03,32 HHPRED33 and PUDGE34 identified the bacterial copper transporter LCopA (PDB ID 3RFU) as the best structural template with statistically significant scores. The pairwise alignment of ATP7B and LCopA was essentially the same in all fold recognition methods, assigning the same regions to the secondary structure elements while slightly differing in the loops. These alignments similarly corresponded to the alignment suggested by Gourdon et al.31 The HHPRED pairwise alignment, consisting of a sequence identity of 36%, was then employed to model ATP7B based on LCopA, covering positions 643–1377 (Fig. S1, ESI†). The PDB file of the model is provided as part of the ESI.†
Conservation analysis and functional patch detection
For both ATP7B and LCopA, collections of 477 and 493 homologues, respectively, were retrieved from a BLAST35 search against the UniRef 90 database,36 with an E-value of 0.0001, and aligned using MAFFT.37 These multiple sequence 7alignments (MSAs) were utilized via the ConSurf webserver (http://consurf.tau.ac.il/38) to compute evolutionary conservation scores for LCopA and ATP7B, mapped on the crystal and model-structure, respectively. Based on the conservation scores, we computed the ConQuass probability scores, reflecting the correspondence of a structure to its conservation profile based on the notion that conserved residues tend to be buried in the protein core, while variable positions exposed to the solvent or membrane.39 The conservation scores were also used to detect the functional site using PatchFinder,40 detecting continuous surface patches of highly conserved residues, corresponding to potential functionally-important sites.
Analysis of clinically relevant mutations
The Wilson Disease Mutation Database describes clinically relevant missense mutations in 228 positions of ATP7B; 197 of the corresponding residues are included in the model-structure.7 We computed the solvent accessibility surface area (SASA) with Naccess using a probe sphere with a radius of 1.4 Å.41 We obtained relative values using comparisons to A–X–A tripeptides, where A represents alanine and X represents any amino acid. The threshold determining whether a position was “exposed” or “buried” was set to 10%. Examining the positions in which mutations were detected, we suggested that evolutionarily conserved positions (grades 8 or 9) that are either buried, or included in the functional patch predicted by PatchFinder, are likely to be highly sensitive-to-mutation. In other words, mutations in these positions are expected to be highly deleterious for protein function. On the other hand, we suggest that mutations in exposed positions that were assigned low conservation scores (grades of 1 and 2) are less likely to have a strong negative effect on the protein’s function and/or structure.
Intracellular copper transport assay
YS fibroblast cells were grown in DMEM with 10% FBS (Fetal Bovine Serum vol/vol), plated on glass coverslips and co-transfected with 2.5 μg of plasmids expressing tyrosinase and either the GFP-tagged wild-type ATP7B or GFP-tagged ATP7B bearing respective mutations. Twelve hours after transfection cells were washed in PBS, fixed in acetone : methanol (1 : 1 v/v), and incubated for 4 h with 0.1 M Na-phosphate (pH 6.8) containing 0.15% (wt/vol) levo-3,4-dihydroxy-L-phenylalanine (L -DOPA) at room temperature. Coverslips were mounted on slides, and formation of the black L-DOPA chrome pigment was detected by phase microscopy. Pigment intensity was quantitated using ImageJ, and the range of values for wild type and each mutant represented graphically.
Results
Overview of the ATP7B model
The model-structure encompasses the C-terminal segment of human ATP7B, consisting of residues 643–1377 of the complete sequence of 1465 amino acid residues. The model presents the core structure shared with LCopA; it includes a TM domain formed by the eight TM helices and a cytosolic portion that represents an ensemble of structures for the N-, P-, and A-domains (Fig. S1, ESI†). The modeled N- and A-domains are very similar to the NMR structures of the apo N-domain and the A-domain of human Cu-ATPases30,42 (PDB IDs 2KMV and 2KIJ, respectively); Fig. S2 (ESI†); minor deviations of the model from the NMR structures may reflect imprecision of the model and/or the fact that the NMR studies were conducted for isolated domains, separated from the rest of the protein. The N-terminal portion of ATP7B was not modeled. Although structures of the individual MBDs in the N-terminal portion of ATP7B have been determined by NMR,26–29 the insufficient number of spatial constrains precludes accurate prediction of packing for the entire N-terminal domain or its interaction with the C-terminal part.
The main difference between the structure of the ATP7B core (represented by the model) and LCopA is the presence of several sequence inserts, which in the model are shown as loops. The luminal TM1,2 loop in ATP7B (Y670–L692) is Ser-rich and hydrophilic. In the structure, the TM1,2 loop is situated in immediate vicinity to the predicted copper release site, where it may regulate the rate of copper release from the transport sites (see below). Another lumenal insert forming the TM5,6 loop (G943–I967) has a strictly conserved length in mammalian pumps. Mutations G943S, V949G, K952R and I967F within this segment are associated with Wilson disease in humans, pointing to its importance. Since in LCopA, the TM5,6 loop is much shorter than in ATP7B, the functional defects caused by mutations in this region of ATP7B likely reflect the role of the TM5,6 loop that is unique for mammalian transporters, such as copper delivery to acceptor proteins or sorting between different intracellular compartments.
The C1104–S1145 segment in the N-domain of ATP7B is exposed into the cytosol. The length of this insert greatly exceeds the corresponding segment of LCopA (G497–H503). This insert is not important for the folding of the N-domain or its ability to bind nucleotides.14 However, mutations C1104F/Y, V1106I/D, V1109M, G1111D, A1140V, Q1142H within this region were reported in Wilson disease patients pointing to a potential role of this insert in ATP7B stability, targeting, or response to regulatory signals in cells.
Conservation and hydrophobicity profiles
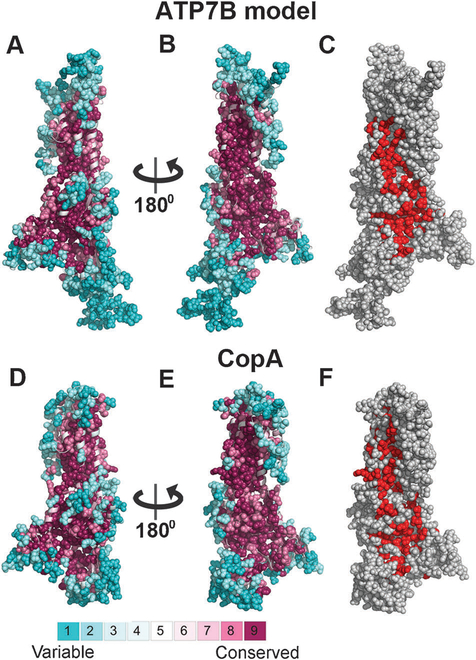
The ATP7B model displays a highly conserved protein core, whereas the most solvent and lipid exposed positions, as well as residues in loops, are variable (Fig. 1A and B). As expected, the model’s conservation profile is similar to that of the template (Fig. 1D and E), and its ConQuass probability score (0.166, Fig. S3A, ESI†) is in the range observed for proteins of known structure of the same size, including the template (0.146, Fig. S3B, ESI†). Notably, both the ATP7B model and the LCopA structure include a large surface patch of highly conserved residues, stretching from the membrane domain to the cytoplasmic region. Many of these residues are included in the functional patch detected by PatchFinder40 for each of the structures (Fig. 1B, C, E and F).
Fig. 1. Conservation analysis of ATP7B and LCopA.
Panels A–B and D–E show side-views of the LCopA structure and ATP7B model in cartoon representation, with the cytoplasm below. The structures are colored according to the ConSurf38 color bar (http://consurf.tau.ac.il), with cyan-to-maroon indicating variable-to-conserved. Positions assigned with the highest or lowest conservation grades (8–9 or 1–2, respectively) are displayed as full atoms spheres, demonstrating the high conservation of the core as opposed to the variability of the peripheral and loop regions. In panels C and F, the LCopA structure and ATP7B model are shown as in panels B and E, respectively, and colored gray, with the functional patches detected by PatchFinder40 in red. Overall, the conservation patterns of the LCopA and ATP7B are remarkably similar, including the location of a highly conserved patch at the proteins’ surface.
The hydrophobicity analysis of the ATP7B model illustrates that most of the charged and strongly polar residues in ATP7B are situated in its extra-membrane regions, as expected for a membrane protein (Fig. S4, ESI†). Nevertheless, the membrane domain contains a significant number of highly conserved, charged and strongly polar residues that are buried in the core, forming the presumed copper translocation pathway, as discussed below. A distinct surface patch of conserved and charged positions is present at the suggested copper entry site31 (Fig. S5, ESI†). Neighboring to this area is a well-defined positively charged pocket (Fig. S6, ESI†), which may serve as an interacting site with the regulatory metal-binding domains, negatively charged C-terminal tail or phospholipids.
Analysis of clinical missense mutations
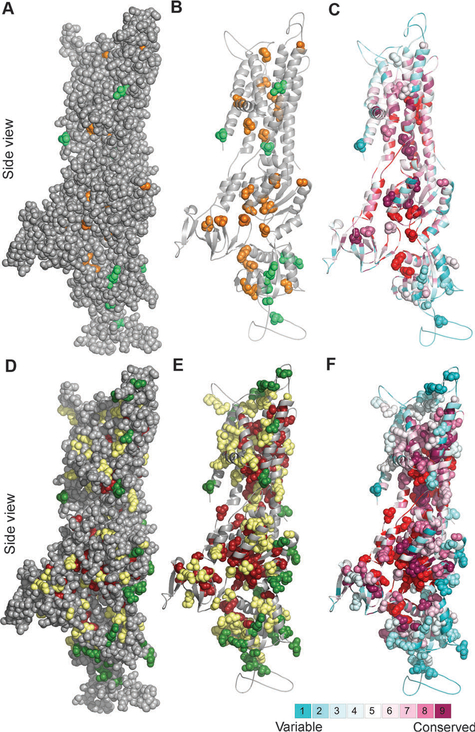
The WD database lists mutations that are detected in patients and therefore are potentially deleterious. However, many WD causing mutations are compound heterozygous, and it is mostly unknown whether each of the contributing mutations has the same consequences. Mapping clinical mutations that have been experimentally characterized (Table S1, ESI†) demonstrates that deleterious mutations, which have a marked negative effect on either protein expression or function (henceforth “positions sensitive to mutation”), tend to map to sites that are buried in the protein core, or belong to the PatchFinder functional patch. In contrast, the least sensitive positions, in which substitutions had no or partial effects on protein stability and/or copper transport rate, occupy exposed regions (Fig. 2A–C), which supports the validity of the model-structure. We thus hypothesized that the structural model and the conservation analysis can be utilized to grade potential sensitivity to mutation of all positions in which missense clinical mutations were detected (Table S2, ESI†). To evaluate this concept, we first graded the sensitivity to mutations of 34 positions in which clinical mutations were detected and characterized experimentally by us or others, employing the relative accessibility of each position and its ConSurf score, as specified in the Methods section. Our analysis indicated that 15 positions were likely to be sensitive to mutation, whereas five positions were probably less sensitive (Table S1, ESI†). The sensitivity of the remaining 14 positions, which did not fall into one of our two categories, was not suggested (Table S1, ESI†). Significantly, all positions that were suggested by our analysis as “sensitive” matched the effect of mutation observed in experiment. We also correctly assessed the effect of mutation in four out of five positions denoted as “less sensitive”. The suggested effect of one position, Pro1052, contradicted the experimental data that showed loss of copper transport activity for the P1052L mutant.43 As this position, situated in the P-domain, is variable (ConSurf score of 2) and exposed (SASA > 59%), we could not resolve this discrepancy. We did observe, however, that this proline is situated at the beginning of a helical segment, and could be important for breaking the secondary structure into a loop.
Fig. 2. Clinically-detected mutations in light of structure and conservation.
Side view of the ATP7B model with the cytoplasm below. (Panels A–C) Location and conservation of clinical missense mutations,7 for which experimental data are available (Table S1, ESI†) (A) Positions sensitive and non-sensitive to mutation are colored orange and green, respectively. The model (in gray) is shown as spheres, demonstrating which of the experimentally assessed positions are buried in the protein core. (B) Same as in panel A, with only the positions sensitive and non-sensitive to mutation in experiment shown as spheres. (C) As in panel B, with the structure colored by ConSurf analysis (Fig. 1A), and the PatchFinder patch (Fig. 1C) colored red. Most sensitive-to-mutation positions map to conserved residues, either buried or part of the functional site, whereas the less sensitive positions tend to be less conserved and more accessible. (Panels D–F) Location and conservation of all clinical missense mutations7 (D) Green and dark red coloring indicate positions suggested by our analysis to be sensitive and less sensitive, respectively, positions for which the effect was not proposed are shown in yellow. (E) Same as panel D, showing only the clinical missense mutations as spheres. (F) Same as panel E, with the coloring scheme corresponding to the ConSurf and PatchFinder analysis.
Since our structure- and conservation-based criteria agreed well with the experimental data (Table S1, ESI†), we next predicted potential effects of mutations for all 197 positions in the model for which missense mutations were detected, but specific consequences remain mostly uncharacterized. According to this analysis, 81 positions were potentially sensitive to mutation, whereas 30 positions were suggested to be “less-sensitive” to substitution (Table S2, ESI†, Fig. 2D–F). We could not suggest the sensitivity of the remaining 86 positions, which resided outside the region covered by the stringent conservation and surface accessibility cutoffs that we used.
Mechanistic interpretation of available experimental data
The above analysis yielded a large-scale overview of the potential severity of clinical mutations. In order to provide better understanding of the molecular characteristics of individual substitutions, we analyzed in more detail several WD mutations, focusing on positions in the membrane domain for which structural data were not available before this study (Fig. 3). Additional mutants are discussed in the ESI.†
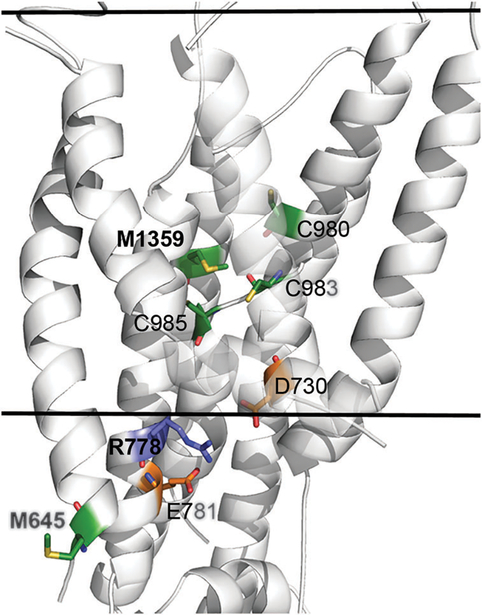
Fig. 3. Structural location and molecular interactions of M645, R778 and M1359.
The model-structure of ATP7B is shown as white cartoon, and viewed from the side with the cytoplasm below. The view is focused on the membrane domain, and the estimated membrane boundaries, calculated by the PPM webserver62 (https://opm.phar.umich.edu/ppm_server.php), are marked by the two solid lines. M645, R778, M1359 and interacting residues of interest are shown as sticks, with Met and Cys in green, Arg in blue and Asp and Glu in orange. Nitrogen, Oxygen and Sulfur atoms of these residues are colored blue, red and yellow, respectively. For clarity, the C-termini of TM2 and TM3 are partly transparent. The variable position M645 is situated at the beginning of TM1, facing outwards in contrast to highly conserved M1359 that is situated at the protein core, facing several other potential highly conserved copper ligands (sticks). The conserved R778 is situated at the region corresponding to the suggested copper entry site of LCopA, in close proximity to highly conserved acidic residues, potentially forming stabilizing interactions.
R778L.
The allele frequency of R778L is 37.9% in Korean patients with WD,44,45 27% in Japanese patients46–49 and 28–44% in Chinese patients.50,51 This mutant has a lower stability9 and an abnormal intracellular localization,10 that can be partially corrected by chemical chaperones. Diminished transport activity of this mutant was suggested based on a whole cell copper export assay.9,10 In fact, even “milder” substitutions to Gly or Gln in this position are associated with the disease phenotype.7 In the ATP7B model, Arg778 is located at the cytoplasm-exposed part of TM4 in the vicinity of the conserved residues Asp730 and Glu781 (Fig. 3). In human cells, replacement of the positively charged Arg with the hydrophobic Leu is associated with the retention of ATP7B in the endoplasmic reticulum,10 indicative of at least partial misfolding. Thus, a charge balance in this region (or a potential ion pairing between Arg778 and Asp730) appears to play an important role in maintaining the ATP7B folding and/or restricting protein motions. This structural feature is characteristic of mammalian transporters in which the charges are highly conserved, whereas in LCopA, the residues equivalent to D730 and R778 are substituted with F149 and Q202, respectively.
M1356V in ATP7B from mice.
Met1356V substitution in mouse ATP7B causes copper accumulation and liver disease.52 Studies of the mutant protein in vitro revealed that the ATP7B-M1356V variant was expressed normally, but lacked a measurable transport activity in a direct copper uptake assay.53 The position of M1356 in mice is equivalent to the human M1359 (Fig. 3). M1359 is a highly conserved residue that is thought to contribute to the intramembrane metal-binding site.31 Given that the intra-membrane copper binding sites in Cu-ATPases are predicted to involve three coordinating ligands,24 substitution of this potential copper ligand is expected to decrease the affnity of ATP7B for copper. In the Cu-ATPase CopA from A. fulgidus, replacement of the equivalent methionine resulted in the loss of one copper binding site and the inability of copper to stimulate catalytic phosphorylation.54 Both these efects would account for the deleterious efect of the mutation on copper transport activity.
M645R.
This substitution, identified in patients from several populations, including Jewish, Spanish, and Italian, was reported by several laboratories.24,55 The M645R variant, when expressed in insect cells using baculovirus system, had significant transport activity that was only 30% lower compared to the wild-type ATP7B (as measured by radioactive copper uptake into vesicles56). Examination of the sequence alignment of ATP7B homologues, constructed for conservation analysis, revealed that M645 is a highly variable position. In the ATP7B model, this residue is located at the cytoplasm-exposed region of TM1, facing the membrane, where it is not expected to have a critical structural or functional role. Furthermore, several ATP7B homologues contain an arginine in the same position, indicating that arginine could be well accommodated in the position 645 (Fig. 3).
These results along with the functional data suggested that the M645R mutation might either be associated with a mild disease phenotype or would require the presence of a more severe mutation in the second allele of ATP7B in order to be deleterious. To verify this prediction, we examined the available clinical data. M645R is a highly prevalent mutation in the Spanish population, where the genotype–phenotype correlations have been investigated.57 In all WD patients with the M645R replacement, this mutation was present only in one allele, supporting our prediction that in a homozygous state this mutation was not deleterious. Furthermore, mutations detected in the second allele involved codon termination (i.e. strong deleterious effect), or the G869R, T977M, H1069Q, V1216M, T1232P mutations. Three of these positions are “sensitive” positions based on our analysis (Table S2, ESI†), i.e. likely to have a significant deleterious effect. For position 869, we could not strongly define sensitivity, as Gly869 was assigned a ConSurf grade of 7 and is solvent accessible. Thus, it is particularly interesting that the combination of M645R with G869R was detected in an individual with an unusually late onset of the disease (at 50 years) reflecting a mild WD phenotype. For comparison, the combination of M645R with a clearly deleterious stop codon in position Q111 in the second allele was associated with disease onset at the age of 7 years.57
Experimental characterization and molecular interpretation of other clinical mutations
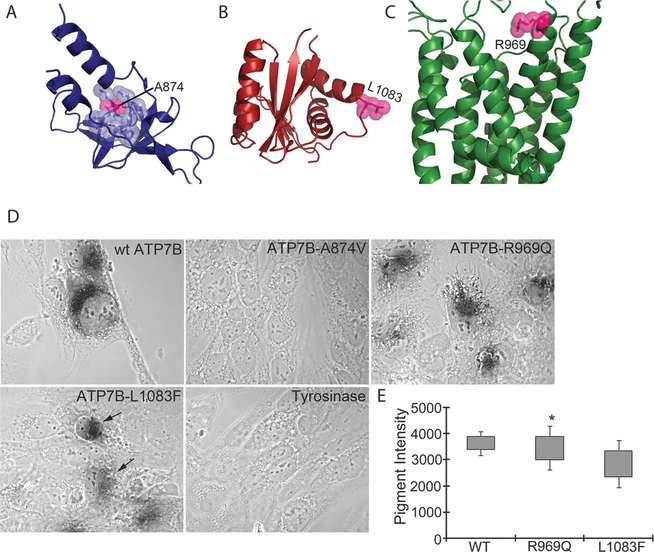
We selected three additional mutations from the WD database, situated in different domains of ATP7B: A874V (A-domain), R969Q (the N-terminal end of TM6 facing the Golgi lumen) and L1083F (N-domain) (Fig. 4A–C, respectively). We could not predict the outcome of substitutions in these positions by our computational analysis, because their evolutionary conservation and degree of exposure were outside the strictly defined categories (Table S1, ESI†). Expression of these variants at lower temperature (in insect cells or in yeast) produces proteins with partial activity.43 To characterize the copper-transport properties of these mutants in mammalian cells at physiologically relevant 37 °C, we examined their ability to deliver copper to a copper-dependent enzyme tyrosinase expressed in fibroblasts derived from a Menkes disease patient. These fibroblasts lack activity of the endogenous Cu-ATPase and the copper-dependent enzymes expressed in these cells (such as tyrosinase) are inactive due to the lack of copper transfer to the TGN. Expression of active ATP7B restores copper transport and activates tyrosinase. This reaction can be visualized by the development of a characteristic black pigment as seen with wt-ATP7B (Fig. 4D). It should be noted that the tyrosinase assay is highly sensitive, and the lack of color usually indicates complete loss of transport activity, whereas decrease in color formation points to a considerable transport impairment.
Fig. 4. Structural location and copper transport activity of the ATP7B mutants.
(A) The A-domain model shown as blue cartoons. A874 is shown as pink atom-spheres, and surrounding residues in blue spheres. (B) The P-domain model in red, with L1083 shown as pink spheres. (C) The membrane domain in green, with R969 as pink spheres. (D) Development of black pigment by tyrosinase is indicative of copper transport by ATP7B. ATP7B-A874V shows no copper transport activity. ATP7B-R969Q shows activity/intensity of color similar to wtATP7B. ATP7B-L1083F shows variable color intensity as indicated by arrows. Cells transfected with tyrosinase plasmids will not develop color and serve as a negative control for the assay. (E). Quantitation of average pigment intensity using ImageJ.
The R969Q-ATP7B variant showed significant activity indicated by intense color and a number of positive cells similar to the wtATP7B. Expression of the A874V-ATP7B variant produced no cells with black pigment. The L1083F-ATP7B mutant showed a range of color intensities with an average that was lower compared to that of the wild-type (Fig. 4D and E).
The results for all mutants were consistent with their structural location of mutated residues and their level of conservation (Fig. 4A–C, Table S1, ESI†). Ala874 is highly conserved (ConSurf grade of 8) and is buried in the core of the A-domain (Fig. 4A). The larger valine might not be properly accommodated in this position, reducing protein stability. In contrast, R969 and L1083 are not conserved (with grades of 4 and 5, respectively) and are situated in the exposed locations. Our prediction that the substitutions in these positions are unlikely to greatly disrupt ATP7B structure and/or function is supported by the ability of these mutants to transport copper for tyrosinase activation (Fig. 4B and C). Overall, the structural location and conservation levels were indeed useful in predicting the functional consequences of these mutations.
Copper entry, binding, and exit sites
The ATP7B model shows that highly conserved amino acids, typically associated with coordination of Cu+ (such as Met, Cys and His), are clustered in three distinct sites, previously described for LCopA as copper entry, binding and exit sites (Fig. 5). When comparing these sites in LCopA and ATP7B, we found that some contributing residues were invariant, whereas other residues differed significantly (Table 1). Clearly, highly conserved amino acids are likely to be associated with common mechanistic steps, whereas dissimilar residues may reflect properties unique for transport by ATP7B, as well as other eukaryotic copper transporters (for example, pH dependencies or regulatory interactions with cytosolic domains or other molecules) (Fig. 5).
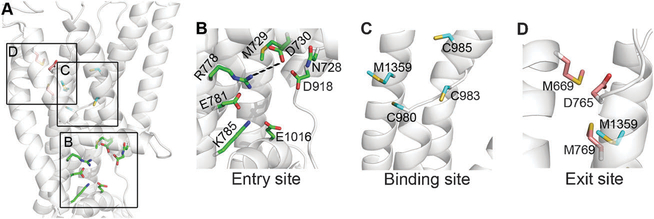
Fig. 5. Potential functional sites in the membrane region.
The ATP7B model is shown as transparent white cartoons, and viewed from the side with the inward side facing below. (A) Focus on the membrane domain, with residues of the functional site (Table 1) shown as colored sticks. The three sites, i.e. entry, intra-membrane binding and exit sites, are marked by squares. (B) The entry site, with suggested functional residues as green sticks. R778 and D730 could be salt-bridged. (C) The binding site, with TM3 removed for clarity. The suggested binding site residues (colored cyan) are C983, C985 and Met1359 with C980 playing an assisting role. (D) Exit site residues in pink, along with Met1359. TM2 and TM3 were extracted for clarity.
Table 1. Potential functional residues in the ATP7B membrane domain (Fig. 5).
Residues in ATP7B, location in model, conservation level according to ConSurf grades (Fig. 1A and B), corresponding LCopA residues and attributed site. Detected clinical mutations, and available experimental evidence are specified as well. Green shades mark potential functional positions that are unique to ATP7B, matched to LCopA positions that are unlikely to possess a similar functional role owing to their different physicochemical nature (shaded in red)
| Experimental evidence | |||||||
|---|---|---|---|---|---|---|---|
| N728 | TM3 | 9 | N147 | 9 | Entry | N728D | - |
| M729 | TM3 | 9 | M148 | 9 | Entry | M729V | - |
| D730 | TM3 | 9 | F149 | 9 | Entry | - | - |
| R778 | TM4 | 8 | Q202 | 9 | Entry | R778G/W/L | Lower stability and abnormal localization9 |
| E781 | TM4 | 9 | E205 | 9 | Entry | - | - |
| K785 | TM4 | 8 | R209 | 8 | Entry | - | - |
| D918 | TM5 | 9 | D337 | 9 | Entry | D918N/A/E | - |
| E1016 | After TM6 | 9 | E415 | 9 | Entry | - | - |
| C980 | TM6 | 9 | 1379 | 9 | Site | C980Y | - |
| C983 | TM6 | 9 | C382 | 9 | Site | - | - |
| C985 | TM6 | 9 | C384 | 9 | Site | C985Y | - |
| Y1331 | TM7 | 9 | Y668 | 9 | Site | Y1331S | - |
| N1332 | TM7 | 9 | N669 | 9 | Site | N1332D/K | - |
| M1359 | TM8 | 9 | M717 | 9 | Site | M1359I | Mice homologue M1356V: copper accumulation and liver disease in mice (toxic milk phenotype),52 no copper transport activity in a copper uptake assay53 |
| S1363 | TM8 | 9 | S721 | 9 | Site | S1363F | Affects folding9,12,17 |
| M668 | TM1 | 7 | E99 | 7 | Exit | - | - |
| M769 | TM4 | 7 | V193 | 7 | Exit | C703Y | Partial transport activity (about 50%), no effect on expression43 |
| D765 | TM4 | 8 | E189 | 9 | Exit | D765N/G | Partial transport activity (about 50%), decreased expression43 |
How copper enters the translocation pathway of copper pumps is presently unknown. In LCopA, the region containing conserved M148, E205, and D337 was suggested as an entry site for copper.31 In ATP7B, the corresponding region has very similar physicochemical properties (the key residues are M729, E781, and D918) (Fig. 5B and Table 1). High evolutionary conservation supports the important role of this site in the basic mechanics of copper pumps. Notably, in ATP7B the region includes four positions in which clinical mutations have been detected, i.e. N728, M729, R778 and D918. Mutation in R778 impairs function,9,10 and the three other positions were denoted by our analysis as sensitive to mutation (Table S2, ESI†).
The proposed role of M729, E781, and D918 as an intermediate copper transfer site is consistent with the location of this triad in the vicinity of the intra-membrane copper-coordinating residues. However, the triad itself is not a favourable site for Cu(I) binding and by itself may not be sufficient to attract copper. The facilitating effect of thiol containing reagents, such as glutathione and/or cysteine on metal-transport rate has been reported for all P1B-ATPases studied so far. Binding of such a cofactor at the entry site may provide additional necessary ligands for guiding copper to the transport sites.
The location of the Cu(I) binding sites in the membrane domain was suggested based on sequence conservation and available functional data for A. fulgidus CopA.24 In ATP7B, several residues at the center of the membrane domain may contribute to copper ligation. Namely, C980, C983, C985 and M1359 (Fig. 5C and Table 1), with C983 and C985 comprising the CPC motif. Of these, only C980 is not conserved (in LCopA, the equivalent position is occupied by a hydrophobic I379 (Fig. 5C). Other residues, identified as functionally important in the bacterial pumps match the identical residues in ATP7B, i.e. Tyr1331, Asn1332 (the invariable YN motif) and Ser1363 (Table 1). Clinical mutations were detected in all positions except C983, and all were suggested by our analysis as deleterious (Table S2, ESI†). The functional significance was experimentally confirmed for the mouse equivalent of M135953 and for the S1363F mutant9,12,17 (Table S1, ESI†).
Finally, we detected significant differences between the putative copper exit site of LCopA and ATP7B. The exit site in LCopA is formed by E99, M100, M711 and E189,31 of which only E189 has a similar, but not identical, residue in the equivalent position of ATP7B, namely D765 (Table 1). The ATP7B model shows that the moderately conserved M665, M669 and M769 are situated in vicinity of Asp765 (Fig. 5D). Of the positions potentially contributing to the ATP7B exit site, clinical mutations were detected in M665, M769 and D765, with the latter two displaying reduced copper transport in experiment (Table 1).43 Comparison of eukaryotic pumps to LCopA revealed another interesting difference in the length and composition of the TM1,2 membrane segment. As described above, the loop connecting TM1 and TM2 is longer in all eukaryotic transporters and has additional Met and His residues (Fig. S7, ESI†, Table 1). In human ATP7A, the recombinant TM1,2 loop binds and stabilizes Cu(I) and mutations within the TM1,2 loop decrease the rate of ATP7A de-phosphorylation (the catalytic step coupled to a copper release into the lumen).58 Thus, in human pumps, the TM1,2 loop may regulate copper release either by attracting copper that exits the transport site and/or by acting as a gate. The availability of ATP7B model provides an opportunity to directly test various mechanistic scenarios for copper release and directly identify residues participating in this process.
Discussion
The human Cu-ATPase ATP7B is of considerable scientific interest, which is further enhanced by its association with Wilson disease. Current lack of structural information, especially on the unique membrane domain hinders characterization of the disease-causing factors and a transport mechanism. In this study, we have developed and validated a structural model for ATP7B, which would aid such investigations. Based on structural data combined with a conservation analysis, we provide a large-scale assessment of clinical mutations included in the WD database, along with a molecular-level interpretation of previous and new experimental and clinical data. We illustrate that integration of computational, structural and experimental data provide new insights into the copper transport mechanism and the molecular basis of WD.
The accuracy of homology models depends on the compatibility of the template structure, and the sequence similarity between the query and template.59 ATP7B has ~ 35% sequence identity to bacterial LCopA, which was used as a template in our studies. Forrest and co-workers showed that with sequence identity above 30%, membrane protein models are expected to deviate at most by 2 Å from the native structure.59 Thus, the model herein should present a reliable structural architecture for ATP7B, appropriate for considering functional implications.
Clinical missense mutations: overview versus detailed analysis
The majority of the 197 clinical mutations included in the ATP7B model and linked to WD7 have not yet been experimentally validated. Our analysis of conservation and location in the structure suggests which positions are likely to be sensitive to mutation, and which positions are probably less affected by a substitution (Fig. 2D and E, Table S2, ESI†). Significantly, when applying this approach to 34 positions for which mutations were examined experimentally, our prediction matched the experimental outcome in 19 cases and only one prediction was in conflict with the experimental data (Table S1, ESI†). We could not firmly propose the sensitivity of the remaining 14 positions due to the stringency of our criteria. These results validate the use of such simple and conservative criteria. Encouraged by the performance of our approach on the evaluated clinical mutations, we suggest that similar analysis can suggest the outcome of other clinically-detected substitutions in ATP7B (Table S2, ESI†), and thus guide the selection of potentially interesting positions and mutations for future experimental studies.
The criteria selected herein for grading the sensitivity to mutation are relatively stringent. Thus, we could suggest the effect of mutation only for a sub-set of residues in the model-structure (Fig. 2D and E, Table S2, ESI†). This conservative criterion was set to reduce errors as much as possible. Nevertheless, the model-structure helps to gain new insights into specific substitutions even for positions that do not conform to the conservative definitions set for the large-scale analysis (Fig. 3 and 4). For instance, R778, sensitive to mutation in experiment,9,10 was not identified as such in our large-scale analysis of mutations (Table S1, ESI†), as this conserved position has surface-accessibility exceeding the selected cutoff. Closer investigation, however, revealed that this residue is positioned at the potential copper entry site, and suggested that a salt bridge could be formed between R778 and D730, providing a potential molecular-level explanation for the effect of the mutation (Fig. 3). Three mutations experimentally examined in this study provide further illustration. Their location in structure and conservation level agree with the experimentally observed effects on function and expression (Fig. 4).
Copper transfer across a membrane
The PatchFinder-detected conserved surface-facing patches, in both the ATP7B model and LCopA structure (Fig. 1C and F), are conspicuous and intriguing; their preservation from bacterial to mammalian transporters points to a general species-independent functional role such as participation in guiding metal ions, possibly through interactions with regulatory domains or molecules such as copper chaperones, glutathione, etc. It is also possible, although less likely, that upon metal-binding, significant structural rearrangements would result in re-orientation of the conserved residues towards the protein core.
In the membrane portion of ATP7B and LCopA, the number of residues typically associated with coordination of Cu+ (such as Met, Cys, and His) is conspicuously small, suggesting that the mechanism of copper passage through the P1B-ATPases differs from a previously reported Met-lined “wire”-type pathway of the bacterial CusA copper efflux system.60 By analogy with CopA from A. fulgidus (the only Cu-ATPase for which copper-binding studies were carried out), ATP7B may have two intra-membrane sites. In ATP7B model, potential copper-ligand C980 is positioned in close vicinity to the established metal-binding CPC motif. This position is not conserved and can be replaced with Ser, Val, or Ile (Table 1). Thus, the Cys residue in this position may assist or stabilize copper binding rather than provide critical interactions. Interestingly, in the apo form depicted by the ATP7B model, C980 and C983 could form a two coordinate binding site, whereas a change of the rotameric state of M729, accompanied by a mild conformational change, would form a three coordinate site between M729 of the entry site and C980 and C983. Similarly, fluctuation of the flexible unwound region comprising the CPC motif could reorient C983 towards C985 and M1359 (Fig. 5C), as suggested for LCopA.31
We propose that the D730 and R778 can form a salt bridge at the proposed entry/gate site. Based on the strategic location and positioning of these charges, we speculate that this electrostatic interaction may contribute to structure stabilization and/or regulation of the copper transport steps. Acidic pH (~4.5) was shown to stimulate Cu+ transport.61 While this pH is not physiological, marked acceleration of transport at this pH suggests that copper transfer to the binding site and/or copper release are controlled by protonation, especially since the pKa values could be highly shifted by local environments. Availability of the ATP7B model facilitates direct testing of this and other mechanistic hypotheses.
The putative copper exit site is especially intriguing. We propose that the luminal end of TM1,2 and the TM1,2 loop (Fig. S7, ESI†) contribute to the copper-exit pathway; the His/Met residues in this region could attract exiting copper and facilitate copper release. In the model, the TM1,2 lumenal end resides in close proximity to the sensitive-to-mutation M1359 and D765 (Fig. 5D and Table 1). Additionally, according to the ATP7B model, a rotamer change of M1359 would result in another three-coordinate Met site with M668 and M769 of the exit site, thus connecting the core transport sites to the exit site (Fig. 5D).
It should be noted that high conservation across different phyla might not be present at the luminal site of the copper pumps, because the eukaryotic transporters (unlike their bacterial orthologues) operate in a different environment, releasing copper into the acidified lumen of intracellular compartments to various acceptor molecules rather than to the extracellular milieu. The TM1,2 transmembrane pair that is characteristic of all P1B-ATPases could be an especially significant player in the copper transport mechanism. It seems plausible that the TM1,2 brackets the rest of the transmembrane portion and in an oscillating motion controls access to the copper entry site from the cytosol (via a conserved flexible hinge in the TM2) and a copper release at the luminal site (vias the TM1,2 loop).
Conclusions
This study has generated a convenient tool for analysis of the ATP7B mechanism and offers insight into the structural architecture of the ATP7B core. Structural data are critical for dissecting the molecular basis of disease, especially in light of the enormous pool of uncharacterized clinical substitutions, and is also useful for deriving hypotheses concerning the copper transport mechanism. Overall, the ATP7B model suggests that a three-coordinate environment of Met-Cys residues, with additional involvement of weaker chelators such as Ser, Tyr and Asn, might be the basis of Cu+ transport along the eukaryotic membrane domain. Emerging structural data of other conformational states is clearly needed to verify the suggestions raised herein. This study highlights the significant functional properties that were well maintained between the bacterial and eukaryotic pumps throughout evolution, while suggesting the molecular basis of functional divergence.
Supplementary Material
Acknowledgements
M.S. was supported by the Edmond J. Safra Bioinformatics program at Tel Aviv University. The work was supported in part by the National Institutive of Health Grant DK071865 to S.L and Israel Science Foundation Grant 1331/11 to N. B-T.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2mt20025b
Notes and references
- 1.Lutsenko S, Barnes NL, Bartee MY and Dmitriev OY, Function and regulation of human copper-transporting ATPases, Physiol. Rev, 2007, 87(3), 1011–1046. [DOI] [PubMed] [Google Scholar]
- 2.El Meskini R, Culotta VC, Mains RE and Eipper BA, Supplying copper to the cuproenzyme peptidylglycine alpha-amidating mono-oxygenase, J. Biol. Chem, 2003, 278(14), 12278–12284. [DOI] [PubMed] [Google Scholar]
- 3.Setty SR, et al. , Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes, Nature, 2008, 454, 1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashino T, et al. , Unexpected role of the copper transporter ATP7A in PDGF-induced vascular smooth muscle cell migration, Circ. Res, 2010, 107(6), 787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das SK and Ray K, Wilson’s disease: an update, Nat. Clin. Pract. Neurol, 2006, 2(9), 482–493. [DOI] [PubMed] [Google Scholar]
- 6.Kaler SG, et al. , Neonatal diagnosis and treatment of Menkes disease, N. Engl. J. Med, 2008, 358(6), 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenney SM and Cox DW, Sequence variation database for the Wilson disease copper transporter, ATP7B, Hum. Mutat, 2007, 28, 1171–1177. [DOI] [PubMed] [Google Scholar]
- 8.Hsi G, et al. , Sequence variation in the ATP-binding domain of the Wilson disease transporter, ATP7B, affects copper transport in a yeast model system, Hum. Mutat, 2008, 29(4), 491–501. [DOI] [PubMed] [Google Scholar]
- 9.van den Berghe PV, et al. , Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin, Hepatology, 2009, 50(6), 1783–1795. [DOI] [PubMed] [Google Scholar]
- 10.Payne AS, Kelly EJ and Gitlin JD, Functional expression of the Wilson disease protein reveals mislocalization and impaired copper-dependent trafficking of the common H1069Q mutation, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huster D, et al. , Defective cellular localization of mutant ATP7B in Wilson’s disease patients and hepatoma cell lines, Gastroenterology, 2003, 124(2), 335–345. [DOI] [PubMed] [Google Scholar]
- 12.Morgan CT, Tsivkovskii R, Kosinsky YA, Efremov RG and Lutsenko S, The distinct functional properties of the nucleotide-binding domain of ATP7B, the human copper-transporting ATPase: analysis of the Wilson disease mutations E1064A, H1069Q, R1151H, and C1104F, J. Biol. Chem, 2004, 279(35), 36363–36371. [DOI] [PubMed] [Google Scholar]
- 13.Dmitriev O, et al. , Solution structure of the N-domain of Wilson disease protein: distinct nucleotide-binding environment and effects of disease mutations, Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dmitriev OY, Bhattacharjee A, Nokhrin S, Uhlemann EM and Lutsenko S, Difference in stability of the N-domain underlies distinct intracellular properties of the E1064A and H1069Q mutants of copper-transporting ATPase ATP7B, J. Biol. Chem, 2011, 286, 16355–16362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsivkovskii R, Efremov RG and Lutsenko S, The role of the invariant His-1069 in folding and function of the Wilson’s disease protein, the human copper-transporting ATPase ATP7B, J. Biol. Chem, 2003, 278, 13302–13308. [DOI] [PubMed] [Google Scholar]
- 16.Iida M, et al. , Analysis of functional domains of Wilson disease protein (ATP7B) in Saccharomyces cerevisiae, FEBS Lett, 1998, 428(3), 281–285. [DOI] [PubMed] [Google Scholar]
- 17.Forbes JR and Cox DW, Functional characterization of missense mutations in ATP7B: Wilson disease mutation or normal variant?, Am. J. Hum. Genet, 1998, 63(6), 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry AN, Shinde U and Lutsenko S, Structural organization of human Cu-transporting ATPases: learning from building blocks, JBIC, J. Biol. Inorg. Chem, 2010, 15(1), 47–59. [DOI] [PubMed] [Google Scholar]
- 19.Raimunda D, Gonzalez-Guerrero M, Leeber BW 3rd and Arguello JM, The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function, BioMetals, 2011, 24(3), 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voskoboinik I, et al. , Functional analysis of the N-terminal CXXC metal-binding motifs in the human Menkes copper-transporting P-type ATPase expressed in cultured mammalian cells, J. Biol. Chem, 1999, 274(31), 22008–22012. [DOI] [PubMed] [Google Scholar]
- 21.Hatori Y, et al. , Intermediate phosphorylation reactions in the mechanism of ATP utilization by the copper ATPase (CopA) of Thermotoga maritima, J. Biol. Chem, 2008, 283(33), 22541–22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsivkovskii R, Eisses JF, Kaplan JH and Lutsenko S, Functional properties of the copper-transporting ATPase ATP7B (the Wilson’s disease protein) expressed in insect cells, J. Biol. Chem, 2002, 277(2), 976–983. [DOI] [PubMed] [Google Scholar]
- 23.Arguello JM, Gonzalez-Guerrero M and Raimunda D, Bacterial Transition Metal P(1B)-ATPases: Transport Mechanism and Roles in Virulence, Biochemistry, 2011, 50(46), 9940–9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Guerrero M, Eren E, Rawat S, Stemmler TL and Arguello JM, Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases, J. Biol. Chem, 2008, 283(44), 29753–29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatori Y, Lewis D, Toyoshima C and Inesi G, Reaction cycle of Thermotoga maritima copper ATPase and conformational characterization of catalytically deficient mutants, Biochemistry, 2009, 48, 4871–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banci L, et al. , The different intermolecular interactions of the soluble copper-binding domains of the menkes protein, ATP7A, J. Biol. Chem, 2007, 282(32), 23140–23146. [DOI] [PubMed] [Google Scholar]
- 27.Banci L, Bertini I, Cantini F, Rosenzweig AC and Yatsunyk LA, Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1, Biochemistry, 2008, 47(28), 7423–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yatsunyk LA and Rosenzweig AC, Cu(I) binding and transfer by the N terminus of the Wilson disease protein, J. Biol. Chem, 2007, 282, 8622–8631. [DOI] [PubMed] [Google Scholar]
- 29.Achila D, et al. , Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake, Proc. Natl. Acad. Sci. U. S. A, 2006, 103(15), 5729–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banci L, et al. , Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins, Biochemistry, 2009, 48(33), 7849–7855. [DOI] [PubMed] [Google Scholar]
- 31.Gourdon P, et al. , Crystal structure of a copper-transporting PIB-type ATPase, Nature, 2011, 475(7354), 59–64. [DOI] [PubMed] [Google Scholar]
- 32.Jaroszewski L, Rychlewski L, Li Z, Li W and Godzik A, FFAS03: a server for profile–profile sequence alignments, Nucleic Acids Res, 2005, 33(Web Server issue), W284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soding J, Biegert A and Lupas AN, The HHpred interactive server for protein homology detection and structure prediction, Nucleic Acids Res, 2005, 33(Web Server issue), W244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norel R, Petrey D and Honig B, PUDGE: a flexible, interactive server for protein structure prediction, Nucleic Acids Res, 2010, 38(Web Server issue), W550–W554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, et al. , Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res, 1997, 25(17), 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzek BE, Huang H, McGarvey P, Mazumder R and Wu CH, UniRef: comprehensive and non-redundant UniProt reference clusters, Bioinformatics, 2007, 23(10), 1282–1288. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Kuma K, Toh H and Miyata T, MAFFT version 5: improvement in accuracy of multiple sequence alignment, Nucleic Acids Res, 2005, 33(2), 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashkenazy H, Erez E, Martz E, Pupko T and Ben-Tal N, ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids, Nucleic Acids Res, 2010, 38(Web Server issue), W529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalman M and Ben-Tal N, Quality assessment of protein model-structures using evolutionary conservation, Bioinformatics, 2010, 26(10), 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimrod G, Schushan M, Steinberg DM and Ben-Tal N, Detection of functionally important regions in “hypothetical proteins” of known structure, Structure, 2008, 16(12), 1755–1763. [DOI] [PubMed] [Google Scholar]
- 41.Hubbard SJ and Thronton JM, Naccess. Computer Program, Department of Biochemistry and Molecular Biology, University College, London, 1993. [Google Scholar]
- 42.Banci L, et al. , The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A, J. Biol. Chem, 2010, 285, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huster D, K. A, Bhattacharjee A, Raines L, Jantsch V, Noe J, Schirrmeister W, Sommerer I, Sabri O, Berr F, Mossner J, Stieger B, Caca K and Lutsenko S, Diverse Functional Properties of Wilson Disease ATP7B Variants, Gastroenterology, 2011, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo HW, Identification of novel mutations and the three most common mutations in the human ATP7B gene of Korean patients with Wilson disease, Genet. Med, 2002, 4(suppl. 6), 43S–48S. [DOI] [PubMed] [Google Scholar]
- 45.Kim EK, et al. , Identification of three novel mutations and a high frequency of the Arg778Leu mutation in Korean patients with Wilson disease, Hum. Mutat, 1998, 11(4), 275–278. [DOI] [PubMed] [Google Scholar]
- 46.Okada T, et al. , Mutational analysis of ATP7B and genotype phenotype correlation in Japanese with Wilson’s disease, Hum. Mutat, 2000, 15(5), 454–462. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu N, et al. , Molecular analysis and diagnosis in Japanese patients with Wilson’s disease, Pediatr. Int, 1999, 41(4), 409–413. [DOI] [PubMed] [Google Scholar]
- 48.Kusuda Y, et al. , Novel mutations of the ATP7B gene in Japanese patients with Wilson disease, J. Hum. Genet, 2000, 45(2), 86–91. [DOI] [PubMed] [Google Scholar]
- 49.Nanji MS, et al. , Haplotype and mutation analysis in Japanese patients with Wilson disease, Am. J. Hum. Genet, 1997, 60(6), 1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee CC, et al. , Molecular analysis of Wilson disease in Taiwan: identification of one novel mutation and evidence of haplotype-mutation association, J. Hum. Genet, 2000, 45(5), 275–279. [DOI] [PubMed] [Google Scholar]
- 51.Chuang LM, et al. , High frequency of two mutations in codon 778 in exon 8 of the ATP7B gene in Taiwanese families with Wilson disease, J. Med. Genet, 1996, 33(6), 521–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theophilos MB, Cox DW and Mercer JFB, The Toxic Milk Mouse is a Murine Model of Wilson Disease, Hum. Mol. Genet, 1996, 5(10), 1619–1624. [DOI] [PubMed] [Google Scholar]
- 53.Voskoboinik I, Greenough M, La Fontaine S, Mercer JF and Camakaris J, Functional studies on the Wilson copper P-type ATPase and toxic milk mouse mutant, Biochem. Biophys. Res. Commun, 2001, 281(4), 966–970. [DOI] [PubMed] [Google Scholar]
- 54.Mandal AK, Yang Y, Kertesz TM and Arguello JM, Identification of the transmembrane metal binding site in Cu+-transporting PIB-type ATPases, J. Biol. Chem, 2004, 279(52), 54802–54807. [DOI] [PubMed] [Google Scholar]
- 55.Deguti MM, et al. , Wilson disease: novel mutations in the ATP7B gene and clinical correlation in Brazilian patients, Hum. Mutat, 2004, 23, 398. [DOI] [PubMed] [Google Scholar]
- 56.Huster D, et al. , Diverse Functional Properties of Wilson Disease ATP7B Variants, Gastroenterology, 2012, 142(4), 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margarit E, et al. , Mutation analysis of Wilson disease in the Spanish population–identification of a prevalent substitution and eight novel mutations in the ATP7B gene, Clin. Genet, 2005, 68(1), 61–68. [DOI] [PubMed] [Google Scholar]
- 58.Barry AN, et al. , The lumenal loop Met672-Pro707 of copper-transporting ATPase ATP7A binds metals and facilitates copper release from the intramembrane sites, J. Biol. Chem, 2011, 286(30), 26585–26594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forrest LR, Tang CL and Honig B, On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins, Biophys. J, 2006, 91(2), 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long F, et al. , Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport, Nature, 2010, 467(7314), 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Safaei R, Otani S, Larson BJ, Rasmussen ML and Howell SB, Transport of cisplatin by the copper efflux transporter ATP7B, Mol. Pharmacol, 2008, 73(2), 461–468. [DOI] [PubMed] [Google Scholar]
- 62.Lomize MA, Pogozheva ID, Joo H, Mosberg HI and Lomize AL, OPM database andPPM web server: resources for positioning of proteins in membranes, Nucleic Acids Res, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.