Abstract
In the Prostate Cancer Prevention Trial (PCPT), genotypes that may modify the effect of finasteride on the risk of prostate cancer have not been identified. Germline Genetic data from 1157 prostate cancer cases in PCPT were analyzed by case-only methods. Genotypes included 357 single nucleotide polymorphisms (SNPs) from 83 candidate genes in androgen metabolism, inflammation, circadian rhythm and other pathways. Univariate case-only analysis was conducted to evaluate whether individual SNPs modified the finasteride effect on the risk of high-grade and low-grade prostate cancer. Case-only classification trees and random forests, which are powerful machine learning methods with resampling-based controls for model complexity, were employed to identify a predictive signature for genotype-specific treatment effects. Accounting for multiple testing, a single SNP in SRD5A1 gene (rs472402) significantly modified the finasteride effect on high-grade prostate cancer (Gleason score >6) in PCPT (family-wise error rate <0.05). Men carrying GG genotype at this locus had a 55% reduction of the risk in developing high-grade cancer when assigned to finasteride (RR=0.45, 95% C.I. [0.27,0.75]). Additional effect-modifying SNPs with moderate statistical significance were identified by case-only trees and random forests. A prediction model built by the case-only random forest method with 28 selected SNPs classified 37% of PCPT men to have reduced risk of high-grade prostate cancer when taking finasteride, while the others have increased risk. In conclusion, case-only methods identified SNPs that modified the effect of finasteride on the risk of high-grade prostate cancer and predicted a subgroup of men who had reduced cancer risk by finasteride.
Introduction
Prostate cancer is the most commonly diagnosed cancer and the second common cause of cancer death in men in United States (1–2). PCPT was launched in the mid-1990s to test the hypothesis whether finasteride, an inhibitor of the enzyme 5α-reductase that is critical to androgen metabolism, can prevent prostate cancer (3). Men were randomized to finasteride or placebo and followed for 7 years. The primary result was published in 2003 and mixed: though finasteride reduced the risk of prostate cancer by 25%, the risk of high-grade prostate cancer (Gleason score >6) was elevated in the finasteride arm (3). Because of this result, finasteride was not approved as a chemoprevention agent for prostate cancer. The finasteride effect on high grade prostate cancer observed in PCPT has since been under considerable debate. Because finasteride increases the sensitivity of PSA, the digital rectal exam, and the needle biopsy for detecting prostate cancer, one plausible explanation for the observed elevation in high grade cancer is a bias due to increased detection in the finasteride arm (4–8). Substantial efforts have been devoted to understanding the biology of finasteride and prostate cancer, including a P01 research program with five projects and a number of single nucleotide polymorphisms (SNPs) genotyped in candidate genes for prostate cancer risk (6). This paper aims to identify SNPs that modified the prevention effect of finasteride in PCPT.
Clinical cancer research is advancing from the empiric approach of “one size fits all” to precision treatment and prevention (9–10). Discovery of predictive biomarkers that delineate subgroup or individual treatment effects is the critical step toward precision medicine. In clinical trials, retrospectively measured biomarkers on stored baseline samples provide high-quality data for determining predictive markers associated with treatment efficacy (11–12). Classical case-control and case-cohort sampling methods have been commonly used to improve cost efficiency of biomarker studies, particularly when study endpoints are rare (13–14). More recently, the case-only method has been advocated as a simple but more efficient means to study gene-treatment interactions and genotype-specific treatment effects, exploiting the independence between randomized treatment assignment and baseline biomarkers that is dictated by randomization (15–16). The main benefit, as we will illustrate using genetic data analyses from the Prostate Cancer Prevention Trial (PCPT), is that only genotypic data in cases are needed, yet it retain statistical efficiency for assessing effect modification by genotypes and developing a multi-genotype predictive marker for treatment selection.
We conducted case-only analyses using genotypic and phenotypic data in PCPT, primarily aiming to identify SNPs that modify the finasteride effect and to determine whether there is a subgroup of men that is more likely to have finasteride prevent both low-grade and high-grade prostate cancer. The PCPT data are ideal for case-only methodologies, because of the large sample size of the trial, a sizable number of prostate cancer cases, the choice of evaluating the finasteride effect on the relative risk scale (17), and the fact that a common case-control study design was used for the P01 program so that SNP data across the five projects can be directly concatenated for case-only analyses. Through this PCPT data analysis, we also seek to illustrate to the clinical research community how case-only methods can be applied in large randomized clinical trials to identify individual predictive genotypes and build a predictive signature for treatment selection.
Materials and Methods
The PCPT trial is registered in ClinicalTrials.gov (NCT00288106) and details of the trial have been described previously (11). Between 1994 and 1997, 18,882 prostate cancer-free men 55 years of age or older were randomly assigned to take finasteride (5 mg per day) or matching placebo daily for a 7-year period. The primary endpoint was the prevalence of prostate cancer in the seven-year period and cancer cases were defined to be biopsy-proven presence of prostate cancer. Men with biopsy-determined prostate cancer were identified either by a for-cause biopsy for cancer diagnosis during follow-up, or end-of-study biopsy for all men without a cancer diagnosis, collected within a time window of 7 years ± 90 days after randomization. The majority of men included in the efficacy analysis were whites (93%) without family history of prostate cancer (81%). The primary results from PCPT were published in 2003, that finasteride reduced the prevalence of prostate cancer by 24.8% (95% confidence interval [18.6%, 30.6%]), though the reduction of cancer risk is only for the low-grade cancer subset (Gleason score < 7). The high-grade prostate cancer, defined as a Gleason score of 7 or higher, was more common in the finasteride arm (relative risk 1.27, 95% confidence interval [1.07,1.50]).
To elucidate the biology underlying the finasteride effect and the risk of prostate cancer, a program project (P01) composed of 5 studies was launched, including efforts to genotype SNPs in candidate genes involved in androgen metabolisms, diet- related factors, insulin-like growth factor axis, inflammation, oxidative damage and DNA repair. Additional genotypes came from other ancillary PCPT projects using genotypes from circadian rhythm genes and others for risk prediction. The control selection scheme has been previously described (18), with frequency matched to cases on distributions of treatment arm, age (in 5-year age groups) and positive family history for first degree relative with prostate cancer. In this analysis, we retrieved genotypic data from 1167 cases and 1365 controls who had sufficient DNA from white blood cells available for genotyping in various projects, which have been published previously with details in genotyping (19–21).This case-control set included additional cases who had end-of-study biopsy between 90 and 180 days after 7 years’ follow-up who were not reported in the primary analysis. Forty-four SNPs were filtered out by the quality-control metric that the false discovery rate for the Hardy-Weinberg test is greater than 0.05. Forty-three SNPs were not included in the case-only analysis because their data were missing in more than 20% cases. This resulted in a total of 357 SNPs from 83 genes for case-only analyses to identify SNPs that modified the effect of finasteride. The minor allele frequencies of these SNPs were similar to those in the HapMap data. Missing genotypic data were imputed by the mean genetic score in cases and controls respectively. The case-only analyses were stratified for 306 high-grade cases and 851 low-grade cases. A small fraction of cases did not have Gleason score available and were excluded from case-only analyses. Cases from whites and other ethnic groups were combined because the effect finasteride did not differ by the ethnic groups and adjustment for the ethnic groups did not yield different results in case-only analyses (3).
The treatment effect of finasteride in a genotype subgroup is defined as the reduction of risk of prostate cancer in the subgroup if receiving finasteride, relative to receiving placebo in the same subgroup. Though typically estimated by a model with a gene-treatment interaction term using data from cases and controls, the gene-treatment interaction and the subgroup-specific treatment effect can also be estimated by case-only methods (7–8). Such approaches have been recently advocated for selecting baseline predictive markers of varying treatment effects and estimating marker-specific treatment effects in randomized clinical trials (7–8). Briefly, let Z denote the treatment assignment (Z=1 if randomized to an investigational treatment or Z=0 if randomized to a standard treatment or placebo), and let D denote the binary study endpoint the trial is designed to ascertain (e.g., 1 for a cancer case or 0 for a control in PCPT). The baseline biomarker that may predict heterogeneous treatments in the trial population is denoted by M, which can be a single candidate marker or a set of high-dimensional markers under investigation. Suppose the marker specific treatment effect in the relative risk scale to be determined is denoted by R(M)=Pr(D=1|Z=1,M)/Pr(D=1|Z=0,M). The case-only approaches to estimating marker-treatment interaction and marker-specific treatment effect are derived by the mathematical expression
| (1) |
The derivation is based on the Bayes theorem and that randomization ensuring Pr(Z|M)=Pr(Z) for any baseline M. The expression in (1) suggests that the marker-specific treatment effect in the relative risk scale can be estimated by the product of the odds of treatment assignment being 1 in cases given the marker M, and the randomization ratio of treatment assignment being 0 and 1. Therefore a simple approach to estimate the treatment effect in relative risk R(M) is to employ a logistic regression model with outcome variable Z and predictor M among cases (D=1) only, adding an offset involving randomization fractions, namely log{Pr(Z=1)/Pr(Z=0)}. Note that the marker-specific treatment effect is interpreted in the relative risk scale, even though a logistic regression is fitted to obtain the odds of the treatment in cases. This case-only logistic regression was applied to the PCPT genetic data to test SNP-treatment interaction for one SNP at a time. Since biomarkers such as genotypes are typically expensive to measure, the case-only approach substantially reduces the cost and saves valuable specimens from controls for other scientific objectives, yet the precision of estimated gene-treatment interaction and marker-specific treatment effect can be comparable to the full cohort analysis where all study participants were assayed for the baseline biomarker (7–8). Furthermore, the genotype-specific treatment effect estimated by case-only approaches is protected from confounding of any baseline characteristic because of randomization (Equation 1).
In addition to estimate marker-specific treatment effect for markers one at a time, case-only methods can be used to select multiple markers, construct a multivariate predictive model, and estimate individualized treatment effect based on the selected markers. Observe that the derivation in (1) implies that investigators can estimate the marker-specific treatment relative risk R(M) using any parametric or nonparametric function to estimate the treatment odds in cases, e.g., LASSO or classification tree methods, with an offset adjusting for the ratio of randomization fractions. In this PCPT study, we will fit case-only classification tree and random forests methods in order to discover effect-modifying SNPs in a multivariate fashion. The R packages rpart and randomForest were used to fit trees and random forests. The rfcv function in randomForest was used to select SNPs to build random forest, by gradually deleting SNPs ranked low in variable importance and evaluating predicting accuracy by cross validation.
Results
Not all prostate cancer cases had blood samples available for genotyping. The collection of white blood cells was introduced after the trial started enrolling men. To establish the interpretability of our analyses, characteristics of 1157 prostate cancer cases (306 high-grade, 851 low-grade) used in our case-only (CO) analyses were compared to all 2003 prostate cancer cases from PCPT. Table 1 shows that there is no difference between the distribution of age, race, family history, body-mass index, diabetes, treatment assignment, or Gleason score between the two case populations. The proportion of for-cause biopsies in cases included in this analysis is less than that in all cases. This is because some earlier cases in PCPT who had for-cause biopsies did not have blood samples for genotyping. Consequently, the follow-up time for the portion of earlier prostate cancer cases included in this analysis is slightly longer than that for all cancer cases: for example, the 20 and 30 quantiles of the distribution of follow-up time are nearly 1 year longer for cases included in this analysis (Table 1).
Table 1:
Characteristics of prostate cancer cases included in case-only analyses in comparison to all cases included in the PCPT primary analysis.
| Cases used in Case-only analyses N=1157 | All cases in PCPT N=2003 |
||
|---|---|---|---|
| Type of diagnosis | For cause | 431 (37%) | 811 (41%) |
| End of study | 726 (63%) | 1163(59%) | |
| Quantile of follow-up time (years) | 10% | 4.1 | 3.3 |
| 20% | 6.0 | 5.1 | |
| 30% | 6.8 | 6.4 | |
| 50% | 7.0 | 7.0 | |
| 75% | 7.1 | 7.1 | |
| Treatment assignment | Finasteride | 477 (41%) | 820 (41%) |
| Placebo | 680 (59%) | 1183 (59%) | |
| Gleason Sum | < 7 | 851 (74%) | 1412 (73%) |
| >=7 | 306 (26%) | 524 (27%) | |
| Age | Median (25%,75%) | 63 (59, 67) |
63 (59, 67) |
| Race | Caucasian | 1081 (93%) | 1868 (93%) |
| Others | 76 (7%) | 135 (7%) | |
| Family history | Yes | 244 (21%) | 415 (21%) |
| No | 913 (79%) | 1588 (79%) | |
| BMI | Median (25%, 50%) | 26.6 (24.7, 29.2) |
26.8 (24.8,29.3) |
| Diabetes | Yes | 51 (4%) | 88 (4%) |
| No | 1106 (96%) | 1914 (96%) |
The 357 SNPs passing quality control were first evaluated one at a time for potential modification of the finasteride effect using a case-only logistic model with an additive genetic effect, stratified by high-grade prostate cancer cases and low-grade prostate cancer cases. Figure 1 shows the quantile-quantile plots of p-values for the SNP-finasteride interactions, separately for high-grade and low-grade prostate cancer. A number of SNPs show evidence for SNP-finasteride interaction as their observed p-values are smaller than expected in the quantile-quantile plot (Figure 1a), while there is clearly no significant SNP-finasteride interaction for low-grade cases when multiple-testing is accounted for (Figure 1b). The top SNP (rs472402, p-value=8 × 10−5) is statistically significant after adjusting for multiple testing. This SNP (C/G) is located in SRD5A1, the gene encodes the enzyme which catalyzes the conversion of testosterone into the more potent androgen, dihydrotestosterone (DHT). The minor allele (G) frequency of rs472402 is 0.48. Table 2 shows the treatment effect in relative risk estimated by case-only methods and stratified by genotype at rs472402, separately shown for high-grade and low-grade cases. Notably, men carrying GG genotype had a 55% reduction of risk to develop high-grade prostate cancer when taking finasteride (RR=0.45, 95% C.I. [0.27,0.75]), contrary to the reported overall hazardous intent-to-treat effect. The SNP-treatment interaction for high-grade prostate cancer is statistically significant (family-wise error rate 0.018 based on the permutation test). These men also had a decreased risk to develop low-grade prostate cancer when taking finasteride (RR=0.69, 95% C.I. [0.51,0.92]), though the estimated case-only interaction between rs472402 and finasteride for low-grade cancer is not significant (p-value=0.08).
Figure 1:
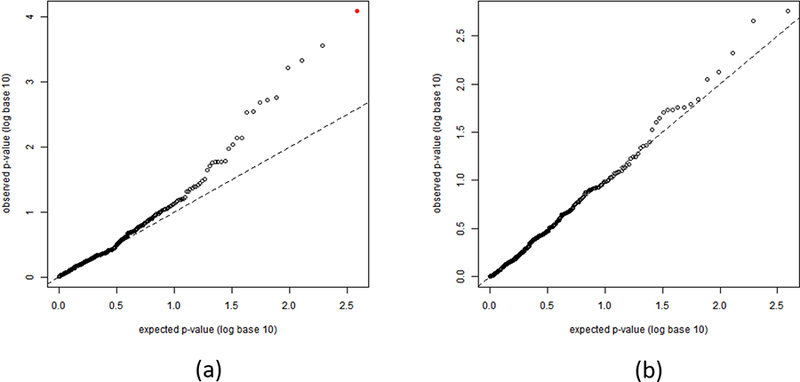
The quantile-quantile plots for p-values for assessing individual SNP-finasteride interactions by the case-only method. (a) SNP-finasteride p-values for interactions for high-grade prostate cancer. The red solid dot represents rs472402. (b) SNP-finasteride p-values for interactions for low-grade prostate cancer.
Table 2:
The finasteride effect on high-grade prostate cancer and low-grade prostate cancer estimated by univariate case-only methods and stratified by rs472402.
| Genotype | Treatment effect (Relative risk comparing finasteride to placebo) | 95%CI | p-value for case-only interaction | |
|---|---|---|---|---|
| High-grade cancer |
All high grade | 1.22 | (0.97,1.52) | 8 × 10−5 |
| CC (n=69) | 1.87 | (1.14,3.08) | ||
| CG(n=169) | 1.52 | (1.11,2.07) | ||
| GG (n=68) | 0.45 | (0.27,0.75) | ||
| Low-grade cancer | All low grade | 0.57 | (0.50,0.66) | 0.08 |
| CC (n=223) | 0.47 | (0.36,0.63) | ||
| CG (n=444) | 0.57 | (0.48,0.71) | ||
| GG (n=184) | 0.69 | (0.51,0.92) |
We investigated whether the multivariate case-only analysis using the classification trees and random forests methods would identify additional SNPs that did not reach univariate statistical significance when evaluated one at a time, but could further refines subgroups determined by rs472402. One SNP (rs1052536) in LIG3, a gene encoding a protein that catalyzes the joining of DNA ends and involves in DNA metabolism, was identified in the best fitting tree selected by cross validation. Figure 2a shows the final classification tree of subgroups with genotypes in the two SNPs. Three nodes were identified in the final tree. In addition to the subgroup defined by GG at rs472402, a small subgroup of men with rs1052536 genotype TT and rs472402 genotype CC or CG have reduced risk by finasteride (RR=0.54), while the third subgroup with at least a C allele in both loci had a substantially increased risk of developing high-grade prostate cancer (RR=2.21). We further investigated whether additional SNPs could be identified to better predict individualized finasteride effects, by generating a large number of case-only trees and perform ensemble learning by random forests. Two thousand trees were randomly generated using bootstrap samples of the case-only data and a prediction model was built from averaging predictions from all trees generated. The feature selection procedure the R packages randomForest resulted in 28 SNPs that were used in the final random forest prediction model (Supplementary Table 1). The cross-validation prediction errors for different numbers of SNPs are shown in Supplementary Figure 1. This prediction model was applied to genotypic data in the case-control sample. Cases and controls were included with different weights to adjust for case-control sampling and generate a distribution of individualized treatment effects corresponding to the population of men in the PCPT trial. Figure 2(b) shows the distribution of the estimated individualized treatment effects in PCPT. The vertical red line indicates zero treatment effect (RR=1). There were estimated 37% of PCPT participants whose risk of high-grade prostate cancer were decreased by finasteride (RR<1), and 26% of participants whose risk of high-grade prostate cancer were decreased by more than 25% (RR<0.75).
Figure 2:
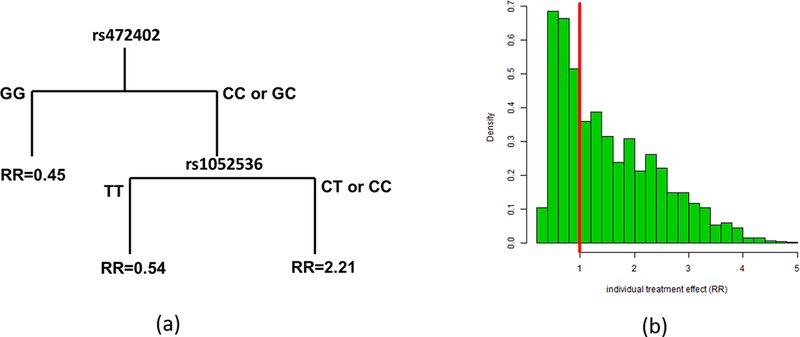
Case-only trees and random forests identified more SNPs collectively predicting genotype specific treatment effects. (a) The classification tree for subgroup treatment effects (relative risk) depending on genotypes in the two SNPs selected by cross validation. (b) The distribution of individual treatment effects for the population of PCPT men based on 28 SNPs identified by random forests. A portion of PCPT men (36%, the left of the red line) did not have increased risk by finasteride.
Table 3 shows the characteristics of the top seven SNPs selected by the feature selection method in the random forest analysis. All of these SNPs are common variants, with the lowest minor allele frequency 0.067 (rs12795870). These SNPs have varying significance levels from univariate case-only interactions, ranging from 0.09 (rs3736544) to 0.00008 (rs472402, the SNP identified in Figure 1a). When using case-control data to estimate SNP-treatment interactions, the case-control interaction p-values for these SNPs are slightly bigger than the case-only interaction p-values. Three out of the seven SNPs came from SRD5A1, including the one SNP (rs472402) already identified by univariate case-only analysis. Three SNPs (rs3736544, rs11689432, rs12795870) are from circadian rhythm genes, CLOCK, PER2 and the intergenic region between RASSF10 and ARNTL (a circadian gene) respectively. One SNP (rs1052536, also found by classification tree in Figure 2a) is from LIG3, a DNA ligase gene involved in DNA repair.
Table 3:
Characteristics, univariate case-only interactions and case-control interactions of the top ten SNPs ranked by the variable importance measure from case-only (high-grade prostate cancer cases) random forests analysis.
| SNP ID | Gene | Minor allele frequency | Genotype | p-value for HWE¶ | Case-only treatment RR estimate | Case-only Interaction p-value |
Case-control interaction p-value |
|---|---|---|---|---|---|---|---|
| rs472402 | SRD5A1 | 0.48 | CC | 0.55 | 1.87(1.14, 3.08) | 0.00008 | 0.0019 |
| CG | 1.52(1.11, 2.07) | ||||||
| GG | 0.45(0.27, 0.75) | ||||||
| rs11689432 | PER2 | 0.45 | GG | 0.02 | 1.80(1.19, 2.72) | 0.0003 | 0.0097 |
| AG | 1.44(0.98, 2.13) | ||||||
| AA | 0.46(0.26, 0.81) | ||||||
| rs1052536 | LIG3 | 0.44 | CC | 0.94 | 1.60(1.05, 2.44) | 0.0017 | 0.0099 |
| CT | 1.42(1.03, 1.96) | ||||||
| TT | 0.47(0.27, 0.83) | ||||||
| rs3756423 | SRD5A1 | 0.20 | TT | 0.90 | 1.71(1.28, 2.28) | 0.002 | 0.0041 |
| TG | 0.63(0.43,0.94) | ||||||
| GG | 2.50(0.48,12.87) | ||||||
| rs12795870 | Intergenic between RASSF10 and ARNTL | 0.067 | TT | 0.16 | 1.36(1.07,1.73) | 0.04 | 0.0612 |
| CT | 0.43(0.21,0.91) | ||||||
| CC | - | ||||||
| rs3736544 | CLOCK | 0.36 | GG | 0.79 | 1.30 (0.91, 1.86) | 0.09 | 0.1149 |
| GA | 1.46 (1.04, 2.04) | ||||||
| AA | 0.56 (0.30, 1.05) | ||||||
| rs248797 | SRD5A1 | 0.50 | CC | 0.78 | 0.66(0.42, 1.04) | 0.003 | 0.0352 |
| CT | 1.41(1.03, 1.93) | ||||||
| TT | 1.79 (1.09,2.95) |
Hardy Weinberg equilibrium
Discussion
Using case-only methods, we identified multiple SNPs that modified the effect of finasteride on high-grade prostate cancer in PCPT. Our results suggest that men carrying specific genotypes in these SNPs may not have increased risk of high-grade prostate cancer when taking finasteride. Discovery of these effect-modifying SNPs have major implications for usage of finasteride. First, finasteride as a chemoprevention agent for prostate cancer may be restricted to the subgroup with the particular genotype that is associated with reduction of risk. Having the GG genotype at rs472402, the single most significant SNP modifying the finasteride effect, is a crude classification rule to define such subgroup, with the population frequency of 25%. Using more sophisticated trees and random forests method, we built a multi-SNP classifier that includes 28 moderately significant SNPs and predicts 36% of men could benefit from finasteride. Second, related to its potential for preventing prostate cancer, finasteride is commonly used to treat benign prostate hyperplasia (BPH). Our analysis suggests that there are men who, if taking finasteride, would have substantially increased risk (e.g. relative risk ~2 or greater) to develop high-grade prostate cancer. For example, men carry the CC genotype at rs472402 had RR=1.87, which raises a cautionary note that these men may not use finasteride as the treatment for BPH.
As the set of SNPs we analyzed were picked from candidate genes related to prostate cancer, some of the identified effect-modifying SNPs have been reported to be associated with risk of prostate cancer in previous PCPT analyses. Five of the six effect-modifying SNPs we detected in SRD5A1 (rs3736316, rs3822430, rs472402, rs1560149, and rs248797) were also significantly associated with risk of high-grade cancer in the placebo arm (19). Interestingly, the GG genotype for SNP rs472402 was associated with increased risk of high-grade cancer (OR=1.7, 95% C.I. [1.05,2.75], reference 19) but not with low-grade cancer, and we found that this GG genotype is also associated with a 55% reduction of high-grade cancer risk by finasteride. This SNP has recently been shown to be associated with d-amphetamine response in a genome-wide association study (22). It has been suggested that SNPs in SRD5A1 could modulate both expression and enzymatic activity of SRD5A1. The SNP rs3736544 from the circadian rhythm gene CLOCK has been found to be located in a 3-SNP haplotype associated with obesity and metabolic syndrome in men (23). Genotypes in the circadian genes including CLOCK and PER2 have been reported to be associated with prostate cancer risk (24). The functional annotation of rs12795870 in the intergenic region between RASSF10 and ARNTL (a circadian gene) is undermined. The SNP rs1052536 from LIG3, a gene encoding a protein involved in mismatch repair, has been previously linked to the risk of young-onset lung cancer (25).
The strengths of this work include a relatively large number of high-grade and low-grade cases from PCPT, and the use of efficient case-only methods for discover SNPs that predict subgroup/individual treatment effects. We have showcased several case-only analyses to estimate marker-specific treatment effects in the relative risk scale, including univariate and multivariate trees and random forests. It is reassuring that the standard case-control estimates yielded slightly less significant p-values (Table 3). As we have shown recently, other machine-learning methods such as LASSO can be adopted in the case-only analysis (8). The simplicity and the efficiency of case-only methods make it ideal for exploring high-dimensional gene-treatment interactions in randomized clinical trials.
Our study has several weaknesses. First, lack of blood samples for some cancer cases (~40% all PCPT high-grade cases) have substantially reduced the sample size and the power of the case-only analysis. Chance of false positives cannot be ruled out even for SNP rs472402 that attains statistical significance in association with high-grade cancers after Bonferroni correction. The statistical significance will be attenuated if accounting for multiple tests incurred by separate analyses for high-grade and low-grade cancers. Second, it is difficult to establish a validation dataset for these newly discovered effect-modifying SNPs since PCPT is the only trial that tests the effect of finasteride for preventing prostate cancer. There are prostate cancer prevention trials for dutasteride (26), which inhibits both SRD5A1 and SRD5A2, however genes interacting with dutasteride may not be the same as genes interacting with finasteride.
Supplementary Material
Acknowledgments
Financial support
This statistical methods work was supported by the National Institute of Health [R01CA233588 to J.Y. Dai, P01CA53996 to J.Y. Dai and M. LeBlanc]. The analysis of the PCPT data was supported by 1UM1CA182883–01 (PCPT and SELECT cohorts: core infrastructure support for cancer research).
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Centers for Disease Control and Prevention (CDC). Cancer among een. http://www.cdc.gov/cancer/dcpc/data/men.htm. Accessed September 27, 2017.
- 2.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2015. CA Cancer J Clin 2015; 65:5–29. [DOI] [PubMed] [Google Scholar]
- 3.Thompson IM, Goodman PJ, Tangen CM, Lucia MS, Miller GJ, Ford LG, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med 2003; 349:215–224. [DOI] [PubMed] [Google Scholar]
- 4.Redman MW, Tangen CM, Goodman PJ, Lucia MS, Coltman CA Jr, Thompson IM. Finasteride does not increase the risk of high-grade prostate cancer: A bias-adjusted modeling approach. Cancer Prev Res (Phila) 2008;1:174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson IM, Chi C, Ankerst DP, Goodman PJ, Tangen CM, Lippman SM, et al. Effect of finasteride on the sensitivity of PSA for detecting prostate cancer. J Natl Cancer Inst 2006; 98: 1128–33. [DOI] [PubMed] [Google Scholar]
- 6.Thompson IM, Tangen CM, Goodman PJ, Lucia MS, Parnes HL, Lippman SM, et al. Finasteride improves the sensitivity of digital rectal examination for prostate cancer detection. J Urol 2007;177:1749–52. [DOI] [PubMed] [Google Scholar]
- 7.Lucia MS, Epstein JI, Goodman PJ, Darke AK, Reuter VE, Civantos F, et al. Finasteride and high-grade prostate cancer in the Prostate Cancer Prevention Trial. J Natl Cancer Inst 2007;99:1375–83. [DOI] [PubMed] [Google Scholar]
- 8.Thompson IM, Tangen CM, Goodman PJ, Lucia MS, Klein EA: Chemoprevention of prostate cancer. J Urol 2009;182:499–507. [DOI] [PubMed] [Google Scholar]
- 9.Roper N, Stensland KD, Hendricks R, Galsky MD. The landscape of precision cancer medicine clinical trials in the United States. Cancer Treatment Reviews 2015; 41: 385–390. [DOI] [PubMed] [Google Scholar]
- 10.Rebbeck TR. Precision prevention of cancer. Cancer Epidemiol Biomarker Prev 2014; 23:2713–5. [DOI] [PubMed] [Google Scholar]
- 11.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009; 101: 1446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nature Review 2013;12:358–369. [DOI] [PubMed] [Google Scholar]
- 13.Breslow NE, Day NE: Statistical methods in cancer research I: the analysis of case-control studies. International Agency for Research on Cancer, Lyon, 1980. [PubMed] [Google Scholar]
- 14.Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika 1986;73: 1–11. [Google Scholar]
- 15.Dai JY, Zhang XC, Wang CY, Kooperberg C. Augmented case-only design for randomized clinical trials with failure time endpoints. Biometrics 2016;72:30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai JY, Liang J, LeBLanc M, Prentice RL, Janes H. Case-only approach to identifying markers predicting treatment effects on the relative risk scale. Biometrics 2018; 74:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman PJ, Thompson IM Jr, Tangen CM, Crowley JJ, Ford LG, Coltman CA Jr. The Prostate Cancer Prevention Trial: Design, biases and interpretation of study results. J Urol 2006;175:2234–2242. [DOI] [PubMed] [Google Scholar]
- 18.Goodman PJ, Tangen CM, Kristal A, Thompson IM, Lucia MS, Platz EA, Figg WD, et al. Transition of a clinical trial into translational research: the prostate cancer prevention trial experience. Cancer Prev Res (Phila) 2010; 3:1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price DK, Chau CH, Till C, Goodman PJ, Leach RJ, Johnson-Pais TL, et al. Association of androgen metabolism gene polymorphisms with prostate cancer risk and androgen concentrations: results from the Prostate Cancer Prevention Trial. Cancer 2016; 122:2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Liu X, Brendler CB, Ankerst DP, Leach RJ, Goodman PJ, et al. Adding genetic risk score to family history identified twice as many high-risk men for prostate cancer: results from the Prostate Cancer Prevention Trial. The Prostate 2016;76:1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu LW, Till C, Yang B, Tangen CM, Goodman PJ, Yu K, et al. Circadian genes and risk of prostate cancer in the Prostate Cancer Prevention Trial. Molecular Carcinogenesis. 2018;57:462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart AB, Engelhardt BE, Wardle MC, Sokoloff G, Stephens M, de Wit H, et al. Genome-wide association study of d-amphetamine response in healthy volunteers identifies putative associations, including cadherin 13 (CAD13). PLoS ONE 2012; 7: e42646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008; 32:658–62. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res 2009; 69:9315–9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landi S, Gemignani F, Canzian F, Gaborieau V, Barale R, Landi D, et al. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Research 2006; 66: 11062–9. [DOI] [PubMed] [Google Scholar]
- 26.Andriole GL, Bostwick DG, Brawley OW, Gomella LG, Marberger M, Montorsi F, et al. Effect of dutasteride on the risk of prostate cancer. NEJM 2010;362: 1192–1202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


