Abstract
Mutations of Hepatocyte-Nuclear-Factor-1-Homeobox-A gene and loss of Liver-Fatty-Acid-Binding-Protein are well documented in hepatocellular adenoma. However, the role of Hepatocyte-NuclearFactor-1-Homeobox-A mutations in hepatocellular carcinoma remains to be determined. In this study, all hepatocellular neoplasms evaluated by our institutional Memorial Sloan Kettering-Integrated Mutational Profiling of Actionable Clinical Targets assay or the Cancer Genome Atlas sequencing, and cases reported in the literature, were queried for Hepatocyte-Nuclear-Factor-1-Homeobox-A mutations. Together, 11 of 672 (1.6%) hepatocellular carcinomas harbored Hepatocyte-Nuclear-Factor-1-Homeobox-A mutations. The single case from our institution (n=153) was extremely well differentiated, arising in a background of adenomatosis. Both the adenoma and carcinoma component contained the same 2 somatic Hepatocyte-Nuclear-Factor-1-Homeobox-A mutations (p. E32* and L214Q), with loss of Liver-Fatty-Acid-Binding-Protein. From the literature, 2 of 146 (1.4%) hepatocellular carcinomas had Hepatocyte-Nuclear-Factor-1-Homeobox-A mutations, and both arose in a background of adenomatosis. Information on pre-existing adenoma for the remaining cases (8/373, from The Cancer Genome Atlas) was not available. Hepatocyte-Nuclear-Factor-1-Homeobox-A mutations in carcinomas were associated with negative viral hepatitis status (p=0.004), mutually exclusive with Catenin-Beta-1 hotspot mutations, and trended to occur more in females (p=0.06) and without cirrhosis (p=0.03). Grade was not associated with Hepatocyte-Nuclear-Factor-1-Homeobox-A status (p=0.28). Somatic Hepatocyte-Nuclear-Factor-1Homeobox-A mutations occur in approximately 1–2% of hepatocellular carcinoma, often in a background of adenomatosis. Our findings suggest that malignant transformation of Hepatocyte-Nuclear-Factor-1Homeobox-A mutated hepatocellular adenoma occurs, albeit infrequently. Hepatocellular adenomas with Hepatocyte-Nuclear-Factor-1-Homeobox-A mutation or adenomatosis with loss of Liver-Fatty-Acid-Binding-Protein warrant thorough sampling and examination.
Keywords: hepatocellular carcinoma, HNF1A, LFABP, adenomatosis
1. Introduction
Hepatocyte nuclear factor 1 homeobox A (HNF1A) encodes a transcription factor that is widely expressed in organs of endoderm lineage. This transcription factor is thought to function as a tumor suppressor because knockdown of HNF1A induces increased proliferation and decreased apoptosis [1]. Mutation of HNF1A is a relatively frequent event in hepatocellular adenomas (HAs) and leads to intralesional steatosis via increased lipogenesis by promoting fatty acid synthesis and down-regulation of expression of liver fatty acid binding protein (LFABP) [2,3].
Germline HNF1A mutations are associated with mature-onset diabetes of the young type 3 (MODY3), and several reports have described liver neoplasia including adenomatosis as well as hepatocellular carcinoma (HCC) in these patients [4–7].
While HNF1A inactivation is thought to be a common feature of HAs and loss of its target LFABP is a frequent event in both HAs and HCC, somatic mutation of HNF1A has only been mentioned in rare cases of HCC. Here, we report the results of an analysis of MSK-IMPACT data, TCGA data, and the available literature on the incidence and analyze the clinicopathologic features of HNF1A mutated HCC.
2. Materials and Methods
The study was approved by our local institutional review board. Two databases and the literature reports were searched for hepatocellular neoplasm with HNF1A mutation. The 2 databases were: 1) the institutional Memorial Sloan Kettering-Integrated Mutational Profiling of Actionable Clinical Targets (MSK-IMPACT) database (MSK-IMPACT is a hybridization based next generation sequencing assay that assesses the coding regions of more than 400 genes, along with select promoters, introns, copy number status, and microsatellite status against a patient’s matched normal)[8–10],and 2) the Cancer Genome Atlas (TCGA)[11] provisional set.
Hepatocellular tumors identified to carry HNF1A mutation, with available clinical data and tissue available were further evaluated. Clinical information was extracted, pathology examination of FFPE sections and histochemical stain for reticulin were performed according to routine procedures. Immunohistochemistry for the following antibodies was performed: Liver Fatty Acid Binding Protein (LFABP) (Polyclonal, Abcam), Beta Catenin (Clone 14, RTU, Cell Marque), Glutamine Synthetase (Clone 6, BD-Transduction Labs), Serum Amyloid A and C reactive protein (the latter 2 antibodies were performed as clinically validated markers in an outside lab). For all pertinent cases, data on viral hepatitis status, background liver fibrosis, age, and grade for HCCs were recorded. Fisher’s exact test using two-tailed p values were calculated for select variable. P<0.0125 was considered significant after correcting for multiple comparison with Bonferroni’s adjustment.
3. Results
3.1. MSK-IMPACT Data Analysis
The data of 153 patients with HCC who underwent MSK-IMPACT findings was reviewed. One case of HCC with HNF1A mutations was identified. This made the frequency of HNF1A mutations in hepatocellular carcinoma at our institution 1 of 153 (0.7%). The case is detailed below.
3.1.1. Clinical History and Findings
A 65 year old female with obesity (BMI= 30.3), hypertension, and remote history of ovarian cystadenocarcinoma presented to the emergency room with severe acute abdominal pain. Family history included father with melanoma and sarcoma, a paternal uncle with a brain tumor, a paternal grandmother with colon cancer, maternal grandmother with breast cancer, a maternal aunt with pancreatic cancer, and a daughter with HAs. Imaging revealed several hepatic lesions with a differential diagnosis of HA vs HCC. Laboratory studies for hepatitis B core and C antibodies were negative. The patient was surgically explored. Lesions in segments II (hepatocellular adenoma), IV (well- differentiated hepatocellular carcinoma), and VI (well differentiated hepatocellular carcinoma) were identified. This was followed by an extended left hepatectomy one month later, which revealed multiple lesions (see Surgical Pathology Findings). Eight months later, a new suspicious lesion in segment VIII was noted, for which the patient underwent a doxorubicin drug-eluting bead (DEB-TACE) embolization. Five months later, to address the concern of further recurrence, the patient underwent a DEB-TACE re-embolization. At last follow-up, 14 months after the initial diagnosis, the patient was alive with stable disease.
3.1.2. Surgical Pathology Findings
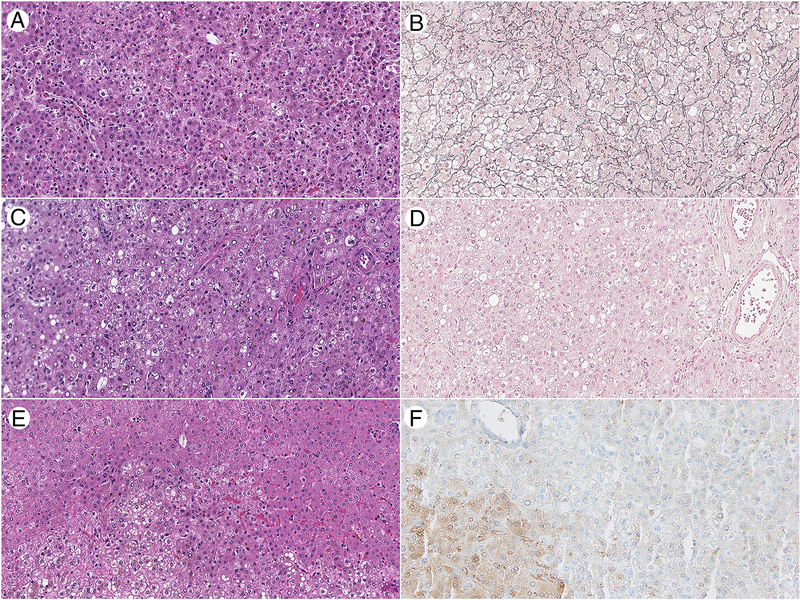
The non-lesional hepatic parenchyma was unremarkable- without significant steatosis, portal or lobular inflammation, fibrosis, cholestasis, or iron deposition. Grossly, the extended left hepatectomy was positive for three yellow, somewhat circumscribed lesions ranging from 0.8 cm to 3.2 cm in greatest dimension. On microscopic examination, 3 well-differentiated hepatocellular neoplasms were identified. The lesions were ill-defined without encapsulation. While the majority of the lesion showed an intact reticulin framework of the cell plates or only slightly decreased reticulin framework in keeping with the diagnosis of HA (Figure 1A and 1B), there were areas where the cell plates were markedly thickened and reticulin framework was significantly reduced or completely lost. These areas were diagnostic of HCC (Figure 1C and 1D).
Figure 1:
Histopathologic and immunohistochemical features of an HNF1A mutated HCC arising in a background of LFABP deficient hepatic adenomatosis. A) and B) An area of hepatic adenoma has retained reticulin framework while C) and D) show an are of carcinoma arising in the hepatic adenoma that has loss of the reticulin framework. E) The borders of the adenoma are ill-defined with the neoplastic hepatocytes on the bottom half of the photo, and F) LFABP is diffusely lost in the adenoma and retained in the non-neoplastic hepatocytes in the lower left corner of the image (A,C, E: H&E 200x original magnification; B, D: reticulin stain 200x original magnification; F: LFABP immunohistochemical stain, Abcam, 200x original magnification)
Immunohistochemical stains showed that LFABP was lost in both the adenoma and the carcinoma components (Figure 1E and 1F). Staining for serum amyloid A (SAA) and C reactive protein (CRP) were also negative in both components. Beta catenin staining demonstrated retained membranous staining without nuclear accumulation. Glutamine synthetase immunohistochemistry was weak and patchy.
3.1.3. Molecular Pathology Results
Two somatic mutations were identified in HNF1A on MSK-IMPACT testing. These 2 mutations: HNF1A p. E32* and L214Q occurred at clonal frequencies (31.3% and 35.8%, respectively). No loss of heterozygosity or copy number losses were detected in HNF1A using FACETs analysis. A third and last mutation identified in the tumor was Mediator Complex Unit 12 (MED12) p. A1133D, with a variant frequency of 29.2%. Other analyses revealed that the tumor was microsatellite stable, diploid without significant copy number gains or losses, and had a tumor mutation burden (2.6 mutations per megabase) while the median for other hepatocellular carcinomas profiled by MSK-IMPACT was 3.5 mutations per megabase.
The patient’s blood was separately analyzed for common pathogenic germline mutations, and no pathogenic alterations were identified on a limited panel. HNF1A was separately reviewed and found to be negative for germline mutations and copy number alterations.
3.2. HNF1A mutations in hepatocellular adenoma and carcinoma: TCGA and the Literature
We then reviewed the provisional TCGA dataset. Interestingly, HNF1A mutations were identified in 8 of 373 patients with mutation data (1.8% of TCGA HCC patients). HNF1A mutations were also identified in the literature in 2 of 146 (1.4%) HCC[12]. In total, 11 of 672 (1.6%) patients with HCC harbor HNF1A mutations were identified between MSK-IMPACT, the provisional TCGA dataset, and the literature.
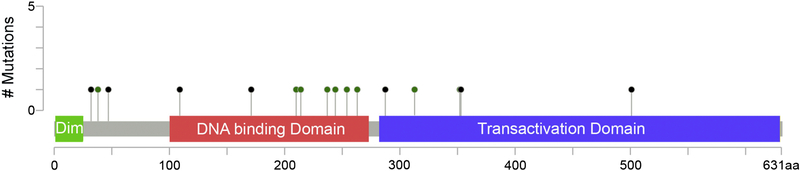
We also reviewed available TCGA data for 30 patients with hepatic adenomas. This analysis revealed that 5 of 30 (17%) of patients with HAs harbored HNF1A mutations. The HNF1A mutations and clinicopathologic features of these cases are summarized in Table 1 and Figure 2.
Table 1.
Clinicopathologic features of HNF1A mutated hepatocellular carcinomas and adenomas.
| Case | Tumor type | HNF1A mutation (amino acid change) | HNF1A mutation (nucleotide change) | Age | Sex | Grade | Stage | Ishak Fibrosis |
|---|---|---|---|---|---|---|---|---|
| MSKCC | HCC | E32*, L214Q | 96G>T, 641T>A | 65 | F | 2 | T1N0MX | 0–1 |
| TCGA-DD-A1EB-01 | HCC | L254q, E48Pfs*106 | 761T>A, 140delGGGA | 72 | F | 2 | T1N0MX | 0 |
| TCGA-DD-A39W-01 | HCC | S210T | 628T>A | 29 | F | 2 | T3NXM0 | 0 |
| TCGA-DD-A3A2-01 | HCC | R171*, N237S | 511C>T, 710A>G | 76 | F | 1 | T1N0MX | 0 |
| TCGA-DD-A73C-01 | HCC | x109_splice | 326+2T>A | 65 | F | 1 | T3aN0MX | 0 |
| TCGA-NI-A4U2-01 | HCC | S352F, R244G, T354Rfs*10 | 1055C>T, 730A>G, 1057delC | 71 | M | 1 | T3N0Mx | 1–2 |
| TCGA-T1-A6J8-01 | HCC | L38P | 113T>C | 68 | M | 2 | T1NXM0 | 0 |
| TCGA-WJ-A86L-01 | HCC | X501_splice | 1502–1G>A | 68 | F | 2 | T1N0Mx | 0 |
| TCGA-CC-A3M9–01 | HCC | R263G | 787C>G | 45 | M | 3 | T2NXM0 | - |
| Inoue et al13 | HCC | P309L | 926C>T | - | - | - | - | - |
| Inoue et al13 | HCC | P291fs | - | - | - | - | - | - |
| CHC462/463T | HA | Q211P, P224A | 632A>C, 670C>G | 35 | F | 0–1 | ||
| Q211P, W206L | 632A>C, 617G>T | |||||||
| CHC464/465T | HA | I27_i30del, q250_g255del | 71del AGGCACTGATCC, 747del ACAGGCACAGGGGCTGGG | 53 | F | 0–1 | ||
| I27_i30del, q250_g255del | 71del AGGCACTGATCC, 747del ACAGGCACAGGGGCTGGG | |||||||
| CHC575/578T | HA | x319_splice, X176_splice, | 956–1delG, 526del CGTAAGTAATGAC, 1107+1G>T, | 34 | F | 0–1 | ||
| x369_splice, R263L | 788G>T, | |||||||
| CHC687/689T | HA | R272S | 814C>A | 47 | F | 0–1 | ||
| S19* | 56C>G | |||||||
| CHC1425/1428T | HA | P447L, R271delinsPG | 1340C>T, 811insCGG | 27 | F | 0–1 | ||
| Q460*, w206L | 1378C>T, 617G>T |
Figure 2:
HNF1A mutations detected across 11 HCCs. The mutations consist of missense (green dots) and truncating (black dots) mutations, without recurrent hotspot, as is seen with many tumor suppressors. No mutations were seen in the dimerizeration domain (dim) while numerous mutations were seen in the DNA binding and transactivation domains.
In the provisional TCGA HCC set, HNF1A mutations were mutually exclusive with hotspot CTNNB1 mutations, the latter occurring in 93 of 366 HCC. Four of the 8 (50%) TCGA HCC with HNF1A mutations also harbored TP53 mutations. In the TCGA HA dataset, HNFA1 mutated HAs (n=5) were mutually exclusive with CTNNB1 HAs (n=13), while no HAs had TP53 mutations.
The Ishak fibrosis score was available for 211 TCGA HCC patients with wt HNF1A (Ishak score 0–1: n=70; Ishak score 1–2: n=30; Ishak score 3–4: n=30; Ishak score 5: n=9; Ishak score 6: n=72). The grade was available for 358 TCGA HCC patients with wt HNF1A (G1: n=55; G2; n=180; G3; n=123). Patient sex was available for 369 TCGA HCC patients with wt HNF1A (female: n=117). T stage was available for 374 TCGA HCC patients with wt HNF1A (T1:n=185; T2: n=95; T3:n=81; T4: n=13). The viral hepatitis status was available for 250 TCGA HCC patients with wt HNF1A (Hepatitis B core positive: n=107, Hepatitis C positive: n=57). In comparison to other TCGA patient with HCC, patients with HNF1A mutations were significantly less likely to have viral hepatitis (p=0.004), and there was a trend for patients to lack cirrhosis (p=0.03) and be predominantly female (p=0.06). No association was seen with grade or stage. These findings are summarized in Table 2.
Table 2.
Clinicopathologic characteristics of HNF1A mutated vs wild type HCC in the TCGA dataset
| Cirrhosis (p=0.03) | Hepatitis B or C (p=0.004) | Grade 3 (p=0.28) | Female % (p=0.06) | |
|---|---|---|---|---|
| HNF1A mutated | 0% | 0% | 11% | 67% |
| HNF1A wild type | 34% | 47% | 34% | 32% |
4. Discussion
We analyzed the prevalence and features of HNF1A mutated hepatocellular neoplasms within institutional (MSK-IMPACT) and TCGA NGS data, including an in depth description of an institutional patient with two somatic mutations in HNF1A in a hepatocellular carcinoma arising in a background of LFABP-deficient hepatocellular adenomatosis.
While the average age of HNF1A mutated HCC was 62 years, the average age of HNF1A mutated HAs was 39 years. These data and the fact that the only 2 HNF1A mutated HCC with described histology patterns have carcinoma arising in an adenoma suggest that HNF1A mutations occur early in hepatocellular neoplasia and that, albeit rarely, HAs with HNF1A mutations may progress to HCC over time. The case sequenced by MSK-IMPACT, which was detected in a 65 year old woman with foci of carcinoma arising within areas of HA, is an example of this malignant transformation.
Molecular sequencing (TCGA data) reveals that approximately 20% of HAs have somatic mutations. Over 90% of HNF1A mutated HAs occur in women, with 67% of these HAs occurring in patients who have used oral contraceptives. Similar to HNF1A mutated adenomas, HNF1A mutated carcinomas trend to occur at a higher frequency in women. This higher prevalence in women, in addition to the link to oral contraceptives suggests that HNF1A-mutated carcinomas may be related to higher levels of estrogen. While HCC classically occurs at a higher incidence in men compared to women, the particularity of HNF1A mutations occurring more frequently in female HCC patients stands out.
Recent research has shown that HNF1A inactivated HAs are a subcategory of HA that account for almost half of HAs. These HAs generally lack abnormalities in other IHC biomarkers that define other HA subcategories such as beta catenin, C-reactive protein, serum amyloid A, and glutamine synthetase (map like in HAs) [12], further supporting the important role of HNF1A activation in tumor development. The mutual exclusivity of CTNNB1 mutations with HNF1A mutations is well known in HAs [2]. Here we find that this mutually exclusive pattern holds true for HCC, supporting the idea that HNF1A mutations play a role in HCC development and occur via a different pathway than HCC with CTNNB1 mutations.
Searching the literature, two case reports in HCC arising in individuals with germline HNF1A mutations [4,7], 1 study reporting 2 HNF1A mutant HCC out of 16 LFABP deficient HAS [13], and 1 study reporting carcinoma arising of 1 of 44 HNF1A mutant HAs were identified.13 Both of the 2 case reports of HCC arising in patients with germline HNF1A mutations occurred in patients with Maturity Onset Diabetes of the Young 3 (MODY3). The patient reported by Willson et al had a germline deletion of HNF1 exons 2–3 and presented at 59 years old with biliary colic. A 4.4 cm mass, originally though to be focal nodular hyperplasia, was identified. When the mass increased in size, biopsy revealed HCC that was unresectable by the time of biopsy, and the patient died at the age of 65 years old [4]. The patient reported by Stueck et al was a 23 year old woman with MODY3 who developed a pathologically distinct HCC arising within an LFABP deficient hepatic adenomatosis [7]. In a report by Zucman-Rossi et al, 1 of 44 (2%) of HNF1A mutated adenomas had carcinoma arising in it; and the specifics of that case were not described [14]. Separately, of the 9 HNF1A mutated HCC identified with clinical history available, none occurred in a patient with viral hepatitis or cirrhosis.
The patient from our institution had only 1 somatic mutation on MSK-IMPACT in addition to the HNF1A mutations, which was a MED12 p. A1133D mutation. While this mutation occurred at a clonal frequency, it has not been described in other HAs or HCCs and is not in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. No other solid malignancies out of over 25,000 tumors sequenced on MSKIMPACT harbored this MED12 mutation. MED12 mutations have mainly been described in benign uterine leiyomomas, and the mutations described occur at hotspots different from the one detected in the hepatocellular neoplasm [15]. Thus, the biologic significance of the non-hotspot missense mutation detected in the HCC is unclear.
There are several limitations to this study. We did not have access to the TCGA tissues to review the histology or thoroughly analyze those cases with IHC and special stains such as reticulin. Additionally, due to the rarity of HNF1A mutations in HCC, the number of cases is very small.
While the number of described cases of HCC with HNF1A mutation remains small (1–2% of all HCCs), we have shown that HNF1A mutated HCC share specific background features and often arise in a background of adenomatosis without significant background liver disease. This information suggests that thorough examination and sampling of HNF1A mutated/ LFABP deficient HAs is necessary. The carcinoma may be very well differentiated and focal. While carcinoma arising in LFABP deficient HAs remains rare, it is a well described event that is important to diagnose.
Highlights.
HNF1A mutated hepatic adenomas can harbor focal areas of malignant transformation
HNF1A mutations occur in 1–2% of hepatocellular carcinomas. Similar to hepatic adenomas, HNF1A mutations are mutually exclusive with CTNNB1 mutations.
HNF1A mutations in HCC are associated with negative viral status and tend to occur more commonly in women and without a background of cirrhosis
Acknowledgments
This study was funded by the National Cancer Institute (NCI) under the MSK Cancer Center Support Grant/Core Grant (P30 CA008748).
This study was approved by the institutional review board at Memorial Sloan Kettering Cancer Center.
Footnotes
The authors declare no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Luo Z, Li Y, Wang H, Fleming J, Li M, Kang Y, Zhang R, Li D. Hepatocyte nuclear factor 1A (HNF1A) as a possible tumor suppressor in pancreatic cancer. PLoS One 2015;10:e0121082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dhingra S, Fiel MI. Update on the new classification of hepatic adenomas: clinical, molecular, and pathologic characteristics. Arch Pathol Lab Med 2014;138:1090–7. [DOI] [PubMed] [Google Scholar]
- [3].Rebouissou S, Imbeaud S, Balabaud C, et al. HNF1 alpha inactivation promotes lipogenesis in human hepatic adenoma independently of SREBP-1 and carbohydrate-response element-binding protein (ChREBP) activation. J Biol Chem 2007;282:14437–14446. [DOI] [PubMed] [Google Scholar]
- [4].Willson JS, Godwin TD, Wiggins GA, Guilford PJ, McCall JL. Primary hepatocellular neoplasms in a MODY3 family with a novel HNF1A germline mutation. J Hepatol 2013;59:904–7. [DOI] [PubMed] [Google Scholar]
- [5].Bacq Y, Jacquemin E, Balabaud C, et al. Familial liver adenomatosis associated with hepatocyte nuclear factor 1alpha inactivation. Gastroenterology 2003;125:1470–5. [DOI] [PubMed] [Google Scholar]
- [6].Reznik Y, Dao T, Coutant R, et al. Hepatocyte nuclear factor-1 alpha gene inactivation: cosegregation between liver adenomatosis and diabetes phenotypes in two maturity-onset diabetes of the young (MODY)3 families. J Clin Endocrinol Metab 2004;89:1476–80. [DOI] [PubMed] [Google Scholar]
- [7].Stueck AE, Qu Z, Huang MA, Campreciós G, Ferrell LD, Thung SN. Hepatocellular Carcinoma Arising in an HNF-1α-Mutated Adenoma in a 23-Year-Old Woman with Maturity-Onset Diabetes of the Young: A Case Report. Semin Liver Dis 2015;35:444–9. [DOI] [PubMed] [Google Scholar]
- [8].Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO PO 2017;1:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nault JC, Paradis V, Cherqui D, Vilgrain V, Zucman-Rossi J. Molecular classification of hepatocellular adenoma in clinical practice. J Hepatol 2017;67:1074–83. [DOI] [PubMed] [Google Scholar]
- [13].Inoue M, Takahashi Y, Fujii T, Kitagawa M, Fukusato T. Significance of downregulation of liver fatty acid-binding protein in hepatocellular carcinoma. World J Gastroenterol 2014;20:17541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zucman-Rossi J, Jeannot E, Nhieu JT, et al. Genotype-phenotype correlation in hepatocellular adenoma: new classification and relationship with HCC. Hepatology 2006;43:515–24. [DOI] [PubMed] [Google Scholar]
- [15].Mäkinen N, Mehine M, Tolvanen J, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011;334:252–5. [DOI] [PubMed] [Google Scholar]