Abstract
Background/Objectives
Obesity is an important risk factor for the development of diseases such as diabetes mellitus, hypertension, and dyslipidemia; however, a small number of individuals with long-standing obesity do not present with these cardiometabolic diseases. Such individuals are referred to as metabolically healthy obese (MHO) and potentially represent a subgroup of the general population with a protective genetic predisposition to obesity-related diseases. We hypothesized that individuals who were metabolically healthy but significantly obese (BMI ≥35 kg/m2) would represent a highly homogenous subgroup, with which to investigate potential genetic associations to obesity. We further hypothesized that such a cohort may lend itself well to investigate potential genotypes that are protective with respect to the development of cardiometabolic disease.
Subjects/Methods
In the present study, we implemented this novel selection strategy by screening 892 individuals diagnosed as Class 2 or Class 3 obese and identified 38 who presented without any manifestations of cardiometabolic disease. We then assessed these subjects for single nucleotide polymorphisms (SNPs) that associated with this phenotype.
Results
Our analysis identified 89 SNPs that reach statistical significance (p<1×10−5), some of which are associated with genes of biological pathways that influences dietary behavior; others are associated with genes previously linked to obesity and cardiometabolic disease as well as neuroimmune disease. This study, to the best of our knowledge, represents the first genetic screening of a cardiometabolically healthy but significantly obese population.
Keywords: MHO, metabolic syndrome, comorbidity, SNP, GWAS, neuroimmune
INTRODUCTION
In the United States, two of every three adults are considered obese and 1 in 20 are severely obese 1. A number of generally modifiable proclivities contribute to these statistics such as sedentary lifestyle, inadequate sleeping practices, and poor eating habits 2. Other factors that are less modifiable, including socioeconomic status and the availability of proper nutrition, also contribute 3. Additionally, some factors, which are not modifiable, including age, gender, and genetic background, likewise contribute 1. While the contributions of age and gender to the development of obesity are largely understood, the contributions of genetics are more complex. For instance, it is generally believed that gene-environment interactions are obligatory in most instances 4. Furthermore, while a few specific genetic polymorphisms are well documented to associate with obesity, the frequency of most are not sufficient in the general population to account for the observed prevalence. In fact, it is likely that polygenic contributions are necessary to define a genetic predisposition to the development of obesity.
The majority of studies that have investigated potential genetic predispositions to the development of obesity have included subjects with obesity-related comorbidities, such as type 2 diabetes or heart disease 5. While obesity is a known risk factor for these comorbidities, their presence may introduce potential confounding factors when conducting strict investigations into the genetic underpinnings of obesity. This is because these diseases may also have a genetic basis independent of obesity, as they impact the non-obese as well. Alternatively, these diseases may have a genetic component that may actually favor the onset of obesity. For those studies exclusively focused on obesity, to the best of our knowledge, none have specifically excluded these potentially confounding comorbidities.
It is believed that certain genetic predispositions exist that are protective with respect to cardiometabolic diseases. Consistent with this supposition, some individuals with long-standing obesity do not develop these cardiometabolic diseases. This phenomenon, originally coined by Sims in 2001 as a metabolically normal subgroup of obese (OBMN) 6 is now more commonly referred to as metabolically healthy obese (MHO) 7. Notwithstanding, the concept of MHO has been the subject of ongoing debate 8, 9. While those who are MHO may not present with cardiometabolic disease at the time of their evaluation; previous studies contend that these individuals are at increased risk of other adverse outcomes, such as cardiovascular disease 10. Conversely, other studies support that an MHO phenotype may be associated with a protective benefit. For instance, recently, Iglesias Molli et al. reported that MHO subjects presented similar levels of chronic inflammation but less insulin-resistance than obese subjects with metabolic syndrome 11.
Although a consensus regarding the definition of MHO has not been reached, most studies define MHO as a body mass index (BMI) ≥ 30 kg/m2 and without the presence of metabolic diseases such as type 2 diabetes, dyslipidemia, or hypertension 6, 12. To the best of our knowledge, an MHO Class 2 or Class 3 (BMI of 35 to < 40 or BMI of 40 or higher, respectively) cohort has not been investigated. With this in mind, we hypothesized that such individuals would represent a profoundly homogenous subgroup, to investigate potential genetic associations to obesity. We further hypothesized that such a cohort may allow potential genotypes, that are protective with respect to the development of cardiometabolic disease, to be identified.
In the present study, we screened 892 individuals classified as either Class 2 or 3 obese and identified 38 who presented without any manifestations of metabolic diseases. When the MHO cohort was compared to 32 lean healthy controls, we identified 89 SNPs that reach statistical significance. In spite of the small sample size of the study cohort, our highly specific selection criteria allowed us to identify novel polymorphisms in obesity-related biological pathways, previously implicated in larger obese genetic studies. Our data provides additional validation of these previous studies as well as identifies novel obesity-related candidate genes.
MATERIALS AND METHODS
Ethics statement
This study was conducted under the guidelines of the Declaration of Helsinki. All subjects provided written informed consent before participation under a protocol approved by the University of Nevada, Reno, Institutional Review Board (Protocol # 2013B027).
Study subjects
Subjects for this study were recruited from patients who visited the Wellness and Weight Management Clinic and Internal Medicine Clinic at the University of Nevada, Reno, between 2009 and 2012. Cases and controls were generally matched with respect to age (Table 1). In order to remove any confounding variables that resulted from gender bias, we elected to include only females in our study. Potential participants were phenotyped using a clinical algorithm to identify cases with the following characteristics: self-identified as European American (Caucasian), female, at least 40 years old, BMI ≥ 35 kg/m2, with normal blood pressure, normal fasting plasma glucose (less than 100 mg/dL), and a desirable fasting lipid panel based on Adult Treatment Panel III recommendations 13. Potential participants who, at any time, had used antidiabetic, lipid-lowering, or antihypertensive drugs were excluded. Participants with a past history of diabetes mellitus, hypertension, metabolic syndrome, coronary artery disease (angina, myocardial infraction, abnormal cardiac stress test), thyroid dysfunctions, Cushing’s disease, polycystic ovary disease, bulimia/anorexia, and family history of premature coronary artery diseases were also excluded. Any potential subjects with a history of diet-associated weight loss within six months of enrollment were further excluded. Additionally, all participants were non-smokers (current or past history of smoking) and all subjects self-reported their diet as “regular”. Lastly, all obese subjects reported as having been obese their entire adult life.
Table 1.
Clinical characteristics of study subjects
| Cases (mean±SD) | Controls (mean±SD) | p-value* | |
|---|---|---|---|
| Age | 51.1±7.4 | 53.1±8.8 | 0.37 |
| Height (cm) | 163.0±7.2 | 166.2±6.8 | 0.08 |
| Weight (kg) | 115.0±19.5 | 62.1±6.4 | 7.90×10−13 |
| BMI (kg/m2) | 42.9±6.4 | 22.4±1.8 | 3.90×10−13 |
| Systolic blood pressure (mmHg) | 123.3±8.7 | 117.0±10.5 | 0.014 |
| Diastolic blood pressure (mmHg) | 75.8±9.0 | 70.1±6.2 | 0.002 |
| Fasting blood glucose (mg/dL) | 92.3±11.0 | 89.2±6.7 | 0.17 |
| Total Cholesterol (mg/dL) | 192.6±32.8 | 191.1±24.3 | 0.80 |
| HDL-cholesterol (mg/dL) | 58.6±17.7 | 70.8±16.2 | 0.02 |
| LDL-cholesterol (mg/dL) | 109.7±28.4 | 105.1±23.6 | 0.63 |
| Triglycerides (mg/dL) | 127.0 ±45.6 | 80.2±26.9 | 2.30×10−6 |
Non-parametric t-tests were performed on all variables to be consistent, as some were not representative of a normally distributed dataset. Note that although there are statistically significant differences in blood pressure, HDL cholesterol, and triglyceride levels, the differences are from the clinical perspective minor and in part due to the small sample size. All subjects meet the criteria for normal blood pressure and lipid panel as per ATPIII and have fasting glucose less than 100 mg/dL.
Sample size justification
Following the study of Smith et al. 14, that reported 64 statistically significantly associated SNPs of effect size (allelic odds ratio) ranging between 0 and 15.7, this study enrolled 38 cases and 32 controls. Post hoc power computations using the QUANTO v. 1.24 power calculator 15 indicate that the 89 SNPs reported here have at least 80% power to detect effect sizes (allelic odds ratios) as small as 2.8, depending on minor allele frequencies (MAFs) of these SNPs and the genetic model used. It is noteworthy that the log-additive model is the least conservative in these calculations. MAF values ranged from 0.043 to 0.50 in this set of 89 SNPs. QUANTO was set to compute power of allelic associations based on the additive, recessive, and dominant models of MAFs between 0.043 and 0.50, significance level alpha = 0.05 and population prevalence 0.351%, following power calculations of similar studies 16, 17. Results from these calculations are available in Supplementary Table 1 and the corresponding odds ratio at each allele frequency are highlighted for clarity.
Our control subjects were comprised of healthy, self-identified European-American females, at least 40 years of age with a BMI ≤ 24.9 kg/m2. In addition, the control subjects who underwent any form of weight loss treatment, including lifestyle interventions, medications, or surgery, were excluded. Anthropometric measurements (weight and height) and blood pressure were obtained using calibrated, professional clinical instruments in the respective clinics described above. Laboratory testing (fasting lipid panel, fasting blood glucose) was performed at nationally standardized, CLIA (Clinical Laboratory Improvement Amendments) certified laboratories as part of routine patient care. A list of the study subjects’ clinical characteristics is presented in Table 1.
Single nucleotide polymorphism analysis
DNA was extracted from approximately 2 ml of saliva, collected using the Oragene®DNA 500 Saliva Collection Kit (DNA Genotek Inc., Ottawa, ON, Canada) according to the manufacturer’s instructions. SNPs were identified using the Affymetrix Genome-Wide SNP Array 6.0 according to methods previously described by us 17.
Data analysis
The Affymetrix Genome-Wide Human SNP Array 6.0 was used to identify potential associations of 905,027 SNPs with the MHO diagnosis. Affymetrix CEL files were first processed using the Corrected Robust Linear Model with maximum Likelihood Classification (CRLMM) genotyping algorithm using the R package crlmm 18. Standard quality control measures were used to assess the chip and sample reliability. Specifically, only SNPs having a minor allele frequency of at least 3.5% in our sample set were considered (193,184 SNPs were excluded due to having a minor allele frequency less than or equal to 3.5%). All SNPs were examined for low call rates (below 95%), and none were excluded because of this. All samples were verified to have an SNP call rate of 95% or greater, and sample genders were verified with heterozygosity of the X chromosome. A total of 1,836 markers were excluded because their genotype frequencies were inconsistent with Hardy-Weinberg equilibrium (Chi-Squared FDR values with p<0.2). There were 710,007 SNPs on the array passing this standard quality control protocol across all 70 samples.
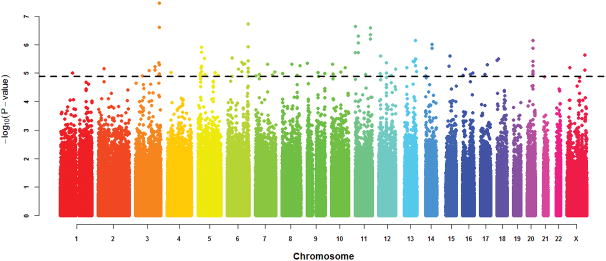
Three single-location association tests were performed on these 710,007 SNPs. A genome-wide test for association was performed on each SNP via a simple logistic regression and computation of the p-value of the likelihood ratio test upon comparison with the null model 19. As the mode of inheritance is currently unknown in obesity, the study was based on the codominant (additive) model, which represents the most general model available 20. Association was also performed using the log-additive, recessive, dominant, and overdominant genetic models. The p-values of the likelihood ratio test were adjusted for multiple testing using the False Discovery Rate (FDR) method (Figure 1) 21. Both raw and adjusted p-values are shown in Supplementary Table 2. SNPs with association tests resulting in any of the five models with a raw p-value of p<1×10−5 were selected for further study. These values correspond to the eligibility criteria of published SNPs in the NHGRI-EBI Catalog of published genome-wide association studies [https://www.ebi.ac.uk/gwas/docs/methods].
Figure 1.
Manhattan plot of genome-wide association raw p-values. The horizontal line corresponds to single SNP associations with p-values of p=1×10−5.
Additionally, a simple Chi-squared hypothesis test (two degrees of freedom) for association between the three possible genotypes of each SNP and the disease trait was also performed on each of the 710,007 SNPs, which we refer to here as the genotypic association test. Next, a standard Fisher’s exact test (one degree of freedom) was performed on the allelic distribution between cases and controls of each SNP. A standard, conservative genomic control method was used to test and control for the overall inflation of the allelic association test statistic (inflation factor γ= 1.03) 22.
SNP positions are consistent with the 2013 human genome assembly (GRCh38/hg38) and are assigned to a gene if the respective SNP is within 40 kb of the gene 23. SNPs that are not within 40 kb of any gene are referred to as intergenic in our tables 23.
RESULTS
Identification of SNPs that associate with MHO
Of the 710,007 candidate SNPs that passed quality control measures, 89 statistically significant SNPs were identified to associate with our MHO cohort (Supplemental Table 2). In order to assess the possibility that these SNPs were associated with a predisposition to the development of obesity or cardiometabolic disease, we conducted a manual search of the medical literature and databases of genotypes, phenotypes, and protein function. Our search determined that 12 genes that were associated with our significant SNPs were also associated with obesity or metabolic disease (Table 2). For example, two SNPs were identified in the 3-prime untranslated region of RTP4; the chemosensory receptor chemosensory transporter protein 4 (p =3×10−8 and p =5×10−6). RTP4 expression has been reported to be predictive of future hypertension incidence in type 2 diabetic patients 24. Additionally, we identified an intron variant of the Potassium Voltage-Gated Channel Subfamily Q Member 1 gene (KCNQ1) that associated with the MHO phenotype (p=1×10−5). Several studies have reported KCNQ1 polymorphism associations for obesity, type 2 diabetes, or cardiovascular risk 25, 26.
Table 2.
Genes with significant SNPs that are associated with obesity or metabolic disease
| Gene | Gene associations with Obesity/Metabolic Phenotype | Ref. |
|---|---|---|
| DPYD | Associated with syndromic obesity | 31 |
| KLHL6 | Associated with acute insulin response to glucose | 32 |
| RTP4 | Associated with future incidence of hypertension; regulates bitter taste receptor | 24, 33 |
| PCDH7 | Associated with waist-to-hip ratio on total cholesterol | 34 |
| FSTL4 | Associated with increased risk of stroke | 35 |
| CSMD1 | Associated with metabolic syndrome | 36 |
| ITIH5 | Expression correlates with body mass index | 37 |
| KCNQ1 | Associated with type 2 diabetes | 38, 39 |
| NBEA | Associated with body mass index | 40 |
| TOX2 | Associated with increased diastolic BP | 40 |
| BBX | Associated with type 1 diabetes mellitus | 41 |
| NTRK2 | Associated with obesity and depression | 28, 29 |
In contrast to the aforementioned examples, which were previously associated with increased risk of obesity or metabolic disease, we also observed a significant polymorphism in the Inter-Alpha-Trypsin Inhibitor Heavy Chain Family Member 5 gene ITIH5. Anveden et al. reported that ITIH-5 is highly expressed in adipose tissue and is increased in obesity, but is down regulated after weight loss 27. Therefore, an ITIH5 polymorphism that decreases transcription may represent a protective phenotype with respect to metabolic disease. We also observed three SNPs in the TOX High Mobility Group Box Family Member 2 gene, TOX2. TOX2 is a putative transcriptional activator involved in the hypothalamo-pituitary-gonadal system. SNPs in TOX2 have been reported to associate with increased, as well as decreased, diastolic blood pressure. Other obesity-related genes are presented in Table 2.
Two significant SNPs were identified in our study have been reported to associate with human disease. The SNP rs984430 is an intron variant in the Neurotrophic Receptor Tyrosine Kinase 2 gene (NTRK2). Polymorphisms in this gene have been identified as a genetic risk factor for the development of severe obesity 28 and this specific SNP was identified as a risk factor for depression 29. The other SNP (rs9736016) is an intergenic variant between the chemokine (C-X-C motif) receptor 5 and the DEAD (Asp-Glu-Ala-Asp) box helicase 6 (CXCR5 and DDX6, respectively) and is identified as a risk factor for multiple sclerosis 30.
Multiple SNPs in proximity to specific genes
In order to identify genotypic differences and patterns in regions near an SNP or SNPs of statistical significance, we utilized the Open Source plotting tool, GenotypePlotter, which was specifically designed to view genotypic patterns of cases and controls simultaneously as well as to organize both phased and unphased chromosomes in regions around potential causative SNPs of interest 17. GenotypePlotter uses a novel clustering scheme to organize samples into similar patterns based on their genotypes across a region, providing a user-friendly overview of differences between cohorts. After organizing the samples over a selection of SNPs, genotypes are portrayed in different colors to represent a type of heatmap: red cells indicate sample genotypes that are homozygous with respect to the minor allele for that SNP; blue cells indicate sample genotypes that are homozygous with respect to the major allele; and yellow cells represent heterozygous genotypes. GenotypePlotter was developed by coauthor Schlauch and is available upon request.
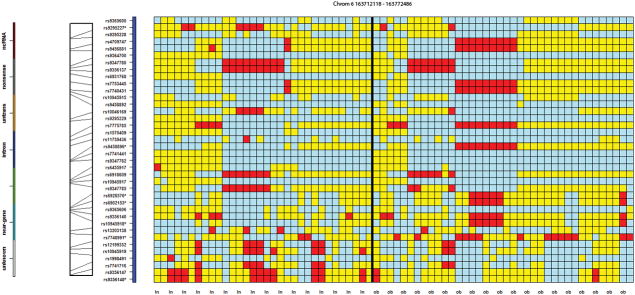
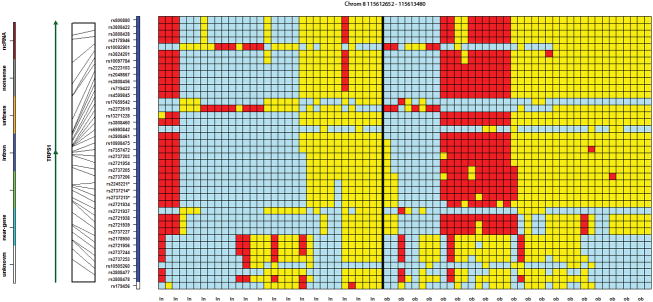
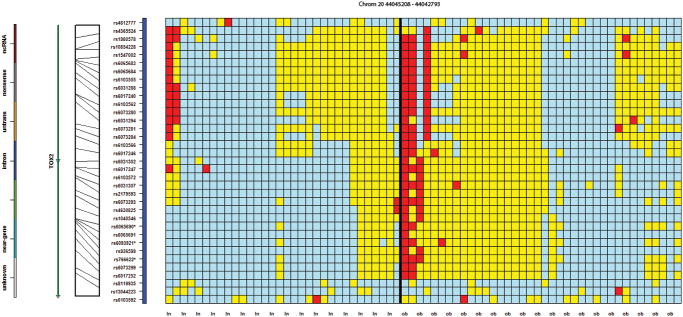
Of the 89 statistically significant SNPs, 31 were observed to be in proximity to at least one other SNP of the same gene or intergenic region, across twelve different loci, suggesting these genes or chromosomal regions may have special relevance. For example, Figure 2 presents a 50 kb region containing seven statistically significant SNPs on chromosome 6 (rs9295227, rs9458896, rs6928576, rs6902153, rs10945918, rs7748991 and rs9356148). Figure 3 presents three SNPs (rs2245221, rs2737214, and rs2737215), within 500 base pairs of each other, on chromosome 8, in an intronic region of the Transcriptional Repressor GATA Binding 1 (TRPS1), and three SNPs (rs766622, rs6065690 and rs6093921) were observed within 3 kb of each other on chromosome 20 in an intronic region of TOX2 (Figure 4). Eight additional regions were observed to have at least two SNPs in proximity to each other (Supplementary Table 3; colored region).
Figure 2.
A genotypic organization of 32 controls (first 32 columns) and 38 MHO cases (last 38 columns) on chromosome 6 between 163,712,118 and 163,772,486, containing the LOC107986666 gene. This region contains seven SNPs found to be statistically significantly associated with the MHO cohort. The first seven cases show a haplotypic pattern not shared by the control cohort: the red cells represent the homozygous genotype of the minor allele. The light blue cells denote the homozygous genotype of the major allele. Also of note is the rather distinct region that the last eight control samples share. These are relatively large haplotypic regions not typically shared between many individuals. The color bar, directly to the left of the heat map, shows the intragenic and near-gene nature of most of the SNPs in the region.
Figure 3.
A genotypic organization of 32 controls (first 32 columns) and 38 MHO cases (last 38 columns) on chromosome 8 between 115,612,652 and 115,613,480 containing the TRPS1 gene. The distinct genotypic pattern shared by the ten cases is seen in only three controls.
Figure 4.
A genotypic organization of 32 controls (first 32 columns) and 38 MHO cases (last 38 columns) on chromosome 20 between 44,045,208 and 44,042,793, containing the TOX2 gene. The genotypic pattern shared by the three MHO cases at the top left of the second panel does not occur in any of the controls.
DISCUSSION
The purpose of this study was to identify novel polymorphisms that contribute to a predisposition to the development of obesity. We also investigated our unique MHO cohort to potentially identify candidate polymorphisms that may represent a protective genotype to the development of cardiometabolic disease. Although our sample size was relatively small for a genome wide association study (GWAS), if one considers the selectivity of our screening process, the power of this method adequately compensates for the small sample size.
A BMI is a ratio of an individual’s weight-to-height and is customarily calculated by dividing one’s weight (in kilograms) by the square of one’s height (in meters). Recent guidelines from the U.S. Centers for Disease Control and Prevention (CDC) and the World Health Organization define a healthy BMI range as 18.5 to 24.9 kg/m2. These guidelines define overweight range as a BMI of 25.0 to 29.9 kg/m2 and obesity as a BMI over 30.0 kg/m2. CDC guidelines further subdivide obesity into three categories: Class 1: BMI of 30 to < 35; Class 2: BMI of 35 to < 40; and Class 3: BMI of 40 or higher. In the present study, we investigated subjects who presented with either Class 2 or Class 3 obesity (mean BMI of 42.9±6.2) and who also met the added criteria of being MHO (Table 1).
As of 2010, it was estimated that approximately 27.2% of the U.S. population was obese with a BMI>30, whereas 3.7% were severely obese (Class 3) with a BMI>40 31. Because the prevalence of a female Caucasian MHO cohort with a BMI>40 not been previously reported, we estimated the prevalence as follows: Although the vast majority of our cases had BMI>40, a small number had BMI between 35 and 40 and thus our inclusion criteria were set at BMI>35 (actual mean BMI was 42.9±6.2). Flegal et al. estimates the prevalence for Caucasian females between the ages of 40 and 59 years to have a BMI > 35 as 15.6% and for BMI>40 as 7.3% 32. Thus, a conservative estimate for the prevalence of Caucasian females in our cohort with mean BMI 42.9 is 7.3%. It was necessary to screen 892 Class 2 Caucasian individuals to identify 38 MHO Caucasian females for this study. As 38/890 = 4.27%, we estimated the prevalence of metabolically healthy but severely obese individuals (BMI>40) in the general US female Caucasian population as 0.315% using Bayes Theorem: P(MHO|BMI>40) = P(MHO ∩ BMI>40)/P(BMI>40). Thus, P(MHO ∩ BMI>40) = (.0427)(.073) = .00351or 0.351%. Consequently, if one were to conduct a GWAS that included subjects who were merely obese, it would be necessary to include over 12,000 obese subjects in order to capture 40 severely obese MHO subjects. This calculation is not meant to be a rigorous power analysis, but to underscore the rigor of our selection method in identifying a very specific subset of the obese.
In our study, we identified 89 SNPs that reached statistical significance. In order to identify candidate SNPs in genes that may lead to a predisposition to obesity or those that may be protective from the development of obesity-related metabolic disease, we conducted a manual search of the medical literature and databases of genotypes, phenotypes, and protein function. Our search determined that 12 genes associated with significant SNPs were also associated with obesity or cardiometabolic disease (Table 2). For instance, we identified two significant SNPs in the (chemosensory) receptor transporter protein 4 gene, RTP4. The product of this gene facilitates trafficking and expression of some G-protein coupled receptors and promotes functional expression of the bitter taste receptor TAS2R1633. Although RTP4 has not been previously associated with obesity or metabolic disease, variation in the bitter-taste receptor genes have been linked to obesity 34, altered glucose and insulin homeostasis35, and circulating levels of thyroid hormones 36. We also observed three SNPs in the TOX High Mobility Group Box Family Member 2 gene, TOX2. Previous studies have reported that the expression of TOX2 is associated with regulation of the transcription factor TBX21 37. Although such an association is indirect, other studies have reported a lack of TBX21 expression in the context of obesity. For instance, Stolarczyk et el. reported that TBX21-deficient mice are fatter but more insulin sensitive than their wild-type counterparts 38, suggesting that decreased TBX21 expression may be associated with obesity on a metabolically healthy background. Therefore, polymorphisms that lead to deceased TOX2 expression may indirectly influence obesity via a TBX21-dependent mechanism; however, the TOX2 polymorphisms identified in the present study will need to be evaluated at the expression level in order to support such an assertion.
A single SNP was identified in the Potassium Voltage-Gated Channel Subfamily Q Member 1 gene, KCNQ1. Several investigators have reported associations between KCNQ1 and obesity, as well as obesity-related metabolic diseases including diabetes mellitus, type 2 diabetes, and reduced insulin release39, 40. As a final example, we observed an SNP in the Inter-Alpha-Trypsin Inhibitor Heavy Chain Family Member 5 gene, ITIH5. Significant correlations between BMI and mRNA expression have been reported for ITIH5. For instance, Anveden et al. reported that ITIH-5 expression in human adipose tissue is increased, suggesting that an SNP that results in decreased transcription may represent a protective genotype with respect to the development of obesity27.
All of the SNPs identified in our study were observed in non-coding regions in their respective gene or intergenic regions. Nevertheless, it is accepted that SNPs residing within introns, or those upstream or downstream of genes, also have the capacity to be causal 41, 42. Indeed, Farh et al. implemented a fine-mapping algorithm to analyze GWAS data sets for 21 autoimmune diseases and showed that approximately 90% of all causal SNPs map to noncoding regions 43. They also reported that only 10–20% of causal SNPs directly alter recognizable transcription factor binding motifs. Additional, studies will be necessary to determine if the SNPs identified in this study lead to altered transcription of their respective genes, and thus are causal.
In this study, we identified nine regions with two statistically significant SNPs in proximity to specific genes, two with three statistically significant SNPs and one with seven statistically significant SNPs. For example, the LOC107986666, which contained seven statistically significant SNPs, represents an uncharacterized RNA gene affiliated with the non-coding RNA class. Although the probability of seven SNPs being found to be statistically significant in a relatively small region is remote, the identification of multiple SNPs within a single gene that are associated with disease is not without precedence. Indeed, multiple SNPs within the RNASEL gene are associated with prostate cancer 44 and several SNPs in the CDKN2B and ANRIL genes are associated with cardiovascular disease 45. Notwithstanding, functional characterization of this gene will be required before the significance of this SNP can be determined.
Simple Mendelian inheritance, resulting from changes of a single gene, is observed in some instances of obesity; however, these occurrences are rare. For instance, mutations in the gene that encodes the melanocortin 4 receptor (MC4R) are a cause of autosomal dominant obesity 46. The melanocortin 4 receptor plays an essential role in energy homeostasis and somatic growth. With a prevalence of 1.0–2.5% in those with a BMI>30, it is the most commonly known genetic predisposing to obesity 46. Polymorphisms in the first intron of the alpha-ketoglutarate dependent dioxygenase gene (FTO) are also commonly associated with increased body mass index and risk of obesity47. Notwithstanding, we did not observe any polymorphisms for these genes in our study cohort. One potential reason for this is that our subjects were specifically selected to exclude any individuals with cardiometabolic disease. In addition to a predisposition to obesity, polymorphisms in MC4R and FTO are also associated with cardiometabolic diseases, including type 2 diabetes and elevated triglycerides 48, 49. This observation suggests that our results implicate an alternative, and perhaps more complicated, mechanism; possibly the results of polygenetic influences.
Although we identified SNPs in several genes associated with metabolic disease or a predisposition to the development of obesity, all but two of the statistically significant SNPs observed in our study were novel with respect to human disease. One of these SNPs (rs984430) is an intron variant in the Neurotrophic Receptor Tyrosine Kinase 2 gene (NRTK2) and is associated with depression and obesity 28, 29. The other (rs9736016) is an intergenic variant between the chemokine (C-X-C motif) receptor 5 and the DEAD (Asp-Glu-Ala-Asp) box helicase 6 (CXCR5 and DDX6, respectively), and was identified by the International Multiple Sclerosis Genetics Consortium as one of 48 new non-MHC variants associated with multiple sclerosis at a genome-wide significance level 30. It is noteworthy that a number of genes we found to contain significant SNPs have also been associated with neurological function. For instance, Suzuki et al. reported that FSTL4 negatively regulates brain derived neurotrophic factor (BNDF) maturation 50, and KCNQ1 levels have been shown to affect neuronal action potentials 51. Further studies will be necessary to definitively ascertain any potential connection between neurological function and predisposition to obesity or cardiometabolic disease.
In summary, to the best of our knowledge, the present study is the first to investigate severely obese subjects who present without metabolic disease. Using an ultra-high density SNP genotyping array, we screened cases and controls to identify 89 potential loci that associate with obesity or obesity-related metabolic disease. The SNPs identified and reported here may help direct future research efforts in a more specific manner by identifying biological pathways and genes that may be associated with, the development of obesity or that are protective with respect to the development of metabolic.
Supplementary Material
Acknowledgments
We are grateful to the Coriell Genotyping and Microarray Center at Coriell Institute for Medical Research in Camden, NJ, and to its former Director, Dr. Norman P. Gerry, for their assistance in processing our specimens. This work was funded in part by the National Institute of General Medical Sciences (GM103440).
Footnotes
The authors declaim no conflicts of interest.
References
- 1.Ogden CL, Carroll MD, Fryar CD, Flegal KM. Prevalence of Obesity Among Adults and Youth: United States, 2011–2014. Centers for Disease Control and Prevention; Nov, 2015. [PubMed] [Google Scholar]
- 2.Hruby A, Manson JE, Qi L, Malik VS, Rimm EB, Sun Q, et al. Determinants and Consequences of Obesity. American journal of public health. 2016;106(9):1656–62. doi: 10.2105/AJPH.2016.303326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stunkard AJ, Sorensen TI. Obesity and socioeconomic status--a complex relation. N Engl J Med. 1993;329(14):1036–7. doi: 10.1056/NEJM199309303291411. [DOI] [PubMed] [Google Scholar]
- 4.Reddon H, Gueant JL, Meyre D. The importance of gene-environment interactions in human obesity. Clinical science. 2016;130(18):1571–97. doi: 10.1042/CS20160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodarzi MO. Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 2017 doi: 10.1016/S2213-8587(17)30200-0. [DOI] [PubMed] [Google Scholar]
- 6.Sims EA. Are there persons who are obese, but metabolically healthy? Metabolism. 2001;50(12):1499–504. doi: 10.1053/meta.2001.27213. [DOI] [PubMed] [Google Scholar]
- 7.Karelis AD, St-Pierre DH, Conus F, Rabasa-Lhoret R, Poehlman ET. Metabolic and body composition factors in subgroups of obesity: what do we know? The Journal of clinical endocrinology and metabolism. 2004;89(6):2569–75. doi: 10.1210/jc.2004-0165. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Moll X. Obesity and prognosis: Time to forget about metabolically healthy obesity. Eur Heart J. 2017 doi: 10.1093/eurheartj/ehx535. [DOI] [PubMed] [Google Scholar]
- 9.Munoz-Garach A, Cornejo-Pareja I, Tinahones FJ. Does Metabolically Healthy Obesity Exist? Nutrients. 2016;8(6) doi: 10.3390/nu8060320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinosa De Ycaza AE, Donegan D, Jensen MD. Long-term metabolic risk for the metabolically healthy overweight/obese phenotype. International journal of obesity. 2017 doi: 10.1038/ijo.2017.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iglesias Molli AE, Penas Steinhardt A, Lopez AP, Gonzalez CD, Vilarino J, Frechtel GD, et al. Metabolically healthy obese individuals present similar chronic inflammation level but less insulin-resistance than obese individuals with metabolic syndrome. PLoS One. 2017;12(12):e0190528. doi: 10.1371/journal.pone.0190528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bluher M. The distinction of metabolically ‘healthy’ from ‘unhealthy’ obese individuals. Current opinion in lipidology. 2010;21(1):38–43. doi: 10.1097/MOL.0b013e3283346ccc. [DOI] [PubMed] [Google Scholar]
- 13.National Cholesterol Education Program Expert Panel on Detection E, Treatment of High Blood Cholesterol in A. Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–421. [PubMed] [Google Scholar]
- 14.Smith AK, Fang H, Whistler T, Unger ER, Rajeevan MS. Convergent genomic studies identify association of GRIK2 and NPAS2 with chronic fatigue syndrome. Neuropsychobiology. 2011;64(4):183–94. doi: 10.1159/000326692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauderman WJMJ. QUANTO 1.2.4: A computer program for power and sample size calculations for genetic–epidemiology studies. University of Southern California; Los Angeles, CA: 2009. [Google Scholar]
- 16.den Hoed M, Luan J, Langenberg C, Cooper C, Sayer AA, Jameson K, et al. Evaluation of common genetic variants identified by GWAS for early onset and morbid obesity in population-based samples. International journal of obesity. 2013;37(2):191–6. doi: 10.1038/ijo.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlauch KA, Khaiboullina SF, De Meirleir KL, Rawat S, Petereit J, Rizvanov AA, et al. Genome-wide association analysis identifies genetic variations in subjects with myalgic encephalomyelitis/chronic fatigue syndrome. Translational psychiatry. 2016;6:e730. doi: 10.1038/tp.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8(2):485–99. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez JR, Armengol L, Sole X, Guino E, Mercader JM, Estivill X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23(5):644–5. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 20.Jorgenson E, Witte JS. Genome-wide association studies of cancer. Future Oncol. 2007;3(4):419–27. doi: 10.2217/14796694.3.4.419. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;(57):289–300. Series B. [Google Scholar]
- 22.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 23.Lehne B, Lewis CM, Schlitt T. From SNPs to genes: disease association at the gene level. PLoS One. 2011;6(6):e20133. doi: 10.1371/journal.pone.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radkowski P, Wator G, Skupien J, Bogdali A, Wolkow P. Analysis of gene expression to predict dynamics of future hypertension incidence in type 2 diabetic patients. BMC proceedings. 2016;10(Suppl 7):113–117. doi: 10.1186/s12919-016-0015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Shammari MS, Al-Ali R, Al-Balawi N, Al-Enazi MS, Al-Muraikhi AA, Busaleh FN, et al. Type 2 diabetes associated variants of KCNQ1 strongly confer the risk of cardiovascular disease among the Saudi Arabian population. Genetics and molecular biology. 2017;40(3):586–590. doi: 10.1590/1678-4685-GMB-2017-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Vliet-Ostaptchouk JV, van Haeften TW, Landman GW, Reiling E, Kleefstra N, Bilo HJ, et al. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS One. 2012;7(3):e32148. doi: 10.1371/journal.pone.0032148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anveden A, Sjoholm K, Jacobson P, Palsdottir V, Walley AJ, Froguel P, et al. ITIH-5 expression in human adipose tissue is increased in obesity. Obesity. 2012;20(4):708–14. doi: 10.1038/oby.2011.268. [DOI] [PubMed] [Google Scholar]
- 28.Yeo GS, Connie Hung CC, Rochford J, Keogh J, Gray J, Sivaramakrishnan S, et al. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat Neurosci. 2004;7(11):1187–9. doi: 10.1038/nn1336. [DOI] [PubMed] [Google Scholar]
- 29.Dong C, Wong ML, Licinio J. Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Molecular psychiatry. 2009;14(12):1105–18. doi: 10.1038/mp.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Multiple Sclerosis Genetics C. Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nature genetics. 2013;45(11):1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturm R, Hattori A. Morbid obesity rates continue to rise rapidly in the United States. International journal of obesity. 2013;37(6):889–91. doi: 10.1038/ijo.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307(5):491–7. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 33.Behrens M, Bartelt J, Reichling C, Winnig M, Kuhn C, Meyerhof W. Members of RTP and REEP gene families influence functional bitter taste receptor expression. The Journal of biological chemistry. 2006;281(29):20650–9. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- 34.Tepper BJ, Banni S, Melis M, Crnjar R, Tomassini Barbarossa I. Genetic sensitivity to the bitter taste of 6-n-propylthiouracil (PROP) and its association with physiological mechanisms controlling body mass index (BMI) Nutrients. 2014;6(9):3363–81. doi: 10.3390/nu6093363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dotson CD, Zhang L, Xu H, Shin YK, Vigues S, Ott SH, et al. Bitter taste receptors influence glucose homeostasis. PLoS One. 2008;3(12):e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark AA, Dotson CD, Elson AE, Voigt A, Boehm U, Meyerhof W, et al. TAS2R bitter taste receptors regulate thyroid function. FASEB J. 2015;29(1):164–72. doi: 10.1096/fj.14-262246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vong QP, Leung WH, Houston J, Li Y, Rooney B, Holladay M, et al. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood. 2014;124(26):3905–13. doi: 10.1182/blood-2014-06-582965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stolarczyk E, Vong CT, Perucha E, Jackson I, Cawthorne MA, Wargent ET, et al. Improved insulin sensitivity despite increased visceral adiposity in mice deficient for the immune cell transcription factor T-bet. Cell metabolism. 2013;17(4):520–33. doi: 10.1016/j.cmet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasuga M. KCNQ1, a susceptibility gene for type 2 diabetes. Journal of diabetes investigation. 2011;2(6):413–4. doi: 10.1111/j.2040-1124.2011.00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavira B, Coto E, Diaz-Corte C, Ortega F, Arias M, Torres A, et al. KCNQ1 gene variants and risk of new-onset diabetes in tacrolimus-treated renal-transplanted patients. Clinical transplantation. 2011;25(3):E284–91. doi: 10.1111/j.1399-0012.2011.01417.x. [DOI] [PubMed] [Google Scholar]
- 41.Al-Haggar M, Madej-Pilarczyk A, Kozlowski L, Bujnicki JM, Yahia S, Abdel-Hadi D, et al. A novel homozygous p. Arg527Leu LMNA mutation in two unrelated Egyptian families causes overlapping mandibuloacral dysplasia and progeria syndrome. Eur J Hum Genet. 2012;20(11):1134–40. doi: 10.1038/ejhg.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordovado SK, Hendrix M, Greene CN, Mochal S, Earley MC, Farrell PM, et al. CFTR mutation analysis and haplotype associations in CF patients. Mol Genet Metab. 2012;105(2):249–54. doi: 10.1016/j.ymgme.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.San Francisco IF, Rojas PA, Torres-Estay V, Smalley S, Cerda-Infante J, Montecinos VP, et al. Association of RNASEL and 8q24 variants with the presence and aggressiveness of hereditary and sporadic prostate cancer in a Hispanic population. Journal of cellular and molecular medicine. 2014;18(1):125–33. doi: 10.1111/jcmm.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Judge AD, Zhang X, Fujii H, Surh CD, Sprent J. Interleukin 15 controls both proliferation and survival of a subset of memory-phenotype CD8(+) T cells. J Exp Med. 2002;196(7):935–46. doi: 10.1084/jem.20020772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocrine reviews. 2006;27(7):710–18. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 47.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yako YY, Guewo-Fokeng M, Balti EV, Bouatia-Naji N, Matsha TE, Sobngwi E, et al. Genetic risk of type 2 diabetes in populations of the African continent: A systematic review and meta-analyses. Diabetes research and clinical practice. 2016;114:136–50. doi: 10.1016/j.diabres.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Manriquez V, Aviles J, Salazar L, Saavedra N, Seron P, Lanas F, et al. Polymorphisms in Genes Involved in the Leptin-Melanocortin Pathway are Associated with Obesity-Related Cardiometabolic Alterations in a Southern Chilean Population. Molecular diagnosis & therapy. 2017 doi: 10.1007/s40291-017-0306-8. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki R, Matsumoto M, Fujikawa A, Kato A, Kuboyama K, Yonehara K, et al. SPIG1 negatively regulates BDNF maturation. J Neurosci. 2014;34(9):3429–42. doi: 10.1523/JNEUROSCI.1597-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abbott GW, Tai KK, Neverisky DL, Hansler A, Hu Z, Roepke TK, et al. KCNQ1, KCNE2, and Na+-coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability. Sci Signal. 2014;7(315):ra22. doi: 10.1126/scisignal.2005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.