Abstract
Colorectal cancer occurs throughout the world but is most common in developed countries. Cancer progression is believed to be driven by genetic mutations in this complex condition. Risk factors for developing colorectal cancer include a genetic family history, long-term ulcerative colitis, and colonic polyps. The use of baicalin has been reported to be clinically efficacious against colon tumors in Asian countries despite an unclear mechanism of action. Several cancers have been found to be biologically dependent on the specificity protein 1 (sp1) transcription factor family. We hypothesized that baicalin may exert its chemotherapeutic effects by sp1 downregulation. Using the SW480 human colorectal cancer cell line, we investigated the physiological properties of baicalin. Our experiments were designed toward clarifying three goals: (a) to determine the mRNA expression profile of transcription factors in colorectal cancer patients using a microarray-based analysis; (b) to determine the effects of baicalin on the sp1 transcription factor with western blotting and reporter cell assays; and (c) to contrast the effects of mithramycin-A (an sp1 transcription factor inhibitor) and baicalin using western blotting and reporter cell assays. Both baicalin and mithramycin-A downregulated sp1 expression, attenuated SW480 cell proliferation, and increased cell apoptosis. Baicalin inhibited sp1 expression and led to SW480 apoptosis, thus clarifying the effect of this traditional Chinese medicine compound in the treatment of colon cancer.
Keywords: baicalin, cell apoptosis, colorectal cancer, specificity protein 1, SW480 cells
Introduction
Colorectal cancer causes the death of about 700 000 patients each year, and is thus the fourth-most deadly cancer worldwide, with the incidence being the highest in developed countries. Countries such as China, currently in the midst of rapid economic development, are likely to witness a spike in the number of colorectal cancer diagnoses in the near future 1. Traditional and complementary medicine may offer an alternative therapeutic approach for this deadly condition.
Traditional Chinese medicine (TCM) has its roots deep in philosophy, which are reflected in its treatment principles, making it uniquely suited for the management of complex conditions. TCM uses a diverse range of medicinal plants for clinical treatment and has been subjected to rigorous scientific scrutiny. These medicines form an untapped pool of potential chemotherapeutic agents for a variety of conditions 2. Well-documented ethno-pharmacological information in addition to robust knowledge on these herbs may serve as guideposts for modern pharmacological research.
Specificity protein 1 (sp1) is a transcription factor expressed throughout the body that drives the activation of oncogenes necessary for tumor progression, metastasis, and survival 3,4. A myriad of cancers, including colorectal cancer, have been found to have high levels of sp1, a finding that is also associated with worse clinical outcomes 5. Therefore, it can be hypothesized that sp1 can be targeted for new treatment options with respect to colorectal cancer 6. In addition, baicalin is a key TCM ingredient that has been used for centuries in China for its anti-tumor and anti-inflammatory properties.
This study examines the in-vitro effects of baicalin on a human colorectal cancer cell line (SW480 cells), with a special focus on the sp1 transcription factor. The following results provide scientific evidence to support the development of baicalin as a chemoprophylactic and chemotherapeutic agent in managing colorectal cancer.
Materials and methods
Baicalin and mithramycin-A were procured from Sigma-Aldrich (St Louis, Missouri, USA). The annexin V-FITC and CCK-8 kits for detecting apoptosis were obtained from Beyotime (Shanghai, China), the RNAiso Plus reagent kit and the PrimeScript RT reagent kit (Perfect Real Time) were obtained from TAKARA, BIO (Kusatsu, Japan), whereas anti-sp1, anti-C-PARP, anti-C-caspase-3, and tubulin antibodies were obtained from Cell Signaling Technology (Danvers, Massachusetts, USA).
Gene expression profiling and network analysis of colorectal cancer patients
The gene expression profiles of patients with colorectal cancer were identified from the PubMed GEO datasets using the keywords ‘colorectal cancer’ and ‘gene expression profiling’. Four datasets were finally selected among the Pubmed GEO datasets (GSE4107, GSE24514, GSE32323, GSE73883). Subsequent GEO2R analysis was carried out to enable inter-dataset comparisons. Gene expression differences were expressed as fold-change ratios that were calculated from values obtained from both the colorectal cancer and healthy control groups. Gene symbols (IDs) and fold-change values were entered into the FunRich software 7 to identify pathways related to canonical signal transduction. Gene promoter sequences were searched for transcription profiles using the Binding Motif Search. The FunRich analytic software was also used to carry out a transcription factor binding site analysis.
Cell cultures
SW480 cells were obtained from Guangzhou Medical University, which in turn sourced the cell line from the American Type Culture Collection (Manassas, Virginia, USA). SW480 cells were maintained under standard conditions (37°C in 5% atmospheric CO2) and cultured in DMEM medium (Gibco, Grand Island, New York, USA) containing 10% fetal bovine serum (Gibco). Cell passage was performed every 2–3 days.
Cell proliferation measurement by the CCK-8 assay
Cells were quantified before inclusion in a suspension containing 5×104 cells/ml. Then, 100 μl of cell suspension was stored in a 96-well culture plate at a concentration of 5×103 cells per well. The plate was then placed in an incubator with 5% CO2 for 24 h at 37°C. In the meantime, baicalin solutions were diluted with complete medium to concentrations of 50, 100, 200, and 400 μg/ml. Then, 100 μl of these baicalin concentrations were added to their allocated wells. An equal amount of dimethyl sulfoxide was used for the control group. The 96-well cell culture plate was then placed back in the 5% CO2 incubator at 37°C for an additional 48 h. Cell staining was performed by first adding 10 μl of CCK-8 to each well, before allowing the cells to grow for 3 h. The cells were then mixed gently for 10 min with a shaker. To calculate the rate of cell inhibition, each well was read with a microplate reader at an absorbance wavelength of 450 nm. Experiments to determine cell proliferation were conducted using similar methods.
Annexin V-FITC/PI double-staining method to detect cell apoptosis
To determine the rate of cell apoptosis, cells were harvested during the logarithmic phase, and then dissociated and inoculated into a six-well plate. The cells were then left to incubate overnight. The respective baicalin concentrations were added to the wells the following day, with the addition of either 50 μg/ml of mithramycin-A or a control solution. The cells were then allowed to incubate further for 48 h before being rinsed twice with PBS and centrifuged for 5 min at 2000 rpm. Collected cells were standardized at a concentration of 5×105 cells per sample. Specifically, 500 μl of binding buffer, 10 μl of propidium iodide, and 5 μl of annexin V-FITC were mixed into the cells and blended evenly. The resultant cell mixture was then allowed to stand in a dark room for 10–20 min. The rate of cell apoptosis was quantified with flow cytometry.
Western blot analysis
For the preparation of western blot analysis, 3×104 cells were plated into six-well plates and left to stand for 24 h to allow the cells to adhere to the well membranes. Cells were then immersed in different baicalin concentrations before adding 50 μg/ml of mithramycin-A or a control solution. The plates were then left to incubate for 48 h. A pyrolysis buffer was added to the protein samples to dilute them to a standardized concentration.
SDS-PAGE electrophoresis was carried out with the protein samples mixed with an equal amount of sample loading buffer first mixed in a test tube and cooled on ice after heating for 5 min at 95–100°C. The protein samples were electrophoresed for 30 min at 80 V in a spacer gel and for 80 min at 60 V in the separation gel. The separated proteins were then transferred onto a polyvinylidene fluoride membrane, which was then exposed to a solution containing 5% skim milk powder for 1 h at room temperature. Next, primary antibodies (1 : 1000) were added to the membranes at a concentration of 0.1 ml/cm2, before they were incubated overnight on a shaker at 4°C. The next day, the proteins were washed with tris-buffered saline and tween 20 and subjected to four 10 min long filtrations. Secondary antibodies with horseradish peroxidase were then added to the membranes, which were shaken and incubated for 1 h at room temperature. The membranes were subjected to four final TBST washings, each 10 min long. Finally, electro-chemi-luminescence solution was added to the membranes in a dark room and the exposure time was adjusted to achieve high-quality images.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from SW480 cells using the RNAiso Plus reagent kit (Takara). Total RNA (500 ng) was used for reverse transcription using a PrimeScript RT reagent kit (Takara). The resulting complementary DNA was analyzed by qPCR performed with SYBR reagent using the LightCycler 480 PCR system (Roche, Rotkreuz, Switzerland). GAPDH expression was used for normalization. The sequences of used primers were as follows: sp1 (forward: 5′-tggcagcagtaccaatggc-3′, reverse: 5′-ccaggtagtcctgtcagaactt-3′), GAPDH (forward: 5′-ctgggctacactgagcacc-3′, reverse: 5′-aagtggtcgttgagggcaatg-3′).
Statistical analysis
SPSS software (IBM, Armonk, USA) was used to carry out all statistical analyses. Results are presented as mean±SD and were calculated using the two-tailed t-test method.
Results
Specificity protein 1 is a central mediator of colorectal cancer
The top ten transcription factors were selected from each of the four gene expression datasets derived from the FunRich analysis (Fig. 1).
Fig. 1.
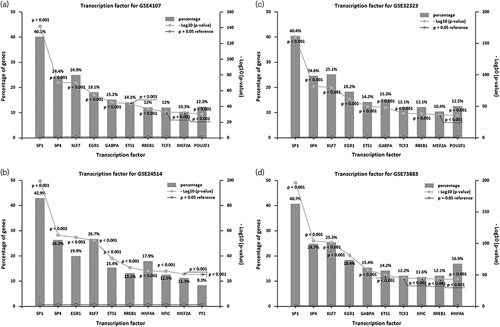
sp1 as a determinant transcription factor of colorectal cancer during the analysis of four datasets. (a) GSE4107 gene expression according to; (b) GSE24514 gene expression according to; (c) GSE32323 gene expression according to; (b) GSE73883 gene expression according to. P<0.001. sp1, specificity protein 1.
sp1 was found to be a central mediator of colorectal cancer.
Baicalin inhibits growth and downregulates specificity protein 1 in SW480 cells
In this study, different concentrations (50–400 µg/ml) of baicalin inhibited the growth of SW480 cells over 48 h (Fig. 2a and b). Both western blot analysis and qPCR data showed that baicalin could attenuate sp1 expression in a dose-dependent manner in contrast to the control group (Fig. 2c and d).
Fig. 2.
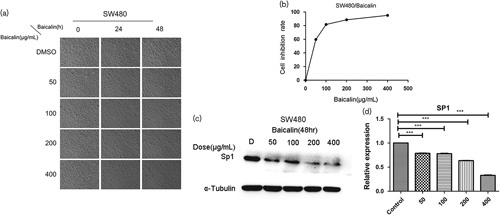
Baicalin inhibits SW480 cells growth and decreases the expression of sp1 in SW480 human colon cancer cells. Cells were treated with DMSO or 50–400 µg/ml baicalin for 24 and 48 h. (a) The photograph of inhibition effects of baicalin on the growth of SW480 cells. (b) Inhibition rate of baicalin, cell proliferation was measured using the CCK-8 kit, as described in ‘Materials and methods’ section. (c) Baicalin decreased the protein expression of sp1 in SW480 human colon cancer cells. Each bar represents the mean±SD of three independent experiments. P<0.05 for comparison with the control cells without baicalin treatment. (d) qPCR analysis of sp1 mRNA expression in different SW480 cells groups. Data are presented as the mean±SD. ***P<0.001 for indicated comparison. sp1, specificity protein 1.
Baicalin induces apoptosis in SW480 cells
The apoptotic effect of baicalin (100 and 400 µg/ml) was investigated in SW480 cells by flow cytometry analysis (Fig. 3a and b). Caspase-3 cleavage and poly ADP-ribose polymerase cleavage are markers of apoptosis; baicalin could increase C-caspase-3 and C-PARP protein expressions in a dose-dependent manner (Fig. 3c).
Fig. 3.
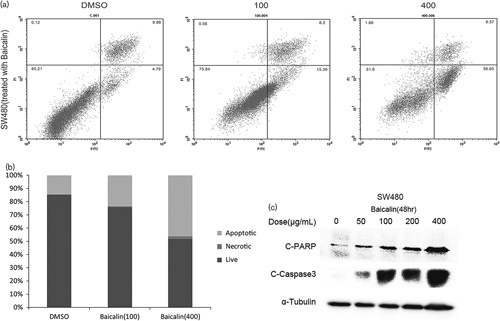
Baicalin induces SW480 human colon cancer cell apoptosis. (a) Baicalin induces SW480 human colon cancer cell apoptosis by flow cytometry methods. (c) The expression of apoptosis proteins PARP cleavage and caspase-3 cleavage was determined by western blot analysis of whole-cell lysates. Each bar represents the mean±SD of three independent experiments. P<0.05 for comparison with the control cells without baicalin treatment.
Specificity protein 1 inhibitor induces apoptosis in SW480 cells
As an sp1 transcription factor inhibitor, mithramycin-A was confirmed by western blot analysis (Fig. 4b). Moreover, treatment of SW480 cells with 50 µg/ml mithramycin-A for 48 h also induced apoptosis by flow cytometry analysis (Fig. 4c and d).
Fig. 4.
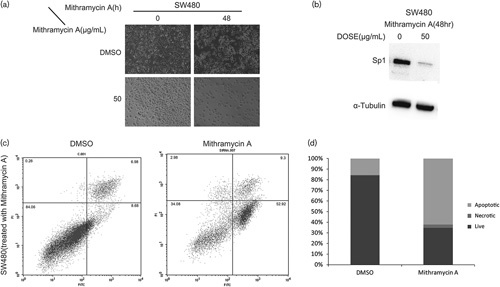
(a, b) sp1 inhibitor mithramycin-A inhibits SW480 cells growth and decreases the expression of sp1, (c, d) mithramycin-A induces SW480 human colon cancer cell apoptosis. Each bar represents the mean±SD of three independent experiments. P<0.05 for comparison with the control cells without baicalin treatment. sp1, specificity protein 1.
These data confirm that the baicalin-mediated apoptotic effect of SW480 cells was accompanied by downregulation of the sp1 transcription factor.
Discussion
The existing literature suggests that sp1 is responsible for the induction of several genes implicated in a variety of cell proliferation and survival mechanisms 8. Past sp1 siRNA experiments have shown that suppression of sp1 could induce apoptosis and attenuate the growth of colon cancer stem cells 9.
Thus, sp1 appears to be a central mediator that determines the progression of colorectal cancer 10. Patients with this malignancy were observed to have abnormally elevated sp1 protein levels 11. This is reflected in the current study 12, where we showed that colon cancer tissues had markedly higher sp1 levels in contrast to healthy tissues. In addition, our findings highlight that sp1 transcription factor targeting may serve to guide the synthesis of novel approaches in treating colon cancer.
Interestingly, baicalin is a Chinese herbal medicine used commonly for its anti-inflammatory and anticancer properties. Our study showed that baicalin could induce apoptosis and downregulate sp1 expression in SW480 colon cancer cells. Moreover, mithramycin-A, an sp1 inhibitor, was also found to induce apoptosis and downregulate sp1 expression in SW480 cells. Apoptosis is a common phenomenon across multicellular organisms and represents the process of programmed cell death.
Apoptosis may be triggered by intrinsic mitochondrial-mediated factors or extrinsic death receptor pathway-facilitated factors 13. These two mechanisms ultimately converge, resulting in caspase-3 cleavage, the formation of apoptotic bodies, and DNA fragmentation. The lifecycle of a cell is determined by the balance between proapoptotic and antiapoptotic protein expressions 14. The protein survivin functions as an inhibitor of apoptosis in cells, and its aberrant overexpression is a common feature of several malignancies, including colorectal cancer 15. The transcription factor sp1 can bind to GC-rich SP sites in the survivin gene promoter region, allowing it to regulate any remaining expression 16.
Taken together, our findings suggest that baicalin-induced apoptosis may be achieved by inhibiting the expression of sp1 in SW480 cells. Such a mechanism supports the development of baicalin as a chemoprophylactic and chemotherapeutic agent in managing colorectal cancer.
Acknowledgements
This work was financially supported by Guangzhou Medical University, (project number: L1751002).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Holmes D. A disease of growth. Nature 2015; 521:S2–S3. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Lv SS, Fu ZY, Hou LL. Baicalein enhances migration and invasion of extravillous trophoblasts via activation of the NF-kappaB pathway. Med Sci Monit 2018; 24:2983–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, SP1. Science 1986; 234:47–52. [DOI] [PubMed] [Google Scholar]
- 4.Bajpai R, Nagaraju GP. Specificity protein 1: its role in colorectal cancer progression and metastasis. Crit Rev Oncol Hematol 2017; 113:1–7. [DOI] [PubMed] [Google Scholar]
- 5.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor SP1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 1997; 89:619–628. [DOI] [PubMed] [Google Scholar]
- 6.Takami Y, Russell MB, Gao C, Mi Z, Guo H, Mantyh CR, et al. SP1 regulates osteopontin expression in SW480 human colon adenocarcinoma cells. Surgery 2007; 142:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015; 15:2597–2601. [DOI] [PubMed] [Google Scholar]
- 8.Black AR, Black JD, Azizkhan-Clifford J. SP1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol 2001; 188:143–160. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y, et al. Inhibition of the transcription factor SP1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol Rep 2013; 30:1782–1792. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol 2013; 229:12–24. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Qian J, Wang F, Ma Z. Cellular prion protein accelerates colorectal cancer metastasis via the Fyn-SP1-SATB1 axis. Oncol Rep 2012; 28:2029–2034. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez JM, Farma JM, Coppola D, Hakam A, Fulp WJ, Chen DT, et al. Expression of the antiapoptotic protein survivin in colon cancer. Clin Colorectal Cancer 2011; 10:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathi S, Jutooru I, Chadalapaka G, Nair V, Lee SO, Safe S. Aspirin inhibits colon cancer cell and tumor growth and downregulates specificity protein (Sp) transcription factors. PLoS One 2012; 7:e48208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha T, Lou Z, Baek SJ, Lee SH. Tolfenamic acid downregulates beta-catenin in colon cancer. Int Immunopharmacol 2016; 35:287–293. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Pathi SS, Safe S. Sulindac sulfide inhibits colon cancer cell growth and downregulates specificity protein transcription factors. BMC Cancer 2015; 15:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chintharlapalli S, Papineni S, Lei P, Pathi S, Safe S. Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer 2011; 11:371. [DOI] [PMC free article] [PubMed] [Google Scholar]


