Abstract
Background:
DNA methylation is involved in numerous biologic events and associates with transcriptional gene silencing, playing an important role in the pathogenesis of endometrial cancer. ESR1/PGR frequently undergoes de novo methylation and loss expression in a wide variety of tumors, including breast, colon, lung, and brain tumors. However, the mechanisms underlying estrogen and progesterone receptors (ER/PR) loss in endometrial cancer have not been studied extensively. The aims of this study were to determine the expression of DNA (cytosine-5)-methyltransferase 3A/3B (DNMT3A/3B) in endometrial cancer to investigate whether the methylation catalyzed by DNMT3A/3B contributes to low ER/PR expression.
Methods:
The clinicopathologic information and RNA-Seq expression data of DNMT3A/3B of 544 endometrial cancers were derived from The Cancer Genome Atlas (TCGA) uterine cancer cohort in May 2018. RNA-Seq level of DNMT3A/3B was compared between these clinicopathologic factors with t-test or one-way analysis of variance.
Results:
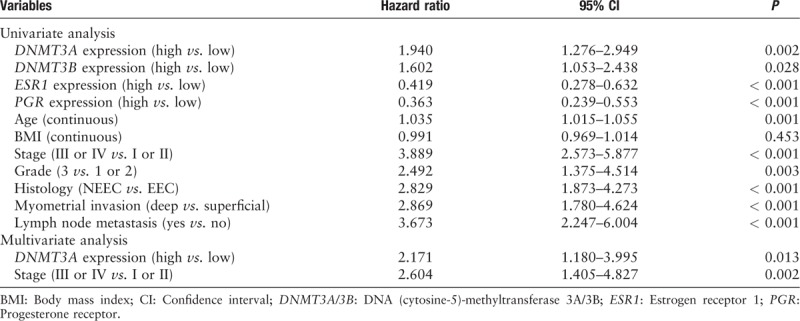
DNMT3A/3B was overexpressed in endometrioid carcinoma (EEC) and was even higher in non-endometrioid carcinoma (NEEC) (DNMT3A, EEC vs. NEEC: 37.6% vs. 69.9%, t = −7.440, P < 0.001; DNMT3B, EEC vs. NEEC: 42.4% vs. 72.8%, t = −6.897, P < 0.001). In EEC, DNMT3A overexpression was significantly correlated with the hypermethylation and low expression of the ESR1 and PGR (P < 0.05). The same trend was observed in the DNMT3B overexpression subgroup. In the ESR1/PGR low-expression subgroups, as much as 83.1% of ESR1 and 59.5% of PGR were hypermethylated, which was significantly greater than the ESR1/PGR high-expression subgroups (31.3% and 11.9%, respectively). However, the above phenomena were absent in NEEC, while DNMT3A/3B overexpression, ESR1/PGR hypermethylation, and low ER/PR expression occurred much more often. In univariate analysis, DNMT3A/3B overexpressions were significantly correlated with worse prognosis. In multivariate analysis, only DNMT3A was an independent predictor of disease-free survival (P < 0.05).
Conclusions:
DNMT3A/3B expression increases progressively from EEC to NEEC and is correlated with poor survival. The mechanisms underlying low ER/PR expression might be distinct in EEC vs. NEEC. In EEC, methylation related to DNMT3A/3B overexpression might play a major role in ER/PR downregulation.
Keywords: DNA (cytosine-5)-methyltransferase 3A/3B, Estrogens receptor, Progesterone receptor, Endometrial carcinoma, The Cancer Genome Atlas
Introduction
Endometrial carcinoma is the most common malignancy of the female genital tract. Histologically, it has long been categorized into two subtypes.[1] Type I tumors (approximately 80%) are endometrioid carcinomas (EECs) with estrogen and progesterone receptors (ERs and PRs) and usually, have a favorable prognosis. However, type II tumors (10–20%), the non-endometrioid carcinomas (NEEC), are not associated with estrogen excess and usually have a poor prognosis. However, approximately 20% of the cases do not fit within this dualistic model; some EECs are aggressive and have poor clinical outcomes.[2] In fact, endometrial carcinoma is a clinically heterogeneous disease, and it is now well recognized that this heterogeneity may be the result of various underlying molecular alterations.
The DNA methylation is involved in numerous biologic events and it concerns approximately 70% to 80% of CpGs in mammalian DNA and associated with transcriptional gene silencing when it occurs in promoters.[3] Studies have indicated that the silencing of the tumor suppressor genes by DNA methylation plays an important role in the pathogenesis of endometrial cancer. Methylation changes to the genome are catalyzed by DNA methyltransferases (DNMT). DNMT3A/3B is de novo methyltransferases with high activity on unmethylated substrates, while DNA methylation is maintained by DNMT1 after the methylation pattern has been established.[4,5] In recent years, our knowledge on the roles of DNMT in DNA methylation has increased substantially. The ESR1/PGR frequently undergoes de novo methylation in a wide variety of tumors, including breast, colon, lung, and brain tumors.[6-9] However, the mechanisms underlying the loss of expression in endometrial cancer have not been studied extensively. Previously, using immunohistochemistry, we found that DNMT3B protein expression was negatively correlated with ER and PR expression in EEC in a small cohort.[10]
The Cancer Genome Atlas (TCGA) provides a wealth of information concerning DNA (somatic mutation, copy number alteration [CNA], and methylation), RNA (transcript level), and particular proteins from thousands of tumor- and case-matched normal tissues.[11] Therefore, in the present study, we collected data from the TCGA database and analyzed the associations between DNMT3A/3B mRNA expression and the methylation and expression patterns of the ESR1/PGR in endometrial carcinomas. We hypothesized that methylation catalyzed by DNMT3A/3B might take part in the ESR1/PGR downregulation in endometrial carcinoma and might possess prognostic utility.
Methods
Study time and design
The TCGA data acquisition Level 3 RNA-Seq, protein RPPA (reverse-phase protein array) and clinical data for uterine cancer patients were obtained from the TCGA data portal (http://cancergenome.nih.gov/). Detailed information concerning the data processing, quality control, and normalization is available on the TCGA open access download directories (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). RSEM (RAN-Seq by Expectation-Maximization) expression values were log2 transformed for statistical analysis.
The TCGA adopted the illumina infinium human methylation 450 (HM450) bead array to assay over 480K CpG sites, and about 99% of RefSeq genes and 96% of CpG islands from UCSC database are included. DNA methylation data were provided as beta values calculated using the formula M/(M+U+100), with M and U representing fully methylated and fully unmethylated intensities, respectively.[12] Mean value was used to stratify these biomarkers as high expression or low expression, hypermethylation or hypomethylation.
The CNA non-linear data were provided as putative copy number variation using GISTIC 2.0 with −2, −1, 0, 1, and 2 representing homozygous deletion, hemizygous deletion, neutral or no change, gain, and high-level amplification, respectively. Mutation data were provided in a mutation annotation manner derived from whole-exome sequencing.
Statistical analysis
Data were analyzed using SPSS19 (SPSS Inc., Chicago, IL, USA). The Kolmogorov-Smirnov method was used to test the normal distribution of measurement data. An unpaired two-sample t-test was performed to compare mRNA or protein expression values. The Chi-squared test or Fisher's exact test were performed to compare categorical variables. A Bonferroni correction was applied for multiple comparisons. Bivariate correlation analysis was performed to investigate the relationship between gene expression values with Pearson's method for data following a normal distribution and Spearman's method for data with an abnormal distribution. Survival analysis was performed using Kaplan-Meier curves, with P-values calculated by the log-rank test. A Cox-proportional hazard regression model was used to perform multivariate analysis. All tests were two-sided, and a P-value <0.05 was considered statistically significant.
Results
Clinical and pathologic characteristics
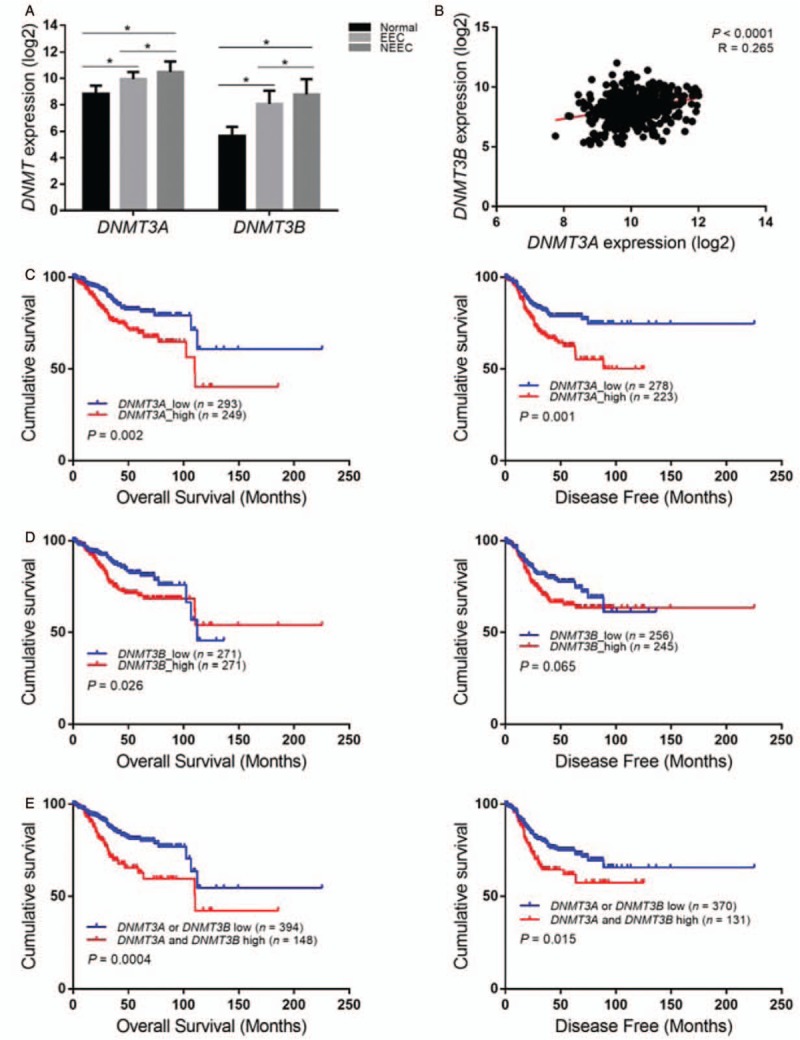
A total of 544 endometrial cancers with both clinical and gene expression data were obtained from the TCGA database. The clinicopathologic features of all of the patients are summarized in Table 1. A total of 408 EEC (75.0%) and 136 NEEC (25.0%, 114 serous carcinomas and 22 mixed carcinomas) were included in this cohort. The median age of the patients was 64 years (range, 31–90 years). A total of 72.9% of the patients were White, 20.7% were African American, and 3.9% were Asian. The ER and PR methylation data were obtained for 430 cases, and 246 cases had mutation data. CNA data were available for 537 cases. Follow-up information was provided for 542 (99.6%) patients (406 EEC, 136 NEEC), and the median follow-up was 29.0 months (range, 0.1–225.3 months).
Table 1.
Clinicopathologic features of 544 endometrial carcinomas
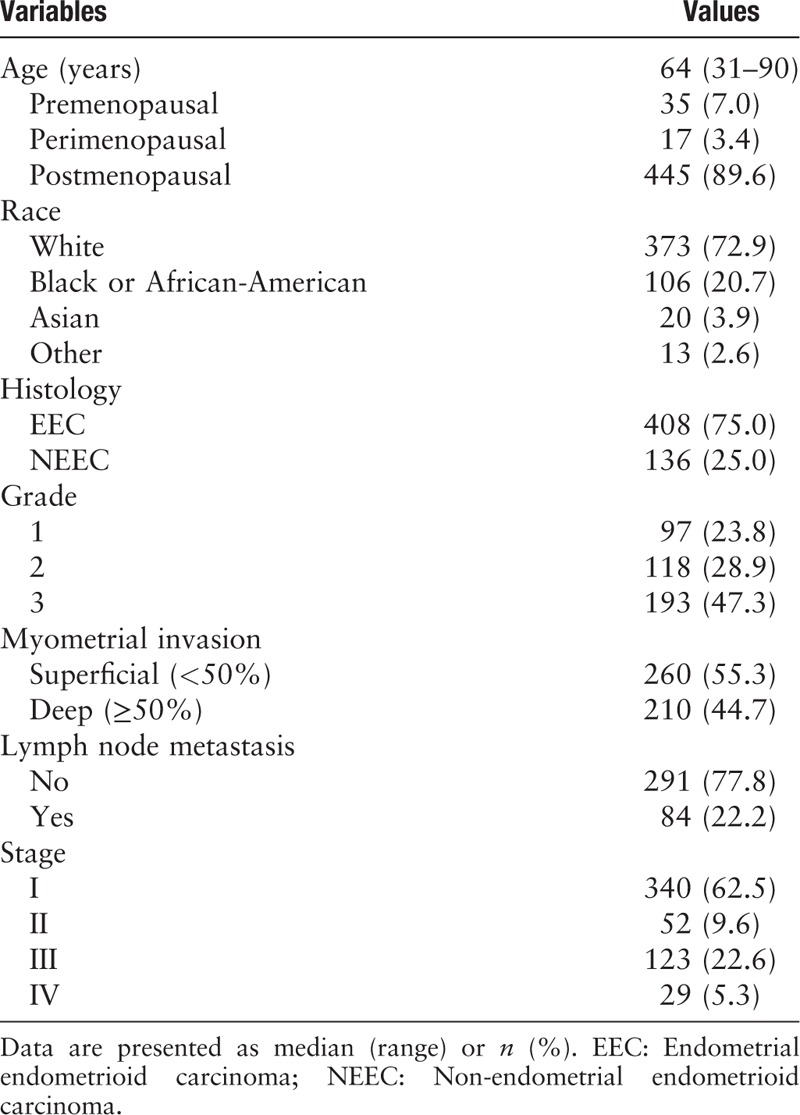
DNMT3A/3B is overexpressed in EEC and NEEC and correlated with poor patient survival
The DNMT3A/3B was overexpressed in EEC and was even higher in NEEC (DNMT3A, EEC vs. NEEC: 37.6% vs. 69.9%, t = −7.440, P < 0.001; DNMT3B, EEC vs. NEEC: 42.4% vs. 72.8%, t = −6.897, P < 0.001). Similarly, the expression levels of DNMT3A and DNMT3B increased in EEC (log2RSEM = 9.94 ± 0.55 and 8.07 ± 1.01) and became even higher in NEEC (log2RSEM = 10.49 ± 0.81 and 8.79 ± 1.16) compared with normal control tissues (log2RSEM = 8.85 ± 0.60 and 5.66 ± 0.69), and all the differences reached significance (all P < 0.001) [Figure 1A]. Among the DNMT3A overexpressed subgroup, as many as 60.5% (150/248) of cases also had DNMT3B overexpression, and the correlation between these two biomarkers was statistically significant (R = 0.265, P < 0.0001; Figure 1B).
Figure 1.
DNMT3A/3B overexpressed in endometrial cancers and indicated a poor prognosis. (A) DNMT3A/3B overexpressed in EEC and was much higher in NEEC compared with normal controls. ∗P < 0.001 vs. normal controls or EEC. (B) The expression of DNMT3A and 3B was significantly positively correlated. (C and D) DNMT3A or DNMT3B overexpression correlated with poor survival and the combined two markers also implied unfavorable prognosis (e). DNMT3A/3B: DNA (cytosine-5)-methyltransferase 3A/3B; EEC: Endometrial endometrioid carcinoma; NEEC: Non-endometrial endometrioid carcinoma.
In addition, higher DNMT3A/3B expression was associated with poor clinicopathologic variables, including tumor type, tumor grade, lymph node metastasis, and advanced stage with statistical significance [Table 2]. Survival analyses demonstrated reduced overall survival and disease-free survival for patients with DNMT3A overexpression (P = 0.002 and P = 0.001) [Figure 1C]. Likewise, DNMT3B overexpression indicated poor overall survival and reduced disease-free months, but the latter was on the borderline of significance (P = 0.026 and P = 0.065) [Figure 1D]. The combined DNMT3A and DNMT 3B overexpression was significantly correlated with poor patient survival [Figure 1E]. However, in the multivariate analysis, only DNMT3A remained as an independent prognostic factor (P = 0.013; Table 3).
Table 2.
Comparison of clinicopathologic features and expression levels of DNMT3A/3B in 544 endometrial cancers
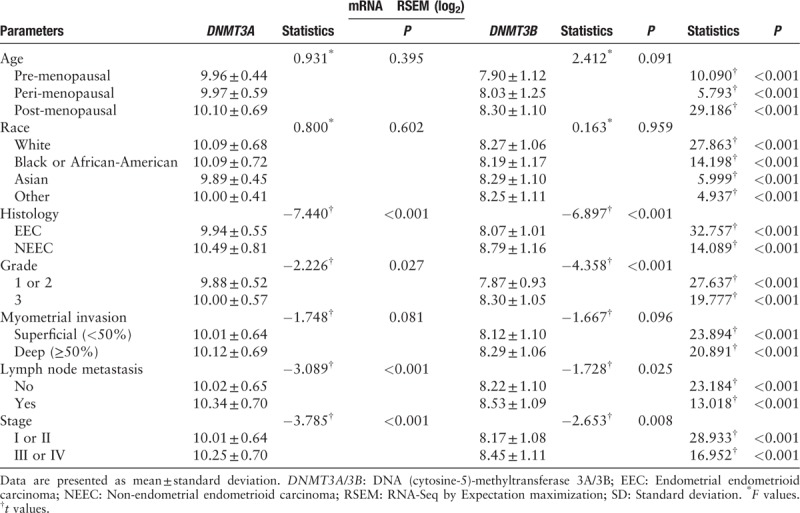
Table 3.
Univariate and multivariate analyses for disease free survival
DNMT3A/3B overexpression was correlated with hypermethylation and reduced expression of the ESR1 and PGR in EEC
Studies have indicated that the ESR1/PGR frequently undergoes de novo methylation and lost expression in a wide variety of tumors, including breast, colon, and lung cancer.[7,9] However, the mechanisms underlying their loss of expression in endometrial cancer have not been studied extensively. Therefore, we next analyzed the relationship between the expression of DNMT3A/3B and the methylation and expression of the ESR1/PGR in EEC and NEEC to assess whether methylation catalyzed by DNMT3A/3B contributes to the low ER/PR expression in endometrial cancer.
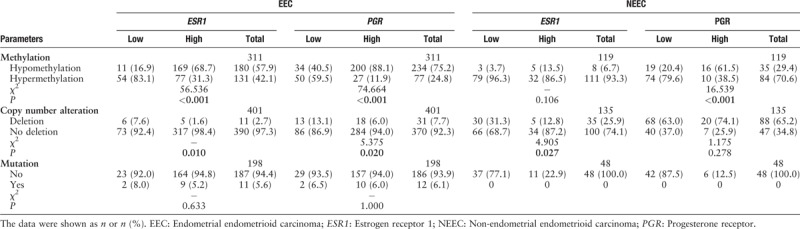
In the current study, the mean value was used to stratify ESR1 or PGR (genes encoding ER-α and PR, respectively) as hypermethylation or hypomethylation. The mean value of ESR1 and PGR methylation was 0.68 and 0.36, respectively. Using these criteria, in EEC, 41.1% of ESR1 and 24.8% of PGR were hypermethylated, and in NEEC, the rate increased drastically up to 93.3% and 70.6%, respectively [Table 4].
Table 4.
Associations between methylation, copy number alteration, and mutation of ESR1, PGR, and their expression status
Further analyses revealed that in EEC DNMT3A overexpression was significantly correlated with the hypermethylation of the ESR1 and PGR (P < 0.001) and its low expression, both at the transcript and protein level (P < 0.05) [Table 5]. The hypermethylation (52.1% and 35.5%) and reduced expression (29.4%, 37.7%) of ER/PR occurred more frequently in tumors with DNMT3A overexpression than in those without DNMT3A overexpression (35.8%, 18.4%; 14.1%, 16.9%). The same trend was observed in the DNMT3B overexpression subgroup, and almost all the associations approached statistical significance, except for the correlation between DNMT3B overexpression and ESR1 mRNA downregulation [Figure 2A and 2B].
Table 5.
Correlation between DNMT3A/3B expression and the methylation and expression status of ESR1/PGR
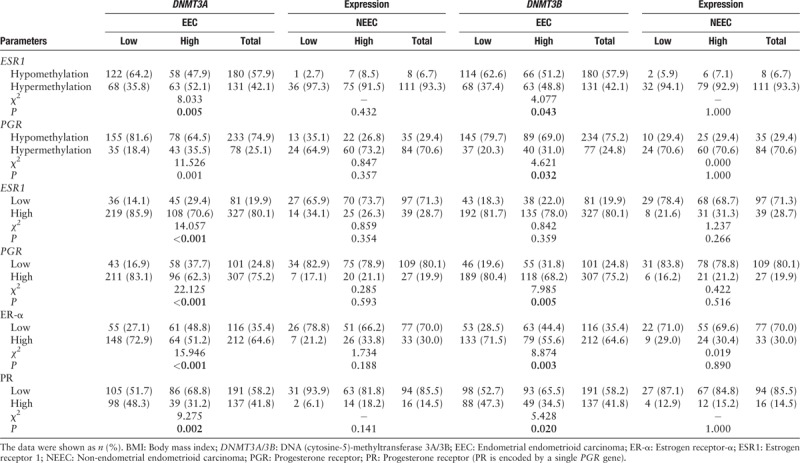
Figure 2.
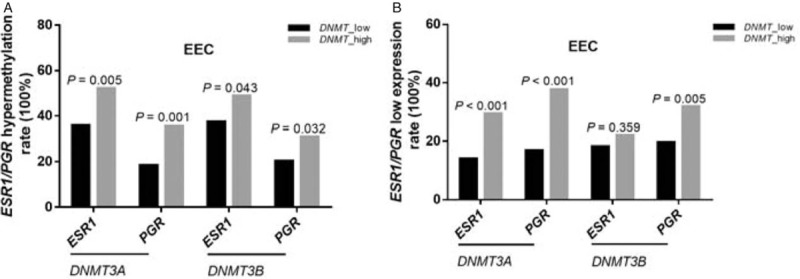
Hypermethylation (A) and reduced expression (B) of ESR1/PGR occurred more frequently in tumors with DNMT3A/3B overexpression compared with tumors without DNMT3A/3B overexpression in EEC; however, the phenomena were not present in NEEC. DNMT3A/3B: DNA (cytosine-5)-methyltransferase 3A/3B; EEC: Endometrial endometrioid carcinoma; ESR1: Estrogen receptor 1; NEEC: Non-endometrial endometrioid carcinoma; PGR: Progesterone receptor.
However, the above phenomena were not present in NEEC. The DNMT3A/3B expression had no relationship to either ESR1/PGR methylation or expression status; however, DNMT3A/3B overexpression, ESR1/PGR hypermethylation, and ER/PR low expression occurred more often in NEEC than that in EEC [Table 5].
Hypermethylation was the dominant mechanism resulting in low ESR1/PGR expression in EEC
We found that DNA methyltransferase 3A/3B overexpression was associated with hypermethylation and reduced ER/PR expression in EEC. In addition, limited studies have reported that the downregulation of the estrogen and progesterone receptor in endometrial carcinoma was associated with the methylation of these two genes. For these reasons, we next explored the relationship between ER/PR expression and methylation, CNA and mutation status of these two genes to investigate whether and to what extent methylation contributes to the reduced ER/PR expression in endometrial cancer.
The reduced ER/PR expression occurred in approximately 20% of EEC cases (also stratified by mean value). Among these tumors with low ER/PR expression, as much as 83.1% (54/65) of ESR1 and 59.5% (50/84) of PGR were hypermethylated, while only 7.6% (6/79) of ESR1 and 13.1% (13/99) of PGR was deleted. Both hypermethylation and copy number deletion were significantly correlated with reduced ER/PR expression (P < 0.05) [Table 4]. Obviously, hypermethylation played a major part in the low expression of the ESR1 and PGR, while copy number deletion played a relatively minor role. The minority of EEC cases had mutations in the ESR1 or PGR; however, it seemed that these mutations had little effect on their expression (P > 0.05).
In NEEC, approximately 70% to 80% of tumors had reduced ER or PR expression. Hypermethylation and copy number deletion of ESR1 and PGR were frequently observed among these tumors (96.3%, 79.6%; 31.3%, and 63.0%, respectively) and were even higher compared with EEC (83.1%, 59.5%; 7.6%, and 13.1%, respectively). However, the mechanism behind low ER or PR expression seemed to be different. The low PR expression was still significantly correlated with the methylation of the PGR (P < 0.001) but this association was lost in the low ER expression subgroup (P = 0.106). By contrast, deletion of the ESR1 was more effective in causing ER low expression. In the low ER expression subgroup, 31.3% of cases showed a deletion in ESR1, which was significantly higher than that in the ER high-expression subgroup (12.8%; P = 0.027). No mutation of ESR1 or PGR was found in NEEC [Table 4].
Combined DNMT3A/3B overexpression and ESR1/PGR methylation or expression were correlated with poor survival
In univariate analysis, DNMT3A/3B overexpression, ESR1/PGR hypermethylation, and low expression alone or in combination were correlated with poor survival in the whole cohort [Figures 1C, 3 and 4]. However, as mentioned above, in multivariate analysis, only DNMT3A was an independent prognostic factor of disease-free survival [Table 3].
Figure 3.
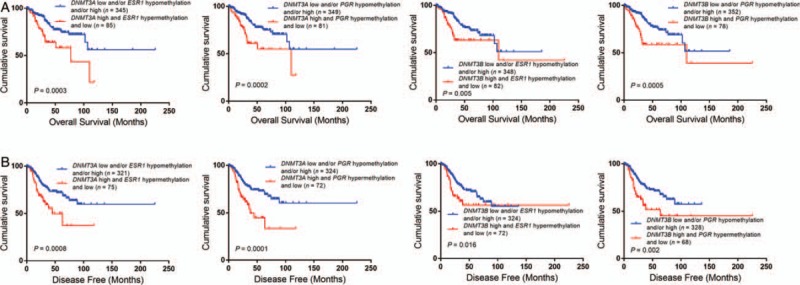
Hypermethylation or low expression of ESR1/PGR was correlated with poor survival. DNMT3A/3B: DNA (cytosine-5)-methyltransferase 3A/3B; EEC: Endometrial endometrioid carcinoma; ESR1: Estrogen receptor 1; NEEC: Non-endometrial endometrioid carcinoma; PGR: Progesterone receptor.
Figure 4.
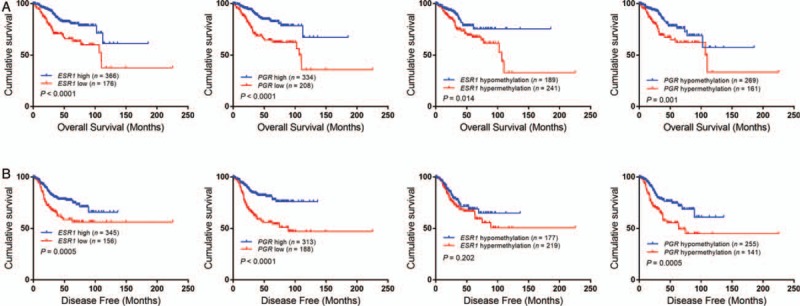
Combination of DNMT3A/3B overexpression with ESR1/PGR low expression and/or hypermethylation indicated poor prognosis in endometrial cancers. DNMT3A/3B: DNA (cytosine-5)-methyltransferase 3A/3B; ESR1: Estrogen receptor 1; PGR: Progesterone receptor.
Discussion
Taking advantage of the large-scale cancer data sets of TCGA, for the first time, we examined the DNMT3A/3B mRNA expression in 544 endometrial carcinomas. Our data set represents the largest series of endometrial cancer cases assessed for DNMT3A/3B alterations. We found that DNMT3A/3B mRNA was overexpressed progressively from EEC to NEEC compared with normal controls and was significantly correlated with a poor prognosis. The results of this study were not completely consistent with the previous study. Previously, using immunohistochemistry, we found in a small cohort that DNMT3B overexpression was more often associated with EEC than NEEC and was significantly correlated with high tumor grade.[10] Two other groups also observed that DNMT3B expression was higher in EEC than in NEEC.[13] In addition, Xiong et al[14] reported no differences of DNMT3A expression among normal endometrium, EEC, and serous endometrial carcinoma. Together, these discrepancies could be explained by two reasons. One might be due to the limited sample size and different testing methods or score criteria used by different studies. The other might be because the DNMT expression at the transcript level was not completely parallel with the protein level.[13] However, regarding the prognostic value, our findings from the present study were in line with others in that poorly differentiated endometrioid cells expressed higher DNMT3B and high DNMT3A or DNMT3B expression implied a poor prognosis.[14-17] In addition, in the present study, we found that DNMT3A and DNMT3B overexpression coexisted in most cases and the combination of these two biomarkers correlated well with poor prognosis. A study has shown that the structures and functions of DNMT3A and DNMT3B are very similar.[3] Thus, our findings were consistent with others that showed that DNMT3A and DNMT3B may cooperate and function synergistically in endometrial cancer.
More interestingly, our data indicated that methylation catalyzed by DNMT3A/3B might be the major mechanism resulting in ER/PR downregulation in EEC. First, our results support that ESR1/PGR silencing occurred primarily at the epigenetic level. Among the ER and PR low expression subgroups, 83.1% of ESR1 and 59.5% of PGR were hypermethylated with statistical significance, with a minority of cases showing deletion (7.6% for ESR1, 13.1% for PGR) or mutation (8.0% for ESR1, 6.5% for PGR). The former was also significantly correlated with ER/PR downregulation, whereas mutations of ESR1 and PGR seemed to have little effect. Second, in EEC, DNMT3A overexpression was significantly associated with ESR1 and PGR hypermethylation and their low expression. The low ER/PR expression (29.4%, 37.7%) and hypermethylation (52.1%, 35.5%) occurred more frequently in the DNMT3A overexpression subgroup compared with the DNMT3A normal expression subgroup (14.1%, 16.9%; 35.8%, and 18.4%, respectively). Similar phenomena were observed in the DNMT3B overexpression subgroups. Third, the combined DNMT3A/3B and ER/PR status or these biomarkers alone were all correlated with shorter survival, and DNMT3A was an independent prognostic factor in multivariate analysis.
To date, studies concerning the basis of ER/PR downregulation in endometrial cancer are not extensive; however, a link between ESR1/PGR methylation and expression loss in breast carcinoma has been established.[6,18] Early studies reported that the ESR1/PGR was de novo methylated in some ER/PR-negative endometrial cancers,[19-22] while the other two groups found that the ESR1 was highly refractory to de novo methylation.[23,24] Considering that TCGA adopted the HM450 bead assay to assess over 480K CpG sites covering approximately 96% of CpG islands from the UCSC database, the findings of the present study are robust and support that in EEC methylation-associated transcriptional silencing may account for the great majority of cases of ER/PR low expression, and this may be related to DNMT3A/3B overexpression. A recent study indicated that DNMT could be specifically targeted on particular loci and take part in specific de novo methylation.[3] From this point of view, our findings were consistent with this notion to some extent.
In addition, our data also demonstrated that the mechanism underlying ER or PR low expression in NEEC was distinct from that in EEC. In NEEC, the ESR1 deletion might play a more important role in the low expression of ESR1 compared with hypermethylation. NEEC showed many more ESR1 deletions (25.9%) than EEC (2.7%), and ESR1 deletion occurred more often in the ER low expression subgroup (31.3%) than in the ER high-expression subgroup (12.8%); the difference showed statistical significance. Furthermore, although DNMT3A/3B overexpression (69.9%, 72.8%), hypermethylation (93.3%) and low expression (68.9%) of ER the gene occurred more often in NEEC than those in EEC, their associations were not statistically significant. ESR1 was consistently hypermethylated in the ER low expression subgroup (96.3%) as well as in the ER high-expression subgroup (86.5%). Together, our findings suggest that low ER expression in NEEC might be related to multiple aberrations, and the contribution of methylation catalyzed by DNMT3A/3B could be shielded by other more dominant factors.
For low PR expression in NEEC, our data support that methylation still exerted great effects, while CNA and mutation had little influence. In the low PR expression subgroup, the hypermethylation rate of PGR (79.6%) was significantly higher than in the PR high-expression subgroup (59.5%). However, such methylation was not correlated with DNMT3A/3B overexpression. Recently, one study indicated that PR expression is downregulated at four different levels.[25] In well-differentiated endometrial carcinomas, ligand-induced receptor activation and downregulation are intact. miRNAs mediate the fine tuning of the PR level. As differentiation is lost, PR silencing is mainly at an epigenetic level. Initially, recruitment of the polycomb repressor complex 2 to the PR promoter suppresses transcription. Subsequently, DNA methylation prevents PR expression. Together, all these findings might imply that although methylation is the primary mechanism of PR downregulation in EEC as well as in NEEC, other regulators may be different between endometrial cancer subtypes.
A major limitation of this study is that we used the RNA-Seq expression count of DNMTs from the TCGA to perform the analysis of EC samples without available corresponding protein data to confirm the result; thus, our findings should be interpreted with caution. In addition, data regarding lymphovascular space invasion, recurrence sites, and treatment modalities were not recorded in the TCGA data set, thus limiting the clinical outcome analysis of patients with EC. Nonetheless, the RNA-Seq and its large-scale, uniform accuracy, robust methodology, and high throughput nature, strengthen this study to some extent.
In summary, our findings suggest that DNMT3A/3B overexpression was associated with poor survival in endometrial cancer. DNMT3A/3B might function synergistically. The mechanisms underlying low ER/PR expression may be distinct in EEC vs. NEEC. In EEC, methylation induced by DNMT3A/3B overexpression might play a major role in ER/PR downregulation. Thus, advances in the understanding of the molecular mechanisms of endometrial carcinomas will facilitate the development of novel anticancer therapeutic strategies.
Funding
This study was supported by a grant from the National Natural Science Foundation of China (No. 81360381).
Conflicts of interest
None.
Footnotes
How to cite this article: He D, Wang X, Zhang Y, Zhao J, Han R, Dong Y. DNMT3A/3B overexpression might be correlated with poor patient survival, hypermethylation and low expression of ESR1/PGR in endometrioid carcinoma: an analysis of the Cancer Genome Atlas. Chin Med J 2019;00:00–00. doi: 10.1097/CM9.0000000000000054
References
- 1.Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol 2014;15:e268–e278. doi: 10.1016/s1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 2.Geels YP, Pijnenborg JM, van den Berg-van Erp SH, Bulten J, Visscher DW, Dowdy SC, et al. Endometrioid endometrial carcinoma with atrophic endometrium and poor prognosis. Obstet Gynecol 2012;120:1124–1131. [DOI] [PubMed] [Google Scholar]
- 3.Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenet 2018;10:17 doi: 10.1186/s13148-018-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 5.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirza S, Sharma G, Prasad CP, Parshad R, Srivastava A, Gupta SD, et al. Promoter hypermethylation of TMS1, BRCA1, ERalpha and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sci 2007;81:280–287. doi: 10.1016/j.lfs.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Harder J, Engelstaedter V, Usadel H, Lassmann S, Werner M, Baier P, et al. CpG-island methylation of the ER promoter in colorectal cancer: analysis of micrometastases in lymph nodes from UICC stage I and II patients. Br J Cancer 2009;100:360–365. doi: 10.1038/sj.bjc.6604859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bello MJ, Aminoso C, Lopez-Marin I, Arjona D, Gonzalez-Gomez P, Alonso ME, et al. DNA methylation of multiple promoter-associated CpG islands in meningiomas: relationship with the allelic status at 1p and 22q. Acta Neuropathol 2004;108:413–421. doi: 10.1007/s00401-004-0911-6. [DOI] [PubMed] [Google Scholar]
- 9.Tekpli X, Skaug V, Baera R, Phillips DH, Haugen A, Mollerup S. Estrogen receptor expression and gene promoter methylation in non-small cell lung cancer - a short report. Cell Oncol (Dordr) 2016;39:583–589. doi: 10.1007/s13402-016-0295-3. [DOI] [PubMed] [Google Scholar]
- 10.Dong Y, Zhou M, Ba XJ, Si JW, Li WT, Wang Y, et al. Characteristic and clinical significance of DNA methyltransferase 3B overexpression in endometrial carcinoma (in Chinese). J Peking Univ Health Sci 2016;48:788. [PubMed] [Google Scholar]
- 11.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y, Dowdy SC, Xue A, Shujuan J, Eberhardt NL, Podratz KC, et al. Opposite alterations of DNA methyltransferase gene expression in endometrioid and serous endometrial cancers. Gynecol Oncol 2005;96:601–609. doi: 10.1016/j.ygyno.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 15.Liao X, Siu MK, Chan KY, Wong ES, Ngan HY, Chan QK, et al. Hypermethylation of RAS effector related genes and DNA methyltransferase 1 expression in endometrial carcinogenesis. Int J Cancer 2008;123:296–302. doi: 10.1002/ijc.23494. [DOI] [PubMed] [Google Scholar]
- 16.Jin F, Dowdy SC, Xiong Y, Eberhardt NL, Podratz KC, Jiang SW. Up-regulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol Oncol 2005;96:531–538. doi: 10.1016/j.ygyno.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 17.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res 2003;9:4415–4422. [PubMed] [Google Scholar]
- 18.Pirouzpanah S, Taleban FA, Mehdipour P, Sabour S, Atri M. Hypermethylation pattern of ESR and PgR genes and lacking estrogen and progesterone receptors in human breast cancer tumors: ER/PR subtypes. Cancer Biomark 2018;21:621–638. doi: 10.3233/cbm-170697. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki M, Dharia A, Oh BR, Tanaka Y, Fujimoto S, Dahiya R. Progesterone receptor B gene inactivation and CpG hypermethylation in human uterine endometrial cancer. Cancer Res 2001;61:97–102. [PubMed] [Google Scholar]
- 20.Ren Y, Liu X, Ma D, Feng Y, Zhong N. Down-regulation of the progesterone receptor by the methylation of progesterone receptor gene in endometrial cancer cells. Cancer Genet Cytogenet 2007;175:107–116. doi: 10.1016/j.cancergencyto.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Shiozawa T, Itoh K, Horiuchi A, Konishi I, Fujii S, Nikaido T. Down-regulation of estrogen receptor by the methylation of the estrogen receptor gene in endometrial carcinoma. Anticancer Res 2002;22:139–143. [PubMed] [Google Scholar]
- 22.Maeda K, Tsuda H, Hashiguchi Y, Yamamoto K, Inoue T, Ishiko O, et al. Relationship between p53 pathway and estrogen receptor status in endometrioid-type endometrial cancer. Human Pathol 2002;33:386–391. doi: 10.1053/hupa.2002.124720. [DOI] [PubMed] [Google Scholar]
- 23.Hori M, Iwasaki M, Shimazaki J, Inagawa S, Itabashi M. Assessment of hypermethylated DNA in two promoter regions of the estrogen receptor alpha gene in human endometrial diseases. Gynecol Oncol 2000;76:89–96. doi: 10.1006/gyno.1999.5662. [DOI] [PubMed] [Google Scholar]
- 24.Navari JR, Roland PY, Keh P, Salvesen HB, Akslen LA, Iversen OE, et al. Loss of estrogen receptor (ER) expression in endometrial tumors is not associated with de novo methylation of the 5′ end of the ER gene. Clin Cancer Res 2000;6:4026–4032. [PubMed] [Google Scholar]
- 25.Yang S, Jia Y, Liu X, Winters C, Wang X, Zhang Y, et al. Systematic dissection of the mechanisms underlying progesterone receptor downregulation in endometrial cancer. Oncotarget 2014;5:9783–9797. doi: 10.18632/oncotarget.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]


