Abstract
Non-alcoholic fatty liver disease (NAFLD) is one of the most common liver disorders. Possible links have been recently found between the gut-microbiota and the host metabolism in development of NAFLD and obesity. Therefore, understanding the changes in intestinal microbiota during the progression of NAFLD, is important. In this study, the effect of Kombucha tea (KT), obtained by microbial fermentation of sugared black tea, was investigated on gut-microbiota during the progression of NAFLD. The results indicated a decrease in Erysipelotrichia class by treatment with KT in comparison to the methionine/choline-deficient (MCD)-fed db/db mice. Allobaculum, Turicibacter, and Clostridium genera, were only detected in MCD-fed db/db mice and were decreased after treatment with KT, whereas Lactobacillus was more abundant in MCD + KT-fed mice than in MCD only-fed mice and Mucispirillum, was found only in the MCD + KT-fed mice group. Our results demonstrated that the change of intestinal microbiota was influenced by KT intake, contributing to combat NAFLD.
Keywords: Gut-microbiota, Kombucha tea, Liver protection, Non-alcoholic fatty liver disease
Introduction
The human gastrointestinal (GI) tract is the habitat for a myriad of bacteria. In an adult man, the number of gut microbiota is approximately 1014, which is more than 10 times as much as the total human cells (Luckey, 1972). Though the gut microbes are composed of a variety of anaerobic bacteria, ranging over at least 500–1000 different species, almost 80% of them are not cultivable (Guarner and Malagelada, 2003). Culture-independent high-throughput sequencing method is preferable to reveal the roles and structures of massive bacterial communities in GI tract, rather than culture-based method. In the past few years, several projects, such as Human Microbiome Project (Turnbaugh et al., 2007) and MetaHIT consortium (Qin et al., 2010), were launched to understand gut microbiota composition and roles. Through these continuous projects and studies of gut microbiome, bacterial community living in the GI tract has been revealed to be associated with several metabolic and physiological functions in the host organisms. Recent reports show that disruption of the gut microbiota influences various metabolic phenotypes and inflammatory human diseases, such as obesity (Ley, 2010; Turnbaugh et al., 2009), type 2 diabetes (Serino et al., 2012), and fatty liver disease (Toye et al., 2007). In particular, fatty liver disease has increased widely due to changes in lifestyle over time, such as dietary changes, widespread metabolic syndrome, and sedentary lifestyle (Nseir et al., 2014).
Non-alcoholic fatty liver disease (NAFLD) is a continuous spectrum of liver diseases characterized by ectopic accumulation of fat in the liver, which is unrelated to excessive alcohol consumption (Machado and Diehl, 2016). Non-alcoholic steatohepatitis (NASH) is defined by the presence of steatosis coexisting with hepatocellular death and inflammation. NAFLD/NASH has a strong association with metabolic abnormalities such as obesity, type 2 diabetes mellitus and dyslipidemia (Birkenfeld and Shulman, 2014). Because of its growing worldwide prevalence, NAFLD is becoming the most common cause of chronic liver disease and it indicates the urgent need for a better understanding of NAFLD/NASH progression (Loomba and Sanyal, 2013). Although the pathogenesis of NAFLD cannot be completely comprehended, several studies suggested that dysbiosis in the gut, caused by obesity and metabolic syndromes, can be related to the development of NAFLD (Abu-Shanab and Quigley, 2010; Compare et al., 2012; Schnabl and Brenner, 2014). In 1980s, early evidence for the relationship between derangement of gut microbiota and fatty liver disease was derived from the pathogenesis of NASH, related to small intestinal bacterial overgrowth (Drenick et al., 1982; Wigg et al., 2001). Since then, through continuous studies in animal models and humans, the microbiota has been shown to play a major role in the pathogenesis of NAFLD (Abu-Shanab and Quigley, 2010).
Recently, the relationship between gut microbiota and fatty liver disease has been clarified to some extent. At an anatomical location, liver is directly supplied with about 70% of blood flow through the portal vein. This gut-liver axis serves as a pathway for exposing liver to antigens or toxic factors, produced by gut microbiota (Compare et al., 2012). Bacterial byproducts such as ammonia, phenols and ethanol, are potentially hepatotoxic and can contribute to the development of liver diseases (Abu-Shanab and Quigley, 2010). Moreover, bacterial lipopolysaccharides (LPS) are mainly involved with NAFLD. The LPS absorbed during lipid absorption induce a cascade of intracellular processes by binding to specific protein and induce inflammation by enhancing expression of target genes (Machado and Cortez-Pinto, 2012). Several studies suggested the association between gut microbiota and intestinal permeability with hepatic steatosis severity. Thus, interventions for modulation of the gut microbiota (e.g. probiotics, prebiotics and diet standardization) are expected to be useful for prevention or treatment of such conditions (Daubioul et al., 2000; Machado and Cortez-Pinto, 2012; Spencer et al., 2011).
Kombucha tea (KT) is a non-alcoholic fermented beverage that is sparkling and has a sour taste. KT is produced by fermentation of a mixture of black tea, sucrose, and tea fungus. The fermentation is performed by microbial metabolism using the symbiosis between bacteria and yeast (Jarrell et al., 2000). The beverage has acquired popularity around the world for its health benefits, such as anti-microbial (Sreeramulu et al., 2000), anti-oxidant (Chu and Chen, 2006), anti-carcinogenic (Jayabalan et al., 2011), and anti-diabetic (Aloulou et al., 2012) effects. Although KT has emerged as a protective substance against liver damage, the effect of KT on NAFLD still remains unclear. In a previous study, we investigated whether KT influenced hepatic steatosis, and found it effective for attenuation of lipid accumulation in liver, thereby protecting the liver from damage (Hyun et al., 2016). Therefore, the objective of this study was to investigate, through metagenomic analyses, whether the gut-microbiota changed during the progression of NAFLD, upon induction by KT and its effect on fat accumulation in liver.
Materials and methods
Preparation of Kombucha extract
Six grams of black tea (type of tea bag, Lipton, Yellow Label Tea) were added to 600 mL of boiling distilled water, allowed to infuse for 5 min, after which the tea bag was removed. Then, 10% (w/v) of sucrose was added and stirred to dissolve into the black tea. After cooling to room temperature, the tea was poured into 1 L glass beaker that had been previously sterilized at 121 °C for 20 min. Finally, the freshly prepared tea was inoculated with freshly grown Kombucha mat that had been cultured in the same medium for 14 days and 30% of previously fermented liquid tea broth. The beaker was covered with clean cheese cloth and fixed with rubber bands. The fermentation (at 25 ± 3 °C) continued for 14 days. During fermentation, a daughter mat (new Kombucha mat) developed over the tea surface. The fermented Kombucha was centrifuged at 7000×g for 20 min and the supernatant was filtered through 0.45 μm (pore size) syringe filter (Sartorius, Minisart syringe filters). Later, the filtrated extracts were frozen at − 80 °C before subjecting to lyophilization. The lyophilized extract of KT was stored at − 20 °C till further use.
Animals and treatments
Six-week-old male C57BLKS db/db mice were purchased from Korea Research Institute of Bioscience and Biotechnology (KRIBB, Daejeon, Korea). During acclimatization, mice were fed normal diet and water, and housed with a 12-h light/dark cycle for 2–3 weeks. Twelve db/db 8-week-old mice (average body weight 38 g) were fed either the control diet (n = 4) or the methionine/choline-deficient (MCD) diet (n = 8; cat no 518810; Central Lab. Animal Inc., Seoul, Korea) for 4 weeks. Among the eight MCD diet-fed mice, four were also fed with Kombucha powder 2 g/kg by oral administration, every 24 h for 3 weeks; the rest were treated with water instead. Kombucha powder, kept at − 80 °C, was completely dissolved in water and used for animal treatment.
Liver histology
Each liver tissue was collected for histological analysis. Liver specimens were fixed in 10% neutral buffered formalin, embedded in paraffin 133, and cut into 4 μm sections. Specimens were de-waxed, hydrated, and stained with standard hematoxylin and eosin staining protocol (H&E).
Collection of stool samples
Stool samples from each group of db/db mice (Group 1: Control, group 2: MCD, group 3: MCD + KT) were collected every week (for 4 weeks). Stool samples were stored at 4 °C for further experiments.
Metagenomic DNA extraction
Metagenomic DNA was extracted from approximately 300 mg of stool using the PowerSoil®DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) with modifications. Specifically, bead tubes were heated to 65 °C for 10 min, and then vortexed horizontally, fixed with tape, on a flat-bed vortex pad for 10 min at the maximum speed. The remaining steps were performed as directed by the manufacturer’s instructions. Extracted DNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and frozen at − 80 °C till further use.
Metagenome sequencing
Extracted DNA samples were sent to National Instrumentation Center for Environmental Management (NICEM, Seoul National University, Seoul, Republic of Korea) for metagenome sequencing. The extracted DNA was used for PCR-amplification of the 16S ribosomal RNA gene. The PCR products were used for metagenome sequencing by NICEM on 454 GS-FLX Titanium platform (454 Life Sciences, Branford, CT, USA).
Microbiome analysis
The filtered read sequences were matched with the taxonomic identifications using the reference database (Silva.bacteria, http://www.arb-silva.de), reference mapping tools (ncbi-blast-2.2.30+, ftp://ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/), and taxonomic classification software (MEGAN5—MEtaGenome ANalyzer, http://www-ab.informatik.uni-tuebingen.de/software/megan5/).
Results and discussion
Kombucha tea (KT) protects against fat accumulation in MCD-fed db/db mice
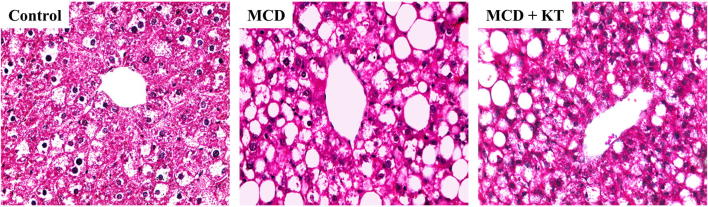
To confirm the previously reported lipoprotective effects of KT on the liver (Hyun et al., 2016), liver histological analysis was performed. The Histological analysis showed severe steatosis with excessive accumulation of macro- and microvesicular fats in livers of MCD-fed db/db mice compared to chow-fed db/db mice (Fig. 1). In comparison to MCD-fed mice without KT treatment, mild steatosis with decreased fat droplets was observed in livers of MCD + KT-fed db/db mice. These data suggest that KT reduced fat accumulation in MCD-fed db/db mice.
Fig. 1.
Kombucha reduces fat accumulation in MCD-fed db/db mice. Histomorphology of the liver sections using H&E staining from representative (Control) normal-fed db/db mice, (MCD) MCD-fed db/db mice, and (MCD + KT) and kombucha-treated MCD-fed db/db mice (× 40). Excessive accumulation of macro- and microvesicular fats were observed in livers of MCD-fed db/db mice compared to normal-fed db/db mice and KT treated mice
Sequence analysis of the 16S rRNA region
Previous histopathological analysis showed that the level of fat accumulation, induced by MCD-feeding, was decreased by KT treatment. In order to investigate whether the alleviation of NAFLD symptoms was due to changes in intestinal microbial communities, gut microbial community of the tested groups (db/db control, MCD and MCD + KT-fed mice) was observed over time post- KT treatment. The gut microbial composition was evaluated following the analysis of 16S rRNA sequences in stools extracted metagenomes. After sequencing the 16S rRNA region, 100,301 raw reads were generated from 12 stool samples (Table 1). The resulting 100,301 read sequences were subjected to quality control standards and the ones that did not meet quality criteria (low-quality read sequences; average quality value < 20, or read sequence length < 300 bp), were filtered out from the data set. The remaining 80,658 read sequences were assigned to the taxonomic identifications using the reference database, reference mapping tools, and taxonomic classification software.
Table 1.
Multiplex identifier (MID)-sorted reads
| Sample | Raw | Q ≥ 20, bp ≥ 300 | Filtered | Clean (%) |
|---|---|---|---|---|
| Control 0Wa | 11,355 | 9073 | 2282 | 79.90 |
| Control 1W | 15,900 | 11,912 | 3988 | 74.92 |
| Control 2W | 9782 | 7601 | 2181 | 77.70 |
| Control 3W | 11,317 | 9092 | 2225 | 80.34 |
| MCD 1Wb | 7286 | 6074 | 1212 | 83.37 |
| MCD 2W | 7568 | 6389 | 1179 | 84.42 |
| MCD 3W | 8782 | 7335 | 1447 | 83.52 |
| MCD + KT 1Wc | 10,836 | 8828 | 2008 | 81.47 |
| MCD + KT 2W | 9499 | 7776 | 1723 | 81.86 |
| MCD + KT 3W | 7976 | 6578 | 1398 | 82.47 |
| Total | 100,301 | 80,658 | 19,643 | 81.00 |
aControl group are normal-fed db/db mice for 0, 1, 2 and 3 weeks
bMCD group are MCD-fed db/db mice for 1, 2 and 3 weeks
cMCD + KT group are Kombucha-treated MCD-fed db/db mice for 1, 2 and 3 weeks
Analysis of the intestinal microbiota between MCD- and MCD + KT-fed db/db mice
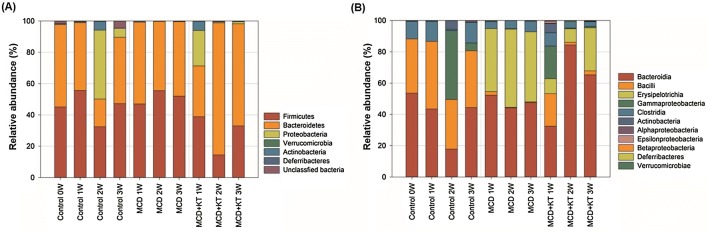
At the phylum level, Firmicutes and Bacteroidetes were present at a high proportion in all the groups (Fig. 2A). The ratio of Firmicutes and Bacteroidetes was almost 1:1 in control and MCD-fed groups, whereas the proportion of Bacteroidetes in MCD + KT-fed group increased over time. Mouzaki et al. (2013) found that patients with NASH had significantly low levels of Bacteriodetes compared to those with simple steatosis and healthy controls. Raman et al. (2013) also reported that the number of Firmicutes was increased in patients with NAFLD versus non-obese controls. However, the results of intestinal microbiome on the relation of NASH with healthy controls in children group, were contrasting (Zhu et al., 2013). Microbiome studies are difficult to establish for such relationships because they are variable depending on the experimental design (Wieland et al., 2015). However, the continuously accumulated results help to understand the relationship between fatty liver disease and gut microbiome.
Fig. 2.
Relative bacterial abundance (A) at phylum level, and (B) at class level in each stool sample of (Control) normal-fed db/db mice, (MCD) MCD-fed db/db mice, and (MCD + KT) kombucha-treated db/db mice on MCD diet. Gut microbial compositions were assessed by sequencing of the 16sr RNA region from the metagenomes of stool samples, followed by taxonomic identifications
At the class level, the Erysipelotrichia class belonging to Firmicutes phylum, shows an interesting aspect (Fig. 2B). The Erysipelotrichia class did not exist in db/db control mice. However, it increased approximately 45% just 1 week from the start of MCD diet and dominated the community. Interestingly, high levels of Erysipelotrichia in fecal microbiota are associated with hepatic steatosis (Spencer et al., 2011). The level of Erysipelotrichia is suggested to be important bacterial marker of susceptibility to choline deficiency-induced fatty liver disease. Moreover, Allobaculum and Turicibacter, belonging to Erysipelotrichia class, are known to produce short-chain fatty acids (SCFAs) (Schwiertz et al., 2010). SCFAs, produced by intestinal microbes, are considered as an additional source of energy to humans (Schwiertz et al., 2010). Therefore, metabolites of various microbes within this family are thought to directly or indirectly affect NAFLD induction.
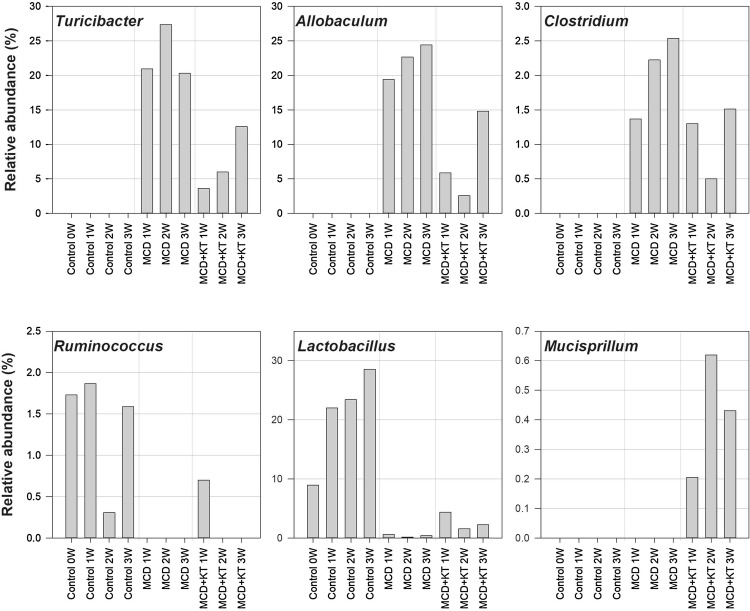
Insight into the changes in intestinal microbiota associated with MCD-fed db/db mice treated with KT
Although there was no significant difference in the composition of dominant genera of MCD or MCD + KT group, a difference in the proportion of dominant genera was noted (Fig. 3). It is noteworthy that Allobaculum and Turicibacter, involved in the pathogenesis of NAFLD, were significantly decreased in MCD-fed db/db mice after KT treatment (Fig. 3). Although the mechanism by which Erysipelotrichia class causes NAFLD is still unclear, we supposed that the microbial structural or chemical components in the fermented KT help to bring NAFLD under control. Clostridium genus was not seen in control-fed db/db mice group and only existed in MCD-fed groups and was under-represented in the intestinal microbiota samples from MCD + KT-fed groups (Fig. 3). The genus is able to produce several SCFAs and alcohol. In addition, some of these bacteria have a possible obesogenic potential, the underlying mechanism of which in mice is still unclear. This microbe was suggested to help reinforce sugar and fat absorption and the associated higher intake of energy supplying nutrients facilitating body fat deposition (Woting et al., 2014). Therefore, Clostridium may be involved in obesity and NAFLD development. Patients with NASH were shown to have a significantly higher percentage of Clostridium than those with NAFLD (Schnabl and Brenner, 2014).
Fig. 3.
The changes in relative abundance of six bacterial genera (Turicibacter, Allobaculum, Clostridium, Ruminococcus, Lactobacillus, and Mucispirillum) whose proportions were altered by MCD or MCD + KT-treatment. Observed changes in the proportion of these genera are suggested to be associated with affecting fat accumulation in liver and consequently non-alcoholic fatty liver disease
In contrast, Ruminococcus did not exist in the intestinal microbiota from MCD-fed groups with NAFLD (Fig. 3). However, these bacteria were maintained in control-fed db/db mice group. Ruminococcaceae was significantly under-represented in the fecal microbiota samples from patients with NAFLD than that from healthy subjects (Mouzaki et al., 2013). While Lactobacillus genus grew 28.6% in control db/db mice group over time, it was significantly under-represented in the intestinal microbiota samples from MCD-fed groups (Fig. 3). In the MCD + KT-fed mice, Lactobacillus was more abundant than in MCD only-fed mice. Several lactobacilli have been known for their beneficial probiotic effect, such as anti-inflammatory actions, and several strains of Lactobacillus have shown protective effect against NAFLD (Li et al., 2014). Wong et al. (2013) investigated the longitudinal microbiome changes in adult patients with biopsy-proven NASH, caused by probiotic supplement treatment consisting of mixed Lactobacillus species. After 6 months, Lactobacillus-treated patients with NASH, reduced hepatic fat simultaneously with a decrease in Firmicutes and an increase in Bacteroidetes. These results summarized that beneficial probiotic bacteria, including Lactobacillus in the intestine, may be related to alleviation of NAFLD.
Although a small percentage, Mucispirillum genus was found only in the MCD + KT-fed mice group. Mucispirillum is known to correlate positively with circulating leptin (Ravussin et al., 2012). Leptin is a hormone made by adipose cells that help to regulate energy balance by inhibiting hunger and maintaining constant body fat. When leptin reaches the brain, it decreases body fat, food intake, level of blood sugar, etc., and increases metabolic efficiency or activity, reducing weight gradually (Brennan and Mantzoros, 2006).
This study showed that KT treatment resulted in changes in the composition of mouse gut microbiota. The proportion of Bacteroidetes increased over time only in MCD + KT-fed group. Bacteria belonging to Erysipelotrichia class increased from 1-week post MCD-feed, eventually dominating the community, but were significantly decreased by KT treatment on MCD-fed db/db mice. Bacteria belonging to Allobaculum, Turicibacter, and Clostridium were selectively decreased, whereas bacteria belonging to Mucispirillum decreased only after treatment with KT. Taken together, the current study demonstrated an indirect relationship between intestinal microbiome changes, caused by KT in MCD-fed db/db mice and the progress of NAFLD. Such microbial changes may help in the suppression of fat accumulation in the liver and consequently reduced NAFLD. The longitudinal study on gut microbiome suggested that continuous KT treatment might help to ameliorate the gut microbiome conditions of patients with NAFLD.
Acknowledgements
This work was supported by a 2-year Research Grant of Pusan National University (Y.-S.S.).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepat. 2010;7:691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- Aloulou A, Hamden K, Elloumi D, Ali MB, Hargafi K, Jaouadi B, Ayadi F, Elfeki A, Ammar E. Hypoglycemic and antilipidemic properties of Kombucha tea in alloxan-induced diabetic rats. BMC Complement. Altern. Med. 2012;12:63–71. doi: 10.1186/1472-6882-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenfeld AL, Shulman GI. Nonalcoholic fatty liver disease, hepatic insulin resistance, and type 2 diabetes. Hepatology. 2014;59:713–723. doi: 10.1002/hep.26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan AM, Mantzoros CS. Drug Insight: the role of leptin in human physiology and pathophysiology—emerging clinical applications. Nat. Rev. Endocrinol. 2006;2:318–327. doi: 10.1038/ncpendmet0196. [DOI] [PubMed] [Google Scholar]
- Chu S-C, Chen C. Effects of origins and fermentation time on the antioxidant activities of Kombucha. Food Chem. 2006;98:502–507. doi: 10.1016/j.foodchem.2005.05.080. [DOI] [Google Scholar]
- Compare D, Coccoli P, Rocco A, Nardone O, De Maria S, Cartenì M, Nardone G. Gut–liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 2012;22:471–476. doi: 10.1016/j.numecd.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Daubioul CA, Taper HS, Laurent D, Delzenne NM. Dietary oligofructose lessens hepatic steatosis, but does not prevent hypertriglyceridemia in obese Zucker rats. J. Nutr. 2000;130:1314–1319. doi: 10.1093/jn/130.5.1314. [DOI] [PubMed] [Google Scholar]
- Drenick EJ, Fisler J, Johnson D. Hepatic steatosis after intestinal bypass—prevention and reversal by metronidazole, irrespective of protein-calorie malnutrition. Gastroenterology. 1982;82:535–548. [PubMed] [Google Scholar]
- Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- Hyun J, Lee Y, Wang S, Kim J, Kim J, Cha J, Seo Y-S, Jung Y. Kombucha tea prevents obese mice from developing hepatic steatosis and liver damage. Food Sci. Biotechnol. 2016;25:861–866. doi: 10.1007/s10068-016-0142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell J, Cal T, Bennett JW. The Kombucha consortia of yeasts and bacteria. Mycologist. 2000;14:166–170. doi: 10.1016/S0269-915X(00)80034-8. [DOI] [Google Scholar]
- Jayabalan R, Chen P-N, Hsieh Y-S, Prabhakaran K, Pitchai P, Marimuthu S, Thangaraj P, Swaminathan K, Yun SE. Effect of solvent fractions of Kombucha tea on viability and invasiveness of cancer cells—characterization of dimethyl 2-(2-hydroxy-2-methoxypropylidine) malonate and vitexin. Indian J. Biotechnol. 2011;10:75–82. [Google Scholar]
- Ley RE. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- Li C, Nie S-P, Zhu K-X, Ding Q, Li C, Xiong T, Xie M-Y. Lactobacillus plantarum NCU116 improves liver function, oxidative stress and lipid metabolism in rats with high fat diet induced non-alcoholic fatty liver disease. Food Funct. 2014;5:3216–3223. doi: 10.1039/C4FO00549J. [DOI] [PubMed] [Google Scholar]
- Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013;10:686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- Luckey T. Introduction to intestinal microecology. Am. J. Clin. Nutr. 1972;25:1292–1294. doi: 10.1093/ajcn/25.12.1292. [DOI] [PubMed] [Google Scholar]
- Machado MV, Cortez-Pinto H. Gut microbiota and nonalcoholic fatty liver disease. Ann. Hepatol. 2012;11:440–449. doi: 10.1016/S1665-2681(19)31457-7. [DOI] [PubMed] [Google Scholar]
- Machado MV, Diehl AM. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology. 2016;150:1769–1777. doi: 10.1053/j.gastro.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- Nseir W, Hellou E, Assy N. Role of diet and lifestyle changes in nonalcoholic fatty liver disease. World J. Gastroenterol. 2014;20:9338–9344. doi: 10.3748/wjg.v20.i28.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2013;11:868–875. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, Waget A, Klopp P, Iacovoni J, Klopp C. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreeramulu G, Zhu Y, Knol W. Kombucha fermentation and its antimicrobial activity. J. Agric. Food Chem. 2000;48:2589–2594. doi: 10.1021/jf991333m. [DOI] [PubMed] [Google Scholar]
- Toye A, Dumas M, Blancher C, Rothwell A, Fearnside J, Wilder S, Bihoreau M, Cloarec O, Azzouzi I, Young S. Subtle metabolic and liver gene transcriptional changes underlie diet-induced fatty liver susceptibility in insulin-resistant mice. Diabetologia. 2007;50:1867–1879. doi: 10.1007/s00125-007-0738-5. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland A, Frank D, Harnke B, Bambha K. Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2015;42:1051–1063. doi: 10.1111/apt.13376. [DOI] [PubMed] [Google Scholar]
- Wigg A, Roberts-Thomson I, Dymock R, McCarthy P, Grose R, Cummins A. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor α in the pathogenesis of non-alcoholic steatohepatitis. Gut. 2001;48:206–211. doi: 10.1136/gut.48.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, Yeung DK, Law PT, Kwan HS, Yu J, Sung JJ. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis—A longitudinal study. PLoS One. 2013;8:e62885. doi: 10.1371/journal.pone.0062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woting A, Pfeiffer N, Loh G, Klaus S, Blaut M. Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. MBio. 2014;5:e01530-01514. doi: 10.1128/mBio.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]