Abstract
Glioblastoma is an aggressive primary brain tumor with bad prognosis. On the other hand, oncolytic measles virus (MeV) therapy is an experimental glioma treatment strategy with clinical safety and first evidence of anti-tumoral efficacy. Therefore, we investigated the combination of MeV with conventional therapies by cytotoxic survival assays in long-term glioma cell lines LN229, LNZ308, and glioma stem-like GS8 cells, as well as the basal viral infectivity in primary glioblastoma cultures T81/16, T1094/17, and T708/16. We employed Chou-Talalay analysis to identify the synergistic treatment sequence chemotherapy, virotherapy, and finally radiotherapy (CT-VT-RT). RNA sequencing and immunopeptidome analyses were used to delineate treatment-induced molecular and immunological profiles. CT-VT-RT displayed synergistic anti-glioma activity and initiated a type 1 interferon response, along with canonical Janus kinase-signal transducers and activators of transcription (JAK-STAT) signaling, and downstream interferon-stimulated genes were induced, resulting in apoptotic cascades. Furthermore, antigen presentation along with immunostimulatory chemokines was increased in CT-VT-RT-treated glioma cells, indicating a treatment-induced pro-inflammatory phenotype. We identified novel treatment-induced viral and tumor-associated peptides through HLA ligandome analysis. Our data delineate an actionable treatment-induced molecular and immunological signature of CT-VT-RT, and they could be exploited for the design of novel tailored treatment strategies involving virotherapy and immunotherapy.
Keywords: glioblastoma, immunopeptidome, oncolytic measles virus, personalized medicine, BATF2, SAMD9
Introduction
Glioblastoma is an aggressive primary tumor of the CNS with an overall median survival of approximately 1.5 years.1, 2, 3, 4, 5 Glioblastomas are immunosuppressive tumors with high levels of regulatory T cell (Treg) infiltration6 and B7H1 upregulation in tumor-associated macrophages.7 Thus, a reshaping of the glioma-associated microenvironment toward a pro-inflammatory signature might be a promising strategy to improve conventional therapeutic strategies. Oncolytic virotherapy using tumor-lytic viruses provides an interesting opportunity in this regard. Such viruses can be genetically engineered and are capable of inducing an oncolytic cascade. Lysed tumor cells release virions, viral components, and cellular debris encompassing highly immunostimulatory danger- and pathogen-associated motif patterns (DAMPs and PAMPs),8, 9, 10 which can serve as a strong induction stimulus of immune responses. The clinical evidence of complete remission in a patient treated with oncolytic measles virus (MeV) in relapsing drug-refractory myeloma further is a strong indicator for the oncolytic efficacy of MeV.11, 12
Moreover, MeV has already also been clinically tested for the treatment of glioblastoma patients. Based on first evidence of the anti-tumoral efficacy of MeV monotherapy against glioblastoma in vitro and in subcutaneous or orthotopic, immunodeficient animal models,13 this treatment option has been tested in a phase I clinical trial to treat patients with glioblastoma.14 While this trial provided clear evidence for the safety of oncolytic MeV applied in significant doses directly to the CNS of human patients, several pre-clinical studies provided evidence that MeV can be modified or combined with other treatment modalities. Among these analyses, it could be shown that MeV can be directly re-targeted to typical tumor markers of glioma, such as epidermal growth factor receptor (EGFR) and/or EGFRvIII,15 or even against glioma stem cells.16, 17 On the other hand, the first indication of a fruitful combination of MeV with radiotherapy became evident;18 but, most interestingly, the combination of MeV with PD-1 checkpoint blocking in immunocompetent animal models indicated a significant immunotherapeutic component of oncolytic MeV in its anti-tumoral efficacy.9
In any case, MeV is not the only virus species that is developed for its use as an anti-glioma entity. Desjardins et al.19 investigated convection-enhanced intratumoral delivery of recombinant nonpathogenic polio-rhinovirus chimera in progressive glioblastoma patients with efficacy analyzed as a secondary endpoint. Treatment with this chimera did not induce neurotoxicity and resulted in higher survival rates at 24 and 36 months post-treatment compared with historic controls. A gamma-retroviral replicating vector encoding cytosine deaminase (Vocimagene amiretrorepvec, Toca 511) was also investigated in a phase I trial in recurrent high-grade glioma patients.20 Infection by this virus becomes cytolytic after administration of the pro-drug 5-fluorocytosine, which is then locally converted into the chemotherapeutic agent 5-fluorouracil. Indeed, durable complete responses were observed in a subgroup of patients.20 As a fourth example, replication-competent oncolytic adenovirus DNX-2401 (tasadenoturev), which had demonstrated pre-clinically anti-glioma efficacy,21 was tested in a recent phase I trial in patients with progressive high-grade glioma. These patients received a single intratumoral injection of DNX-2401, but tumors became resected afterward to acquire post-treatment tissue.21 Also this trial observed a subgroup of long-term responders. DNX-2401-induced oncolysis was observed in post-treatment tissues, and histologies revealed treatment-induced tumor infiltration by CD8+ and T-bet+ T cells, while the transmembrane immunoglobulin mucin-3 hinted at a treatment-induced anti-tumoral immune reaction.22
These selected recent examples highlight the potential of this therapeutic modality, in general, but especially MeV, with special focus on immunotherapy. Next steps include to question how their efficacy might be further enhanced. One option is the combination of virotherapy with other immunotherapeutic modalities, i.e., oncolytic virus therapy might help to alter the immunosuppressive glioma-associated microenvironment and thereby pave the way for the efficacy of subsequent immunotherapies, including peptide vaccinations strategies.
Based on these considerations, we investigated (1) how oncolytic MeV could be synergistically incorporated as a part of a sequential combination treatment with standard treatment modalities, i.e., radiotherapy (RT), or the chemotherapeutics temozolomide (TMZ) or lomustine (CCNU); (2) whether MeV-containing treatments induce usable therapy-induced molecular and immunological signatures; and (3) whether immunopeptidome analysis can reveal treatment-induced presentation of peptides that might be utilized therapeutically.
Results
Expression of Oncolytic MeV Receptor CD46 Is Modulatd by Hypoxia or TMZ
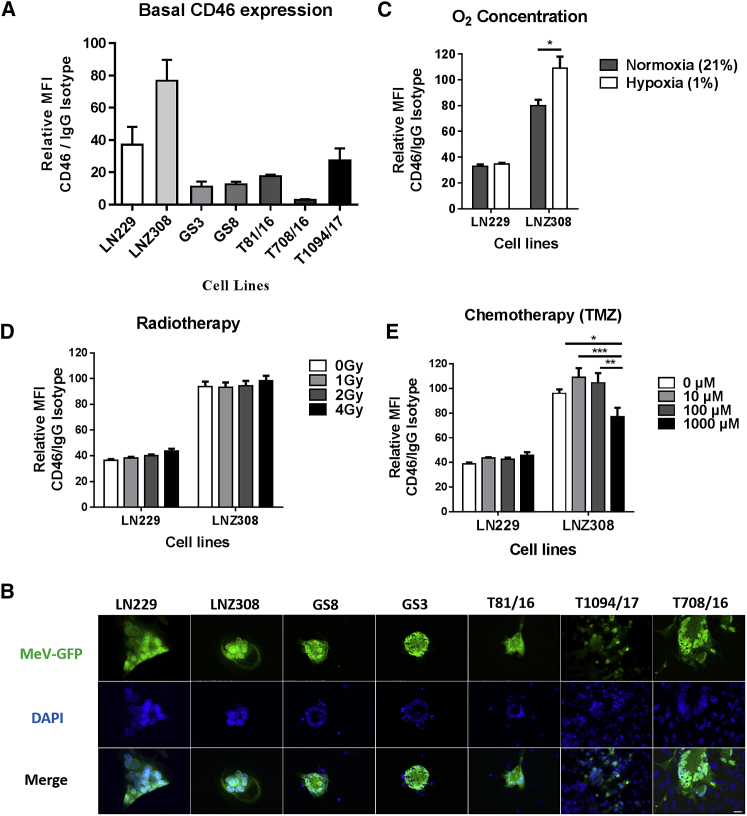
Membrane cofactor protein or CD46 serves as receptor for cell entry of vaccine strain MeVs.23 Wild-type strains also use signaling lymphocyte activation molecule SLAM-F1 expressed in immune cells24 or nectin-4, an epithelial receptor.25, 26 All glioma cells except primary GBM T708/16 expressed CD46 with mean fluorescence intensity (MFI) coefficient (CD46 to immunoglobulin G [IgG]) > 10 (Figure 1A). No expression of nectin-4 was detected on the same panel of tumor cells (Figure S1). In any case, all cell lines were susceptible to MVNSe-GFP (P), an Edmonston strain vaccine virus, which was developed for oncolysis (Figure 1B).
Figure 1.
MeV Receptor Expression and Infectivity in Glioma Cells
(A) Basal level of CD46 receptor expression in glioma cells, glioma stem-like cells, and primary glioblastoma cells determined via flow cytometry and depicted as relative MFI compared to isotype control. (B) MeV infectivity observed via GFP expression along with characteristic syncytial morphology as giant multinuclear aggregates visualized by fluorescence microscopy. Scale bar, 20 μm. (C) Significant (p = 0.011) increase in CD46 surface expression on LNZ308 cells subjected to hypoxia, with no effect of hypoxia in LN229 cells. (D) No change in CD46 expression after γ-irradiation of LN229 or LNZ308 cells. (E) A low dose (10 or 100 μM) of TMZ tendentially increases CD46 surface expression, while a high dose of TMZ (1,000 μM) significantly impairs CD46 expression in LNZ308 cells. Multiple t test with Holm-Sidak post hoc test in (C) and two-way ANOVA with Tukey’s multiple comparison test in (E); *p < 0.05; **p < 0.01; ***p < 0.001. Data are expressed as mean ± SEM, n = 3 (A) and n = 9 (C–E).
To check for receptor expression under more physiologic or standard treatment-related conditions, we assessed CD46 expression after the cultivation of tumor cells at 1% O2 or after treatment with irradiation or TMZ (Figures 1C–1E). The expression of CD46 increased after hypoxia in LNZ308 (Figure 1C), but it remained unchanged after RT (Figure 1D). Low doses of TMZ, i.e., 10 or 100 μM, increased the expression of CD46 on LNZ308 cells, while treatment with 1,000 μM TMZ significantly reduced the surface expression of CD46 (Figure 1E). These treatment-induced changes in CD46 expression correlated with moderately increased MeV replication using low TMZ doses and decreased MeV replication with 1,000 μM TMZ (Figure S2). These results prompted us to check systematically for potential synergism of the standard glioma treatment regimen with MeV oncolysis.
Combined Treatments Are Only Synergistic When Radiation Therapy Is Administered after Virotherapy
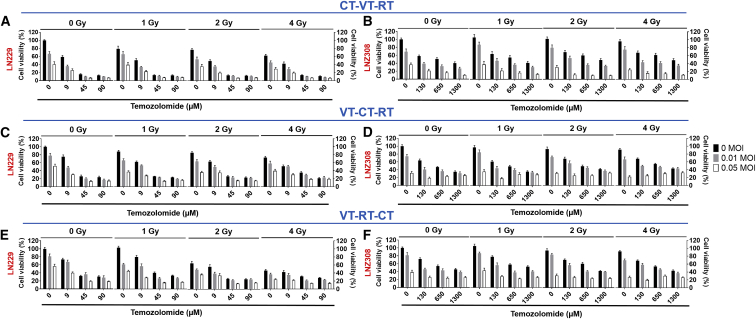
For synergism of different treatment modalities, the ways the tumor cells are respectively killed may be relevant. MeV can induce autophagy ensued by mitophagy to sustain an anti-apoptotic environment through the prevention of cytochrome c release to aid its replication.27, 28, 29 On the other hand, glioma cell killing by MeV has been described to have also apoptotic features.18 We verified MeV-induced autophagy in our studies via western blot analysis and co-localization studies utilizing a GFP-tagged LC3 (LC3-GFP), demonstrating cleavage and co-localization pattern expected for autophagy (Figure S3). Due to their relevance for the current clinical standard of care in glioma therapy, we combined MeV virotherapy (VT) with RT and chemotherapy (CT) to seek synergistic treatment sequences. RT induces apoptosis.30, 31 Accordingly, for MeV-containing combination treatments starting with RT, a rather antagonistic antiviral effect became evident. In contrast the VT-RT regimen sequence resulted in lower cell viability (Figures S4A–S4C), mirroring previous reports.18 Consequently, we only considered combination treatments for the design of triple combinations of MeV, RT, and CT, where VT preceded RT. This resulted in the following combination regimens in our investigations: (1) VT-RT-CT, (2) VT-CT-RT, and (3) CT-VT-RT.
Triple Therapies Are Synergistic if the Treatment Algorithm Starts with Alkylating CT and Is Followed by VT and RT
We carried out cytotoxic survival assays utilizing TMZ dosed according to respective EC5032 in combination with VT and RT (Figure S5). All triple regimens revealed dose-dependent killing, with synergistic effects mainly observed in CT-VT-RT (Figures 2A and 2B). The effect of VT was most prominent in CT-VT-RT (Figures 2A and 2B) compared with VT-RT-CT and VT-CT-RT (Figures 2C–2F). Given the clinical routine for treatment of glioblastoma with fractionated individual doses up to 2-Gy irradiation, we identified CT-VT-RT as the only regimen to exhibit maximal synergism at 2 Gy (Tables S1–S6).
Figure 2.
Anti-glioma Activity of CT-VT-RT as Synergistic Triple Regimen Using TMZ as Chemotherapeutic Agent
The treatment sequence CT-VT-RT is outlined at the indicated concentrations in (A) LN229 and (B) LNZ308 cells. Non-synergistic regimen VT-CT-RT showed poor combinatorial efficacy in (C) LN229 and (D) LNZ308 cells. Similarly, regimen VT-RT-CT exhibited poor combinatorial efficacy in (E) LN229 and (F) LNZ308 cells. Data are expressed as mean ± SEM, n = 9. Calculations of synergy are outlined in Tables S1–S6.
To employ a more relevant system, we used the glioblastoma stem-like cell line GS8. We first determined the EC50 of TMZ in GS8 cells to be 250 μM (Figure S6), and we continued with the investigations of triple therapies. Again, CT-VT-RT elicited synergy with doses of just 2-Gy radiation, 10% EC50 TMZ, and 0.05 MOI of MeV-GFP (Figure S7). To also switch the chemotherapeutic agent, we then determined the EC50 of another clinically relevant alkylating agent, CCNU (Figure S8), in LN229 and LNZ308 cells. The CT-VT-RT-initiated synergistic effect was retained when substituting TMZ with CCNU (Figures S9 and S10). Moreover, the synergistic effect in CT-VT-RT was sustained in the TMZ-resistant cell line R-LN229 (Figure S11, red line), while these cells displayed a comparable susceptibility to MeV as the parental LN229 cells.
Activation of Type I Interferon Response, Canonical JAK-STAT-Signaling Pathway, and MHC Class I Expression
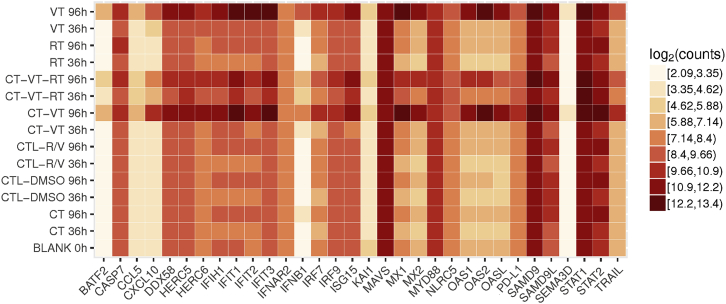
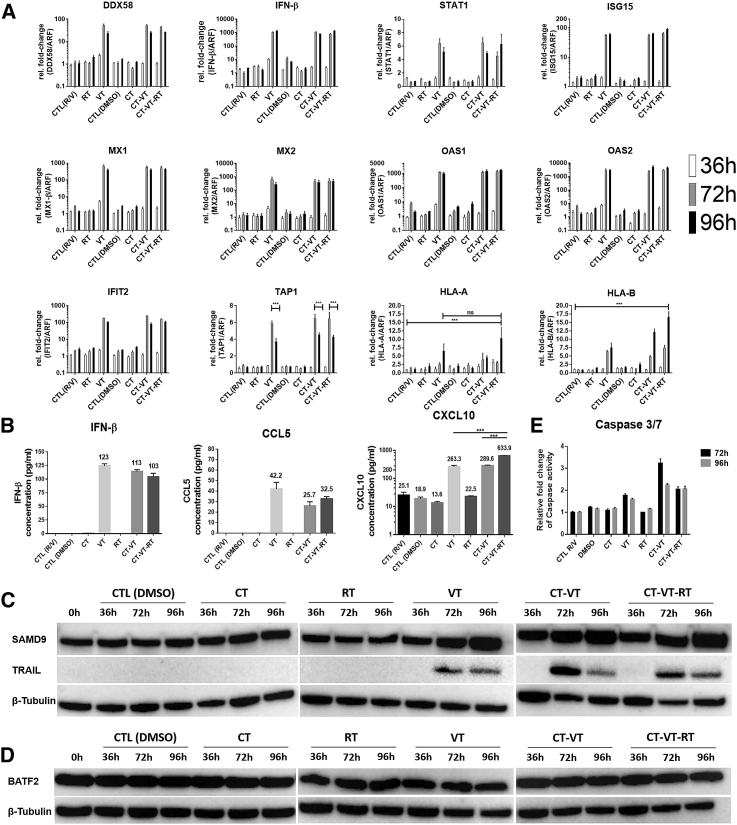
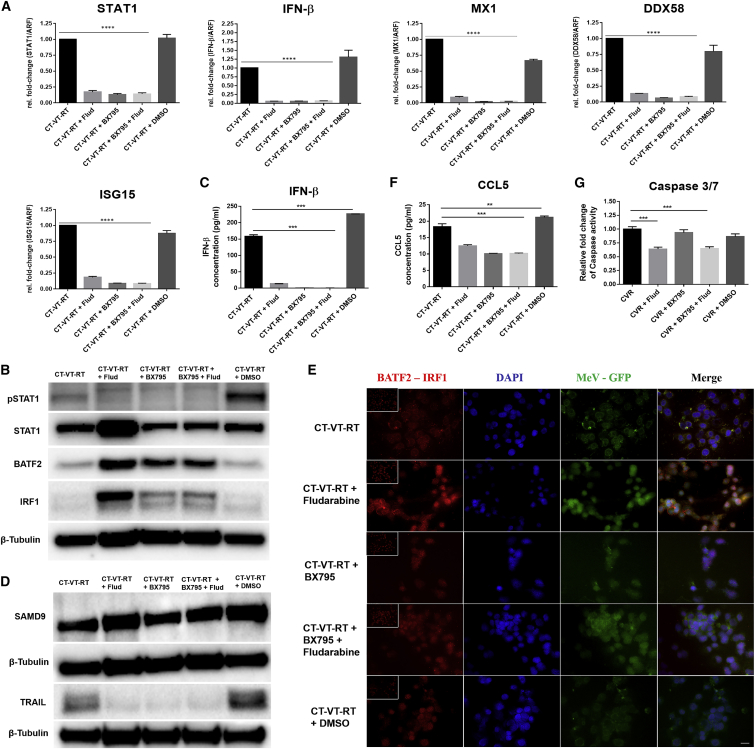
We next analyzed the treatment-induced molecular cascade by next-generation sequencing (NGS)-based RNA sequencing (RNA-seq) and subsequent validation by qPCR and ELISA. We included different time points for RNA sequencing analysis, as outlined schematically in Figure S12. We identified a total of 2,592 genes with significant expression profile changes, and we continued our analysis with a selected subset of the identified genes of interest (Figure 3). All MeV-containing regimens (VT, CT-VT, and CT-VT-RT) upregulated RIG-I-like receptor (RLR)-signaling pathway genes, presumably upon detection of MeV RNA genomes (Figure 3). The innate RNA sensor DEAD box protein-58 (DDX58) showed an approximately 100-fold increase in mRNA levels at 72 h post-initial treatment (hpt) in the VT, CT-VT, and CT-VT-RT regimens (Figure 4A), resulting in roughly 1,000-fold increased transcription of interferon-beta (IFN-β) at 96 hpt within these groups (Figure 4A), which was reflected by increased amounts of secreted IFN-β protein (Figure 4B).
Figure 3.
Transcriptional Signature Heatmap of Selected Genes Identified by RNA-Seq
Depicted genes (x axis) were identified as main factors, with their transcriptional profiles in all treatments at 36 and 96 hpt (y axis) represented as log2 counts (mRNA copy numbers) in a heatmap. Blank data (0 hpt) served as the basal expression control for further analysis.
Figure 4.
Molecular Profile in CT-VT-RT-Treated LNZ308 Cells
(A) Fold change in mRNA expression levels of selected genes identified by RNA-seq, validated at sequential time points post-treatment (36, 72, and 96 hpt) by qPCR. The fold changes in gene expression were significantly higher at 72 and 96 h in VT, CT-VT, and CT-VT-RT when compared to other regimens and with non-significant differences among the VT-containing regimen. The significance values are not depicted to avoid confusing graph outline, unless differing from the above statement. (B) ELISA revealed production of IFN-β along with secretion of pro-inflammatory chemokines CCL5 and CXCL10 measured at 96 hpt. (C) Immunoblot analysis revealed overexpression of SAMD9 (upper panel) in MeV-containing regimen and the induction of TRAIL (middle panel). β-tubulin (lower panel) served as the loading control. (D) Immunoblot analysis showed no change in BATF2 protein expression (upper panel). β-tubulin (lower panel) served as the loading control. (E) Increased effector caspase 3 or 7 activity detected in MeV-containing regimen over respective control, CTL (R/V) regimen. Two-way ANOVA with Tukey’s multiple comparison test in (A) and one-way ANOVA with Dunnett’s multiple comparison test in (B); ns, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001. Data are expressed as mean ± SEM.
IFN-β binding to the IFN α-β receptor (IFNAR) triggers downstream canonical JAK-STAT signaling with an inherent positive feedback loop.33 Accordingly, we indeed observed a 6-fold increase in STAT1 mRNA in VT and CT-VT regimens at 72 hpt (Figure 4A). Delayed STAT1 signaling might allow for efficient viral proliferation, as documented by the expression of MeV nucleocapsid protein in the CT-VT-RT regimen at 72 hpt (Figure S13). STAT1 signaling resulting in the transcription of IFN-stimulated genes (ISGs) indicated a distinct antiviral state, with a 100-fold increase in IFN-induced 15-kDa protein (ISG15) and IFN-induced protein with tetratricopeptide-repeats 2 (IFIT2) mRNA levels (Figure 4A). Moreover, myxoma resistance proteins 1 (MX1) and 2 (MX2), as well as 2′-5′-oligoadenylate synthase 1 (OAS1) and 2 (OAS2) mRNAs (Figure 4A), were increased in all virus-containing regimens.
Upon activation by IFN-β, the ISGF3 complex can bind the IFN-sensitive response element (ISRE) in respective promoter regions and upregulate, e.g., major histocompatibility complex (MHC) class I expression.34 CT-VT-RT increased the cellular capacity of antigen presentation, with 1.5- to 2-fold elevated expression of MHC-I, type A (HLA-A) or B (HLA-B), mRNA at 96 hpt compared to CT-VT or VT treatment schedules (Figure 4A). Moreover, peptide transporter TAP1 protein was upregulated in all virus-containing regimens (Figure 4A), with a lack of MHC class I expression in other treatments (Figure 4A). These data might indicate a dependence on IFN-β signaling. Moreover, MeV infection induced the secretion of immunostimulatory chemokines. The synergistic regimen CT-VT-RT upregulated C-X-C motif chemokine-10 (CXCL10), with significantly increased protein expression (Figure 4B) of 633.9 pg/mL/μg total protein detected by ELISA, in comparison to just 289.6 pg/mL using only CT-VT, as had been suggested by the RNA sequencing data (Figure 3). Interestingly, CCL5 was uniquely detected in MeV-containing regimens (Figure 4B), confirming the transcriptional signature detected by RNA sequencing (Figure 3). VT monotherapy elicited a maximal secretion of CCL5, reaching a concentration of up to 45.2 pg/mL/μg total protein, while CT-VT and CT-VT-RT regimens triggered lower expression of CCL5, with 25.7 and 32.5 pg/mL, respectively (Figure 4B).
We identified several upregulated IFN-related genes upon MeV infection by RNA sequencing. The sterile alpha motif domain-containing protein 9 (SAMD9) was expressed at basal levels (0 h), with no change in expression upon TMZ, DMSO, or radiation treatments (Figure 4C, upper panel). However, SAMD9 protein was significantly increased upon MeV infection at 72 and 96 hpt in all virus-containing regimens, with CT-VT-RT showing higher levels in comparison to VT alone (Figure 4C, upper panel). Similarly, we observed an increase of anti-tumoral host factor basic leucine zipper transcriptional factor ATF2-like protein (BATF2) in all virus-containing regimens (Figure 3). Yet, the increased mRNA expression did not correlate with BATF2 protein levels (Figure 4D, upper panel).
Apart from their innate antiviral responses, ISGs possess the ability to initiate apoptosis.35 We observed an activation of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) triggering an apoptotic cascade in all virus-containing regimens. TRAIL protein was higher in CT-VT and CT-VT-RT compared to VT at 72 and 96 hpt (Figure 4C, middle panel). Furthermore, TRAIL corresponded to caspase 3 or 7 activity, with a 3-fold increase observed in CT-VT at 72 hpt and a more than 2-fold increase in the CT-VT-RT regimen at 72 and 96 hpt in comparison to a 1.5-fold increase for VT alone (Figure 4E).
Taken together, we observed key roles for IFN and STAT1 signaling as well as coherent increases in MHC-I upregulation and immunostimulatory chemokines during oncolytic virus (OV) treatment. Consequently, we continued our investigations on the respective molecular mechanisms (1) using inhibitors of IFN-β production (BX795) and STAT1 signaling (fludarabine); and (2) with an immunopeptidome analysis.
Selective Inhibition of Type I IFN Signaling Triggers an Interaction of BATF2 with IRF1
When downregulating STAT1 transcription and phosphorylation by fludarabine and by BX795, alone or in combination (Figures 5A and 5B), IFN-β secretion was abolished in CT-VT-RT plus BX795, in CT-VT-RT plus BX795 and fludarabine, and also in CT-VT-RT plus fludarabine (Figures 5A and 5C). Inhibition of the IFN-β-STAT1 axis resulted in a complete loss of ISG induction and an absence of TRAIL expression (Figure 5D, lower panel). Yet, the stimulation of SAMD9 expression was not impaired upon inhibition of the IFN-β-STAT1 pathway (Figure 5D, upper panel). Most interestingly, BATF2 was highly upregulated upon the inhibition of IFN-β or STAT1 signaling when compared to the CT-VT-RT regimen without any additional manipulation or the CT-VT-RT + DMSO control treatment (Figure 5B). Also IFN regulatory factor 1 (IRF1) expression was increased (Figure 5B). Subsequent proximity ligation assays identified the potential molecular interaction of BATF2 with IRF1 (Figure 5E) upon treatment with CT-VT-RT plus fludarabine. Interestingly, CCL5 is a known BATF2-IRF1-induced gene.36 Indeed, CCL5 expression was lower in the CT-VT-RT plus fludarabine (Figure 5F) regminen. Thus, the observed correlation of CCL5 expression in accordance with BATF2 and IRF1 expression in our setting might suggest a BATF2-IRF1 interaction-dependent transcription of CCL5. Moreover, we observed caspase activity in CT-VT-RT plus BX795 similar to CT-VT-RT without any inhibitor, despite the abrogation of TRAIL expression by BX795 (Figure 5G).
Figure 5.
Selective Inhibition of Canonical JAK-STAT Signaling Might Lead to an Alternative Antiviral Signaling Network following CT-VT-RT Treatment
(A) Transcriptional profiles determined via qPCR reveal the downregulation of IFN-β and STAT1 upon inhibitor treatment, also affecting downstream ISGs (MX1 and ISG15) and DDX58. (B) Immunoblot analysis after CT-VT-RT regimen with or without signaling cascade inhibitors monitoring levels of STAT1 and phosphorylated STAT1, along with BATF2, IRF1, and β-tubulin as loading controls as indicated. (C) Loss of IFN-β production upon inhibitor treatment determined by ELISA. (D) Immunoblot analysis after CT-VT-RT regimen with inhibitors. There was no change in SAMD9 expression (upper panel), but abrogation of TRAIL expression (lower panel) is shown upon inhibition. β-tubulin served as the loading control. (E) Proximity ligation assay reveals potential molecular interaction of BATF2 with IRF1 observed through increased red amplification signal in cells treated with CT-VT-RT plus fludarabine. Basal signal was observed in CT-VT-RT and CT-VT-RT plus DMSO alongside IgGs (inset) in all treatment regimens. Scale bar, 20 μm. (F) Decreased but detectable CCL5 expression in samples treated with CT-VT-RT plus molecular inhibitors determined via ELISA. (G) Effector caspase 3 or 7 activity in samples treated with CT-VT-RT plus molecular inhibitors indicate alternative host machinery initiating apoptosis. One-way ANOVA with Dunnett’s multiple comparison test (A, C, F, and G); *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Flud, fludarabine.
MHC Class I Upregulation Correlates with Increased Presentation of Tumor and Viral Antigens by Glioma Cells
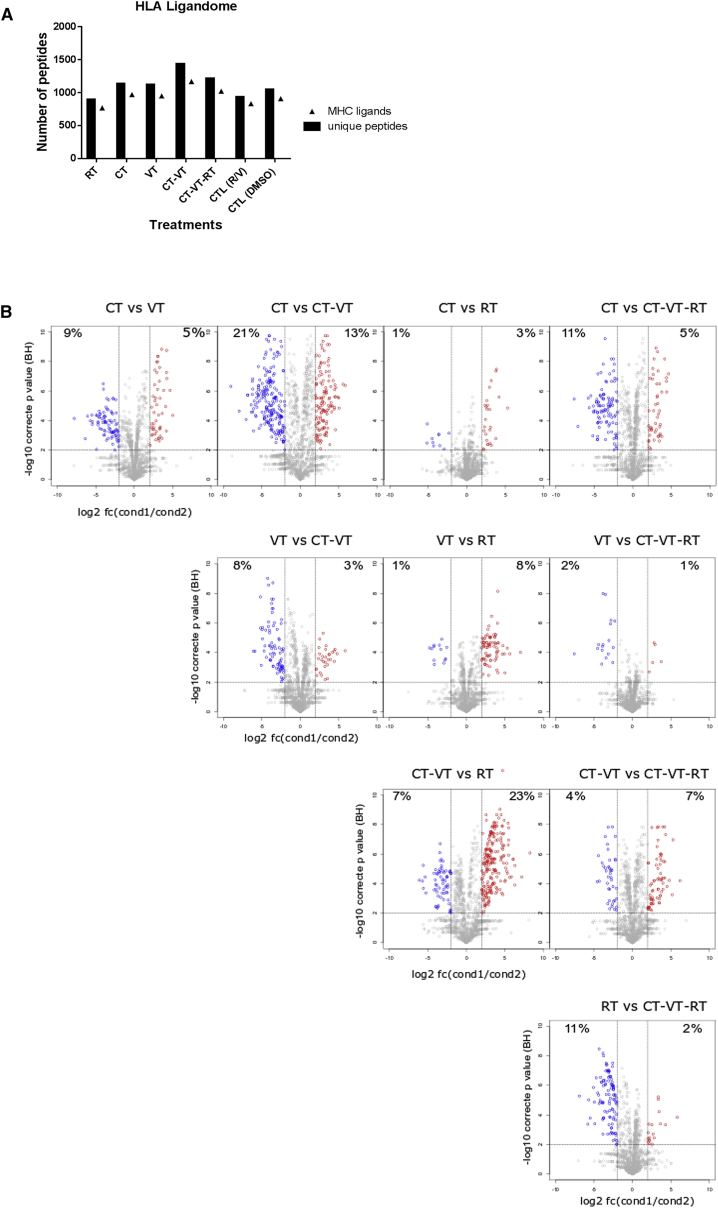
Based on the observed MHC-I upregulation and an increase of immunostimulatory chemokines, we hypothesized that the CT-VT-RT regimen might stimulate the presentation of tumor antigens by MeV-infected glioma cells. Thus, we performed an immunopeptidome analysis. Indeed, both CT-VT and CT-VT-RT distinctively increased antigen presentation, with 1,430 and 1,222 peptides isolated from MHC molecules after treatment, respectively (Figure 6A). The differential expression of antigenic ligands was higher in the VT-containing regimens (i.e., VT, CT-VT, and CT-VT-RT) when compared with CT or RT, and there were fewer differences among them (Figure 6B). In particular, CT-VT seemed to exhibit a significantly differential profile compared with CT and RT, respectively, and in complete correlation with their quantitative increase in isolated antigenic ligands. All treatment regimens revealed the presentation of tumor-associated antigenic peptides of Ephrin type-A receptor 2 (EPHA2) and Tenascin (TNC), which had been found associated with tumors, including glioblastoma.37, 38 We isolated a novel tumor-associated peptide (LYTDRTEKL) processed from transforming growth factor-beta-induced (TGFBI), which had been exclusively presented in CT-VT. TGFBI is associated with anti-adhesive metastatic properties in cancers, including melanoma and prostate cancer,39, 40 and it was significantly overexpressed in glioblastoma based on The Cancer Genome Atlas (TCGA) dataset interrogation (Figure S14). Moreover, other TGFBI-derived peptides from glioblastoma sample have been previously discovered and revealed immunogenicity.41
Figure 6.
HLA Ligandome Analysis Revealed Increased Peptide Presentation upon MeV-Containing Combinatorial Treatments
(A) Graph enumerating peptides and source proteins isolated from LNZ308 cells after individual treatments using HLA ligandomics, with CT-VT showing maximal presentation of peptides with 1,430 peptides, followed by CT-VT-RT with 1,222 peptides post-normalization. (B) Semiquantitative volcano plot analysis visualizing modulations in HLA ligands presented on LNZ308 cells upon different treatment regimens. Each dot represents an HLA ligand, with log2-fold changes in HLA ligand number indicated on the x axis and the respective Benjamini-Hochberg-corrected significance levels on the y axis. Significantly upregulated or downregulated HLA ligands (>4-fold difference in number; p < 0.01) are highlighted in red and blue, respectively. The significantly modulated HLA ligand percentages are mentioned in the quadrants.
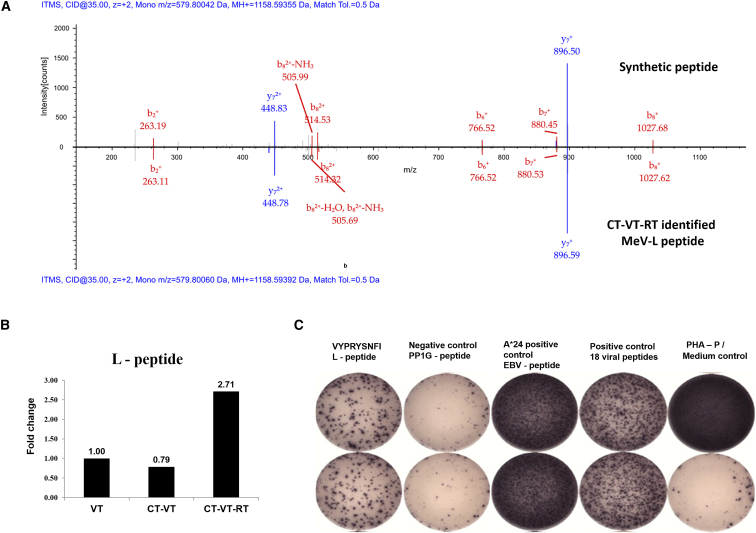
In addition to the endogenous peptides, we observed the presentation of a novel HLA-A*24-restricted MeV peptide (Figure 7A), VYPRYSNFI, in all virus-containing regimens. This peptide is processed from a highly conserved viral region, the MeV-L polymerase, and it was particularly increased in the triple regimen CT-VT-RT (2.7-fold increase) compared to VT alone (Figure 7B). Presentation of this MeV-L peptide triggered significant IFN-γ secretion from donor peripheral blood mononuclear cells (PBMCs) in enzyme linked immunospot (ELISpot) assays, with a mean spot count of 264.5, measured after a 12-day recall (Figure 7C, positive control with strongest signal, negative controls without signal), demonstrating that the identified viral peptide is highly immunogenic.
Figure 7.
MeV Polymerase Peptide (L) Identified from HLA Ligandome Analysis Is Immunogenic and Highly Enriched in CT-VT-RT
(A) Mass spectrometry spectra match between synthesized peptide (upper axis, above 0) and CT-VT-RT-identified MeV-L peptide (lower axis, below 0) from LNZ308 cells. (B) Semiquantitative analysis of ligandome (n = 5) indicates a 2.7-fold increase in MeV L-peptide, VYPRYSNFI, upon CT-VT-RT treatment over virotherapy alone. (C) VYPRYSNFI-stimulated healthy donor PBMCs secrete IFN-γ in ELISpot analyses, indicating the immunogenic potential of the peptide to elicit CD8+ T cell responses with appropriate signal-generating positive and signal-absent negative controls.
Discussion
We identified a synergistic anti-glioma strategy based on oncolytic MeV in combination with standard treatment options, and we investigated the treatment-induced molecular and immunological signature. The combination of VT and RT was indeed synergistic and in line with previous results.18 Upon incorporation of TMZ or CCNU as standard chemotherapeutic agents (CT) in the clinical care, we identified the sequence of a triple regimen, CT-VT-RT, eliciting synergistic therapeutic effect.
We investigated the temporal molecular signature by transcriptomic (RNA sequencing) at different time points, and we detected an MeV-induced pro-inflammatory molecular signature in tumor cells (Figure 3), including increased transcription of SAMD9, BATF2, KAI1, and SEMA3D. These possess anti-tumoral characteristics, complementing results describing a role of IFN for oncolytic MeV infectivity.42 Contrary to reports suggesting downregulation of SAMD9 in response to IFN inhibition in Sendai virus-treated cells,43 SAMD9 expression remained unchanged by the inhibition of IFN-β or STAT1 signaling here (Figure 5D, upper panel). Furthermore, we observed similar levels of effector caspase 3 or 7 activity in CT-VT-RT, when compared to CT-VT-RT plus BX795, using a small molecule inhibitor of IFN-β production and the downstream JAK-STAT pathway. The consistent caspase activity accompanied by the loss of TRAIL expression in CT-VT-RT plus BX795 suggests that SAMD9 does not influence TRAIL-dependent apoptosis. Besides its anti-tumorigenic properties,44, 45 SAMD9 has been suggested as an innate antiviral host factor in response to IFN stimulation and interaction with Sendai virus, vaccinia virus, myxomavirus, and others.43, 46, 47, 48 Our data might indicate SAMD9 as an innate antiviral host factor, whose expression might even be independent of STAT1 signaling.
Despite increased abundance of the BATF2 mRNA in MeV-containing regimens (according to RNA sequencing), the amount of expressed BATF2 protein remained unchanged (Figures 3 and 4D). Strikingly, the inhibition of IFN-β and STAT1 signaling through BX795 or fludarabine, respectively, led to an increase in BATF2 protein levels corresponding to elevated IRF1 protein expression (Figure 5B). Roy et al.36 demonstrated co-immunoprecipitation of BATF2 with IRF1 in response to mycobacterial infection, thereby revealing this interaction. The concomitant increase of BATF2 and IRF1 protein in response to the inhibition of type 1 IFN signaling was confirmed here also for MeV infection, and their molecular interaction was detected via proximity ligation assay (Figure 5E). Moreover, we detected in cells treated with CT-VT-RT, in combination with fludarabine and/or BX795, the secretion of a BATF2-IRF1 transcriptionally induced chemokine, CCL5,36 while IFN-β was not detectable (Figures 5C and 5F). This is particularly relevant, as both CCL5 and IFN-β are usually secreted in response to viral infection via the IRF3-signaling pathway, which was specifically inhibited upon BX795 treatment in our experimental setting.49, 50 Thus, the expression of CCL5 and the activity of effector caspase 3 or 7 indicate a potential role for BATF2-IRF1 as an alternative antiviral host machinery.
The profound increase in chemokines regulating immune responses was consistent with upregulation of the antigen-processing machinery (Figure 4A). This prompted us to investigate the immunopeptidome. We detected increased numbers of antigenic peptides presented on HLA molecules (quantified through extrinsic ligandome enrichment), with maximal presentation detected in CT-VT and CT-VT-RT. All VT-containing regimens presented a specific novel MeV polymerase (L) peptide (VYPRYSNFI) on HLA-A*24, while the CT-VT regimen induced presentation of a tumor-associated antigenic peptide TGFBI (LYTDRTEKL). The MeV-L peptide was particularly presented at high levels in the triple regimen CT-VT-RT (Figure 7B), and it is highly immunogenic, as outlined by IFN-γ ELISpot data (Figure 7C). This treatment-induced immunologic signature and the presentation of the VYPRYSNFI peptide might be clinically relevant in two ways: First, MeV-infected cells might reshape the tumor-associated microenvironment toward an immunogenic signature by the attraction of antiviral immune cells, generating an inflammatory milieu. Second, the peptide could be exploited for a tailored vaccination strategy that enhances immune responses toward the remaining MeV-infected cells. Of note, the treatment-induced immunogenic signatures were accompanied by increased PD-1L transcription (Figure 3) that might explain the previously reported benefit of combining MeV with immune checkpoint blockade inhibitors.9, 51
Speranza et al.52 combined a chemo-virotherapeutic approach of non-replicating adenovirus encoding thymidine kinase along with anti-PD1 antibodies, resulting in the activation of proinflammatory type 1 IFN response in vitro and in vivo. Such a regimen enhances immunogenic cell death effectuated in combination with checkpoint blockade inhibitors due to upregulated PD-L1 expression, as can be seen with the increased percentage of long-term survivors when compared to either treatment alone in mice as well as the lack of tumor growth upon rechallenge in such long-term-surviving animals.52 Similarly, our combinatorial oncolytic approach using MeV could be exacerbated through the cloning of cytotoxic genes in clinical use, such as super cytosine deaminase (SCD) or human thyroidal sodium iodide symporter (NIS), and the latter also utilized for live imaging of viral dynamics through non-invasive radioiodine single-photon emission computed tomography (SPECT).11, 53
Kurokawa et al.42 identified IFN pathway activation as a key determinant for efficient oncolytic measles viral infectivity in human glioblastoma specimens. While this study is in the same research field, the driving questions are different and the results are complementary. First, we present a comprehensive analysis of a combination therapy enhancing the immunogenic signature of MeV-based treatments, as suggested by Kurokawa et al.42 Second, we present novel data on the treatment-induced immunopeptidome by applying mass spectrometry (MS)-based ligandomics. Third, our results characterizing the role of IFN signaling upon STAT inhibition further support their results, despite the use of different inhibitors (JAK inhibition).42 Fourth, uncovering the role of BATF2-IRF1 as a potential antiviral host machinery in the absence of IFN signaling is a novel feature in our study. Finally, our molecular model delineates further key players in IFN signaling determining oncolytic MeV infectivity for successful implementation in clinical trials, as well as understanding MeV biology.
In conclusion, a sequential triple combination of TMZ (or CCNU), MeV, and radiation therapy has synergistic anti-glioma activity, and it leads to an actionable treatment-induced molecular and immunological signature. Thus, a CT-VT-RT regimen could be combined with tailored peptide vaccinations with our newly identified peptide sequences, potentially in combination with checkpoint blockade antibodies. Our data might thus lead to the conception of new tailored immuno-virotherapeutic strategies.
Materials and Methods
Cell Lines and Primary Tissue
The human long-term glioma cell lines LN229 and LNZ308, human colorectal adenocarcinoma cell line HT29, and the cell line HEK293T were obtained from American Type Culture Collection (ATCC), and they were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum and 50 μg/mL gentamycin (Thermo Fisher Scientific). The MeV producer cell line 293-3-4654 was a kind gift by Dr. Roberto Cattaneo, and African green monkey kidney Vero B4 cells were obtained from Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Cultures (DSMZ) and cultured as described. The glioma stem-like (GS) cell lines GS3 and GS8 were kindly provided by Katrin Lamszus.55 The primary tumor cell lines T81/16, T708/16, and T1094/17 were harvested from fresh glioblastoma tissue obtained from patients undergoing surgery at the Department of Neurosurgery, University Hospital Tübingen (ethical approval 456/2009B02), and they were cultured similar to GS cells in Neurobasal medium (Thermo Fisher Scientific) supplemented with 20 ng/mL recombinant human fibroblast growth factor (PeproTech), 20 ng/mL recombinant human epidermal growth factor (PeproTech), 2 mM L-glutamine (Thermo Fisher Scientific), 10% B-27 supplement (Thermo Fisher Scientific), and 50 μg/mL gentamycin (Thermo Fisher Scientific). The TMZ-resistant cell line R-LN229 was generated by repetitive exposure of parental LN229 to TMZ, as previously described.56
Flow Cytometry for Basal CD46 Receptor Expression and Post-treatments In Vitro
Cells were stained with 0.5 μg antibody (either CD46-PE [8E2] or IgG1κ-PE, Thermo Fisher Scientific; and nectin-4-APC or IgG2B-APC, R&D Systems), and receptor expression was measured in a MACSQuant Analyzer 10 (Miltenyi Biotec). The ratios of MFIs of receptor antibody versus isotype control were calculated, and graphs generated using GraphPad Prism 6 software.
CD46 Receptor Expression after Treatments In Vitro
LN229 and LNZ308 cells were treated with TMZ (0, 10, 100, or 1,000 μM), γ-irradiation (0, 1, 2, or 4 Gy), or under different oxygen conditions (21% O2 [normoxia] or 1% O2 [hypoxia]) for 48 h. Then, levels of CD46 receptor expression were analyzed via flow cytometry along with viability staining using 7-aminoactinomycin D (7-AAD, Thermo Fisher Scientific), as described above.
MeV Production
The recombinant Edmonston strain of MeV (NSe) encoding GFP or the rapidly maturing variant of Discosoma sp. red fluorescent protein (DsRed) in the post-P position was rescued as described previously.54 The viruses were propagated in Vero cells, and titers were determined by 50% tissue culture infective dose (TCID50) titration by the Kärber method.57
Measles Viral Infectivity Post-TMZ Treatment
105 LNZ308 cells were seeded in 6-well plates 1 day before treatment initiation with TMZ at doses of 0, 10, 100, and 1,000 μM for 48 h. Post-TMZ treatment, cells were infected with MeV-GFP at doses of 0.01 and 0.05 MOI for 48 h to measure viral infectivity. The virus-treated cells were harvested along with supernatants to calculate viral titers by TCID50 titration using the Kärber method by propagation in Vero cells, as described above.
Fluorescence Microscopy
Cells were seeded on glass chamber slides coated with poly-L-lysine (PLL, Sigma-Aldrich) alone for adherent cells or along with laminin (Sigma-Aldrich) for suspension cultures 1 day before infection with MeV-Nse-GFP at 1 MOI for 72 h at 37°C. Cells were fixed with 4% paraformaldehyde followed by nuclear staining with DAPI (Thermo Fisher Scientific). Images were captured with a Zeiss microscope using Apotome (Carl Zeiss), and image analysis was carried out with AxioVision software (Carl Zeiss).
LC3-GFP Puncta Assay
Cells were treated with different treatment regimens (blank, RT, and VT) in serum-free medium followed by serum supplementation. Cells were stained for LC3-GFP 12 hpt along with nuclear Hoechst 33342 using CYTO-ID Autophagy detection kit 2.0 (Enzo Life Sciences), as per the manufacturer’s instructions. Images were captured with a Zeiss microscope using Apotome (Carl Zeiss), and image analysis was carried out with AxioVision software (Carl Zeiss).
Acute Cytotoxicity Assay
LN229 and LNZ308 were seeded at 5,000 cells/well in 96-well plates 1 day before treatment initiation. All treatments were carried out in serum-free DMEM along with their respective controls, and cell viability was measured at 72 hpt using Cell titer blue solution (Promega), as per the manufacturer’s instructions. The measured values were converted to percentage cell viability by normalizing each treatment to its respective control within the individual regimen.
Cytotoxic Survival Assay
We designed a cell viability assay to elicit and determine the oncolytic cascade effect initiated by MeV in combination with cytotoxic agents such as TMZ. Herein, measurement of cytotoxicity associated with recurrent ineffective mismatch repair or lysis of MeV-induced syncytia requires longer readout periods of 144 hpt as opposed to a classical 72 hpt acute cytotoxicity assay. The predicted values of combinations used to determine synergy were calculated from the individual monotherapies, RT, CT, and VT, by the Chou-Talalay fractional product method.58
LN229, R-LN229, and LNZ308 cells were seeded at 5,000 cells/well in 96-well plates 1 day before treatment initiation with γ-irradiation, alkylating CT, or MeV VT, alone or sequentially as combinatorial regimens, in serum-free DMEM at the indicated concentrations or doses. The percentage cell viability at 144 hpt was measured using Cell titer blue solution (Promega), as per the manufacturer’s instructions. The measured values were converted to percentage cell viability by normalizing each treatment to its respective control within the individual regimen. The predicted values of combinations used to determine synergy were calculated from the individual monotherapies, RT, CT, and VT, by the Chou-Talalay fractional product method,58 defined as the product of the viable fractions after treatment with all agents alone. If the observed value of the co-treatment is less than that of the calculated predictive value, the combination of agents was deemed to be synergistic and not only additive. This is in accordance with an alternative method of synergy calculation using the coefficient of drug interaction without conversion to ratios.59
GS8 cells were seeded at 50,000 cells/35-mm dish 1 day before treatment initiation, and all treatments were performed in serum-free neurobasal medium. The measurement of cell viability was performed in triplicates with 5,000 cells at 144 hpt using Cell titer blue solution (Promega), as described previously. The graphs were generated using GraphPad Prism 6 with percentage cell viability post-treatments.
Immunoblotting
Cells of interest were lysed in lysis buffer containing 25 mM Tris hydrochloric acid (Tris-HCl), 120 mM sodium chloride (NaCl), 5 mM EDTA, 0.5% NP-40, and phosphatase inhibitor cocktails (Sigma-Aldrich). Proteins were separated on 4%–12% polyacrylamide gels (Thermo Fisher Scientific) followed by transfer to polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific). The blots were blocked in Tris-buffered saline containing 5% skim milk (Becton Dickinson) and 0.05% Tween 20 (Sigma-Aldrich) for 1 h. Protein blots were probed with primary antibodies overnight at 4°C and, after thorough washing, for 1 h at room temperature with horseradish peroxidase-coupled secondary antibodies (Abcam) the following day. Protein bands were visualized using Pierce chemiluminescent substrate solutions (Thermo Fisher Scientific), and image capture and analysis were carried out with ChemiDoc MP imaging system (Bio-Rad) using Image Lab software (Bio-Rad).
Antibodies and Reagents
Antibodies against SAMD9 (EPR13603), BATF2 (EPR10667), STAT1-α (EPYR2154), STAT1 phospho-Y701 (M135), and IRF1 (EPR18301) were purchased from Abcam. Antibodies against TRAIL (C92B9), LC3A (D50G8), and β-tubulin (9F3) were purchased from Cell Signaling Technology. Antibody against MeV- nucleoprotein (N) was purchased from Novus Biologicals. Antibody against IRF1 (13H3A44) was purchased from BioLegend for the proximity ligation assay. Anti-rabbit and anti-mouse horseradish peroxidase-coupled secondary antibodies were purchased from Abcam. TBK1 and IKKε inhibitor BX795 was purchased from InvivoGen and used at a concentration of 10 μM.60 Fludarabine (Selleckchem) was used as a functional STAT1 inhibitor at a concentration of 100 μM.61
Real-Time qPCR
RNA extractions were performed using the RNeasy mini kit (QIAGEN) and reversely transcribed to cDNA using the High-Capacity RNA-to-cDNA kit (Thermo Fisher Scientific), according to the manufacturers’ instructions. For each qPCR reaction, 20 ng cDNA was amplified with custom-designed primers (Table S7) and qPCR Mastermix Plus for SYBR Green (Eurogentec) using the 7500 Fast Real time PCR system (Thermo Fisher Scientific). Relative gene expression was determined using the ΔΔ-CT method versus the housekeeping gene ARF1.
ELISA
The respective chemokine or cytokine concentrations were determined enzymatically per microgram of total protein using the respective DuoSet ELISA kits, according to the manufacturer’s instructions (R&D Systems).
RNA Sequencing and Analysis
LNZ308 cells were seeded at 3 × 106 cells/dish and treated with monotherapies (RT, 2 Gy; CT, 130 μM; VT, 0.05 MOI), double regimen (CT-VT, 130 μM-0.05 MOI), and synergistic triple regimen (CT-VT-RT, 130 μM-0.05 MOI-2 Gy), along with DMSO as control for CT-initiated regimens (CTL-DMSO) and controls for RT-alone and VT-alone regimens (CTL [R/V]) under serum-free conditions. Blank (0 hpt) served as basal control for expression. RNA was isolated at 36 and 96 hpt using an RNeasy Mini kit (QIAGEN), according to the manufacturer’s instructions, and quality was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies). Samples with high RNA integrity number (RIN > 8) were selected for library construction.
Using the TruSeq Stranded RNA Sample Prep Kit (Illumina) and 400 ng total RNA for each sequencing library, poly(A)-selected single-read sequencing libraries (68-bp read length) were generated according to the manufacturer’s instructions. All libraries were sequenced on an Illumina HiSeq2500 platform at a depth of 18–20 million read search. Library preparation and sequencing procedures were performed by the same individual to minimize technical batch effects. Raw fastq files were pre-filtered using the chastity filter to remove reads that contained a “Y” flag. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used to determine quality of the resulting fastq files. Subsequently, an adaptor trimming or removal process was conducted with Cutadapt (https://pypi.org/project/cutadapt/), version 1.8.3.62 This step used the FastQC output to identify reads that showed a match to some typical overrepresented (Illumina) sequences or adapters. TopHat2 was used as the aligner to map the quality-controlled remaining reads to the human genome.63 Read counting to features (genes) in the genome was performed with HTSeq (https://htseq.readthedocs.io/en/release_0.11.1/), version 0.6.0.64 Counting was performed using “union” mode on the feature “gene_id,” where each gene is considered here as the union of all its exon counts. The stranded option was also set to “stranded=no” to count features on both strands. For differential expression analysis, the raw read count table provided by HTSeq was used in the R package DESeq2 (version 1.10.1).65 Adjusted p values were used at an FDR (false discovery rate) <0.05 to account for multiple hypothesis testing. The RNA sequencing results have been deposited and are accessible at GEO: GSE111247.
Functional Inhibition of IFN-β and JAK-STAT Pathway
LNZ308 cells were seeded at 3 × 106 cells/dish, and they were treated with the synergistic CT-VT-RT regimen as described previously (RNA sequencing). At 36 hpt upon completion of CT-VT-RT, IFN-β inhibitor (BX795) and STAT1 inhibitor (fludarabine) were added alone or in combination along with a DMSO control to obtain five regimens. The RNA, protein, and supernatants were harvested at 96 hpt for further analysis.
Proximity Ligation Assay
The cells were subjected to proximity ligation assay using Duolink In Situ red starter kit Mouse/Rabbit (Sigma-Aldrich) along with suitable IgG controls, as per the manufacturer’s instructions. Images were captured with a Zeiss microscope using Apotome (Carl Zeiss), and image analysis was carried out with AxioVision software (Carl Zeiss).
HLA Ligandome Analysis
Cells subjected to treatment regimens as mentioned above (RNA sequencing) were harvested at 96 h, washed twice with cold PBS (Lonza), and stored frozen at −80°C, with subsequent isolation of HLA class I molecules using standard immunoaffinity purification, as described previously.66, 67 The MS proteomics data have been deposited via the PRIDE partner repository, and they are accessible at ProteomeXchange Consortium: PXD008984.
Database Search and Spectral Annotation of HLA Ligandome
Data were processed against the LNZ308 proteome derived from RNA sequencing and the oncolytic MeV proteome (UniProt) applying the SequestHT algorithm68 in the Proteome Discoverer 1.3 (Thermo Fisher Scientific) software. Precursor mass tolerance was set to 5 ppm, fragment mass tolerance was set to 0.5 Da, and oxidized methionine was allowed as a dynamic modification. Percolator69-assisted FDR calculation was set at a target value of q ≤ 0.05 (5% FDR). Peptide spectrum matches with q ≤ 0.05 were filtered according to additional orthogonal parameters to ensure spectral quality and validity. Peptide lengths were limited to 8–12 amino acids (aa). HLA annotation was performed using NetMHCpan-3.0,70 based on the HLA class I typing of LNZ308.71
Analysis of LNZ308 Ligandomes in Different Conditions
For label-free quantification of the relative HLA ligand abundances under the different conditions (CT, CT-VT, CT-VT-RT, VT, and RT), the injected peptide amounts of paired samples were normalized, and liquid chromatography-tandem MS (LC-MS/MS) analysis was performed in five technical replicates for each sample.66 In brief, relative amounts of substance of paired samples were calculated from average precursor ion intensities determined in dose-finding MS runs and adjusted accordingly by dilution. Relative quantification of HLA ligands was performed by calculating the area under the curve of the corresponding precursor-extracted ion chromatograms using Proteome Discoverer 1.3. The ratios of the mean areas of the individual peptides in the five label-free quantification MS runs of each sample were calculated, and two-tailed t tests were performed using an in-house MATLAB script (version [v.]8.2, MathWorks).
Peptide Synthesis
The automated peptide synthesizer Liberty Blue (CEM) was used to synthesize peptides using 9-fluorenylmethyl-oxycarbonyl/tert-butyl (Fmoc/tBu) strategy. The identity and purity of peptides were validated by reversed-phase LC (nanoUHPLC, UltiMate 3000 RSLCnano, Dionex) and online coupled LTQ Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific) system. Synthesized peptide was used in the validation of LC-MS/MS identification as well as in functional experiments.
TCGA Dataset Analysis
We downloaded the gene expression RNA sequencing data of low grade gliomas (LGGs) and glioblastomas (GBMs) (Illumina) and the associated clinical data from the NIH National Cancer Institute GDC portal (https://portal.gdc.cancer.gov) released by TCGA. The survival data from TCGA were merged with gene expression data and other associated clinical information using corresponding sample IDs.
T Cell Culture
Blood samples of healthy donors matched for HLA-A*24 serotype were kindly provided by the Institute for Clinical and Experimental Transfusion Medicine at the University Hospital Tübingen after obtaining written informed consent. PBMCs were isolated by standard Ficoll-Hypaque (Biocoll, Biochrom) density gradient centrifugation.72 Cells were stored at −80°C in fetal calf serum (FCS) (Sigma-Aldrich) and 10% DMSO (Merck, Whitehouse Station, NJ, USA). After thawing, the cells were rested overnight prior to stimulation with culture conditions of 37°C and 7.5% CO2 in humidified incubators.
IFN-γ ELISpot Assay
The IFN-γ ELISpot assay after 12-day stimulation was performed as described previously73 along with suitable controls. Briefly, cells were stimulated 24 h after thawing with 1 μg/mL candidate MeV peptide or control peptides. IL-2 (R&D Systems) was added on days 2, 5, and 7 with a final concentration of 20 U/mL or 1,000 U/mL for PBMCs of healthy donors or tumor infiltrating lymphocytes (TILs), respectively. On day 12, cells were harvested and IFN-γ ELISpot was performed. Phytohaemagglutinin (PHA, Sigma-Aldrich), the HLA-A*24:02-restricted Epstein Barr virus (EBV) epitope TYPVLEEMF (EBV BRLF1_198-206), or a pool of 18 viral peptides of different HLA restrictions was used as the positive control to generate a strong positive signal. HLA-A*24:02-restricted PP1G peptide KYPENFFLL (HUMAN PP1G_113-121) or medium alone served as the negative control. Spot counts were determined using an ImmunoSpot S6 Analyzer (Cellular Technology), with T cell responses considered to be positive if the mean number of spots per well was at least 10 and more than three times the mean number of spots of the negative controls.
Statistical Analysis
Statistical significance was calculated using two-way ANOVA followed by Tukey’s multiple comparison test, one-way ANOVA with Dunnett’s multiple comparison test, or with multiple t test, as suitable and respectively indicated. p values less than 0.05 were considered statistically significant and all values are expressed as mean ± SEM.
Author Contributions
G.T. was responsible for the design of the entire study. S.R., S.N., M.G., S.S., and H.-G.R. contributed important aspects for the design and conduction of transcriptomics and immunopeptidome analysis. S.R. conducted all experiments with contributions and support for data collection from D.C., M.G., M.C.C., R.S., F.W., and M.K. Data analysis was performed by S.R., M.C.C., M.T., S.N., H.-G.R., M.D.M., S.S., and G.T. S.R. and G.T. wrote the first draft of the manuscript. S.R. designed the first drafts of the figures. All authors edited and approved the final version of the manuscript.
Conflicts of Interest
G.T. served on Advisory Boards of BMS, MSD, and AbbVie; received research and/or travel grants from Roche Diagnostics, Novocure, and Medac; and received speaking fees from Medac and Novocure. A patent has been filed at the European Patent Office for immunotherapeutic peptides, TGFBI (LYTDRTEKL) and MeV-L polymerase (VYPRYSNFI) with G.T., S.R., M.G., and S.S. as inventors.
Acknowledgments
We thank Sarah Hendel, Heike Pfrommer, and Yeliz Donat for excellent technical assistance. We also thank Susanne Berchtold for assistance with viral infections, Jan Hanauer for assistance with virus production, and Maren Lübke for her assistance with the ELISpot assay. Parts of this work were funded by the Wilhelm Sander Foundation and the Else Kröner Forschungskolleg “Therapieresistenz solider Tumore” (EKFS_Kolleg_2015_14 to G.T.). S.N. and M.C.C. acknowledge funding from the Deutsche Forschungsgemeinschaft (core facilities initiative KO-2313-2). M.D.M. received funding from the Deutsche Krebshilfe (109614).
Footnotes
Supplemental Information includes fourteen figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.omto.2018.12.010.
Supplemental Information
References
- 1.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups. National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert M.R., Dignam J.J., Armstrong T.S., Wefel J.S., Blumenthal D.T., Vogelbaum M.A., Colman H., Chakravarti A., Pugh S., Won M. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert M.R., Wang M., Aldape K.D., Stupp R., Hegi M.E., Jaeckle K.A., Armstrong T.S., Wefel J.S., Won M., Blumenthal D.T. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J. Clin. Oncol. 2013;31:4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinot O.L., Wick W., Mason W., Henriksson R., Saran F., Nishikawa R., Carpentier A.F., Hoang-Xuan K., Kavan P., Cernea D. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N. Engl. J. Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R., Taillibert S., Kanner A., Read W., Steinberg D., Lhermitte B., Toms S., Idbaih A., Ahluwalia M.S., Fink K. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA. 2017;318:2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fecci P.E., Mitchell D.A., Whitesides J.F., Xie W., Friedman A.H., Archer G.E., Herndon J.E., 2nd, Bigner D.D., Dranoff G., Sampson J.H. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 7.Bloch O., Crane C.A., Kaur R., Safaee M., Rutkowski M.J., Parsa A.T. Gliomas promote immunosuppression through induction of B7-H1 expression in tumor-associated macrophages. Clin. Cancer Res. 2013;19:3165–3175. doi: 10.1158/1078-0432.CCR-12-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly O.G., Errington-Mais F., Steele L., Hadac E., Jennings V., Scott K., Peach H., Phillips R.M., Bond J., Pandha H. Measles virus causes immunogenic cell death in human melanoma. Gene Ther. 2013;20:7–15. doi: 10.1038/gt.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardcastle J., Mills L., Malo C.S., Jin F., Kurokawa C., Geekiyanage H., Schroeder M., Sarkaria J., Johnson A.J., Galanis E. Immunovirotherapy with measles virus strains in combination with anti-PD-1 antibody blockade enhances antitumor activity in glioblastoma treatment. Neuro-oncol. 2017;19:493–502. doi: 10.1093/neuonc/now179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauvrit A., Brandler S., Sapede-Peroz C., Boisgerault N., Tangy F., Gregoire M. Measles virus induces oncolysis of mesothelioma cells and allows dendritic cells to cross-prime tumor-specific CD8 response. Cancer Res. 2008;68:4882–4892. doi: 10.1158/0008-5472.CAN-07-6265. [DOI] [PubMed] [Google Scholar]
- 11.Russell S.J., Federspiel M.J., Peng K.W., Tong C., Dingli D., Morice W.G., Lowe V., O’Connor M.K., Kyle R.A., Leung N. Remission of disseminated cancer after systemic oncolytic virotherapy. Mayo Clin. Proc. 2014;89:926–933. doi: 10.1016/j.mayocp.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dispenzieri A., Tong C., LaPlant B., Lacy M.Q., Laumann K., Dingli D., Zhou Y., Federspiel M.J., Gertz M.A., Hayman S. Phase I trial of systemic administration of Edmonston strain of measles virus genetically engineered to express the sodium iodide symporter in patients with recurrent or refractory multiple myeloma. Leukemia. 2017;31:2791–2798. doi: 10.1038/leu.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phuong L.K., Allen C., Peng K.W., Giannini C., Greiner S., TenEyck C.J., Mishra P.K., Macura S.I., Russell S.J., Galanis E.C. Use of a vaccine strain of measles virus genetically engineered to produce carcinoembryonic antigen as a novel therapeutic agent against glioblastoma multiforme. Cancer Res. 2003;63:2462–2469. [PubMed] [Google Scholar]
- 14.Msaouel P., Dispenzieri A., Galanis E. Clinical testing of engineered oncolytic measles virus strains in the treatment of cancer: an overview. Curr. Opin. Mol. Ther. 2009;11:43–53. [PMC free article] [PubMed] [Google Scholar]
- 15.Paraskevakou G., Allen C., Nakamura T., Zollman P., James C.D., Peng K.W., Schroeder M., Russell S.J., Galanis E. Epidermal growth factor receptor (EGFR)-retargeted measles virus strains effectively target EGFR- or EGFRvIII expressing gliomas. Mol. Ther. 2007;15:677–686. doi: 10.1038/sj.mt.6300105. [DOI] [PubMed] [Google Scholar]
- 16.Bach P., Abel T., Hoffmann C., Gal Z., Braun G., Voelker I., Ball C.R., Johnston I.C., Lauer U.M., Herold-Mende C. Specific elimination of CD133+ tumor cells with targeted oncolytic measles virus. Cancer Res. 2013;73:865–874. doi: 10.1158/0008-5472.CAN-12-2221. [DOI] [PubMed] [Google Scholar]
- 17.Kleinlützum D., Hanauer J.D.S., Muik A., Hanschmann K.M., Kays S.K., Ayala-Breton C., Peng K.W., Mühlebach M.D., Abel T., Buchholz C.J. Enhancing the Oncolytic Activity of CD133-Targeted Measles Virus: Receptor Extension or Chimerism with Vesicular Stomatitis Virus Are Most Effective. Front. Oncol. 2017;7:127. doi: 10.3389/fonc.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C., Sarkaria J.N., Petell C.A., Paraskevakou G., Zollman P.J., Schroeder M., Carlson B., Decker P.A., Wu W., James C.D. Combination of measles virus virotherapy and radiation therapy has synergistic activity in the treatment of glioblastoma multiforme. Clin. Cancer Res. 2007;13:7155–7165. doi: 10.1158/1078-0432.CCR-07-1306. [DOI] [PubMed] [Google Scholar]
- 19.Desjardins A., Gromeier M., Herndon J.E., 2nd, Beaubier N., Bolognesi D.P., Friedman A.H., Friedman H.S., McSherry F., Muscat A.M., Nair S. Recurrent Glioblastoma Treated with Recombinant Poliovirus. N. Engl. J. Med. 2018;379:150–161. doi: 10.1056/NEJMoa1716435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cloughesy T.F., Landolfi J., Vogelbaum M.A., Ostertag D., Elder J.B., Bloomfield S., Carter B., Chen C.C., Kalkanis S.N., Kesari S. Durable complete responses in some recurrent high-grade glioma patients treated with Toca 511 + Toca FC. Neuro-oncol. 2018;20:1383–1392. doi: 10.1093/neuonc/noy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai B., Roife D., Kang Y., Gumin J., Rios Perez M.V., Li X., Pratt M., Brekken R.A., Fueyo-Margareto J., Lang F.F., Fleming J.B. Preclinical Evaluation of Sequential Combination of Oncolytic Adenovirus Delta-24-RGD and Phosphatidylserine-Targeting Antibody in Pancreatic Ductal Adenocarcinoma. Mol. Cancer Ther. 2017;16:662–670. doi: 10.1158/1535-7163.MCT-16-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang F.F., Conrad C., Gomez-Manzano C., Yung W.K.A., Sawaya R., Weinberg J.S., Prabhu S.S., Rao G., Fuller G.N., Aldape K.D. Phase I Study of DNX-2401 (Delta-24-RGD) Oncolytic Adenovirus: Replication and Immunotherapeutic Effects in Recurrent Malignant Glioma. J. Clin. Oncol. 2018;36:1419–1427. doi: 10.1200/JCO.2017.75.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dörig R.E., Marcil A., Richardson C.D. CD46, a primate-specific receptor for measles virus. Trends Microbiol. 1994;2:312–318. doi: 10.1016/0966-842x(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 24.Tatsuo H., Ono N., Tanaka K., Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 25.Mühlebach M.D., Mateo M., Sinn P.L., Prüfer S., Uhlig K.M., Leonard V.H.J., Navaratnarajah C.K., Frenzke M., Wong X.X., Sawatsky B. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noyce R.S., Bondre D.G., Ha M.N., Lin L.T., Sisson G., Tsao M.S., Richardson C.D. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia M., Meng G., Jiang A., Chen A., Dahlhaus M., Gonzalez P., Beltinger C., Wei J. Mitophagy switches cell death from apoptosis to necrosis in NSCLC cells treated with oncolytic measles virus. Oncotarget. 2014;5:3907–3918. doi: 10.18632/oncotarget.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joubert P.E., Meiffren G., Grégoire I.P., Pontini G., Richetta C., Flacher M., Azocar O., Vidalain P.O., Vidal M., Lotteau V. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Richetta C., Grégoire I.P., Verlhac P., Azocar O., Baguet J., Flacher M., Tangy F., Rabourdin-Combe C., Faure M. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 2013;9:e1003599. doi: 10.1371/journal.ppat.1003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seki H., Kanegane H., Iwai K., Konno A., Ohta K., Yachie A., Taniguchi N., Miyawaki T. Ionizing radiation induces apoptotic cell death in human TcR-gamma/delta+ T and natural killer cells without detectable p53 protein. Eur. J. Immunol. 1994;24:2914–2917. doi: 10.1002/eji.1830241150. [DOI] [PubMed] [Google Scholar]
- 31.Fulda S., Scaffidi C., Pietsch T., Krammer P.H., Peter M.E., Debatin K.M. Activation of the CD95 (APO-1/Fas) pathway in drug- and gamma-irradiation-induced apoptosis of brain tumor cells. Cell Death Differ. 1998;5:884–893. doi: 10.1038/sj.cdd.4400419. [DOI] [PubMed] [Google Scholar]
- 32.Hermisson M., Klumpp A., Wick W., Wischhusen J., Nagel G., Roos W., Kaina B., Weller M. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J. Neurochem. 2006;96:766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]
- 33.Uddin S., Chamdin A., Platanias L.C. Interaction of the transcriptional activator Stat-2 with the type I interferon receptor. J. Biol. Chem. 1995;270:24627–24630. doi: 10.1074/jbc.270.42.24627. [DOI] [PubMed] [Google Scholar]
- 34.Leeuwenberg J.F., van Damme J., Jeunhomme G.M., Buurman W.A. Interferon beta 1, an intermediate in the tumor necrosis factor alpha-induced increased MHC class I expression and an autocrine regulator of the constitutive MHC class I expression. J. Exp. Med. 1987;166:1180–1185. doi: 10.1084/jem.166.4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chawla-Sarkar M., Lindner D.J., Liu Y.F., Williams B.R., Sen G.C., Silverman R.H., Borden E.C. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- 36.Roy S., Guler R., Parihar S.P., Schmeier S., Kaczkowski B., Nishimura H., Shin J.W., Negishi Y., Ozturk M., Hurdayal R. Batf2/Irf1 induces inflammatory responses in classically activated macrophages, lipopolysaccharides, and mycobacterial infection. J. Immunol. 2015;194:6035–6044. doi: 10.4049/jimmunol.1402521. [DOI] [PubMed] [Google Scholar]
- 37.Neidert M.C., Schoor O., Trautwein C., Trautwein N., Christ L., Melms A., Honegger J., Rammensee H.G., Herold-Mende C., Dietrich P.Y., Stevanović S. Natural HLA class I ligands from glioblastoma: extending the options for immunotherapy. J. Neurooncol. 2013;111:285–294. doi: 10.1007/s11060-012-1028-8. [DOI] [PubMed] [Google Scholar]
- 38.Hatano M., Eguchi J., Tatsumi T., Kuwashima N., Dusak J.E., Kinch M.S., Pollack I.F., Hamilton R.L., Storkus W.J., Okada H. EphA2 as a glioma-associated antigen: a novel target for glioma vaccines. Neoplasia. 2005;7:717–722. doi: 10.1593/neo.05277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nummela P., Lammi J., Soikkeli J., Saksela O., Laakkonen P., Hölttä E. Transforming growth factor beta-induced (TGFBI) is an anti-adhesive protein regulating the invasive growth of melanoma cells. Am. J. Pathol. 2012;180:1663–1674. doi: 10.1016/j.ajpath.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 40.Chen W.Y., Tsai Y.C., Yeh H.L., Suau F., Jiang K.C., Shao A.N., Huang J., Liu Y.N. Loss of SPDEF and gain of TGFBI activity after androgen deprivation therapy promote EMT and bone metastasis of prostate cancer. Sci. Signal. 2017;10:eaam6826. doi: 10.1126/scisignal.aam6826. [DOI] [PubMed] [Google Scholar]
- 41.Singh H., Walter S., Weinschenk T., Hilf N., Schoor O., Trautwein C. Google Patents; 2016. Immunogenic Epitopes for Immunotherapy. [Google Scholar]
- 42.Kurokawa C., Iankov I.D., Anderson S.K., Aderca I., Leontovich A.A., Maurer M.J., Oberg A.L., Schroeder M.A., Giannini C., Greiner S.M. Constitutive Interferon Pathway Activation in Tumors as an Efficacy Determinant Following Oncolytic Virotherapy. J. Natl. Cancer Inst. 2018;110:1123–1132. doi: 10.1093/jnci/djy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka M., Shimbo T., Kikuchi Y., Matsuda M., Kaneda Y. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int. J. Cancer. 2010;126:1982–1991. doi: 10.1002/ijc.24965. [DOI] [PubMed] [Google Scholar]
- 44.Topaz O., Indelman M., Chefetz I., Geiger D., Metzker A., Altschuler Y., Choder M., Bercovich D., Uitto J., Bergman R. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am. J. Hum. Genet. 2006;79:759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Q., Yu T., Ren Y.Y., Gong T., Zhong D.S. Overexpression of SAMD9 suppresses tumorigenesis and progression during non small cell lung cancer. Biochem. Biophys. Res. Commun. 2014;454:157–161. doi: 10.1016/j.bbrc.2014.10.054. [DOI] [PubMed] [Google Scholar]
- 46.Liu J., McFadden G. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J. Virol. 2015;89:1925–1931. doi: 10.1128/JVI.02262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J., Wennier S., Zhang L., McFadden G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J. Virol. 2011;85:3270–3282. doi: 10.1128/JVI.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivan G., Ormanoglu P., Buehler E.C., Martin S.E., Moss B. Identification of Restriction Factors by Human Genome-Wide RNA Interference Screening of Viral Host Range Mutants Exemplified by Discovery of SAMD9 and WDR6 as Inhibitors of the Vaccinia Virus K1L-C7L- Mutant. MBio. 2015;6:e01122. doi: 10.1128/mBio.01122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 50.Schröder M., Baran M., Bowie A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engeland C.E., Grossardt C., Veinalde R., Bossow S., Lutz D., Kaufmann J.K., Shevchenko I., Umansky V., Nettelbeck D.M., Weichert W. CTLA-4 and PD-L1 checkpoint blockade enhances oncolytic measles virus therapy. Mol. Ther. 2014;22:1949–1959. doi: 10.1038/mt.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speranza M.C., Passaro C., Ricklefs F., Kasai K., Klein S.R., Nakashima H., Kaufmann J.K., Ahmed A.K., Nowicki M.O., Obi P. Preclinical investigation of combined gene-mediated cytotoxic immunotherapy and immune checkpoint blockade in glioblastoma. Neuro-oncol. 2018;20:225–235. doi: 10.1093/neuonc/nox139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lange S., Lampe J., Bossow S., Zimmermann M., Neubert W., Bitzer M., Lauer U.M. A novel armed oncolytic measles vaccine virus for the treatment of cholangiocarcinoma. Hum. Gene Ther. 2013;24:554–564. doi: 10.1089/hum.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radecke F., Spielhofer P., Schneider H., Kaelin K., Huber M., Dötsch C., Christiansen G., Billeter M.A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen R., Nishimura M.C., Bumbaca S.M., Kharbanda S., Forrest W.F., Kasman I.M., Greve J.M., Soriano R.H., Gilmour L.L., Rivers C.S. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 56.Perazzoli G., Prados J., Ortiz R., Caba O., Cabeza L., Berdasco M., Gónzalez B., Melguizo C. Temozolomide Resistance in Glioblastoma Cell Lines: Implication of MGMT, MMR, P-Glycoprotein and CD133 Expression. PLoS ONE. 2015;10:e0140131. doi: 10.1371/journal.pone.0140131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 1931;162:480–483. [Google Scholar]
- 58.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 59.Wang D., Wang Z., Tian B., Li X., Li S., Tian Y. Two hour exposure to sodium butyrate sensitizes bladder cancer to anticancer drugs. Int. J. Urol. 2008;15:435–441. doi: 10.1111/j.1442-2042.2008.02025.x. [DOI] [PubMed] [Google Scholar]
- 60.Clark K., Plater L., Peggie M., Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase ε: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanafi L.A., Gauchat D., Godin-Ethier J., Possamaï D., Duvignaud J.B., Leclerc D., Grandvaux N., Lapointe R. Fludarabine downregulates indoleamine 2,3-dioxygenase in tumors via a proteasome-mediated degradation mechanism. PLoS ONE. 2014;9:e99211. doi: 10.1371/journal.pone.0099211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 63.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalewski D.J., Stevanović S. Biochemical large-scale identification of MHC class I ligands. Methods Mol. Biol. 2013;960:145–157. doi: 10.1007/978-1-62703-218-6_12. [DOI] [PubMed] [Google Scholar]
- 67.Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 68.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 69.Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 70.Nielsen M., Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J.G., Eguchi J., Kruse C.A., Gomez G.G., Fakhrai H., Schroter S., Ma W., Hoa N., Minev B., Delgado C. Antigenic profiling of glioma cells to generate allogeneic vaccines or dendritic cell-based therapeutics. Clin. Cancer Res. 2007;13:566–575. doi: 10.1158/1078-0432.CCR-06-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Böyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of mononuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Invest. Suppl. 1968;97:77–89. [PubMed] [Google Scholar]
- 73.Peper J.K., Bösmüller H.C., Schuster H., Gückel B., Hörzer H., Roehle K., Schäfer R., Wagner P., Rammensee H.G., Stevanović S. HLA ligandomics identifies histone deacetylase 1 as target for ovarian cancer immunotherapy. OncoImmunology. 2015;5:e1065369. doi: 10.1080/2162402X.2015.1065369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.