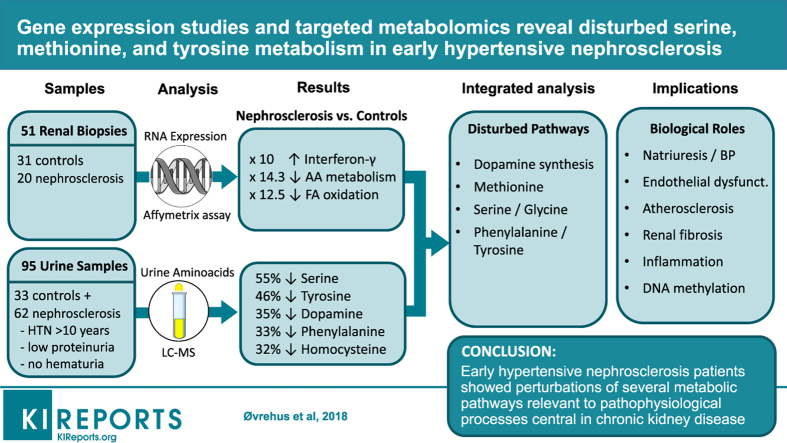
Abstract
Introduction
Hypertensive nephrosclerosis is among the leading causes of end-stage renal disease, but its pathophysiology is poorly understood. We wanted to explore early metabolic changes using gene expression and targeted metabolomics analysis.
Methods
We analyzed gene expression in kidneys biopsied from 20 patients with nephrosclerosis and 31 healthy controls with an Affymetrix array. Thirty-one amino acids were measured by liquid chromatography coupled with mass spectrometry (LC-MS) in urine samples from 62 patients with clinical hypertensive nephrosclerosis and 33 age- and sex-matched healthy controls, and major findings were confirmed in an independent cohort of 45 cases and 15 controls.
Results
Amino acid catabolism and synthesis were strongly underexpressed in hypertensive nephrosclerosis (13- and 7-fold, respectively), and these patients also showed gene expression patterns indicating decreased fatty acid oxidation (12-fold) and increased interferon gamma (10-fold) and cellular defense response (8-fold). Metabolomics analysis revealed significant distribution differences in 11 amino acids in hypertensive nephrosclerosis, among them tyrosine, phenylalanine, dopamine, homocysteine, and serine, with 30% to 70% lower urine excretion. These findings were replicated in the independent cohort. Integrated gene-metabolite pathway analysis showed perturbations of renal dopamine biosynthesis. There were also significant differences in homocysteine/methionine homeostasis and the serine pathway, which have strong influence on 1-carbon metabolism. Several of these disturbances could be interconnected through reduced regeneration of tetrahydrofolate and tetrahydrobiopterin.
Conclusion
Early hypertensive nephrosclerosis showed perturbations of intrarenal biosynthesis of dopamine, which regulates natriuresis and blood pressure. There were also disturbances in serine/glycine and methionine/homocysteine metabolism, which may contribute to endothelial dysfunction, atherosclerosis, and renal fibrosis.
Keywords: amino acids, gene expression, hypertensive nephropathy, metabolomics, nephrosclerosis
Graphical abstract
See Commentary on Page 194
Hypertensive nephrosclerosis is one of the leading causes of end-stage renal disease in industrialized countries.1 Renal parenchymal loss is believed to occur as a consequence of antecedent hypertension-induced pre-glomerular microvascular alterations. The disease is often diagnosed on clinical criteria only, typically in patients with chronic kidney disease (CKD) with longstanding hypertension or signs of blood pressure–related organ damage, low proteinuria, and no signs of other kidney diseases like hematuria, diabetes, and glomerulonephritis.2 However, current diagnostic criteria have low accuracy,3, 4, 5 the pathogenesis is incompletely understood, and no specific treatment is available. More data on the underlying pathophysiological mechanisms and consequences are needed for this understudied, but still very common, disease.
Human gene expression studies find that chronic hypoxia is a central mechanism contributing to both glomerular and tubulointerstitial damage in hypertensive nephrosclerosis.6 The downstream consequences of ischemia and other risk factors in nephrosclerosis are not well studied. Metabolomics platforms now enable the simultaneous measurement of hundreds of small molecular metabolites describing the downstream effects of genes and proteins. Patients with glomerulonephritis,7, 8, 9 kidney transplantation,10, 11 diabetes nephropathy,12 acute kidney injury,13 and general CKD14, 15, 16, 17 have been studied, and important disturbances in amino acid, glucose, and fatty acid metabolism as well as the central energy metabolism have been described.12, 18, 19 Whether this extends to patients with hypertensive nephrosclerosis is not well studied. Although there is no very good animal model for nephrosclerosis, current experimental data indicate decreased citric acid (TCA)-cycle activity20 and extensive mitochondrial abnormalities and dysfunctions in kidneys exposed to various forms of hypertension.21 Pathway analysis based on gene expression in human nephrosclerosis biopsies indicate that amino acid metabolism (including arginine, serine, and tryptophan metabolisms), TCA-cycle, glycolysis/gluconeogenesis, fatty acid oxidation, peroxisome proliferator–activated receptor signaling, and others were significantly disturbed.22
We therefore used kidney gene expression and targeted urinary metabolomic analyses to explore the metabolic consequences in early stages of hypertensive nephrosclerosis compared with age- and sex-matched controls. The goal of this work was to generate a relevant hypothesis to be further tested in experimental models to elucidate the underlying mechanisms of nephrosclerosis.
Material and Methods
Study Populations
For our biopsy study, we included patients with biopsy-proven hypertensive nephrosclerosis and healthy kidney transplant donors before kidney transplantation from the European Renal cDNA Bank.23
For urine metabolomics studies, we included subjects from the Third Nord-Trøndelag Health Study (HUNT3), a cross-sectional population study performed in the Norwegian county of Nord-Trøndelag between 2006 and 2008. Biological sampling, recording of medical and lifestyle information, and simple physical examinations were done as previously described.24 Blood pressure was measured 3 times in a sitting position by trained nurses at the study venue and recorded as the mean of the last 2 measurements using the oscillometric method (Dinamap 845XT; Criticon, Tampa, FL). Clinical hypertensive nephrosclerosis was defined as individuals with an estimated glomerular filtration rate (eGFR) less than 60 ml/min per 1.73 m2 or eGFR 60–89 ml/min per 1.73 m2 with a decline in eGFR larger than 30 ml/min per 1.73 m2 over the previous 10 years, combined with self-reported hypertension lasting more than 10 years and absence of diabetes mellitus, hematuria, and macroalbuminuria (defined as a urine albumin-to-creatinine ratio >30 mg/mmol). Hypertension was defined as systolic blood pressure >140 mm Hg and/or diastolic blood pressure >90 mm Hg or treatment with antihypertensive medication. The control group included individuals with estimated glomerular filtration rates above 75 ml/min per 1.73 m2, who did not have hypertension, diabetes, hematuria, or proteinuria. Controls were matched to patients with nephrosclerosis regarding sex and age group in age intervals of 20 to 50 years, 50 to 70 years, and older than 70 years.
To replicate the main metabolomics findings of the HUNT study, we also analyzed urine and blood samples from the Study of Glucose and Insulin in Renal Disease (SUGAR).25 This cohort included nondiabetic patients with CKD with eGFR <60 ml/min per 1.73 m2, as well as age- and gender-matched controls. Most patients had a clinical diagnosis of hypertensive nephrosclerosis. See Supplementary Materials for further details on study design and analytical methods.
Metabolomic Analyses
Fresh urine samples were collected midstream at the venue of the HUNT study, immediately put into a −20 °C freezer and transferred to a −80° freezer within 24 hours, without centrifugation or use of additives during storage. Targeted quantitative analysis of amino acids in urine was performed using LC-MS using the commercial EZ:faast LC/MS Physiological (Free) Amino Acids Kit (Phenomenex, Inc, Torrance, CA), which includes the most clinical amino acids and related compounds (n = 47). Urine was diluted, centrifuged, spiked with the internal standard solution, and derivatized following the Phenomenex EZ-faast protocol. Samples were separated and measured with a Waters Acquity UPLC–TQ-S tandem mass spectrometer (Waters Corporation, Milford, MA) with cases and controls in a random order with 1 quality control every 15 samples. Values were normalized to 1 mmol creatinine. We excluded 3 amino acids (sarcosine, citrulline, arginine) because they were not detected, or detected in only near zero amounts, in more than 80% of the samples. We also excluded 13 amino acids because we did not achieve reliable transitions, good peak shapes, or retention times that matched with expected times. Interbatch variability was 11.9%, calculated as the relative SD (= SD/mean). The average variance of the internal standard within the same batch (intrabatch) was 2.1% (range 0.5–4.2%). Between all batches, 71% of these amino acids had relative SDs of less than 35%. All remaining amino acids were included for further analyses. See Supplementary Methods for details.
Transcriptomic Analyses
Samples from the glomerular and the tubulointerstitial compartment were separated by microdissection in kidney biopsies from the European Renal cDNA Bank. Total RNA was isolated, reverse-transcribed and amplified, and then fragmented and hybridized to the Affymetrix (Santa Clara, CA) GeneChip Human Genome U133A 2.0 and U133 Plus 2.0 Array as described in previous publications.6, 23, 26 Quality controls have been integrated in each step of the standard procedures and include, but are not limited to, RNA quality control, hybridization control, and data analysis control. In addition, a minimum of 3 reference controls (Stratagene, San Diego, CA) were used in each batch of hybridization, and living donors or tumor nephrectomy samples were included by design as “healthy” to control for disease conditions. Specific to this selected dataset, confirmatory real-time polymerase chain reaction analyses were performed to replicate the microarray results using micro-dissected glomeruli from biopsy specimens from an independent cohort of patients. Genes with false discovery rate (FDR) <0.05 and more than 1.5-fold change were considered differentially expressed and included for further analysis. We used gene ontology enrichment analysis (PANTHER, supported by the Gene Ontology Consortium) to integrate all the gene expression data making it easier to understand overall biological function. Upregulated and downregulated genes were analyzed separately.27
Statistics
Central tendency for urine metabolites was evaluated as percent differences in median values (patients with nephrosclerosis minus controls), and statistical testing was done with the Mann Whitney U-test (2-sample Wilcoxon rank test). We also tested for distribution differences using the 2-sample Kolmogorov-Smirnov test. FDR was calculated according to the Benjamini Hochberg ranking procedure to adjust for multiple testing,28 and FDR 0.10 was considered significant in this exploratory setting. Principal Component Analysis and Partial Least Squares Discriminant Analysis (PLS-DA) were used to evaluate overall discrimination between patients and controls after normalizing to 1 mmol of urine creatinine, log transformation, and autoscaling (MetaboAnalyst 3.0; www.metaboanalyst.ca). Variable importance in projection (VIP) was used as a measure of the importance of variables from the PLS-DA analysis. The VIP score is based on the sum of variable influence over all model dimensions, looking at the PLS loadings relative to the amount of explained Y-variation,29 and can be used to identify discriminating variables or predictors.30
Bioinformatics
For pathway analysis, we used a combination of enrichment analysis and pathway topology analysis (MetaboAnalyst 3.0). Enrichment analysis, rather than evaluating one metabolite at a time for significance, evaluates groups of functionally related metabolites to see if they occur more often than expected or not. Pathway topology analysis takes into consideration that some metabolites are more central and others more peripheral in a metabolic pathway, using so-called relative betweenness centrality and degree centrality measures.29 We also combined gene and metabolite information into the same pathway analysis (integrated pathway analysis, MetaboAnalyst 3.0). See Supplementary Methods for further details, especially on LC-MS methods.
To further elucidate the differentially expressed genes relevant for our top pathways, we also explored the Nephroseq version 5. This public website displays preanalyzed data on the associations between specific genes of interest and various renal outcomes (eGFR decline, CKD, albuminuria, age, and others) based on selected datasets. The study was approved by the Regional Committee for Medical and Health Research Ethics, Central Norway. All participants gave written informed consent.
Results
Participants
Baseline characteristics of patients with nephrosclerosis and healthy controls with kidney biopsies (n = 51), urine metabolomics in the main cohort (HUNT, n = 95), and the replication cohort (SUGAR, n = 60) are summarized in Table 1. Mean age was close to 60 in all groups except kidney biopsy controls (47 years). By definition, none of the controls had diabetes, hypertension, reduced kidney function, or signs of kidney damage. Patients with nephrosclerosis had higher blood pressure and body mass index combined with lower eGFR (only modestly reduced in the HUNT cohort, mean of 67 ml/min per 1.73 m2, with range 40–89).
Table 1.
Baseline characteristics of participants
| Gene expression studies (European Renal Biopsy Bank) |
Metabolomics studies (HUNT study) |
Metabolomics studies (SUGAR study) |
||||
|---|---|---|---|---|---|---|
| Healthy controls (n = 31) | Hypertensive nephrosclerosis (n = 20) | Healthy controls (n = 33) | Hypertensive nephrosclerosis (n = 62) | Healthy controls (n = 15) | Hypertensive non-DM CKD (n = 45) | |
| Age | 47.2 | 57.1 | 58.1 (11.3) | 59.7 (10.8) | 55.6 (11.8) | 61.5 (14.3) |
| Male gender (%) | 57 | 80 | 64 | 58 | 60 | 53 |
| Systolic BP (mm Hg) | <140 | 146 (22.9) | 126.8 (12.2) | 141.9 (13.8) | 122.4 (14.1) | 134.4 (15.9) |
| Diastolic BP (mm Hg) | <80 | 88 (13.5) | 73.1 (8.9) | 79.3 (10.9) | 77.2 (9.3) | 80.6 (9.7) |
| Cholesterol (mmol/l) | n.a. | 6.3 (1.0) | 5.9 (1.0) | 5.7 (1.0) | 5.2 (1.0) | 4.7 (1.1) |
| Current-smoker (%) | n.a. | n.a. | 25 | 11 | 7 | 20 |
| Body mass index (kg/m2) | n.a. | 29.1 (4.7) | 26.4 (3.5) | 28.6 (3.6) | 27.3 (5.9) | 30.3 (6.2) |
| Cardiovascular disease (%) | 0 | n.a. | 3 | 18 | 7 | 40 |
| DM (%) | 0 | 0 | 0 | 0 | 0 | 0 |
| eGFR (ml/min per 1.73 m2)a | 105.4 (30.9) | 40.9 (23.8) | 94.8 (14.2) | 67.1 (10.8) | 90.4 (18.0) | 36.0 (12.8) |
| eGFR ≥90 (%) | 76 | 0 | 57 | 0 | 47 | 0 |
| eGFR 75–89 (%) | 10 | 20 | 43 | 44 | 27 | 0 |
| eGFR 60–74 (%) | 14 | 0 | 0 | 27 | 27 | 0 |
| eGFR 45–59 (%) | 0 | 13 | 0 | 26 | 0 | 31 |
| eGFR 30–44 (%) | 0 | 27 | 0 | 3 | 0 | 33 |
| eGFR 15–29 (%) | 0 | 40 | 0 | 0 | 0 | 36 |
| u-ACR (mg/mmol) | <3.0 | 57 (56) | 1.3 (0.5) | 2.1 (1.6) | 0.8 (0.7) | 40.2 |
| ACR 0–2.9 (%) | 100 | 0 | 100 | 84 | 93 | 34 |
| ACR 3.0–29.9 (%) | 0 | 44 | 0 | 16 | 7 | 44 |
| ACR ≥30.0 (%) | 0 | 56 | 0 | 0 | 0 | 22 |
ACR, albumin-to-creatinine ratio; BP, blood pressure; CKD, chronic kidney disease; DM, diabetes mellitus; eGFR, estimated glomerular filtration rate; HUNT, Nord-Trøndelag Health Study; SUGAR, Study of Glucose and Insulin in Renal Disease.
eGFR calculated using the CKD-Epidemiology collaboration equation.
Kidney Biopsy Gene Expression
Kidney biopsy gene expression analysis displayed substantial differences between patients with nephrosclerosis and healthy kidney donors. Among 11,936 genes tested, 306 and 267 were significantly (FDR <0.05 and absolute fold-change >1.5) over- or underexpressed, respectively. We carried out gene ontology enrichment analysis to combine the information from these differentially expressed genes into main biological functions (Table 2). Amino acid–related biological processes were substantially underexpressed in patients with nephrosclerosis (7- to 13-fold). Other highly enriched functions indicated decreased fatty acid oxidation (12-fold) and gluconeogenesis (10-fold), whereas several immunological and defense functions were increased (interferon gamma response [10-fold], cellular defense response [8-fold], and cytokine signaling [7-fold]). We also tested the association between main gene expression findings and age due to the 10-year age difference between cases and controls. There were, however, either no significant associations with age (e.g., phenylalanine-hydroxylase, phosphoenolpyruvate carboxykinase, 5,10-methylene-tetrahydrofolate reductase) or the fold-change for the 10-year age difference was <1.1 (e.g., L-dihydroxyphenylalanine [L-DOPA]-decarboxylase, betaine-homocysteine methyl-transferase).
Table 2.
Gene ontology analysis showing the top 10 biological processes up- or downregulated by fold enrichment in hypertensive nephrosclerosis kidney biopsies versus biopsies from healthy kidney donors
| Biological process | Total | No. found | No. expected | Fold enrichment | Raw P value | FDR |
|---|---|---|---|---|---|---|
| Cellular amino acid catabolic process | 56 | 10 | 0.70 | −12.81 | 1.05E-07 | 1.28E-05 |
| Fatty acid beta-oxidation | 20 | 3 | 0.25 | −11.96 | 2.77E-03 | 3.98E-02 |
| Response to interferon gamma | 58 | 9 | 0.87 | +10.40 | 5.80E-07 | 1.57E-05 |
| Gluconeogenesis | 23 | 3 | 0.29 | −10.40 | 3.96E-03 | 4.83–02 |
| Cellular defense response | 105 | 13 | 1.57 | +8.30 | 2.05E-08 | 2.50E-06 |
| Cellular amino acid metabolic process | 230 | 22 | 2.89 | −7.62 | 8.65E-13 | 2.11E-10 |
| Cellular amino acid biosynthetic process | 64 | 6 | 0.80 | −7.47 | 2.31E-04 | 7.04E-03 |
| Cellular calcium ion homeostasis | 115 | 12 | 1.72 | +6.99 | 3.94E-07 | 1.37E-05 |
| Cytokine-mediated signaling pathway | 60 | 6 | 0.9 | +6.70 | 4.17E-04 | 5.36E-03 |
| Monosaccharide metabolic process | 76 | 6 | 0.95 | −6.29 | 5.45E-04 | 1.21E-02 |
Note: Data from separate gene ontology (GO) enrichment analysis of 306 upregulated and 267 downregulated genes. Analyses done with PANTHER application and genes with false discovery rate (FDR) <0.05 and |FC| >1.5 were included. Table shows PANTHER GO-Slim Biological Processes. GO is a framework for modeling biological function using defined concepts/classes to describe gene function and the relationships between these concepts. GO slims are cut-down versions of the full GO ontologies containing a subset of the most important and instructive terms, that is, an output particularly useful for giving a summary of the results when broad classification of gene product function is required.
Urine Metabolomics
Table 3 displays median values and distribution of the 31 amino acids included for further analyses. The relative differences indicated lower urine concentrations for many amino acids in nephrosclerosis, but none had a statistically different central tendency (i.e., median) after correcting for multiple testing. For 11 amino acids, the distribution of concentrations was statistically different in nephrosclerosis versus controls (serine, tyrosine, leucine, phenylalanine, homocysteine, threonine, ornithine, proline, carnosine, glutamic acid, and dopamine) (Table 3). The distribution difference measure includes information on range and skewness in addition to central tendency, which can be important for discriminating between groups. Serine, tyrosine, leucine, phenylalanine, and homocysteine had the largest distribution differences (P < 0.01 for all); however, the distribution of these metabolites in the hypertensive nephrosclerosis group and the age- and sex-matched controls still showed substantial overlap (Figure 1). There was no association between the amino acids and eGFR in multiadjusted regression analysis, so the amino acids do not vary simply with filtration between the 2 groups (data not shown).
Table 3.
Urinary amino acid excretion in healthy controls versus hypertensive patients with nephropathy
| HUNT cohort |
SUGAR cohort |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Central location difference (Nephroscl. – Ctrl., medians) |
Distribution difference (Kolmogorov-Smirnov) |
Discrimination (PLS-DA) |
Central location difference (Nephroscl. – Ctrl., medians) |
||||||
| Δ % | Raw P value | FDR <0.10 | Raw P value | FDR <0.10 | VIP score | Δ % | Raw P value | FDR <0.10 | |
| Carnosine (Car) | −71.1 | 0.15 | N.s. | 0.03 | Sign. | 1.25 | n.a. | n.a. | n.a. |
| Glycine (Gly) | −58.1 | 0.11 | N.s. | 0.06 | N.s. | 1.18 | −35.8 | <0.001 | Sign. |
| Serine (Ser) | −55.4 | 0.06 | N.s. | 0.001 | Sign. | 1.01 | −69.6 | <0.001 | Sign. |
| Tyrosine (Tyr) | −46.9 | 0.03 | N.s. | 0.005 | Sign. | 1.40 | −45.1 | 0.04 | Sign. |
| Threonine (Thr) | −46.7 | 0.19 | N.s. | 0.02 | Sign. | <1.0 | +12.8 | 0.21 | N.s. |
| Histidine (His) | −42.9 | 0.14 | N.s. | 0.24 | N.s. | 1.41 | n.a. | n.a. | n.a. |
| 2.4-diaminobutyric acid | +35.8 | 0.4 | N.s. | 0.7 | N.s. | 1.02 | n.a. | n.a. | n.a. |
| Dopamine (DA) | −35.4 | 0.05 | N.s. | 0.05 | Sign. | 1.0 | −28.2 | 0.05 | Sign. |
| Ornithine (Orn) | −35.2 | 0.04 | N.s. | 0.02 | Sign. | 1.03 | n.a. | n.a. | n.a. |
| Phenylalanine (Phe) | −33.4 | 0.07 | N.s. | 0.009 | Sign. | 1.08 | −51.2 | 0.004 | Sign. |
| Homocysteine (Hcys) | −32.7 | 0.02 | N.s. | 0.009 | Sign. | 1.82 | n.a. | n.a. | n.a. |
| Leucine (Leu) | −32.6 | 0.09 | N.s. | 0.006 | Sign. | <1.0 | n.a. | n.a. | n.a. |
| Kynurenine (Kyn) | −31.9 | 0.06 | N.s. | 0.1 | N.s. | 1.38 | −21.2 | 0.31 | N.s. |
| Lysine (Lys) | −31.8 | 0.11 | N.s. | 0.08 | N.s. | 1.08 | n.a. | n.a. | n.a. |
| Alanine (Ala) | −31.8 | 0.13 | N.s. | 0.1 | N.s. | <1.0 | −63.1 | <0.001 | Sign. |
| α-aminoadipic acid (Aaa) | −31.5 | 0.1 | N.s. | 0.15 | N.s. | 1.07 | |||
| Valine (Val) | −30.3 | 0.18 | N.s. | 0.11 | N.s. | <1.0 | |||
| Cystathionine (Cth) | −29.2 | 0.11 | N.s. | 0.17 | N.s. | 1.15 | |||
| Proline (Pro) | −27.8 | 0.07 | N.s. | 0.03 | Sign. | <1.0 | |||
| Glutamic acid (Glu) | −26.6 | 0.04 | N.s. | 0.05 | Sign. | 1.43 | |||
| Glutamyl-Lysine (Glu-Lys) | −26.1 | 0.17 | N.s. | 0.27 | N.s. | <1.0 | |||
| 5-aminovalerate (5Aval) | −25.5 | 0.48 | N.s. | 0.51 | N.s. | <1.0 | |||
| Methionine (Met) | −24.9 | 0.09 | N.s. | 0.09 | N.s. | 1.20 | |||
| Isoleucine (Ile) | −24.6 | 0.13 | N.s. | 0.15 | N.s. | <1.0 | |||
| 2,6-Aminopimelat (Dapa) | +22.8 | 0.21 | N.s. | 0.2 | N.s. | <1.0 | |||
| Proline-OH-proline (PHP) | −22.1 | 0.68 | N.s. | 0.29 | N.s. | <1.0 | |||
| 1-Met-Histidine (1Mhis) | −22.1 | 0.64 | N.s. | 0.57 | N.s. | <1.0 | |||
| Cystine | −20.9 | 0.05 | N.s. | 0.04 | Sign. | 1.24 | |||
| Hydroxyproline (Hyp) | −15.6 | 0.49 | N.s. | 0.65 | N.s. | <1.0 | |||
| Aspartic acid (Asp) | −9.3 | 0.51 | N.s. | 0.69 | N.s. | <1.0 | |||
| 3-Met-Histidine (3Mhis) | −3.4 | 0.85 | N.s. | 0.45 | N.s. | <1.0 | |||
Note: We intended to replicate the top 15 HUNT metabolites in the SUGAR cohort, but not all metabolites were available and some are therefore marked with “n.a.” Central location is the difference between median values, that is, controls (Ctrl.) subtracted from patients with nephrosclerosis (Nephroscl.).
FDR, false discovery rate; HUNT, Nord-Trøndelag Health Study; N.s, not significant; PLS-DA, partial least squares discriminant analysis; Sign., significant; SUGAR, Study of Glucose and Insulin in Renal Disease; VIP, variable importance in projection.
Figure 1.
Relative frequency plots of central amino acids. Hypertensive nephrosclerosis (red) versus healthy controls (blue). (a) Plot of relative frequency versus relative concentrations of serine. (b) Plot of relative frequency versus relative concentrations of tyrosine. Red line: Nephrosclerosis. (c) Plot of relative frequency versus relative concentrations of homocystein.
Metabolomics Replication
We were able to replicate the main findings from the HUNT cohort in the SUGAR cohort (Table 3). Regarding the serine metabolism, our top hit, we found that cases had 70% lower urine excretion of serine in this cohort compared with their controls (P < 0.001), and glycine excretion was 36% lower (P < 0.001). For the tyrosine metabolism, we found very similar reduction for phenylalanine (−51%, P = 0.004), tyrosine (−45%, P = 0.04), and dopamine (−28%, P = 0.05).
Bioinformatics Analyses
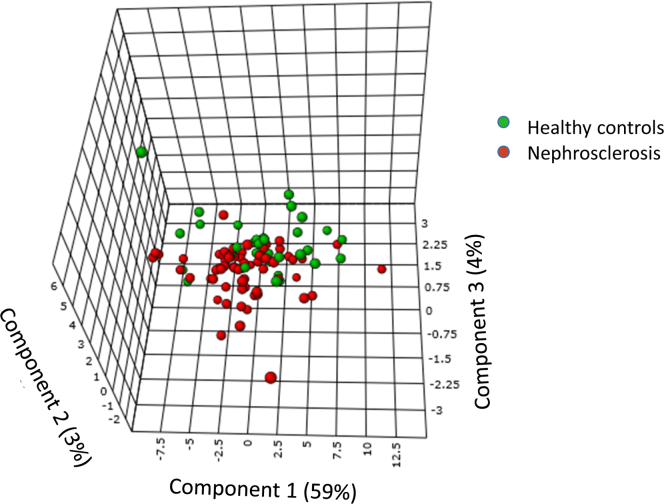
PLS-DA showed that urine amino acid levels moderately separated hypertensive patients with nephrosclerosis from controls and explained 72% of the variance (Figure 2). Eighteen of the amino acids (Hcys, Glu, His, Tyr, Kyn, Car, Cys, Met, Gly, Cth, Lys, Phe, Aaa, Orn, Dap, Ser, DA, Ala) had VIP scores >1.0, and these were considered to contribute significantly to the discrimination (see Supplementary Table 1 for details). The top 5 metabolites were homocysteine (generally known as an independent cardiovascular risk factor), glutamic acid (playing a central role in overall nitrogen homeostasis), histidine (having antioxidant, anti-inflammatory, and antisecretory properties), tyrosine (important for protein and catecholamine biosynthesis), and kynurenine (major metabolite in tryptophan metabolism with influence on production of neurotransmitters, inflammation, and aging).
Figure 2.
Overall ability of urine amino acids to discriminate between patients with early nephrosclerosis (chronic kidney disease stage 2–3) and healthy controls. Partial least squares discriminant analysis shows the overall variance between the groups divided into 3 vectors (principal components 1–3). Eighteen amino acids had variable importance in projection scores ≥1.0 (in decreasing order: Hcys, Glu, His, Tyr, Kyn, Car, Cys, Met, Gly, Cth, Lys, Phe, Aaa, Orn, Dap, Ser, DA, Ala), which is considered a significant contribution to discrimination.
We tested the ability to distinguish between a patient with nephrosclerosis CKD and a healthy control using receiver operating characteristic analysis. We used Principal Component 1 scores as a measure for all metabolites, and we also tested the most significant individual metabolites from the PLS-DA and the Kolmogorov-Smirnov tests in separate receiver operating characteristic analysis (Table 4). Principal Component 1 had an area under the curve of 0.63, which indicates a weak but significant ability (95% confidence interval 0.51–0.75) to correctly diagnose nephrosclerosis versus healthy controls (area under the curve = 0.50 means no information beyond coin tossing). Homocysteine, glutamic acid, and kynurenine also had similar results. When analyzing the various clinical diagnostic criteria of nephrosclerosis separately, the metabolites were able to correctly diagnose only hypertension (PC1, homocysteine, glutamate, and leucine). They contained no relevant information for diagnosing low eGFR, proteinuria, or hematuria (Table 4).
Table 4.
Diagnostic accuracy of important amino acids for nephrosclerosis and its clinical criteria
| PC 1 | Homocysteine | Glutamate | Histidine | Kynurenine | Leucine | Phenylalanine | Serine | Tyrosine | |
|---|---|---|---|---|---|---|---|---|---|
| Nephrosclerosis | 0.63 (0.51–0.75) | 0.65 (0.53–0.77) | 0.63 (0.50–0.75) | 0.59 (0.47–0.72) | 0.62 (0.50–0.74) | 0.61 (0.48–0.73) | 0.62 (0.49–0.74) | 0.61 (0.48–0.75) | 0.64 (0.51–0.0.77) |
| Hypertension | 0.63 (0.50–0.77) | 0.65 (0.52–0.78) | 0.63 (0.51–0.76) | 0.58 (0.45–0.72) | 0.61 (0.48–0.74) | 0.63 (0.51–0.75) | 0.60 (0.48–0.73) | 0.60 (0.47–0.74) | 0.63 (0.50–0.76) |
| eGFR | 0.50 (0.37–0.62) | 0.51 (0.39–0.63) | 0.52 (0.40–0.64) | 0.58 (0.46–0.69) | 0.50 (0.38–0.62) | 0.54 (0.42–0.67) | 0.53 (0.42–0.65) | 0.56 (0.44–0.68) | 0.55 (0.42–0.66) |
| Proteinuria | 0.55 (0.36–0.74) | 0.55 (0.42–0.76) | 0.52 (0.35–0.69) | 0.60 (0.30–0.90) | 0.54 (0.29–0.79) | 0.50 (0.27–0.72) | 0.59 (0.44–0.75) | 0.51 (0.28–0.75) | 0.61 (0.47–0.76) |
| Hematuria | 0.60 (0.41–0.77) | 0.56 (0.37–0.76) | 0.62 (0.40–0.84) | 0.52 (0.32–0.72) | 0.58 (0.39–0.79) | 0.51 (0.30–0.72) | 0.62 (0.45–0.79) | 0.68 (0.49–0.86) | 0.62 (0.45–0.80) |
Note: Principal Component (PC) 1 includes information from all amino acids reduced into 1 variable describing the variation in the dataset irrespective of diagnostic group. The specific amino acids displayed are the top 5 from Table 2 (highest significance for distribution differences) and the top 5 from Figure 1 (highest variable importance in projection score in partial least squares discriminant analysis). Data are area under the receiver operating characteristic curves (95% confidence interval). Negative associations giving values below 0.5, which is the line of indifference, are transformed (1 − x) for ease of comparison. Significant data are highlighted in bold.
eGFR, estimated glomerular filtration rate.
Overrepresentation analysis was used to gain insight into the underlying biological mechanisms and functional implications of the statistically significant metabolites discovered. Methionine metabolism was most significantly different from the background set (P = 0.00001, FDR = 0.0008) and had a 9.8-fold enrichment (6 significant metabolites found while only 0.612 expected by chance). Catecholamine biosynthesis had the highest enrichment (15.6-fold), but there were also other highly enriched metabolite sets: glycine, serine, and threonine metabolism (9.0-fold), ammonia recycling (8.7-fold), glutathione metabolism (11.8-fold), and histidine metabolism (10.6-fold). See Supplementary Table S2 and Supplementary Figure S1 for further details.
Integrated pathway analysis displayed that several metabolic pathways had significant overrepresentation of differentially expressed genes combined with corresponding changes in urine metabolites (Table 5). The glycine, serine, and threonine metabolism had the highest rank based on a combination of enrichment and topology analysis. The enrichment analysis displayed a 2.0-fold enrichment with a P value of 0.0004, showing that it is very unlikely that so many significantly differing metabolites and genes should occur in this pathway by chance. The topology analysis displayed scores of 2.0, indicating that very central metabolites and genes in this pathway were up- or downregulated compared with the other pathways. The phenylalanine, tyrosine, and tryptophan metabolism was also significantly perturbed (3.3-fold enriched with a topology score 2.1), and the methionine metabolism showed similar results (1.8-fold enrichment and topology score 3.3).
Table 5.
Integrated pathway analysis combining significant genes and metabolites in clinical nephrosclerosis versus healthy controls
| Pathway/metabolism | Enrichment analysis |
Topology analysis | Rank | ||||
|---|---|---|---|---|---|---|---|
| Total | No. expected | No. found | P | Fold-change | |||
| Glycine, serine, threonine | 68 | 11.36 | 23 | 0.0004 | 2.02 | 3.70 | 7.50 |
| Phenylalanine, tyrosine, tryptophan | 9 | 1.50 | 5 | 0.0089 | 3.32 | 2.06 | 6.85 |
| Methionine and homocystein | 63 | 10.53 | 19 | 0.0053 | 1.80 | 3.31 | 5.98 |
| Glycerolipid | 72 | 12.03 | 20 | 0.0116 | 1.66 | 2.60 | 4.31 |
| One-carbon pool by folate | 28 | 4.68 | 10 | 0.0116 | 2.14 | 1.71 | 3.65 |
| Arginine and proline | 102 | 17.05 | 30 | 0.0008 | 1.76 | 2.00 | 3.52 |
| Glycolysis/gluconeogenesis | 91 | 15.21 | 23 | 0.0224 | 1.51 | 2.27 | 3.44 |
| N-Glycan biosynthesis | 50 | 8.36 | 15 | 0.0134 | 1.80 | 1.40 | 2.51 |
| Phenylalanine | 29 | 4.85 | 13 | 0.0003 | 2.68 | 0.88 | 2.35 |
| Butanoate | 47 | 7.85 | 17 | 0.0009 | 2.16 | 0.88 | 1.89 |
| Beta-alanine | 50 | 8.36 | 19 | 0.0002 | 2.27 | 0.76 | 1.73 |
| Linoleic acid | 34 | 5.68 | 15 | 0.0001 | 2.64 | 0.57 | 1.51 |
Note: Pathways are ranked according to their combined enrichment and topology using multiplication.84 The analysis is using hypergeometric test for enrichment and betweenness centrality for topology. Enrichment analysis tests if compounds involved in a particular pathway are represented more often than expected by chance, and data are presented as fold enrichment. Topology analysis takes the pathway structure into consideration when determining which pathways are more likely to be involved in the conditions under study with changes in key positions of a network triggering more severe impact on the pathway than changes on marginal or relatively isolated positions. Analyses were done using MetaboAnalyst 3.0.
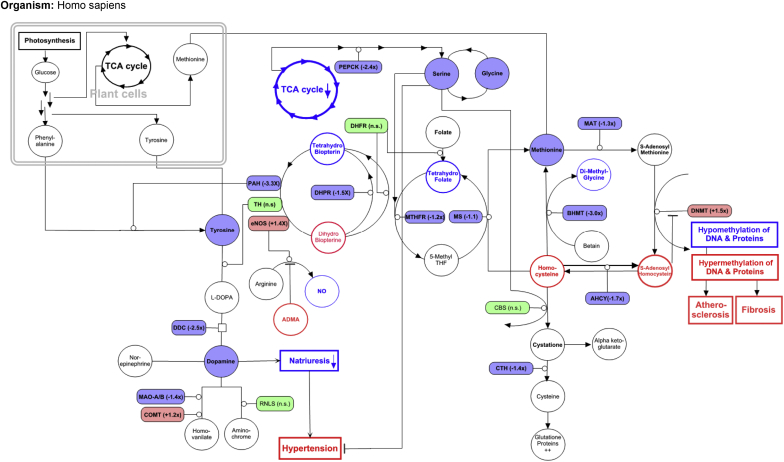
Figure 3 gives an overview of biologically interesting parts of the top-3 pathways and their interactions. Our metabolite and gene expression data are indicated in the figure, and other well-documented relevant findings from the kidney disease literature are also integrated. Parts of the serine and the methionine metabolisms are strongly connected via the folate cycle into the so-called 1-carbon metabolism. Information on metabolites and enzymes indicate that this is downregulated in nephrosclerosis, which could lead to disturbances in the methylation of DNA and protein with correspondingly increased renal fibrosis and atherosclerosis. Finally, the data also show reduced substrate and gene expression consistent with reduced intrarenal dopamine synthesis, which would strongly impair natriuresis and blood pressure control. The activity of main enzymes could even be reduced due to reduced regeneration of their co-enzyme tetrahydrobiopterin, which represents a potential connection with the downregulated folate cycle.
Figure 3.
Major disturbances connecting the major perturbed pathways in nephrosclerosis: tyrosine, serine, and methionine metabolism. Filled figures are data from the current study, circles are metabolites, and rectangles are enzymes. Colored but open figures are based on literature findings in nephrosclerosis. Red is increased/upregulated, green is normal/unchanged, and blue is reduced/downregulated. ADMA, assymetric dimethylarginine; AHCY, adenosylhomocysteinase; BHMT, betaine-homocysteine methyl-transferase; CBS, cystathionine beta-synthase; COMT, catechol-O-methyltransferase; CTH, cystathionase; DHFR, dihydrofolate reductase; DHPR, dihydropteridine reductase; DNMT, DNA methyltransferase; DOC; deoxycorticosterone; eNOS, endogenous nitric oxide synthase; L-DOPA, L-dihydroxyphenylalanine; MAO-A/B, monoamine oxidase A/B; MAT, methionine adenosyl-transferase; MS, methionine synthase; MTHFR, 5,10-methylene-tetrahydrofolate reductase; NO, nitric oxide; n.s., not significant; PAH, phenylalanine-hydroxylase; PEPCK, phosphoenolpyruvate carboxykinase; RNLS, renalase; TCA, citric acid; TH, tyrosine hydroxylase.
Discussion
Patients with nephrosclerosis had lower expression of genes related to amino acid and energy metabolism, whereas there was increased expression of genes related to immune response. Urinary metabolomics combined with gene expression in kidney biopsies displayed perturbations in several relevant pathophysiology pathways in early hypertensive nephrosclerosis, such as serine metabolism (endothelial dysfunction and oxidative stress), methionine metabolism (cardiovascular risk and fibrosis), and tyrosine metabolism (catecholamine biosynthesis and natriuresis).
The diagnosis of nephrosclerosis has been disputed for decades, and some argue that these patients display only normal age-related kidney findings.31 Clearly, the typical histopathological findings of arteriolar hyalinosis, glomerulosclerosis, and tubulointerstitial fibrosis32 also can be found in kidneys from healthy elderly subjects,5, 33 but the findings are more pronounced in nephrosclerosis.34, 35, 36 Several studies indicate that high blood pressure and other cardiovascular risk factors are also involved in the pathogenesis of nephrosclerosis,37, 38, 39 but recent 3-dimensional microscopy shows that nephrosclerosis is a small vessel disease different from the traditional atherosclerotic disease found in arcuate and larger renal arteries.40 The underlying pathophysiological mechanisms are therefore still unresolved. Our findings point toward several amino acid–based dysregulations in central metabolic pathways compared with age- and sex-matched controls, which can both initiate and aggravate nephrosclerosis pathology.
Serine is a nutritionally nonessential amino acid, but it is metabolically indispensable and has received increasing attention over the past years. Twenty-five percent of serine comes from protein breakdown, and the remaining 75% is from de novo synthesis.41 Both rat and human studies show that the kidney is the main site of serine production.42 Although some serine is produced by regeneration from glycine (15%), most (>50%) comes from TCA-cycle intermediates that are converted into phosphoenolpyruvate by phosphoenolpyruvate carboxykinase.41 Serine is required for cellular and tissue growth in general and for the nervous system in particular.43 It acts as neurotransmitters, is important for the catalytic activity of many enzymes, and is an important building block of many lipids. More recently, kidney-relevant properties have also been disclosed; for example, serine is a major methyl group provider,44 and it also has direct blood pressure–lowering effects.45
Methionine is an essential amino acid produced in plants only from TCA-cycle intermediates. It plays, in addition to general protein synthesis, critical roles in important human metabolic processes, including transmethylation reactions, the tetrahydrofolate associated 1-carbon metabolism, and as a precursor for other sulfur compounds. Methionine is converted into homocysteine via a 3-step process in which demethylation of s-adenosyl-methionine to s-adenosyl-homocysteine (SAH).46 This step is the major provider of methyl groups in all human cells and is used for methylation of DNA to regulate transcription (epigenetics) and for methylation of proteins to modify their function (e.g., posttranslational regulation of the folding of proteins). Homocysteine must then be re-methylated to methionine by combining with 5-methyl-tetrahydrofolate, which depends on serine to be regenerated, or with betaine, or it can be metabolized to cysteine for further degradation or urine excretion.
Tyrosine is a nonessential amino acid that can be synthesized from plant-derived phenylalanine and then used for catecholamine synthesis. In general, the first step from tyrosine to L-DOPA, which is catalyzed by tyrosine hydroxylase, is regarded as the rate-limited step in this process. However, for the much higher levels of intrarenal dopamine, expression of L-DOPA-decarboxylase as well as availability of L-DOPA substrate, could be of additional importance.47 Intrarenal dopamine has been found to account for more than 50% of the kidney’s salt excretion ability and is therefore of great importance for blood pressure control.48, 49
Renal fibrosis and atherosclerosis are often found in nephrosclerosis, although the latter is more likely an associated finding and not a cause of the disease. In early nephrosclerosis, we demonstrate pathophysiological perturbations in both serine and methionine metabolism, which are closely connected and could cause both renal fibrosis and general atherosclerosis. Hyperhomocysteinemia has long been studied as a risk factor of cardiovascular disease,50, 51 and it has also been suggested as a risk factor for hypertension52 and incident CKD.53, 54, 55, 56, 57 Oxidative stress58, 59 and upregulated inflammation60, 61 have been suggested as direct effects of the high homocysteine levels. Furthermore, several studies have shown an association between hyperhomocysteinemia and DNA hypomethylation in CKD62; however, it is more likely that the methylation disturbances are due to the accumulation of SAH. Plasma SAH levels are often more strongly increased with reduced kidney function than homocysteine levels,63, 64 and SAH acts as a strong inhibitor of most methylation reactions.65 We have previously found that TCA-cycle activity is downregulated in nondiabetic CKD,66 and in the current study we find reduced renal expression of phosphoenolpyruvate carboxykinase in nephrosclerosis. This combination could lead to reduced serine production and a state of reduced substrate for the tetrahydrofolate cycle. We also demonstrate reduced renal expression of 5,10-methylene-tetrahydrofolate reductase, methionine synthase, and betaine-homocysteine methyl-transferase, which are key enzymes for the remethylation of methionine, and consequently reduce methionine levels. Unfortunately, we do not have data on s-adenosyl-methionine and SAH in nephrosclerosis, and the interpretation of the related epigenome literature is also difficult. Several reports indicate that there is a global hypomethylation in CKD,67 and our data show that patients with nephrosclerosis also have the prerequisites for such an epigenetic change. Global hypomethylation has been found in both blood cells and in vascular lesions of patients with atherosclerosis,68, 69 and it is also associated with aging in general.70 However, whether it is a direct facilitator for harmful effects or merely a marker of a generalized epigenetic dysregulation is not well studied. Site-specific and organ-specific information on methylation in various candidate genes is clearly needed, and several studies show that both hyper- and hypomethylation coexist in various diseases.71, 72
Blood pressure control could be substantially impaired by the dopamine and serine disturbances described in our study. Urine tyrosine has previously been found to be reduced in early CKD16, 73 and in patients with end-stage renal disease,74, 75 and this is caused by reduced phenylalanine-hydroxylase enzyme activity,45 also demonstrated in our study. Even though we found no change in renal expression of tyrosine hydroxylase, which is considered the rate-limiting enzyme in catecholamine synthesis, production of L-DOPA could still be hampered in CKD. Tetrahydrobiopterin is an essential cofactor for all amino acid hydroxylases (e.g., phenylalanine-hydroxylase and tyrosine hydroxylase) and nitric oxide synthases. Regeneration of tetrahydrobiopterin from dihydrobiopterin is reduced in CKD leading to reduced tetrahydrobiopterin-dihydrobiopterin ratios,76 which probably are the best functional measures of the tetrahydrobiopterin. We found a significantly reduced expression of dihydropteridine reductase, the main enzyme for regeneration, in nephrosclerosis. Furthermore, regeneration is also connected with the folate cycle, which we also found to be suppressed. The expression of L-DOPA-decarboxylase for conversion of L-DOPA to dopamine was also strongly downregulated in our patients with nephrosclerosis, and mice with reduced renal expression of L-DOPA-decarboxylase demonstrate reduced renal and urine dopamine concentrations, leading to reduced salt and water excretion, activation of renin-angiotensin system, and increased blood pressure.47
Taken together, our data suggest that enzymatic downregulation and metabolite deficiency in the phenylalanine-tyrosine-dopamine axis is found in early hypertensive nephrosclerosis. Disturbances in renal dopamine are linked to hypertension, both by reduced renal L-DOPA uptake or conversion to dopamine,77, 78 and disturbed dopamine D1-like receptor function.79 In humans, a defect in the tubular D1 receptor has been shown in salt-sensitive hypertension.80 The renal dopamine system has also been shown to interact with the renin-angiotensin-aldosterone system in sodium homeostasis and blood pressure regulation in normotensive individuals.81
There are, however, some limitations that need to be discussed. First, although urine is produced in very close proximity to kidney parenchyma, intracellular metabolic changes are not necessarily reflected in urine concentrations. However, recent experimental and clinical studies found the same metabolic perturbations whether using urine samples or kidney cell extracts, and especially amino acids show good correlation between the 2 media.66, 82, 83 Likewise, urine excretion of small molecules, like creatinine and amino acids, is not influenced by reduced GFR per se until the advanced CKD stages. Second, we have defined hypertensive nephrosclerosis from a set of clinical and laboratory criteria known to be rather unspecific, rather than by histological definition based on renal biopsy findings. Third, we could not decide whether our findings were caused by hypertension, reduced GFR, or the combination, but typically patients with nephrosclerosis display both characteristics. This study was exploratory in its nature, focusing on data-based pathway and gene-metabolite enrichment analysis rather than a predefined hypothesis. As such, its strengths lie in hypothesis generation and elucidation of possible pathophysiological disturbances. For definite mechanism, additional animal experiments with a more focused view are needed.
In conclusion, renal gene expression analysis showed reduced amino acid catabolism and synthesis in patients with nephrosclerosis. Urine samples from clinically diagnosed cases with well-preserved eGFR showed significantly reduced excretion of 11 amino acids, among them tyrosine, phenylalanine, dopamine, homocysteine, and serine. Metabolite pathway enrichment analysis revealed downregulation of the phenylalanine-tyrosine-dopamine axis, which regulates natriuresis and blood pressure, due to enzymatic downregulation and metabolite deficiency. We also found disturbances in methionine/homocysteine and serine metabolism, involved in methylation status, endothelial dysfunction, inflammation, and atherosclerosis. Albeit explorative in nature, our combined genomic and metabolomic analysis highlights pathologically perturbed pathways in early-stage hypertensive nephrosclerosis.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The HUNT study is a collaboration between the HUNT Research Center, Faculty of Medicine, Norwegian University of Science and Technology; the Norwegian Institute of Public Health; Nord-Trøndelag County Council; and Central Norway Regional Health Authority. The authors thank the health service and people of Nord-Trøndelag for their endurance and participation.
The SUGAR study was supported by an investigator-initiated research grant from AbbVie, with additional support from the University of Washington Institute of Translational Health Sciences (UL1TR000423), as well as National Institute of Diabetes and Digestive and Kidney Diseases grants R01DK087726 and R01DK087726-S1. None of the funders had any role in study design, data collection, data analysis, interpretation, or writing of the manuscript.
Footnotes
Supplementary Materials and Methods. Methods for the HUNT cohort and SUGAR replication cohort.
Table S1. Variable importance in projection scores.
Table S2. Pathway enrichment.
Figure S1. Pathway enrichment.
Supplementary material is linked to the online version of the paper at http://www.kireports.org/.
Supplementary Material
Methods for the HUNT cohort and SUGAR replication cohort.
Variable importance in projection scores.
Pathway enrichment.
Pathway enrichment.
References
- 1.Jha V., Garcia-Garcia G., Iseki K. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382:260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 2.Wright J.T., Jr., Kusek J.W., Toto R.D. Design and baseline characteristics of participants in the African American Study of Kidney Disease and Hypertension (AASK) pilot study. Control Clin Trials. 1996;16:3s–16s. doi: 10.1016/s0197-2456(96)00081-5. [DOI] [PubMed] [Google Scholar]
- 3.Weisstuch J.M., Dworkin L.D. Does essential hypertension cause end-stage renal disease? Kidney Int Suppl. 1992;36:S33–S37. [PubMed] [Google Scholar]
- 4.Luft F.C. Hypertensive nephrosclerosis—a cause of end-stage renal disease? Nephrol Dial Transplant. 2000;15:1515–1517. doi: 10.1093/ndt/15.10.1515. [DOI] [PubMed] [Google Scholar]
- 5.Meyrier A. Nephrosclerosis: a term in quest of a disease. Nephron. 2015;129:276–282. doi: 10.1159/000381195. [DOI] [PubMed] [Google Scholar]
- 6.Neusser M.A., Lindenmeyer M.T., Moll A.G. Human nephrosclerosis triggers a hypoxia-related glomerulopathy. Am J Pathol. 2010;176:594–607. doi: 10.2353/ajpath.2010.090268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X., Chen W., Li R. Systematic variations associated with renal disease uncovered by parallel metabolomics of urine and serum. BMC Syst Biol. 2012;6(Suppl 1):S14. doi: 10.1186/1752-0509-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedic M., Gethings L.A., Vissers J.P. Label-free mass spectrometric profiling of urinary proteins and metabolites from paediatric idiopathic nephrotic syndrome. Biochem Biophys Res Commun. 2014;452:21–26. doi: 10.1016/j.bbrc.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Wang C., Feng Y., Wang M. Volatile organic metabolites identify patients with mesangial proliferative glomerulonephritis, IgA nephropathy and normal controls. Sci Rep. 2015;5:14744. doi: 10.1038/srep14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J.-N. Prediction of acute cellular renal allograft rejection by urinary metabolomics using MALDI FTMS. J Proteome Res. 2008;7:3597–3601. doi: 10.1021/pr800092f. [DOI] [PubMed] [Google Scholar]
- 11.Blydt-Hansen T.D., Sharma A., Gibson I.W. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am J Transplant. 2014;14:2339–2349. doi: 10.1111/ajt.12837. [DOI] [PubMed] [Google Scholar]
- 12.Sharma K., Karl B., Mathew A.V. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24:1901–1912. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beger R.D., Holland R.D., Sun J. Metabonomics of acute kidney injury in children after cardiac surgery. Pediatr Nephrol. 2008;23:977–984. doi: 10.1007/s00467-008-0756-7. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi K. Use of serum and urine metabolome analysis for the detection of metabolic changes in patients with CKD stage 1 and 2. Nephrourol Mon. 2011;3:164–171. [Google Scholar]
- 15.Duranton F., Lundin U., Gayrard N. Plasma and urinary amino acid metabolomic profiling in patients with different levels of kidney function. Clin J Am Soc Nephrol. 2014;9:37–45. doi: 10.2215/CJN.06000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molnar G. Urinary ortho tyrosine excretion in diabetes mellitus and renal failure Evidence for hydroxyl radical production. Kidney Int. 2005;68:2281–2287. doi: 10.1111/j.1523-1755.2005.00687.x. [DOI] [PubMed] [Google Scholar]
- 17.Posada-Ayala M., Zubiri I., Martin-Lorenzo M. Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney Int. 2014;85:103–111. doi: 10.1038/ki.2013.328. [DOI] [PubMed] [Google Scholar]
- 18.Barrios C., Spector T.D., Menni C. Blood, urine and faecal metabolite profiles in the study of adult renal disease. Arch Biochem Biophys. 2016;589:81–92. doi: 10.1016/j.abb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Kang H.M., Ahn S.H., Choi P. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanada Y., Okuda J., Kato T. The metabolic profile of a rat model of chronic kidney disease. PeerJ. 2017;5:e3352. doi: 10.7717/peerj.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eirin A., Saad A., Tang H. Urinary mitochondrial DNA copy number identifies chronic renal injury in hypertensive patients. Hypertension. 2016;68:401–410. doi: 10.1161/HYPERTENSIONAHA.116.07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T., Liu M., Shang P. Investigation into the underlying molecular mechanisms of hypertensive nephrosclerosis using bioinformatics analyses. Mol Med Rep. 2018;17:4440–4448. doi: 10.3892/mmr.2018.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasuda Y., Cohen C.D., Henger A. Gene expression profiling analysis in nephrology: towards molecular definition of renal disease. Clin Exp Nephrol. 2006;10(2):91–98. doi: 10.1007/s10157-006-0421-z. [DOI] [PubMed] [Google Scholar]
- 24.Krokstad S., Langhammer A., Hveem K. Cohort profile: the HUNT study, Norway. Int J Epidemiol. 2013;42:968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 25.de Boer I.H., Zelnick L., Afkarian M. Impaired glucose and insulin homeostasis in moderate-severe CKD. J Am Soc Nephrol. 2016;27:2861–2871. doi: 10.1681/ASN.2015070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ju W., Greene C.S., Eichinger F. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23:1862–1873. doi: 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong G., Zhang W., Li H. Separate enrichment analysis of pathways for up- and downregulated genes. J R Soc Interface. 2014;11:20130950. doi: 10.1098/rsif.2013.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 29.Xia J., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palermo G., Piraino P., Zucht H.-D. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics type data. Adv Appl Bioinform Chem. 2009;2:57–70. doi: 10.2147/aabc.s3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glassock R.J., Rule A.D. Aging and the kidneys: anatomy, physiology and consequences for defining chronic kidney disease. Nephron. 2016;134:25–29. doi: 10.1159/000445450. [DOI] [PubMed] [Google Scholar]
- 32.Churg JB J., Glassock R.J. 2nd ed. Igaku-Shoin; New York, NY: 1995. Renal Disease: Classification and Atlas of Glomerular Diseases. [Google Scholar]
- 33.Rule A.D., Amer H., Cornell L.D. The association between age and nephrosclerosis on renal biopsy among healthy adults. Ann Intern Med. 2010;152:561–567. doi: 10.1059/0003-4819-152-9-201005040-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremers W.K., Denic A., Lieske J.C. Distinguishing age-related from disease-related glomerulosclerosis on kidney biopsy: the Aging Kidney Anatomy study. Nephrol Dial Transplant. 2015;30:2034–2039. doi: 10.1093/ndt/gfv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lusco M.A., Najafian B., Alpers C.E. AJKD atlas of renal pathology: arterionephrosclerosis. Am J Kidney Dis. 2016;67:e21–e22. doi: 10.1053/j.ajkd.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 36.Marcantoni C., Fogo A.B. A perspective on arterionephrosclerosis: from pathology to potential pathogenesis. J Nephrol. 2007;20:518–524. [PubMed] [Google Scholar]
- 37.Freedman B.I., Iskandar S.S., Appel R.G. The link between hypertension and nephrosclerosis. Am J Kidney Dis. 1995;25:207–221. doi: 10.1016/0272-6386(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 38.Kopp J.B. Rethinking hypertensive kidney disease: arterionephrosclerosis as a genetic, metabolic, and inflammatory disorder. Curr Opin Nephrol Hypertens. 2013;22:266–272. doi: 10.1097/MNH.0b013e3283600f8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Sullivan E.D., Hughes J., Ferenbach D.A. Renal aging: causes and consequences. J Am Soc Nephrol. 2017;28:407–420. doi: 10.1681/ASN.2015121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uesugi N., Shimazu Y., Kikuchi K. Age-related renal microvascular changes: evaluation by three-dimensional digital imaging of the human renal microcirculation using virtual microscopy. Int J Mol Sci. 2016;17:E1831. doi: 10.3390/ijms17111831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalhan S.C., Hanson R.W. Resurgence of serine: an often neglected but indispensable amino acid. J Biol Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowry M., Hall D.E., Hall M.S., Brosnan J.T. Renal metabolism of amino acids in vivo: studies on serine and glycine fluxes. Am J Physiol. 1987;252:F304–F309. doi: 10.1152/ajprenal.1987.252.2.F304. [DOI] [PubMed] [Google Scholar]
- 43.Metcalf J.S., Dunlop R.A., Powell J.T. L-serine: a naturally-occurring amino acid with therapeutic potential. Neurotox Res. 2018;33:213–221. doi: 10.1007/s12640-017-9814-x. [DOI] [PubMed] [Google Scholar]
- 44.Davis S.R., Stacpoole P.W., Williamson J. Tracer-derived and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am J Physiol Endocrinol Metab. 2004;286:E272–E279. doi: 10.1152/ajpendo.00351.2003. [DOI] [PubMed] [Google Scholar]
- 45.Mishra R.C., Tripathy S., Desai K.M. Nitric oxide synthase inhibition promotes endothelium-dependent vasodilatation and the antihypertensive effect of L-serine. Hypertension. 2008;51:791–796. doi: 10.1161/HYPERTENSIONAHA.107.099598. [DOI] [PubMed] [Google Scholar]
- 46.Mandaviya P.R., Stolk L., Heil S.G. Homocysteine and DNA methylation: a review of animal and human literature. Mol Genet Metab. 2014;113:243–252. doi: 10.1016/j.ymgme.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M.Z., Yao B., Wang S. Intrarenal dopamine deficiency leads to hypertension and decreased longevity in mice. J Clin Invest. 2011;121:2845–2854. doi: 10.1172/JCI57324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris R.C., Zhang M.Z. Dopamine, the kidney, and hypertension. Curr Hypertens Rep. 2012;14:138–143. doi: 10.1007/s11906-012-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang M.Z., Harris R.C. Antihypertensive mechanisms of intra-renal dopamine. Curr Opin Nephrol Hypertens. 2015;24:117–122. doi: 10.1097/MNH.0000000000000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoogeveen E.K., Kostense P.J., Jakobs C. Hyperhomocysteinemia increases risk of death especially in Type 2 diabetes. Circulation. 2000;101:1506–1511. doi: 10.1161/01.cir.101.13.1506. [DOI] [PubMed] [Google Scholar]
- 51.Humphrey L.L., Fu R., Rogers K. Homocysteine level and coronary heart disease incidence: a systematic review and meta analysis. Mayo Clin Proc. 2008;83:1203–1212. doi: 10.4065/83.11.1203. [DOI] [PubMed] [Google Scholar]
- 52.Refsum H., Nurk E., Smith A.D. The Hordaland Homocysteine study: a community based study of homocysteine, its determinants, and associations with disease. J Nutr. 2006;136:1731S–1740S. doi: 10.1093/jn/136.6.1731S. [DOI] [PubMed] [Google Scholar]
- 53.Ninomiya T., Kiyohara Y., Kubo M. Hyperhomocysteinemia and the development of chronic kidney disease in a general population: the Hisayama study. Am J Kidney Dis. 2004;44:437–445. [PubMed] [Google Scholar]
- 54.Fox C.S., Gona P., Larson M.G. A multi-marker approach to predict incident CKD and microalbuminuria. J Am Soc Nephrol. 2010;21:2143–2149. doi: 10.1681/ASN.2010010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie D., Yuan Y., Guo J. Hyperhomocysteinemia predicts renal function decline: a prospective study in hypertensive adults. Sci Rep. 2015;5:16268. doi: 10.1038/srep16268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jager A., Kostense P.J., Nijpels G. Serum homocysteine levels are associated with the development of (micro)albuminuria: the Hoorn study. Arterioscler Thromb Vasc Biol. 2001;21:74–81. doi: 10.1161/01.atv.21.1.74. [DOI] [PubMed] [Google Scholar]
- 57.Marti F., Vollenweider P., Marques-Vidal P.M. Hyperhomocysteinemia is independently associated with albuminuria in the population-based CoLaus study. BMC Public Health. 2011;11:733. doi: 10.1186/1471-2458-11-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi F., Zhang A.Y., Li N. Inhibition of ceramide-redox signaling pathway blocks glomerular injury in hyperhomocysteinemic rats. Kidney Int. 2006;70:88–96. doi: 10.1038/sj.ki.5001517. [DOI] [PubMed] [Google Scholar]
- 59.Yi F., dos Santos E.A., Xia M. Podocyte injury and glomerulosclerosis in hyperhomocysteinemic rats. Am J Nephrol. 2007;27:262–268. doi: 10.1159/000101471. [DOI] [PubMed] [Google Scholar]
- 60.Shastry S., Ingram A.J., Scholey J.W., James L.R. Homocysteine induces mesangial cell apoptosis via activation of p38-mitogen-activated protein kinase. Kidney Int. 2007;71:304–311. doi: 10.1038/sj.ki.5002031. [DOI] [PubMed] [Google Scholar]
- 61.Han H., Wang Y., Li X. Novel role of NOD2 in mediating Ca2+ signaling: evidence from NOD2-regulated podocyte TRPC6 channels in hyperhomocysteinemia. Hypertension. 2013;62:506–511. doi: 10.1161/HYPERTENSIONAHA.113.01638. [DOI] [PubMed] [Google Scholar]
- 62.Dwivedi R.S., Herman J.G., McCaffrey T.A., Raj D.S. Beyond genetics: epigenetic code in chronic kidney disease. Kidney Int. 2011;79:23–32. doi: 10.1038/ki.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wollesen F., Brattstrom L., Refsum H. Plasma total homocysteine and cysteine in relation to glomerular filtration rate in diabetes mellitus. Kidney Int. 1999;55:1028–1035. doi: 10.1046/j.1523-1755.1999.0550031028.x. [DOI] [PubMed] [Google Scholar]
- 64.Jabs K., Koury M.J., Dupont W.D., Wagner C. Relationship between plasma S-adenosylhomocysteine concentration and glomerular filtration rate in children. Metabolism. 2006;55:252–257. doi: 10.1016/j.metabol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 65.Clarke S., Banfield K. S-adenosylmethionine-dependent methyltransferases. In: Carmel R., Jacobsen D.W., editors. Homocysteine in Health and Disease. Cambridge University Press; Cambridge, UK: 2001. pp. 63–78. [Google Scholar]
- 66.Hallan S., Afkarian M., Zelnick L.R. Metabolomics and gene expression analysis reveal down-regulation of the citric acid (TCA) cycle in non-diabetic CKD patients. EBioMedicine. 2017;26:68–77. doi: 10.1016/j.ebiom.2017.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chu A.Y., Tin A., Schlosser P. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8:1286. doi: 10.1038/s41467-017-01297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng Q., Huang W., Peng C. Genomic 5-mC contents in peripheral blood leukocytes were independent protective factors for coronary artery disease with a specific profile in different leukocyte subtypes. Clin Epigenetics. 2018;10:9. doi: 10.1186/s13148-018-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aavik E., Lumivuori H., Leppanen O. Global DNA methylation analysis of human atherosclerotic plaques reveals extensive genomic hypomethylation and reactivation at imprinted locus 14q32 involving induction of a miRNA cluster. Eur Heart J. 2015;36:993–1000. doi: 10.1093/eurheartj/ehu437. [DOI] [PubMed] [Google Scholar]
- 70.Gensous N., Bacalini M.G., Pirazzini C. The epigenetic landscape of age-related diseases: the geroscience perspective. Biogerontology. 2017;18:549–559. doi: 10.1007/s10522-017-9695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zawada A.M., Rogacev K.S., Heine G.H. Clinical relevance of epigenetic dysregulation in chronic kidney disease-associated cardiovascular disease. Nephrol Dial Transplant. 2013;28:1663–1671. doi: 10.1093/ndt/gft042. [DOI] [PubMed] [Google Scholar]
- 72.Smyth L.J., McKay G.J., Maxwell A.P., McKnight A.J. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young G.A., Parsons F.M. Plasma and urine amino acid imbalance in chronic renal failure. Proc Eur Dial Transpl Ass. 1970;7:167–174. [Google Scholar]
- 74.Kopple J. Phenylalanine and tyrosine metabolism in chronic kidney failure. J Nutr. 2007;137:1586S–1590S. doi: 10.1093/jn/137.6.1586S. [DOI] [PubMed] [Google Scholar]
- 75.Pena M.J., Lambers Heerspink H.J., Hellemons M.E. Urine and plasma metabolites predict the development of diabetic nephropathy in individuals with Type 2 diabetes mellitus. Diabet Med. 2014;31:1138–1147. doi: 10.1111/dme.12447. [DOI] [PubMed] [Google Scholar]
- 76.Yokoyama K., Tajima M., Yoshida H. Plasma pteridine concentrations in patients with chronic renal failure. Nephrol Dial Transplant. 2002;17:1032–1036. doi: 10.1093/ndt/17.6.1032. [DOI] [PubMed] [Google Scholar]
- 77.Brismar H., Asqhar M., Carey R.M. Dopamine-induced recruitment of dopamine D1 receptors to the plasma membrane. Proc Natl Acad Sci U S A. 1998;95:5573–5578. doi: 10.1073/pnas.95.10.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gill J.R., Jr., Güllner G., Lake C.R. Plasma and urinary catecholamines in salt sensitive idiopathic hypertension. Hypertension. 1988;11:312–319. doi: 10.1161/01.hyp.11.4.312. [DOI] [PubMed] [Google Scholar]
- 79.Jose P.A., Eisner G.M., Felder R.A. Renal dopamine receptors in health and hypertension. Pharmacol Ther. 1998;80:149–182. doi: 10.1016/s0163-7258(98)00027-8. [DOI] [PubMed] [Google Scholar]
- 80.O'Connell D.P., Ragsdale N.V., Boyd D.G. Differential human renal tubular responses to dopamine type 1 receptor stimulation are determined by blood pressure status. Hypertension. 1997;29:115–122. doi: 10.1161/01.hyp.29.1.115. [DOI] [PubMed] [Google Scholar]
- 81.Natarajan A.R., Eisner G.M., Armando I. The renin-angiotensin and renal dopaminergic systems interact in normotensive humans. J Am Soc Nephrol. 2016;27:265–279. doi: 10.1681/ASN.2014100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao L., Dong M., Liao S. Identification of key metabolic changes in renal interstitial fibrosis rats using metabonomics and pharmacology. Sci Rep. 2016;6:27194. doi: 10.1038/srep27194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Z., Li A., Gao J. Kidney tissue targeted metabolic profiling of unilateral ureteral obstruction rats by NMR. Front Pharmacol. 2016;7:307. doi: 10.3389/fphar.2016.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tofallis C. Add or multiply? A tutorial on ranking and choosing with multiple criteria. INFORMS Transactions on Education. 2014;14:109–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods for the HUNT cohort and SUGAR replication cohort.
Variable importance in projection scores.
Pathway enrichment.
Pathway enrichment.