Abstract
Cranial endocasts, or the internal molds of the braincase, are a crucial correlate for investigating the neuroanatomy of extinct vertebrates and tracking brain evolution through deep time. Nevertheless, the validity of such studies pivots on the reliability of endocasts as a proxy for brain morphology. Here, we employ micro‐computed tomography imaging, including diffusible iodine‐based contrast‐enhanced CT, and a three‐dimensional geometric morphometric framework to examine both size and shape differences between brains and endocasts of two exemplar archosaur taxa – the American alligator (Alligator mississippiensis) and the domestic chicken (Gallus gallus). With ontogenetic sampling, we quantitatively evaluate how endocasts differ from brains and whether this deviation changes during development. We find strong size and shape correlations between brains and endocasts, divergent ontogenetic trends in the brain‐to‐endocast correspondence between alligators and chickens, and a comparable magnitude between brain–endocast shape differences and intraspecific neuroanatomical variation. The results have important implications for paleoneurological studies in archosaurs. Notably, we demonstrate that the pattern of endocranial shape variation closely reflects brain shape variation. Therefore, analyses of endocranial morphology are unlikely to generate spurious conclusions about large‐scale trends in brain size and shape. To mitigate any artifacts, however, paleoneurological studies should consider the lower brain–endocast correspondence in the hindbrain relative to the forebrain; higher size and shape correspondences in chickens than alligators throughout postnatal ontogeny; artificially ‘pedomorphic’ shape of endocasts relative to their corresponding brains; and potential biases in both size and shape data due to the lack of control for ontogenetic stages in endocranial sampling.
Keywords: Alligator, diffusible iodine‐based contrast‐enhanced computed tomography, Gallus, geometric morphometrics, micro‐computed tomography, neuroanatomy
Introduction
Fossils are indispensable resources for elucidating ancient biotas and evolutionary dynamics through deep time (Gauthier et al. 1988a; Donoghue et al. 1989; Raff, 2007; Lee & Palci, 2015; Rabosky, 2015). Nevertheless, fossils are inherently limited in the biological information that they can provide. For example, the rarity of soft‐tissue preservation requires paleontologists to frequently utilize anatomical correlates of preserved hard tissues to infer unpreserved soft‐tissue characteristics of extinct taxa (e.g. Witmer, 1995; Wedel & Sanders, 2002; Watanabe et al. 2015). In the field of paleoneurology (Edinger, 1929; Kochetkova, 1978) internal molds of the cranial cavity, called cranial endocasts (hereafter ‘endocasts’), have provided crucial information on the brain morphology of extinct vertebrates (e.g. Jerison, 1963, 1969; Edinger, 1975; Hopson, 1979; Balanoff & Bever, 2017). More recently, the advent of micro‐computed tomography (μCT) imaging has propelled the use of high‐resolution virtual endocasts to reconstruct the neuroanatomy of fossil and extant taxa (Balanoff et al. 2016; and references therein). Modern comparative studies have harnessed this technology to infer large‐scale trends in brain evolution, including the origins of highly encephalized brains in mammals and birds (e.g. Rowe et al. 2011; Balanoff et al. 2013; Neubauer, 2014).
Despite its capacity to provide valuable information, the validity of any anatomical correlate relies on the degree to which it accurately reflects the soft‐tissue structures of interest. Naturally, endocasts overestimate brain sizes due to the intermediary space between the brain and the internal surface of the skull that contains meninges, arteries, venous sinuses, cerebrospinal fluid, and the roots of cranial nerves. Seminal work based on volumetric measurements of brains and endocasts has shown that size differences between them are negligible in extant mammals (Haight & Nelson, 1987; de Miguel & Henneberg, 1999) and birds (Jerison, 1973; Zusi, 1993; Iwaniuk & Nelson, 2002) because their enlarged brains occupy nearly the entire endocranial space. The neuroanatomical literature has historically referenced these volumetric analyses to justify the use of endocasts in studies of mammals, birds, and closely related extinct clades (e.g. Rowe et al. 2011; Balanoff et al. 2013). In contrast, the endocasts of most other vertebrate clades have been considered to be poor representations of brain size because the brains do not closely occupy the endocranial space (Jerison, 1969; Hopson, 1977, 1979; Kochetkova, 1978).
Although endocasts may be a good proxy for brain volume in certain taxa, quantitative assessments of brain–endocast correspondence in shape have been limited. The development of three‐dimensional (3D) semi‐landmark techniques (Gunz et al. 2005; Gunz & Mitteroecker, 2013) has facilitated the use of landmark‐based geometric morphometric (GM) methods to characterize the relatively featureless surfaces of endocasts (Gómez‐Robles et al. 2018; Pareira‐Pedro & Bruner, 2018). This growing use of GM approaches in paleoneurological research prompts an examination into the degree to which endocranial shape reflects true brain morphology. Such investigations will demonstrate which brain regions are accurately or poorly approximated by the corresponding areas on endocasts.
Furthermore, both size and shape correspondences between brains and endocasts likely change throughout ontogeny. For instance, the volume of cerebrospinal fluid in the endocranial cavity increases with age in humans (Wanifuchi et al. 2002; Sherwood et al. 2011), and differential, non‐linear development of the brain and the skull occurs across hominins (Neubauer et al. 2010; Bruner et al. 2015). In crocodylians, Jirak & Janacek (2017) reported changes to the brain–endocast volumetric correspondence throughout ontogeny, with closer correspondence in younger individuals. These observations imply that the inferential power of endocasts differs across not only taxa but also ontogenetic stages. However, the ontogenetic data analyzed by Jirak & Janacek (2017) covered a mixture of multiple species and sexes, preventing a systematic analysis of brain–endocast differences.
Here, we examine brain–endocast correspondence in size and shape during ontogeny in two exemplar extant archosaurs – the American alligator (Alligator mississippiensis) and the domestic chicken (Gallus gallus). Archosauria sensu Gauthier et al. (1988b) (e.g. crocodylians, birds, non‐avian dinosaurs) is of interest to neurobiologists because birds possess highly encephalized brains that evolved independently from those of mammalian groups, including primates (Jerison, 1973; Northcutt & Kaas, 1995; Nieuwenhuys et al. 1998). We employ a suite of modern techniques, including standard μCT imaging, as well as diffusible iodine‐based contrast‐enhanced CT (diceCT) imaging (Metscher, 2009a,b; Gignac & Kley, 2014; Gignac et al. 2016), to create high‐resolution endocasts and brain reconstructions, respectively. For the first time in archosaurs, we employ a high‐dimensional 3D GM approach to characterize the shape of brains and endocasts and their major functional subdivisions. This dataset was then subjected to statistical methods to assess whether (1) the brain and endocranial shapes are distinct in alligators and chickens, (2) the brain–endocast deviation changes throughout ontogeny, and (3) the magnitude of this deviation could overcome real signals of intra‐ and interspecific variation in archosaurian neuroanatomical shape. In light of the results, we formulate important considerations for future comparative neuroanatomical studies on archosaurs.
Materials and methods
Specimens
We obtained postnatal specimens of A. mississippiensis from the Rockefeller Wildlife Refuge (Grand Chenier, LA, USA): four ‘perinatal’ (< 1 year old) individuals, five ‘yearlings’ (1–2 years old), and two ‘juvenile’ specimens (2–3 years old) (n = 11; Table 1). Specimens were euthanized with an overdose of 150 mg kg–1 body mass solution of sodium pentobarbital (Fatal‐PlusTM, Vortech Pharmaceuticals, Dearborn, MI, USA) injected into the intraperitoneal space. This protocol was approved by the Stony Brook University Institutional Animal Care and Use Committee (IACUC, Protocol #236370‐1) and the Oklahoma State University Center for Health Sciences (OSU‐CHS) IACUC (Protocol #2015‐1). Specimens were then decapitated between the third and fourth cervical vertebrae and immediately fixed in 10% neutral‐buffered formalin to prevent postmortem decomposition of the brain. To minimize shape distortion from tissue fixation (Weisbecker, 2012), we fixed the specimens in formalin for over 8 weeks before imaging.
Table 1.
List of sampled specimens with information on age, sex, staining protocol, and neuroanatomical measurements
| Taxon | Age | Sex | Lugol's iodine concentration (% I2KI, w/v) | Stain duration (days) | Brain volume (mm3) | Endocranial volume (mm3) |
|---|---|---|---|---|---|---|
| Alligator | 0–1 year | ♀ | 11.25 | 28 | 520.37 | 742.42 |
| 0–1 year | ♂ | 11.25 | 14 | 657.90 | 760.89 | |
| 0–1 year | ♂ | 7.50 | 14 | 783.90 | 814.49 | |
| 0–1 year | ♂ | 7.50 | 14 | 963.82 | 964.30 | |
| 1–2 years | ♀ | 11.25 | 21 | 1157.20 | 1393.67 | |
| 1–2 years | ♀ | 11.25 | 21 | 1331.76 | 2507.89 | |
| 1–2 years | ♀ | 11.25 | 14 | 1544.82 | 1767.80 | |
| 1–2 years | ♀ | 11.25 | 14 | 1649.50 | 2147.62 | |
| 1–2 years | ♂ | 11.25 | 14 | 2389.41 | 3678.68 | |
| 2–3 years | ♀ | 11.25 | 36 | 2164.22 | 4154.08 | |
| 2–3 years | ♂ | 11.25 | 36 | 2629.69 | 4627.71 | |
| Gallus | 1 day | ♂ | 5.00 | 14 | 541.66 | 901.88 |
| 1 day | ♂ | 3.00 | 14 | 748.15 | 965.36 | |
| 1 day | ♂ | 3.00 | 14 | 756.69 | 939.77 | |
| 1 day | ♂ | 3.00 | 14 | 830.41 | 1061.83 | |
| 1 week | ♂ | 5.00 | 14 | 1007.63 | 1206.21 | |
| 1 week | ♂ | 5.00 | 14 | 1034.74 | 1285.18 | |
| 3 weeks | ♂ | 7.50 | 14 | 1643.88 | 1958.14 | |
| 3 weeks | ♂ | 7.50 | 14 | 1648.04 | 1891.17 | |
| 6 weeks | ♂ | 10.00 | 14 | 2207.13 | 2512.06 | |
| 6 weeks | ♂ | 10.00 | 14 | 2207.51 | 2487.25 | |
| > 8 weeks | ♂ | 10.00 | 22 | 2852.36 | 3257.38 | |
| > 8 weeks | ♂ | 10.00 | 21 | 2856.40 | 3152.19 | |
| > 8 weeks | ♂ | 10.00 | 21 | 3057.25 | 3277.91 | |
| > 8 weeks | ♂ | 10.00 | 21 | 3501.76 | 3769.36 |
The Charles River Laboratory (North Franklin, CT, USA) supplied male G. gallus specimens at 1 day, 1 week, 3 weeks, 6 weeks, and over 8 weeks of age (Table 1). Two individuals were sampled for each age group, with the exception of four individuals at one‐day and over 8 weeks of age (n = 14). These specimens were euthanized at the Charles River Laboratory via cervical dislocation and decapitation, followed immediately by submersion into 10% neutral‐buffered formalin solution. After 2 weeks, the fixed specimens were transported to the American Museum of Natural History (AMNH; New York, NY, USA) wrapped in formalin‐saturated gauze. Upon arrival, the specimens were again submerged in formalin for over 8 weeks before imaging to minimize distortion in brain morphology (Weisbecker, 2012). Ontogenetic sampling of alligators and chickens did not encompass equivalent developmental stages due to the difficulty of euthanizing and fixing A. mississippiensis specimens at substantially larger body sizes.
Virtual endocasts and brain reconstructions
Specimens were scanned with Phoenix v|tome|x μCT scanner (General Electric Company, Fairfield, CT, USA) at the AMNH Microscopy and Imaging Facility. We varied the scan parameter values in an effort to optimize the contrast and resolution of the X‐ray images (Supporting Information Table S1). The creation of virtual endocasts consisted of scanning formalin‐fixed heads of Alligator and Gallus, then processing and volume‐rendering CT images using the Phoenix datos|x 2 reconstruction software v2.3.2 (GE Sensing & Inspection Technologies, Hürth, Germany). For larger specimens requiring multiple scans, separate image stacks were fused using the ‘3D Stitching’ function in imagej (FIJI) v1.49u (Schindelin et al. 2012). In vgstudio MAX v2.2 (Volume Graphics, Heidelberg, Germany), we imported full X‐ray image stacks of each specimen and digitally segmented the endocranial cavity following the protocol outlined by Balanoff et al. (2016). Any impressions of the cranial nerves were removed digitally from segmented regions in transverse view, except for the trigeminal ganglion in frontal view, which allowed the clearest and most consistent delimitation of the endocast and the ganglion. Segmented endocasts were exported in Polygon file format (PLY) using ‘Precise with simplification’ setting.
DiceCT imaging utilizes Lugol's iodine (iodine potassium‐iodide, I2KI) as a contrast agent, rendering soft tissues more radio‐opaque (Metscher, 2009a,b; Gignac & Kley, 2014; Gignac et al. 2016). We used diceCT to create high‐resolution in situ reconstructions of the brain from the same set of specimens used to create endocasts. The size of the specimens informed both the concentration and duration of iodine stains for optimizing the contrast among soft‐tissue types (Table 1; also see Gignac et al. 2016). Although iodine staining has been associated with soft‐tissue shrinkage (Vickerton et al. 2013; Cox & Faulkes, 2014), our CT images show that the brains are in close proximity to the skulls, suggesting that neither formalin fixation nor iodine staining resulted in substantial soft‐tissue distortions (Fig. 1). This result is consistent with a recent assessment of potential shrinkage artifacts in bat brains, where specimens stained shortly after their collection in the field incurred minimal shrinkage effects compared to museum specimens that had been fixed in ethanol prior to staining (Hedrick et al. 2018).
Figure 1.
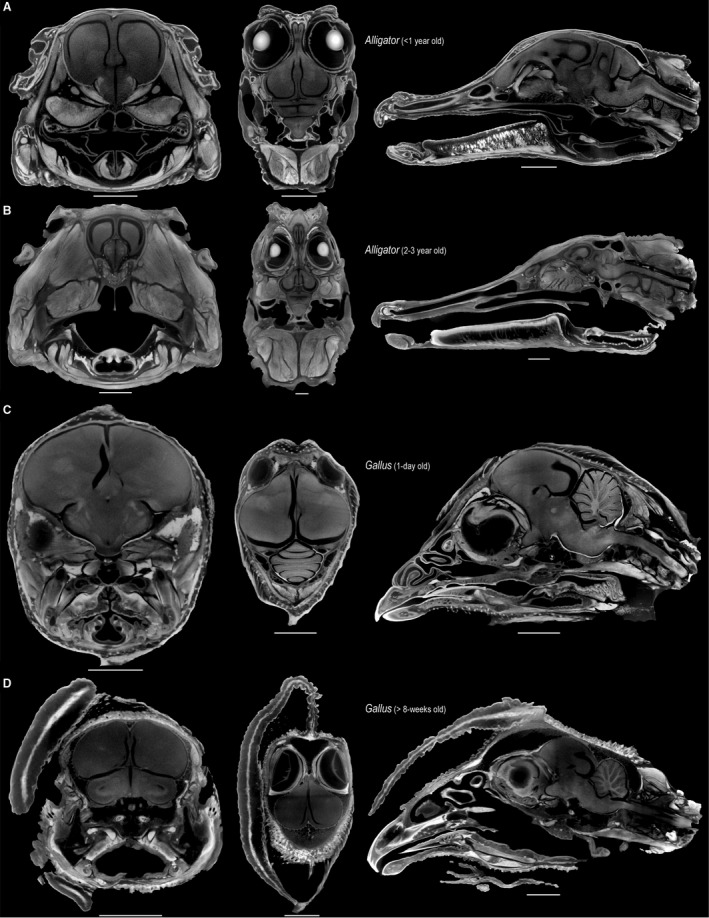
Selected transverse (left), frontal (middle), and sagittal (right) μCT slices through the heads of perinatal Alligator (A), 2‐ to 3‐year‐old Alligator (B), 1‐day‐old Gallus (C), > 8‐week‐old Gallus (D), illustrating the minimal shrinkage artifact from staining neural tissue with high concentrations of Lugol's iodine. Scale: 5 mm (A,C), 10 mm (B,D).
The specimens were submerged in aqueous solutions of Lugol's iodine for certain periods (Table 1) and regularly agitated on a Vortex‐Genie 2 machine (Scientific Industries, Inc., Bohemia, NY, USA) for 60 s every 2–3 days to facilitate the incorporation of iodine into deeper tissue layers. During the staining process, the containers with the specimen and Lugol's iodine solution were stored in the dark to limit loss of stain potency (Gignac et al. 2016). Processing of CT image stacks followed those for unstained specimens, with two exceptions. First, the image stacks were subjected to the enhanced local contrast (CLAHE) script (Saalfeld, 2010) in imagej (FIJI) with the default parameters to increase local grayscale contrast. Secondly, we used the Adaptive Gauss filter in vgstudio MAX with the default parameters to sharpen edges, thus improving edge recognition during digital segmentation. The ventricles were left unfilled in the brain reconstructions for more accurate calculation of brain tissue volume.
Morphological data
We calculated the volume (mm3) of endocasts and brain reconstructions in meshlab v2016.12 (Cignoni et al. 2008; Table 1). To characterize neuroanatomical shape, we used a 3D landmark‐based GM approach. The collection of coordinate data from endocasts is difficult due to the dearth of discrete anatomical landmarks on their surfaces (Neubauer, 2014). For example, landmark configurations in previous studies on avian brain shape included one or two landmarks within major functional subdivisions (e.g. cerebrum, optic lobes, cerebellum), preventing a robust characterization of morphological variation within these structures (e.g. Kawabe et al. 2013, 2015; Marugán‐Lobón et al. 2016). An automated collection and alignment of coordinate data (Boyer et al. 2011, 2015) and the landmark‐free iterative closest point algorithm (Pomidor et al. 2016) are not suitable for this study because the endocasts in both species include surfaces that are interpolations and do not directly represent a structural surface (e.g. unossified regions of the laterosphenoid in immature specimens).
In this study, we established a landmark configuration combining discrete landmarks with semi‐landmarks on curves and surfaces using landmark editor v3.6 (Wiley et al. 2005). The ‘patch’ tool in landmark editor allows the placement of discrete, consistently identifiable landmarks that define the boundaries of major functional brain divisions (i.e. left and right cerebra, left and right optic lobes, cerebellum, medulla) with a specified density of semi‐landmarks sampled within these subdivisions (Table 2). The bilateral landmark data comprised 24 discrete landmarks, 87 curve semi‐landmarks that define the boundaries of major functional divisions (i.e. cerebrum, optic lobes, cerebellum, and medulla), and 114 surface semi‐landmarks that characterize the shape within each division. When placing surface semi‐landmarks on the reconstructions of the cerebella from diceCT data, we visually confirmed that the landmarks were placed on gyri and not within the sulci.
Table 2.
Landmark scheme used in this study. Each major neuroanatomical region was characterized by discrete landmarks, curve semi‐landmarks that define the regional boundaries, and surface semi‐landmarks to characterize the shape within each region. The numbers in parentheses denote number of median and right semi‐landmarks analyzed in the study after performing generalized Procrustes alignment on bilateral data
| Region | No. curve semi‐landmarks | No. surface semi‐landmarks | Discrete landmarks defining each region |
|---|---|---|---|
| Cerebrum | 24 (12) | 78 (39) | Anteriormost median point of the cerebrum on dorsal side. |
| Posteromedial point of the cerebrum on dorsal side. | |||
| Dorsalmost junction point of cerebrum and optic lobe. | |||
| Ventralmost junction point of cerebrum and optic lobe. | |||
| Optic lobe | 28 (14) | 24 (12) | Dorsalmost junction point of cerebrum and optic lobe. |
| Ventralmost junction point of cerebrum and optic lobe. | |||
| Junction point of optic lobe, midbrain, and medulla. | |||
| Junction point of optic lobe, cerebellum, and medulla. | |||
| Cerebellum | 18 (12) | 8 (4) | Anteriormost median point of cerebellum on dorsal side. |
| Anterolateral point of the cerebellum on the dorsal side. | |||
| Posteriormost median point of the cerebellum on dorsal side. | |||
| Posterolateral point of the cerebellum. | |||
| Medulla | 17 (11) | 8 (4) | Anteriormost median point adjacent to midbrain on ventral side. |
| Junction point of optic lobe and medulla. | |||
| Posterolateral point of medulla. | |||
| Posteriormost median point of medulla. |
We used the geomorph r package v3.0.1 (Adams & Otárola‐Castillo, 2013) to perform a generalized Procrustes alignment on the combined Alligator and Gallus data (Gower, 1975; Rohlf & Slice, 1990), with sliding semi‐landmarks minimizing total bending energy (Gunz et al. 2005; Gunz & Mitteroecker, 2013). Procrustes distances, or the sums of squared differences between corresponding landmarks and semi‐landmarks, were calculated to measure shape differences among specimens. In addition to shape, we recorded the centroid sizes of the endocasts and brains from the coordinate data. After alignment, the landmarks and semi‐landmarks on the left side were removed to exclude redundancy in morphological information while avoiding artifacts from aligning one‐sided data of bilaterally symmetric structures (Cardini, 2016a,b). The resulting unilateral shape data comprised 16 landmarks, 49 curve semi‐landmarks, and 59 surface semi‐landmarks (Fig. 2, Table 2, Supporting Information Data S1). We also generated form data (combined shape and size data) by multiplying the shape data with the corresponding centroid size for each specimen. The form difference between brains and endocasts is a metric concomitant with physical distances between each corresponding landmark and semi‐landmark. We computed digitization error by repeatedly collecting landmark data from a 1‐day‐old chicken (10 replications), which accounted for 4.14% of the total shape variation and was considered to be negligible.
Figure 2.
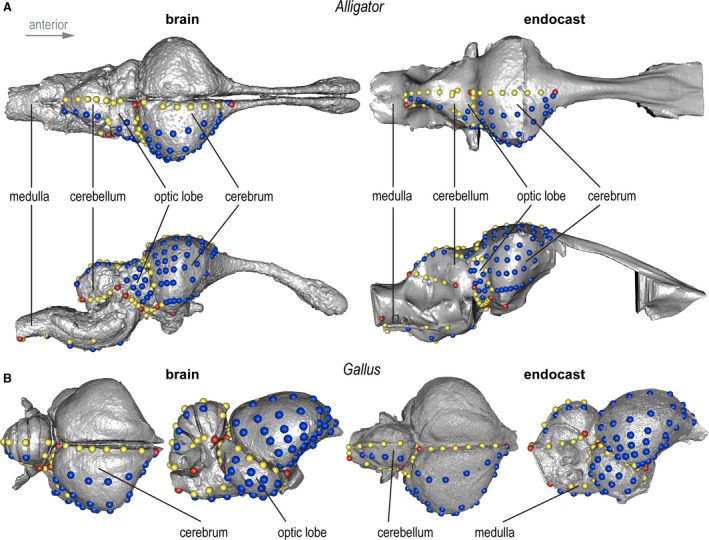
3D landmark configuration used in this study on brains (left) and endocasts (right) on (A) a juvenile Alligator and (B) adult Gallus. Red, yellow, and blue points denote discrete, curve, and surface landmarks and semi‐landmarks, respectively. Images not in scale.
Analysis
All statistical analyses were performed in R v3.2.4 (R Core Development Team, 2018). First, the volumetric correspondence between brains and endocasts was evaluated with a least‐squares regression analysis on brain and endocranial sizes. We then calculated the ratio of brain to endocranial volumes. These ratios were regressed onto log‐transformed brain centroid size (logCS) to identify ontogenetic trends in brain–endocast size differences in Alligator and Gallus.
We employed a series of statistical analyses to investigate brain–endocast shape differences. To visualize the pattern of neuroanatomical variation, we created a morphospace of brains and endocasts using the scores along the first two principal components of shape variation. To test whether endocasts and brains differ significantly in shape, multivariate analysis of variance (manova) was performed on endocranial and brain shapes of chickens and alligators separately, as well as on the combined data, using the procD.lm function in the geomorph package. Localized form and shape differences were visualized by observing the direction and magnitude of changes in landmark positions. The Procrustes distances between corresponding endocasts and brains were plotted against logCS of the brain to examine whether shape differences between brains and endocasts change predictably throughout ontogeny. Least‐squares regression analysis was used to detect the presence of a trend. Next, we used analysis of variance (anova) to evaluate whether the magnitude of brain–endocast shape distances is different from (1) intraspecific variation within Alligator and Gallus separately; (2) interspecific variation between these two taxa; and (3) interspecific differences in endocranial shape among extant birds after conducting a generalized Procrustes alignment on a pooled coordinate dataset. For the latter comparison, we collected shape data from endocasts sampled in previously published studies (Balanoff et al. 2013). In association with these analyses, we constructed box‐and‐whisker plots illustrating the extent of overlap between brain–endocast differences across these hierarchical levels of neuroanatomical shape variation.
Results
Brain‐endocast volume
The brain occupies 52–99% and 60–93% of endocranial volume in Alligator and Gallus, respectively. Despite these volumetric differences, endocranial volume correlates strongly with brain volume in both Alligator and Gallus (Fig. 3A; R 2 > 0.92, P < 0.001). As expected, the regression line for Gallus shows a steeper slope than that for Alligator, indicating that chickens have proportionately larger brains within the endocranial space. Regressing brain‐to‐endocast volume onto logCS of the brain indicates divergent trends in the two taxa (Fig. 3B). Alligator shows greater brain–endocast size deviation in larger specimens (R 2 = 0.395; P = 0.038), implying that the brain occupies a smaller proportion of the endocranial cavity in more mature individuals. Conversely, we find that the proportional brain size within the endocranial cavity increases throughout ontogeny in chickens (R 2 = 0.658; P < 0.001).
Figure 3.
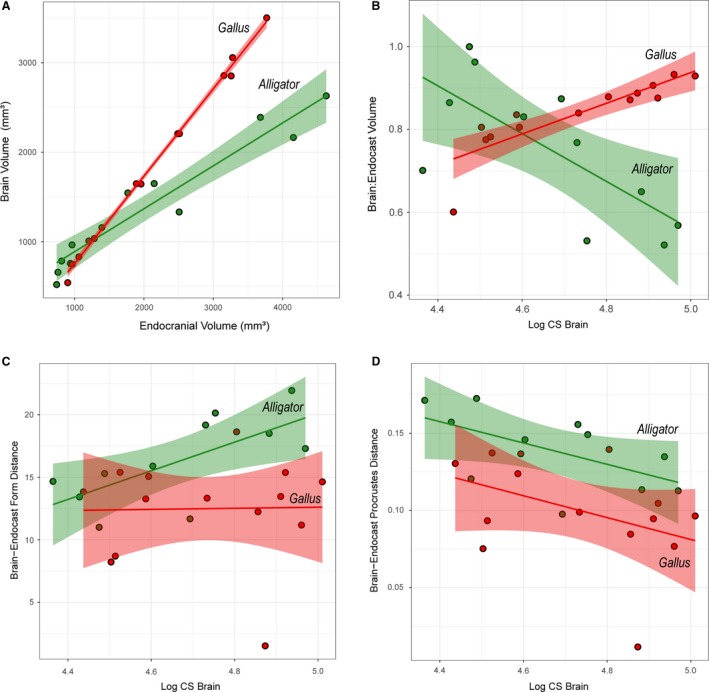
Bivariate plots of brain–endocast correspondence and distances in Alligator (green) and Gallus (red). (A) Brain and endocranial volume (mm3); (B) brain‐endocast volumetric ratio against logCS of brain; (C) form distance against logCS of brain; (D) Procrustes distance (shape difference) against logCS of brain. Solid lines indicate regression lines, and semi‐transparent bands denote 95% confidence intervals.
Brain‐endocast form
Differences in form at each landmark illustrate localized morphological differences between brains and endocasts that reflect true physical distances (Fig. 4). Across taxa and ontogenetic stages, the endocasts generally exhibit less dorsoventral convexity in the cerebrum, greater anterior and ventral extent of the optic lobe region, less posteroventrally flexed cerebellum, and less dorsoventral convexity in the medulla. In Alligator the magnitude of these form differences is generally smaller in larger, more mature individuals, especially in the cerebrum (Fig. 4A). A regression analysis supports the observation that larger Alligator specimens exhibit smaller form differences (Fig. 3C; R 2 = 0.478; P = 0.019). In Gallus, form differences are concentrated in the dorsoventral extent of anterior and posterior margins of the cerebrum, the lateral extent of the cerebrum and optic lobe, and the dorsoventral extent of both the cerebellum and medulla (Fig. 4B). In contrast to Alligator, the magnitude of form differences in chickens does not seem to change throughout postnatal ontogeny (Fig. 3C; R 2 < 0.001; P = 0.943).
Figure 4.
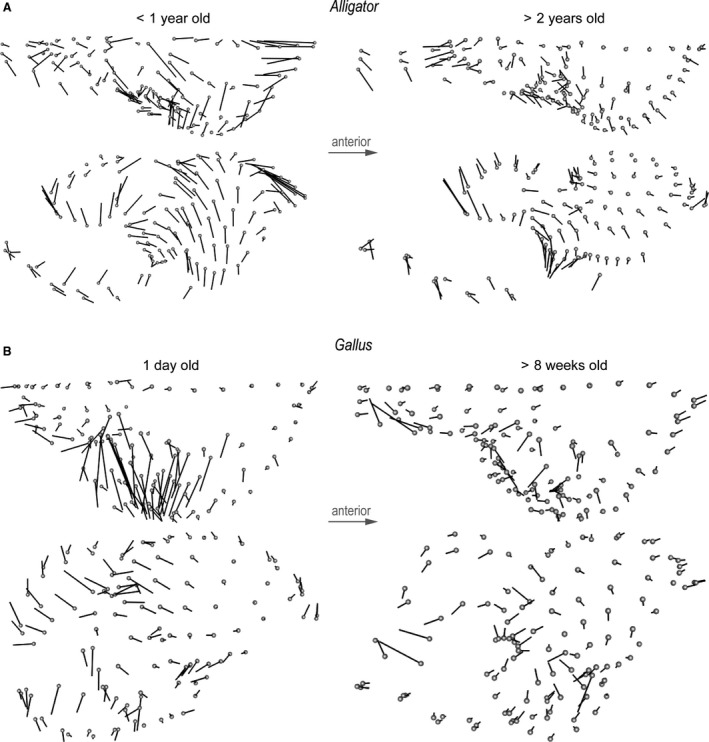
Form differences between brains and endocasts of (A) perinatal and juvenile Alligator and (B) 1‐day‐old and > 8‐week‐old Gallus. Diagrams indicate direction and magnitude of changes in landmark positions from brains to endocasts. The magnitude of vectors reflects true distance.
Brain–endocast shape
A morphospace constructed from the first two principal components (PC) illustrates the taxonomic and ontogenetic trends in neuroanatomical shape (Fig. 5A). PC1 accounts for 59.5% of the total shape variation and separates the neuroanatomical shape between Alligator and Gallus. It is primarily associated with the (1) lateral expansion of the cerebrum; (2) sphericity, relative position, and proportional size of the optic lobe; (3) dorsoventral flexion of the cerebellum; and (4) relative anteroposterior length of the medulla. PC2 accounts for 10.8% of the total shape variation and corresponds to the (1) degree of dorsoventral flexion in the entire brain and endocast; (2) relative size of the cerebrum; (3) position of the optic lobes; and (4) dorsoventral convexity of the cerebellum and medulla along the longitudinal axis. PC2 aligns with ontogenetic changes in brains and endocasts, where the ontogenetic trajectories of Alligator and Gallus are parallel to each other. Brain shape is collectively shifted to occupy areas further along the ontogenetic trajectory than the corresponding endocasts. Endocasts, therefore, exhibit an artifactually ‘pedomorphic’ shape relative to brains of the same age.
Figure 5.
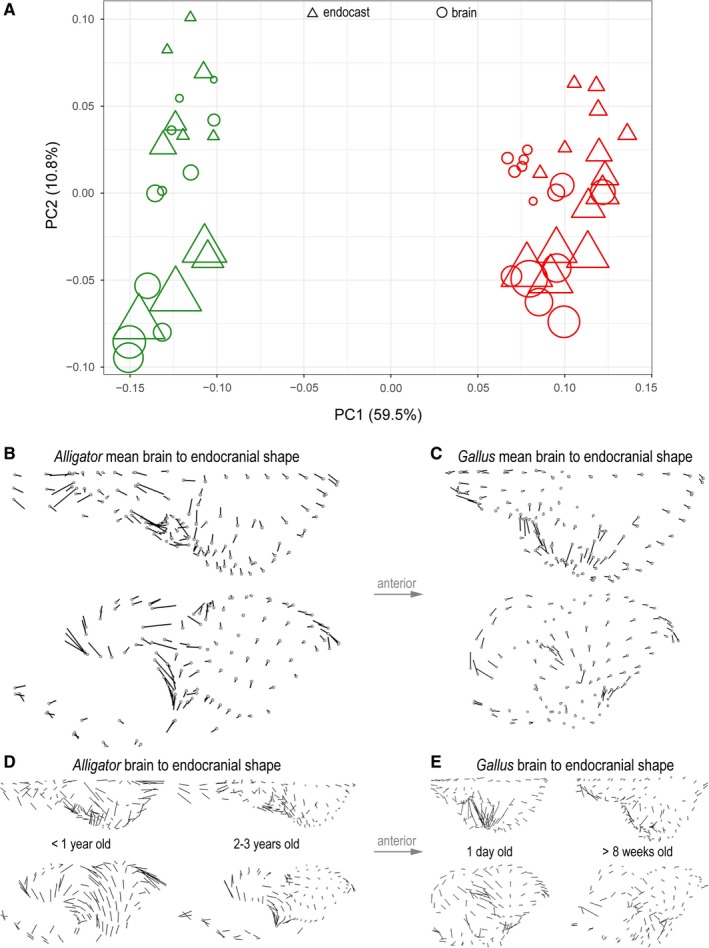
Shape variation in endocranial and brain shape data of Alligator (green) and Gallus (red). (A) Morphospace constructed from PC1 and PC2 of shape. Circles and triangles denote brains and endocasts, respectively. The size of data points corresponds to non‐log‐transformed centroid size. Direction and magnitude of differences in landmark positions from mean brain to mean endocranial shape in Alligator (B) and Gallus (C). Brain‐to‐endocast shape differences in perinatal and more mature specimens in (D) Alligator and (E) Gallus. The magnitude of vectors reflects true shape distance.
In both Alligator and Gallus, the endocasts generally exhibit an anteroposteriorly restricted medulla and less dorsoventral flexion in the hindbrain (Fig. 5B,C). In Alligator the impression of the optic lobe on the mean endocranial shape extends further ventrally than the position of the optic lobe in the mean brain shape (Fig. 5B). Although similar shape divergences remain in the cerebellum and medulla, the divergence in the cerebrum decreases in more mature Alligator specimens (Fig. 5D). In Gallus, the endocasts exhibit greater lateral extent in the cerebrum and optic lobes that are more limited in posterolateral breadth (Fig. 5C). The overall shape divergence seems to decrease in more mature Gallus specimens (Fig. 5E). The manova corroborates that brains and endocasts are significantly different in their mean shapes in Alligator (R 2 = 0.19; P < 0.001) and Gallus (R 2 = 0.15; P < 0.001). The plot of Procrustes distances against logCS of brains and the regression lines (Fig. 3D) suggest that brain–endocast shape differences may gradually decrease throughout ontogeny, but manova fails to reject the absence of a trend in Alligator (R 2 = 0.335; P = 0.261) and Gallus (R 2 = 0.169; P = 0.144).
Comparisons with intra‐ and interspecific variation
We calculated the pairwise Procrustes distances within Alligator and Gallus to represent intraspecific neuroanatomical variation for these taxa, as well as between the brains of Alligator and Gallus specimens to measure the interspecific variation between these two taxa (Table 3). Even with more restricted ontogenetic sampling, the magnitude of brain–endocast shape differences is greater in Alligator than in Gallus (Fig. 6), as confirmed by anova (R 2 = 0.303; P = 0.004). When compared to the intraspecific variation in brain shape within these taxa, the brain–endocast shape differences are comparable (Fig. 6; Alligator: R 2 = 0.026; P = 0.103; Gallus: R 2 = 0.029; P = 0.083). The brain–endocast shape differences are smaller than the interspecific differences between Alligator and Gallus (R 2 = 0.786; P < 0.001) and are generally less than the magnitude of interspecific endocranial variation in crown‐group birds (R 2 = 0.117; P < 0.001), although they partially overlap in the value of Procrustes distances (Fig. 6).
Table 3.
Mean and range of brain–endocast shape differences (Procrustes distance) at multiple levels of variation. To permit comparison of Procrustes distances, the values are based on alignment of pooled data comprising Alligator, Gallus, and interspecific sampling of extant birds. These values correspond to Fig. 6
| Variation type | Mean | Range |
|---|---|---|
| Brain–endocast (Alligator) | 0.142 | 0.097–0.179 |
| Brain–endocast (Gallus) | 0.098 | 0.009–0.137 |
| Brain–endocast (Alligator and Gallus) | 0.118 | 0.009–0.179 |
| Intraspecific variation (Alligator) | 0.122 | 0.071–0.193 |
| Intraspecific variation (Gallus) | 0.113 | 0.047–0.160 |
| Interspecific variation (Alligator, Gallus) | 0.245 | 0.200–0.304 |
| Interspecific variation (Neornithes) | 0.165 | 0.081–0.309 |
Figure 6.
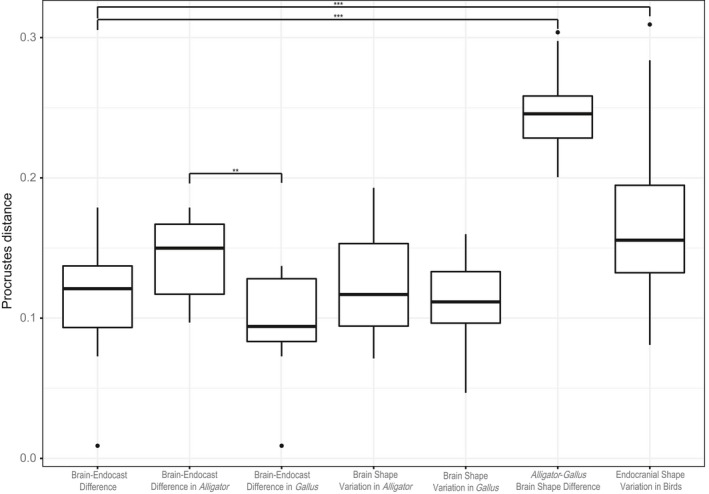
Box‐and‐whisker plots of Procrustes shape distances between corresponding brains and endocasts within Alligator and Gallus (brain‐endocast difference), for intraspecific brain shape variation within Alligator and Gallus (brain‐shape difference), interspecific brain shape variation between Alligator and Gallus (Alligator‐Gallus brain shape difference), and interspecific endocranial shape variation in extant birds (endocranial shape variation in birds). The pooled coordinate data were re‐aligned with data from extant birds to place all specimens within a single shape space to allow comparison of variation. The brackets indicate significant differences in Procrustes distances between two levels (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
Volumetric correspondence
Our study corroborates and extends previous reports on brain–endocast congruence in archosaurs utilizing modern techniques on postnatal ontogenetic series of Alligator and Gallus. In Alligator the brain displays negative allometry relative to endocranial size throughout ontogeny (Fig. 3B), corroborating previous studies (Hopson, 1979; Rogers, 1999; Hurlburt & Waldorf, 2002; Hurlburt et al. 2013). We also find that perinatal alligators exhibit high brain–endocast correspondences in volume (> 90%). These values are consistent with Crocodylus acutus at Stage 28 embryonic stage showing 97.5% brain‐to‐endocast correspondence (Jirak & Janacek, 2017), and are much greater than previously reported for A. mississippiensis (i.e. 67%; Hurlburt & Waldorf, 2002). The brain occupies nearly half of the endocranial volume in the largest Alligator specimens sampled in this study (Fig. 3B), but volumetric correspondence is expected to decrease further in more mature individuals because previous studies have shown that the brain occupies 32 and 29% of the endocranial space in much larger specimens of A. mississippiensis (Hurlburt & Waldorf, 2002; Hurlburt et al. 2013) and Crocodylus niloticus (Jirak & Janacek, 2017), respectively. Although we did not sample somatically mature specimens in Alligator, our study fills a crucial gap in ontogenetic sampling that provides an evidence of consistent reduction in brain–endocast size correspondence during the first few years of life.
In contrast to Alligator, brain size converges towards endocranial size as growth proceeds in Gallus (Fig. 3B). The high proportional brain size in older individuals supports previous studies showing that the brain occupies nearly the entire endocranial space (> 90%) in somatically mature birds (Jerison, 1973; Zusi, 1993; Iwaniuk & Nelson, 2002). We also find that the brain occupies less than 80% of the endocranial cavity in neonatal chickens. Taken together, Alligator and Gallus exhibit divergent allometric trends in brain size relative to endocranial cavity. The precise mechanism for these ontogenetic trends is yet unclear. In the Nile crocodile (C. niloticus), the central nervous system continues to grow indeterminately with body size, where the brain grows more slowly than the spinal cord relative to body size (Ngwenya et al. 2013). This combined effect of indeterminate growth and the brain‐to‐body allometric relationship in crocodylians is consistent with the observed reduction in proportional brain size through ontogeny. Like other birds, Gallus undergoes an abbreviated period of somatic growth that is characterized by a derived allometric relationship between brain and body size (e.g. Jerison, 1973; Tsuboi et al. 2018). Therefore, these contrasting developmental strategies likely underlie these opposing trends in brain–endocast correspondence. Fabbri et al. (2017), for instance, showed that the skull roof closely tracks the shape of adjacent regions on the forebrain and midbrain during early ontogeny that become decoupled later in development. Given this observation, Gallus, and more broadly birds, may maintain high brain–endocast correspondence due to their truncated period of somatic growth. Clarifying the molecular and functional mechanisms that explain the varying degrees of association between skull and brain development will be an important line of research in archosaur neurobiology (e.g. Marcucio et al. 2005, 2011; Young et al. 2010; Hu et al. 2015).
Form and shape correspondence
In both Alligator and Gallus, brain–endocast shape differences are concentrated in the dorsoventral convexity of the cerebellum and medulla, whereas the cerebrum (and optic lobes in Gallus) shows higher correspondences. These non‐uniform deviations in shape across brain regions mirror that in form, indicating that areas with greater physical distances between the brain and endocranial surfaces also represent the regions with greater deviations in shape. The areas of relatively high deviations and close correspondences are consistent with previous studies. In crocodylians, Hopson (1979) noted that the dorsal longitudinal venous sinus and its divisions occupy a substantial portion of the area around the cerebellum and medulla. Conversely, the venous sinus is relatively thin around the cerebrum, allowing a closer correspondence between the cerebrum and adjacent areas of the braincase (Evans, 2005). This anatomical feature may extend to Gallus, where a thicker dorsal longitudinal venous sinus surrounding the hindbrain contributes to the greater morphological deviations in the cerebellum and medulla (Balanoff & Bever, 2017). In many non‐avian dinosaurs, the endocast exhibits a ‘dural peak’ in the cerebellar region (Balanoff & Bever, 2017), which may represent even more size and shape deviations from the actual brain morphology.
Implications for paleoneurology
Our study shows that endocasts are clearly distinct from brains in size, form, and shape, and that the magnitude of this shape difference is comparable to intraspecific variation in brain shape. Although these results seem to suggest that endocasts are poor correlates for brain morphology, the pattern of morphological variation is congruent between brains and endocasts. For instance, endocranial volume is tightly correlated with brain volume within Alligator and Gallus (Fig. 3A), indicating that raw endocranial volume will closely reflect the variation in brain size within a taxon. Whether the same linear relationship can be applied to other archosaur taxa remains to be seen with ontogenetic data from additional species. Similarly, the distribution of endocranial shape variation tightly corresponds to that of brain shape variation. Performing a two‐block partial least‐squares analysis (Rohlf & Corti, 2000) on brain and endocranial shape data indicates strong correlations between them (Alligator: R = 0.965, P < 0.001; Gallus: R = 0.951, P < 0.001). Furthermore, the morphospace shows that the overall distribution of endocranial shape is equivalent to that of brain shape but translated, where endocasts exhibit artifactually ‘pedomorphic’ shapes relative to their corresponding brains (Fig. 5A). Therefore, we expect that principal conclusions drawn from endocranial shape data reflect genuine large‐scale patterns in brain morphology. Nevertheless, we present several considerations and recommendations for mitigating potential artifacts in archosaur paleoneurological research:
Endocasts exhibit relatively poor form and shape correspondences in the hindbrain (cerebellum, medulla) and closer correspondences in the forebrain. Sampling Alligator and Gallus forms an ‘extant phylogenetic bracket’ (sensu Witmer, 1995; Farris, 1983), suggesting that this pattern applies to all archosaurs. In fact, endocasts from non‐avian dinosaurs, such as hadrosaurids, exhibit clear impressions of blood vessels along the lateral surface of the brain anterior to the facial nerve (CN VII), indicating a good correspondence between osteological surface and underlying structures (Evans, 2005). However, these clear features are absent on corresponding surfaces of most of the hindbrain (Evans, 2005). Beyond archosaurs, lungfishes show even more substantial brain–endocast shape differences in the hindbrain (Clement et al. 2015), indicating that the same neuroanatomical regions have poor correspondences across choanates. Taken together, paleoneurological studies should consider the regional differences in the inferential power of endocasts.
The correction factor to convert endocranial volume to brain volume should consider ontogenetic stage. Historically, comparative neuroanatomical studies have used ˜ 50% for converting endocasts of non‐avian reptiles to their corresponding brain volume (Dendy, 1910; Jerison, 1969, 1973; Hopson, 1977, 1979). Although such values are observed in larger alligator specimens, our study indicates that this correction factor severely underestimates brain size for perinatal crocodylians, where the brain occupies ˜ 90% of the endocranial cavity in specimens younger than 1 year of age. Gallus exhibits larger proportional brain volumes, ranging from 60% in neonates up to > 90% in somatically mature specimens. Although endocranial size has been known to reflect brain size in somatically mature birds accurately, the use of raw endocranial volume could overestimate brain size in perinatal specimens by 20% or more.
Merging endocranial with brain data should be avoided. Although we are not aware of any study that combines morphometric data of endocasts and brains, we discourage such a ‘total evidence’ approach to comparative anatomy. Besides exaggerating form and shape variation in the hindbrain, these shape data would artificially incur a developmental signal, where endocasts exhibit ‘pedomorphic’ shapes relative to their corresponding brains.
Ontogenetic stage should be controlled for purely interspecific studies whenever possible. For volumetric data, our results caution against mixing multiple species from various ontogenetic stages because the proportional brain volume varies considerably throughout ontogeny. Merging multiple ontogenetic stages would unintentionally include intraspecific variation that would be spuriously interpreted as interspecific variation. The same issue extends to form and shape data, unless an ontogenetic correction factor could be established through equivalent studies on additional archosaur taxa.
Merging neuroanatomical data from crocodylians and birds should be avoided. Such data will tend to overestimate brain volumes in crocodylians relative to birds, assuming that the results from Alligator and Gallus are applicable to their respective crown groups. As with size, birds have a greater shape correspondence than crocodylians at similar brain sizes. Therefore, endocranial size or shape data from these two groups will distort real variation in brain morphology. To resolve this issue, identifying when particular ontogenetic trends evolved in the archosaur phylogeny is crucial for formulating a clade‐specific correction factor for both size and shape (e.g. Kochiyama et al. 2018; for hominins). Endocasts of stem archosaurian clades seem to show closer associations with external brain structures, suggesting that the patterns observed here in Alligator and previously in Crocodylus (Ngwenya et al. 2013) may be a derived feature of crocodylians (Pierce et al. 2017; and references therein). Until we identify the polarity and evolutionary timing of derived ontogenetic trends in neuroanatomical size and shape, we discourage inferring the ontogenetic stage of extinct archosaur lineages based on an estimated proportional size of the brain (e.g. Hurlburt et al. 2013).
Conclusions
Through the use of μCT imaging and diceCT, we introduce a new neuroanatomical dataset comprising size and shape data from endocasts and brains of the same Alligator and Gallus specimens. By employing a suite of computational methods on 3D GM data, we demonstrate that (1) Alligator and Gallus show discordant ontogenetic trends in volumetric correspondence between brains and endocasts; (2) the brain–endocast shape deviation is greater in Alligator than in Gallus for the ontogenetic stages sampled; (3) brains and endocasts differ significantly in shape, particularly with respect to the dorsoventral flexion of the cerebellum and medulla; and (4) the magnitude of brain–endocast shape difference is comparable to intraspecific variation within these taxa but generally lower than interspecific variation between Alligator and Gallus, as well as among extant birds. While we show that endocasts retain the overall pattern of brain shape variation, we provide several suggestions to mitigate artifacts in neuroanatomical data (see ‘Implications for paleoneurology’). Moving forward, equivalent studies on additional archosaur taxa are necessary to establish clade‐specific ontogenetic trends in brain–endocast correspondence, akin to what has been achieved with research on interspecific allometric trends in brain and body size. Such endeavors will be critical for accurate and precise inferences of brain morphology in paleoneurological studies.
Author contributions
A.W. conceived the study, created the figures, and wrote the manuscript with assistance from all co‐authors. AW., P.M.G., T.L.G., and N.J.K. collected and prepared specimens. A.W. and P.M.G. scanned the specimens and created endocasts and brain reconstructions. A.M.B. provided endocranial reconstructions of modern birds.
Conflict of interest
The authors have no conflict of interest to declare.
Supporting information
Table S1. Parameters used for CT imaging of specimens.
Data S1. Procrustes aligned shape data of brains and endocasts of Alligator and Gallus specimens (plain text file).
Acknowledgements
We thank Ruth Elsey (Rockefeller Wildlife Refuge), Gregory Erickson (Florida State University), David Kay (Oklahoma State University CHS), Broderick Vaughan (Vaughan Gators), and Doug Warner (Charles River Laboratory) for providing and collecting alligator and chicken specimens; Morgan Hill and Henry Towbin (American Museum of Natural History) for assistance with CT imaging; and Isabelle Brenes (Saint Francis Preparatory School; currently Montana State University) for CT data processing. This study was funded by the National Science Foundation (NSF) Graduate Research Fellowship (to A.W.), NSF Division of Environmental Biology (DEB‐1406849 to A.W.; DEB‐1457180 to P.M.G.; DEB‐1801224 to A.W., P.M.G., A.M.B., M.A.N.), EAGER (1450842 to N.J.K., 1450850 to P.M.G.), Mary R. Dawson Predoctoral Fellowship Grant through the Society of Vertebrate Paleontology (to A.W.), Jurassic Foundation (to A.W.), Macaulay Family (to M.A.N.), Richard Gilder Graduate School (to A.W.), the Division of Paleontology at the AMNH (to A.W., M.A.N.), and the Department of Anatomy and Cell Biology at Oklahoma State University, Center for Health Sciences (to P.M.G.)
References
- Adams DC, Otárola‐Castillo E (2013) geomorph: an R Package for the collection and analysis of geometric morphometric shape data. Methods Ecol Evol 4, 393–399. [Google Scholar]
- Balanoff AM, Bever GS (2017) The role of endocasts in the study of brain evolution In Evolution of Nervous Systems: Volume 1, 2nd edn (eds Kaas J, Striedter G.), pp. 223–241. Oxford: Academic Press. [Google Scholar]
- Balanoff AM, Bever GS, Rowe TB, et al. (2013) Evolutionary origins of the avian brain. Nature 501, 93–96. [DOI] [PubMed] [Google Scholar]
- Balanoff AM, Bever GS, Colbert MW, et al. (2016) Best practices for digitally constructing endocranial casts: examples from birds and their dinosaurian relatives. J Anat 229, 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer DM, Lipman Y, St. Clair E, et al. (2011) Algorithms to automatically quantify the geometric similarity of anatomical surfaces. Proc Natl Acad Sci USA 108, 18221–18226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer DM, Puente J, Gladman JT, et al. (2015) A new fully automated approach for aligning and comparing shapes. Anat Rec 298, 249–276. [DOI] [PubMed] [Google Scholar]
- Bruner E, Amano H, de la Cuétara JM, et al. (2015) The brain and the braincase: a spatial analysis on the midsagittal profile in adult humans. J Anat 227, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardini A (2016a) Left, right or both? Estimating and improving accuracy of one‐side‐only geometric morphometric analyses of cranial variation. J Zool Syst Evol Res 55, 1–10. [Google Scholar]
- Cardini A (2016b) Lost in the other half: improving accuracy in geometric morphometric analyses of one side of bilaterally symmetric structures. Syst Biol 65, 1096–1106. [DOI] [PubMed] [Google Scholar]
- Cignoni P, Corsini M, Ranzuglia G (2008) MeshLab: an open‐source 3D mesh processing system. ERCIM News 73, 45–46. [Google Scholar]
- Clement AM, Nysjö J, Strand R, et al. (2015) Brain‐endocast relationship in the Australian lungfish, Neoceratodus forsteri, elucidated from tomographic data (Sarcopterygii: Dipnoi). PLoS ONE 10, e0141277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox PG, Faulkes CG (2014) Digital dissection of the masticatory muscles of the naked mole‐rat, Heterocephalus glaber (Mammalia, Rodentia). PeerJ 2, e448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendy A (1910) On the structure, development and morphological interpretation of the pineal organs and adjacent parts of the brain in the tuatara (Sphenodon punctatus). Philos Trans R Soc Lond B 201, 226–331. [Google Scholar]
- Donoghue MJ, Doyle JA, Gauthier J, et al. (1989) The importance of fossils in phylogeny reconstruction. Annu Rev Ecol Syst 20, 431–460. [Google Scholar]
- Edinger T (1929) Die fossilen Gehirne. Ergebnisse der Anatomische Entwicklungsgeschichte 28, 1–249. [Google Scholar]
- Edinger T (1975) Paleoneurology, 1804–1966: an annotated bibliography. Adv Anat Embryol Cell Biol 49, 12–258. [PubMed] [Google Scholar]
- Evans DC (2005) New evidence on brain‐endocranial cavity relationships in ornithischian dinosaurs. Acta Palaeontol Pol 50, 617–622. [Google Scholar]
- Fabbri M, Mongiardino Koch N, Pritchard AC, et al. (2017) The skull roof tracks the brain during the evolution and development of reptiles including birds. Nature Ecol Evol 1, 1543–1550. [DOI] [PubMed] [Google Scholar]
- Farris JS (1983) Logical basis of phylogenetic analysis In: Advances in Cladistics (eds Platnick NI, Funk VA.), pp. 7–36. New York: Columbia University Press. [Google Scholar]
- Gauthier J, Kluge AG, Rowe T (1988a) Amniote phylogeny and the importance of fossils. Cladistics 4, 105–209. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Kluge AG, Rowe T (1988b) The early evolution of the Amniota In: The Phylogeny and Classification of the Tetrapods, Volume 1: Amphibians, Reptiles, Birds (ed. Benton MJ.), pp. 103–155. Oxford: Clarendon Press. [Google Scholar]
- Gignac PM, Kley NJ (2014) Iodine‐enhanced micro‐CT imaging: methodological refinements for the study of the soft‐tissue anatomy of post‐embryonic vertebrates. J Exp Zool B Mol Dev Evol 322, 166–176. [DOI] [PubMed] [Google Scholar]
- Gignac PM, Kley NJ, Clarke JA, et al. (2016) Diffusible iodine‐based contrast‐enhanced computed tomography (diceCT): an emerging tool for rapid, high‐resolution, 3‐D imaging of metazoan soft tissues. J Anat 228, 889–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Robles A, Reyes LD, Sherwood CC (2018) Landmarking brains In: Digital Endocasts (eds Bruner E, Ogihara N, Tanabe HC.), pp. 115–126. Tokyo: Springer. [Google Scholar]
- Gower JC (1975) Generalized Procrustes analysis. Psychometrika 40, 33–51. [Google Scholar]
- Gunz P, Mitteroecker P (2013) Semilandmarks: a method for quantifying curves and surfaces. Hystrix, Ital J Mammal 24, 103–109. [Google Scholar]
- Gunz P, Mitteroecker P, Bookstein FL (2005) Semilandmarks in three dimensions In: Modern Morphometrics in Physical Anthropology (ed. Slice DE.), pp. 73–98. New York: Kluwer Academic/Plenum Publishers. [Google Scholar]
- Haight JR, Nelson JE (1987) A brain that doesn't fit its skull: a comparative study of the brain and endocranium of the koala, Phascolarctos cinereus (Marsupialia: Phascolarctidae) In: Possums and Oppossums: Studies on Evolution (ed. Archer M.), pp. 331–352. Sydney: Surrey Beatty & Sons. [Google Scholar]
- Hedrick BP, Yohe L, Linden AV, et al. (2018) Assessing soft‐tissue shrinkage estimates in museum specimens imaged with diffusible iodine‐based contrast‐enhanced computed tomography (diceCT). Microsc Microanal 24, 284–291. [DOI] [PubMed] [Google Scholar]
- Hopson JA (1977) Relative brain size and behavior in archosaurian reptiles. Ann Rev Ecol Syst 8, 429–448. [Google Scholar]
- Hopson JA (1979) Paleoneurology In: Biology of the Reptilia, vol. 9 (eds Gans C, Northcutt RG, Ulinski P.), pp. 39–146. New York: Academic Press. [Google Scholar]
- Hu D, Young NM, Xu Q, et al. (2015) Signals from the brain induce variation in avian facial shape. Dev Dyn 244, 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt GR, Waldorf L (2002) Endocast volume and brain mass in a size series of alligators. J Vertebr Paleontol 23 (Supplement to 3), 69A. [Google Scholar]
- Hurlburt GR, Ridgely RC, Witmer LM (2013) Relative size of brain and cerebrum in tyrannosaurid dinosaurs: an analysis using brain–endocast quantitative relationships in extant alligators In: Tyrannosaurid Paleobiology (eds Parrish JM, Molnar RE, Currie PJ, Koppelhus EB.), pp. 134–154. Bloomington: Indiana University Press. [Google Scholar]
- Iwaniuk AN, Nelson JE (2002) Can endocranial volume be used as an estimate of brain size in birds? Can J Zool 80, 16–23. [Google Scholar]
- Jerison HJ (1963) Interpreting the evolution of the brain. Hum Biol 35, 263–291. [PubMed] [Google Scholar]
- Jerison HJ (1969) Brain evolution and dinosaur brains. Am Nat 103, 575–588. [Google Scholar]
- Jerison HJ (1973) Evolution of the Brain and Intelligence. New York: Academic Press. [Google Scholar]
- Jirak D, Janacek J (2017) Volume of the crocodilian brain and endocast during ontogeny. PLoS ONE 12(6), e0178491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe S, Shimokawa T, Miki H, et al. (2013) Variation in avian brain shape: relationship with size and orbital shape. J Anat 223, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe S, Matsuda S, Tsunekawa N, et al. (2015) Ontogenetic shape change in the chicken brain: implications for paleontology. PLoS ONE 10, e0129939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetkova V (1978) Paleoneurology. Washington, DC: VH Winston. [Google Scholar]
- Kochiyama T, Ogihara N, Tanabe HC, et al. (2018) Reconstructing the Neanderthal brain using computational anatomy. Sci Rep 8, 6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MSY, Palci A (2015) Morphological phylogenetics in the genomic age. Curr Biol 25, R922–R929. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, et al. (2005) Molecular interactions coordinating the development of the forebrain and face. Dev Biol 284, 48–61. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Young NM, Hu D, et al. (2011) Mechanisms that underlie co‐variation of the brain and face. Genesis 49, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugán‐Lobón J, Watanabe A, Kawabe S (2016) Studying avian encephalization with geometric morphometrics. J Anat 229, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD (2009a) MicroCT for comparative morphology: simple staining methods allow high‐contrast 3D imaging of diverse non‐mineralized animal tissues. BMC Physiol 9, 1– 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD (2009b) MicroCT for developmental biology: a versatile tool for high‐contrast 3D imaging at histological resolutions. Dev Dyn 238, 632–640. [DOI] [PubMed] [Google Scholar]
- de Miguel C, Henneberg C (1999) Encephalization of the koala, Phascolarctos cinereus . Aust Mammal 20, 315–320. [Google Scholar]
- Neubauer S (2014) Endocasts: possibilities and limitations for the interpretation of human brain evolution. Brain Behav Evol 84, 117–134. [DOI] [PubMed] [Google Scholar]
- Neubauer S, Gunz P, Hublin JJ (2010) Endocranial shape changes during growth in chimpanzees and humans: a morphometric analysis of unique and shared aspects. J Hum Evol 59, 555–566. [DOI] [PubMed] [Google Scholar]
- Ngwenya A, et al. (2013) The continuously growing central nervous system of the Nile crocodile (Crocodylus niloticus). Anat Rec 296, 1489–1500. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R, Donkelaar JHT, Nicholson C (1998) The Central Nervous System of Vertebrates. New York: Springer. [Google Scholar]
- Northcutt RG, Kaas JH (1995) The emergence and evolution of mammalian neocortex. Trends Neurosci 18, 373–379. [DOI] [PubMed] [Google Scholar]
- Pareira‐Pedro AS, Bruner E (2018) Landmarking endocasts In: Digital Endocasts (eds Bruner E, Ogihara N, Tanabe HC.), pp. 127–142. Tokyo: Springer. [Google Scholar]
- Pierce SE, Williams M, Benson RBF (2017) Virtual reconstruction of the endocranial anatomy of the early Jurassic marine crocodylomorph Pelagosaurus typus (Thalattosuchia). PeerJ 5, e3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomidor BJ, Makedonska J, Slice DE (2016) A landmark‐free method for three‐dimensional shape analysis. PLoS ONE 11, e0150368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; Available at https://www.R-project.org/. [Google Scholar]
- Rabosky DL (2015) Challenges in the estimation of extinction from molecular phylogenies: a response to Beaulieu and O'Meara. Evolution 70, 218–228. [DOI] [PubMed] [Google Scholar]
- Raff RA (2007) Written in stone: fossils, genes and evo–devo. Nat Rev 8, 911–920. [DOI] [PubMed] [Google Scholar]
- Rogers SW (1999) Allosaurus, crocodiles, and birds: evolutionary clues from spiral computed tomography of an endocast. Anat Rec 257, 162–173. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, Corti M (2000) Use of two‐block partial least‐squares to study covariation in shape. Syst Biol 49, 740–753. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, Slice D (1990) Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool 39, 40–59. [Google Scholar]
- Rowe TB, Macrini TE, Luo Z‐X (2011) Fossil evidence on origin of the mammalian brain. Science 332, 955–957. [DOI] [PubMed] [Google Scholar]
- Saalfeld S (2010) Enhance Local Contrast (CLAHE) [Software plugin]. Available at http://imagej.net/Enhance_Local_Contrast_(CLAHE) (Accessed 15 November 2015)
- Schindelin J, Arganda‐Carreras I, Frise E, et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood CC, Gordon AD, Allen JS, et al. (2011) Aging of cerebral cortex differs between humans and chimpanzees. Proc Natl Acad Sci USA 108, 13029–13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M, et al. (2018) Breakdown of brain‐body allometry and the encephalization of birds and mammals. Nature Ecol Evol 2, 1492–1500. [DOI] [PubMed] [Google Scholar]
- Vickerton P, Jarvis J, Jeffery N (2013) Concentration‐dependent specimen shrinkage in iodine‐enhanced microCT. J Anat 223, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanifuchi H, Shimizu T, Maruyama T (2002) Age‐related changes in the proportion of intracranial cerebrospinal fluid space measured using volumetric computerized tomography scanning. J Neurosurg 97, 607–610. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Gold MEL, Brusatte SL, et al. (2015) Vertebral pneumaticity in the ornithomimosaur Archaeornithomimus (Dinosauria: Theropoda) revealed by computed tomography imaging and reappraisal of axial pneumaticity in Ornithomimosauria. PLoS ONE 10, e0145168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel MJ, Sanders RK (2002) Osteological correlates of cervical musculature in Aves and Sauropoda (Dinosauria, Saurischia), with comments on the cervical ribs of Apatosaurus . PaleoBios 22, 1–6. [Google Scholar]
- Weisbecker V (2012) Distortion in formalin‐fixed brains: using geometric morphometrics to quantify the worst‐case scenario in mice. Brain Struct Funct 217, 677–685. [DOI] [PubMed] [Google Scholar]
- Wiley DF, Amenta N, Alcantara DA, et al. (2005) Evolutionary Morphing. In Proceedings of IEEE Visualization 2005. Available at http://www.idav.ucdavis.edu/research/EvoMorph.
- Witmer LM (1995) The extant phylogenetic bracket and the importance of reconstructing soft tissue in fossils In: Functional Morphology in Vertebrate Paleontology (ed. Thomason J.), pp. 19–33. New York: Cambridge University Press. [Google Scholar]
- Young NM, Chong HJ, Hu D, et al. (2010) Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development 137, 3405–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusi R (1993) Patterns of diversity in the avian skull In The Skull, Vol. 2: Patterns of Structural and Systematic Diversity (eds Hanken J, Hall BK.), pp. 391–437, Chicago: University of Chicago Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Parameters used for CT imaging of specimens.
Data S1. Procrustes aligned shape data of brains and endocasts of Alligator and Gallus specimens (plain text file).


