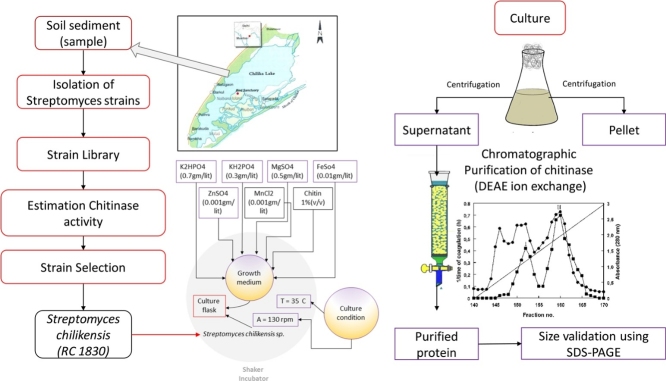
Graphical abstract
Keywords: Streptomyces, Chitinase, Halo tolerant, Chio oligosaccharides
Abstract
An extracellular thermo-alkali stable chitinase was obtained from Streptomyces chilikensis RC1830, a novel actinobacterial strain isolated from the sediments of Chilika lake, India. Purification of the enzyme was carried out by concentrating the enzyme with centrifugal device followed by chromatographic separation by DEAE Sepharose ion exchange resin.The molecular weight of the enzyme was 10.5 kDa as determined by SDS-PAGE. The optimum pH and temperature for the partially purified chitinase was pH 7 and 60 °C. The chitinase showed 40% activity at pH 11 after 24 h exposure at room temperature. The chitinase exhibited Km and Vmax values are 0.02 mM and 3.184 mol/min/mg of enzyme respectively. The 6 residue N-terminal sequence of the enzyme was not found similar to any of the reported chitinase enzyme. Based on the SDS PAGE, zymogram analysis, activity assays and other characteristics, it is proposed that the purified enzyme from S.chilikensis RC1830 is a chitinase.
1. Introduction
Chitin is a polymer and made up of consists of N-acetylglucosamine by β-(1 → 4) glycosidic bond to construct poly(β-(1 → 4)-N-acetyl-d-glucosamine (GlcNAc). It is the second most abundant biopolymer existing in nature. Being produced in 1010-1011 tons every year. It requires chitinase enzymes for its breakdown. Improvement of chitinase production has the potential to broaden the range of practicable biomass conversion and lowering the environmental impact of chemical-based industries. In recent years a lot of knowledge regarding various prospects of chitinase are known while challenges still remain related to economical use of chitinases at industrial scale.
Chitin and its derivatives are of commercial and biotechnological interest because of their wider range of biological activities (Bhushan and Hoondal 1998).The enzymes responsible for chitin degradation and modification are chitinases (3.2.1.14), which are found in a variety of organisms such as bacteria, fungi, actinomycetes, yeasts, plants, protozoans, coelenterates, nematodes, molluscs, arthropods and also in human beings. Chitinases have received increased attention due to their wider range of biotechnological applications, especially in the bio control of fungal phyto pathogens (Mathivanan et al.1998) and harmful insects (Mendonsa et al. 1996; Pinto et al.1997).They have also been used in the preparation of sphaeroplasts and protoplasts from yeast and fungal species (Peberdy 1985 and Mizuno et al. 1997), which can be used further for strain improvement (Kelkar et al. 1990). Other applications of chitinases are bioconversions of chitin waste to single cell protein and ethanol (Vyas and Deshpande 1991), and fertilizer [1]. Speci¢c sized oligo-mers have been synthesized by transglycosylation activites of chitinases (Kadowaki et al. 1997). Chitinase colloidal gold- labelled complexes have been used for immuno-cytochemical and cytochemical localization of chitin in biological sys- tems (Manocha and Zhonghua 1997). Industrial applications of chitinases have been governed mainly by key factors such as cost of production, shelf-life stabilities and improvement in enzyme properties by immobilization. Chitinolysis is performed by three separate enzymes: the endochitinases, which produce multimers of N-acetyl-glucosamine (NAG), the exochitinases, which release soluble low molecular weight dimers, and chitobiase which hydrolyses chitobiose to NAG (Ulhoa and Peberdy 1992). The properties of the immobilized enzyme, the kinetics of chitin degradation and the shelf-life stabilities of chitinase are also reported.
Chitinases are widely distributed in nature and play an important role in the degradation of chitin. They serve a variety of other functions including morphogenesis, defense and pathogenesis [2], and have been reported previously in different micro-organisms, such as fungi (Ulhoa and Peberdy 1992; Fenice et al. 1998; Sakurada et al. 1998) and bacteria (Sakai et al. 1998), and streptomycetes (Neugebauer and Wilson 1967; Gupta et al. 1995; Gomes et al. 1999, 2000a, b).
Enzymes belonging to Chitinase family are classified on the basis of individual mode of hydrolysis of the enzyme. Generally chitinases are random splitting hydrolases that recognise the O-glycosyl linkage between chito-saccharide residues for catalysis. Endochitinases hydrolyse internal O-glycosyl linkages to break chitin polymers into oligosaccharides, while exochitinases function by splitting only terminal GlcNAc residues or dimers from the reducing end of the chitin polymer. Collective action of exochitinases and endochitinases leads to the degradation of Chitin polymers into soluble N-acetyl d-glucosamine units. Bacterial chitinases are primarily produced to digest chitin to utilize it as a carbon and energy source.
Actinomycetes, particularly Streptomyces, are well known to produce various antifungal agents that inhibit several pathogenic fungi [3,4] and antibacterial agents [5]; Zhang et al.
The ability to hydrolyze chitin as the sole carbon source is an important characteristic of Streptomyces sp. and thus these are regarded as the major producers of chitinases in soil. Different chitinase enzymes have been purified from diverse Streptomyces sp. Four chitinases were purified from Streptomyces olivaceoviridis, showing multiplicity of chitinases in Streptomyces [6]. This multiplicity of chitinase gene was reported after cloning of chi A, chi B and chi C from S. lividans (Miyashita et al., 1991). A family 19 chitinase, chi C, was reportedly found in Streptomyces sp, not reported earlier (Fuji and Miyashita, 1993). Both families of chitinases (family 18 and 19) are reported from the members of genus Streptomyces. All reported Streptomyces chitinases have molecular weight ranging from 60-20.5 kDa.
The organism Streptomyces chilikensis RC1830 was reported earlier as a novel halophilic strain from Chilika lake of Odisha, India (Ray et al., 2013). In the present study, purification and characterization of a low molecular weight chitinase is being reported for the first time from this strain of Actinobacteria.
2. Material and methods
2.1. Media, chemicals, reagents
All microbiological media, chemicals for buffers and components of TLC (thin layer chromatography) like spray reagents (aniline diphenyl amine) were purchased from Hi Media, Pvt. Ltd. Mumbai, India; Merck, Germany and SD Fine, India. All the chromatography resins were purchased from Sigma-Aldrich, USA. Chito oligo sachharides were purchased from Sigma-Aldrich, USA and MEGAzyme, Ireland. Centrifugal devices for concentration of protein were purchased from PALL life sciences, USA. Substrates such as glycol chitin, N-acetyl glucosamine, Di N-acetylchitobiose, Tri-N-acetyl-chitotriose were obtained from Sigma-Aldrich, Germany. Penta-N- acetyl chito pentose, Hexa-N-Acetylchitohexaose were obtained from Megazyme (Megazyme International, Ireland). All the substrates were dissolved in deionized water and stored at −20 °C.
2.2. Preparation of colloidal chitin media
5 g of powdered chitin (Hi Media) was acidified with 60 ml of conc. HCl (12 N) for 1–2 h followed by addition of ice-cold distilled water till a white precipitate was obtained. The filtered chitin precipitate on filter paper was washed with ice-cold water several times (20 times) until the pH of the suspension was 5. At pH 5, the suspension was filtered and dried in hot air oven at 50-60 °C. Dried chitin precipitate was weighed and suspended in distilled water to make 4–5% (w/v) of colloidal solution and autoclaved at 121 °C at 15 lb for 15 min. This solution after sterilization was preserved for several months and was added to the growth media
2.3. Growth medium of the organism
Culture strain of Streptomyces chilikensis RC 1830 (= JCM 18411T = DSM 42072T) was isolated from the sediments of Chilika lake. The organism was identified using polyphasic taxonomical methods (Ray et al., 2013). For chitinase production, S. chilikensis was cultivated in 500 ml Erlenmeyer flasks containing 250 ml of production medium consisting of (g/L) K2HPO4, KH2PO4, MgSO4, MnCl2, FeSO4, and ZnSO4 at pH 7 under shaking condition at 30 °C. Chitinase activity in RC1830 was induced by the addition of colloidal chitin (0.5% w/v) to the growth medium (mineral salt medium; MSM). 50 ml of mineral salt medium with and without chitin were prepared separately. Pre-culture of RC 1830 was grown in 50 ml of seawater (50% v/v) LB (50% v/v) for 48 h. 1% (w/v) (containing approx.1.2–1.8 × 106 cells/ml) of pre-culture was inoculated to flasks containing media with and without chitin, separately and incubated at 30 °C in shaking condition. Aliquots of 2 ml were taken out at different time intervals and analyzed for enzyme activity, protein content and viable cell count as well.
2.4. Chitinase enzyme production
Pre-culture of RC 1830 was inoculated into 2 L flasks containing 500 ml minimal media with 0.5% colloidal chitin and incubated for 48–72 hours at 30 °C for with continuous agitation. Cells were harvested by centrifugation at 6500 g for 20 min at 4 °C. The pellet was discarded and the supernatant (approx. 750 ml) was used for enzyme purification.
2.5. Enzyme assay
Chitinase enzyme activity was measured according to a modified protocol originally developed by [7] 400 μl of enzyme solution was added to 400 μl of colloidal chitin and incubated at 37° for 2 h. The reaction mixture was centrifuged and the supernatant was collected. 100 μl of 0.8 M potassium tetraborate was added to 400 μl of supernatant and boiled for 3 min. To this, 1 ml of Dimethyl amino benzaldehyde solution was added and incubated at room temperature for 30 min to 1 h till pink colour develops. Different concentrations of NAG were used for standard curve. Absorbance was measured at 585 nm on UV Vis Spectrophotometer (Shimadzu 1800). 1 Unit of chitinase enzyme activity was measured as described by Reissig et al. [7]
2.6. Protein estimation
The protein was estimated by Bradford method (Bradford et al., 1976). 100ul of enzyme solution was added to 900ul of Bradford reagent. Different concentrations of BSA (bovine serum albumin) were used for standard curve. Absorbance was measured at 595 nm on UV Vis Spectrophotometer (Shimadzu 1800).
2.7. Partial purification of chitinase
For concentration of the chitinase centrifugal device of 3 kD size (Centrifugal Devices, PALL corporation, USA) were used. Culture supernatant was added to the upper chamber of the centrifugal device and centrifuged at 1500 g for 3–4 h. 750 ml of culture supernatant was concentrated 10 times.
The concentrated enzyme extract was then subjected to ion exchange chromatography with DEAE Sephacel and DEAE Sepharose (Sigma-Aldrich, USA) separately. After loading the enzyme concentrate, 2 ml of flow through was collected in 10 fractions in micro centrifuge tubes. Column was washed again with 10 ml of 50 mM Tris HCl, pH 7 without NaCl. Five fractions of 2 ml aliquots were collected and designated as wash. The enzyme bound in column was eluted with a step wise increase of NaCl concentration in 5 ml with 100, 200, 300, 400, 500, 600 and 700 mM NaCl. Total 30 fractions of 2 ml each were collected and stored at 4 °C until further use. The eluted fractions were then checked for total protein estimation (Bradford method) and chitinase enzyme activity at 585 nm. The fractions were also checked on SDS PAGE (by Coomasie Brilliant Blue).
2.8. SDS-PAGE and activity staining of chitinase enzyme
Detection of chitinase activity on gel electrophoresis was carried out according to the method described by Liau and Lin [8]. The enzyme solution (containing 5 μg protein) was loaded on to 12% SDS- PAGE prepared by Laemmli’s method [9], containing 0.01% (w/v in water) glycol chitin added during preparation of the stacking and resolving gel. After electrophoresis the gel was immersed in 0.1 M sodium acetate buffer (pH 5) containing 1% (w/v) deionized TritonX-100 for 10–15 min. The gel was then washed with distilled water and incubated overnight at 37 °C with 0.1 M sodium acetate buffer (pH 5). After incubation, the gel was transferred to a staining solution containing CBB G-250 (0.0.025% w/v) in 10% (v/v) acetic acid and 25% (v/v) isopropanol. The gel was then de stained with 15% (v/v) methanol and 10% (v/v) acetic acid.
2.9. N–terminal amino acid sequence analysis of purified 10.5 kDa chitinase
For N-terminal amino acid sequencing, purified chitinase was run on SDS-PAGE and electro transferred to poly vinylidene- difluoride (PVDF) membrane (GE healthcare, India) using electro blotting apparatus (Bio-Rad, Richmond, CA) with Tris-Glycine buffer (TGB) transfer buffer (pH 8.3) at 100v/0.01amp for 30 min. The membrane was stained with staining solution for 10 min and distained with de staining solution with several changes. The blotted band was cut and then sent for sequencing.
3. Characterization of the enzyme
3.1. Effect of temperature on chitinase enzyme activity and stability
The optimal temperature for chitinase enzyme activity was determined by incubating 400ul of partially purified chitobiosidase and chitin deacetylase enzyme solutions with 0.5% (w/v) colloidal chitin and NAG (50 μmole), respectively. The reaction mixtures were incubated at 20, 30, 40, 50, 60, 70, 80 °C separately. The mixtures were then assayed for chitinase activity.
3.2. Effect of pH on chitinase enzyme activity and stability
The optimum pH for chtinase activity was determined by using buffers with pH ranging between 3.0 and 11.0. 400ul of partially purified chitinase was incubated with 0.5% (w/v) colloidal chitin. The reaction mixtures were incubated with 400ul of potassium phosphate buffer for pH 7, Tris Cl buffer for pH 8, citrate buffer for pH 5 and 6 for 2 h, separately. The mixtures were assayed for chitinase activity by standard assay methods as detailed earlier.
3.3. Effect of metal ion, and surfactants on chitinase enzyme activity and stability
The effect of metal ion and other inhibitors were studied by incubating the enzyme solution with different metal ion solution (10 mM).
3.4. Effect of different substrates on chitinase enzyme activity
Different substrates including carboxymethyl cellulose, colloidal chitin, starch, xylan were tested to determine the substrate binding of the chitinase (chitobiosidase) enzyme. 5 ml of partially purified chitobiosidase enzyme was added to 5 ml of 1% (w/v) colloidal chitin, 1% (w/v) starch, 1% (w/v) CMC and 1% (w/v) xylan (5 mg substrate/ mg protein) (Chiba et al., 2008). The mixture of enzyme-substrate was incubated at 4 °C with gentle agitation for 24 h to allow for binding of the enzyme to the substrate. After centrifugation at 11,000 x g for 20 min, the supernatant was discarded and the pellet water washed to remove any unbound contaminating proteins. The chitinase enzyme was eluted by incubation at 30 °C on an orbital shaker). The sample was centrifuged (11 000x g, 20 min) and the pellet was discarded. Aliquots of 200ul were taken out at 0 h, 30mn, 1 h, 2 h, 6 h and 24 h time interval and assayed for chitinase enzyme (NAG release assay).
3.5. Analysis of the hydrolytic product using thin layer chromatography (TLC)
Chitinase activity was checked by using chito-oligosaccharide substrates Diacetyl chitobiose (GlcNAc)2 (stock, 2.5 mM), N,N,N-Triacetyl chitotriose (GlcNAc)3 (stock, 2.5 mM), OF N,N,N,N,N-Pentacetyl chitopentaose (GlcNAc)5 (stock, 1 mM), and N,N,N,N,N,N-Hexacetyl chitohexaose (GlcNAc)6 (stock, 1 mM) (Sigma Aldrich, Germany).
The reaction mixture consists of 200 μl (0.2 mM working conc.) and 200ul of enzyme solution (2 Units/mg protein). The enzyme substrate mixture was incubated at 37 °C for 0, 5 s, 15 s, 30 s, 1 min, 5 min, 10 min, 15 min, 30 min and 1 h separately. Enzyme mixture with and without substrate served as negative control. The reaction was stopped by boiling the mixture at 100 °C and then concentrated using a vacuum evaporator (Eyela, Japan). TLC analysis was carried according to the method of Tanaka et al., 1999. 20 μl of reaction mixture was spotted on TLC plates (Silica gel 60, F 254 (20 x 20 cm); E. Merck, Darmstadt, Germany) using micro-capillary (Drummond, Scientific company, USA), alongwith respective chito-oligosaccharide standards. The plates were air dried. Mobile phase (butanol:methanol:ammonia:water (5:4:2:1)) was allowed to run along the TLC plate till solvent front reached more than 3/4th of its length after which the plate was marked and dried. The plates were then developed by spraying with developing reagent: aniline diphenylamine reagent (Appendix I) and heated at 120 °C for 10–15 minutes. Dark brown-greenish spots were identified and Rf value was compared to that of standards.
3.6. Kinetic characterization
Kinetic experiments were conducted with substrate colloidal chitin 1 mM dissolved in sodium phosphate buffer at 37 °C. The Michaelis-Menten constant (Km) and maximum velocity (Vmax) were determined by Lineweaver Burk plot analysis.
4. Results
4.1. Protein purification and molecular weight determination
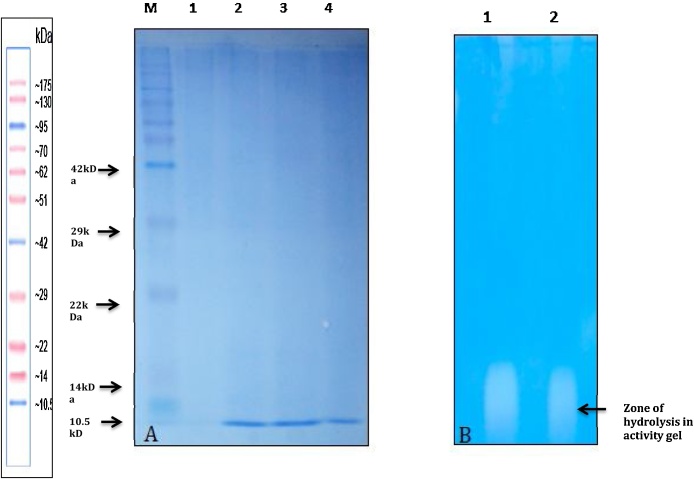
The chitinase enzyme from S. chilikensis RC1830 was concentrated by using centrifugal device (3 kD) to yield the sample that was used as starting material for column purification. The concentrated enzyme sample was further purified using DEAE-Sephacel ion exchange chromatography. A 5.3 fold purification was achieved with 29% recovery of chitinase enzyme activity of 1210 (±2) IU (Table 1). The partially purified chitinase preparation was represented in the form of a single band on SDS-PAGE (Fig). The molecular weight of the same was estimated to be 10.5 kDa by SDS-PAGE. Activity staining revealed the presence of a zone of hydrolysis that corresponded with the SDS-PAGE result (Fig. 1)
Table 1.
Purification table for the partial purification of chitinase (10.5 kD) from Strepromyces chilikensis RC1830.
| Purification steps | Total activity (U) | Protein (mg) | Specific activity (U/mg) | Yeild (%) |
Purification (fold) |
|---|---|---|---|---|---|
| Crude supernatant | 6053.4 | 540 | 11.21 | 100 | |
| Concentrated by molecular cutter (3 kD) | 4035.6 | 180 | 22.52 | 66 | 2 |
| DEAE Sephacel | 1210.68 | 20 | 60.53 | 29 | 5.3 |
Fig. 1.
(A) SDS-PAGE (15%) showing the protein band at 10.5 kD. and (B) showing activity staining of the enzyme at 10.5 kD The samples collected from DEAE Sephacel column chromatography were analysed on a SDS-PAGE. 80 μl of sample was mixed with 5X SDS PAGE loading buffer with beta mcp and boiled for 3 mintues followed by storing on ice. The samples (70 μl) were then loaded onto the gel. The samples were electrophoresed overnight at 50 V. The gels were then stained with CBB-R-250 for 4 h and destained. Lane 1: crude sample, lane 2: concentrate sample, lane 3 and lane 4: partially purified sample by DEAE Sephacel. Samples showed prominent bands .10.5 kD. These samples also showed significant chitinase activity. Thus these bands may be probable the smallest chitinase (chitobiosidase) enzyme.
RC1830 grown in Mineral salt medium with colloidal chitin. The culture supernatant was concentrated and loaded on SDS-PAGE with 0.1% glycol chitin. After electrophoresis gel was washed with triton X-100 and incubated with potassium phosphate buffer (pH6) overnight, Stained in CBB G-250 and distained. The formation of clear zones ascertained the presence of chitinolytic enzyme.
4.2. Temperature and pH optima and stability
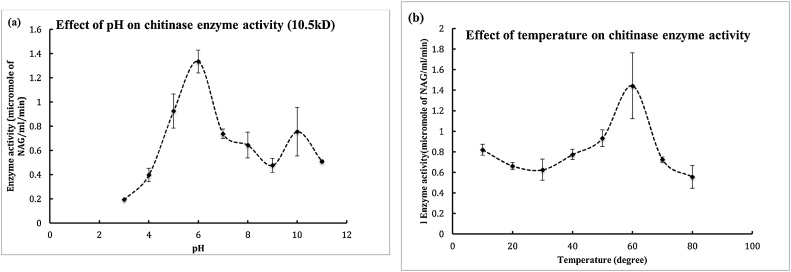
The purified chitinase was active between pH 5–8 with an optima of 7. The enzyme was stable till pH 11 with retention of 40% activity after overnight exposure of the enzyme to various pH buffers at room temperature (Fig. 2 (a) and (b)). The enzyme exhibited temperature optima of 60 °C. Optimum temperature for activity and stability of the chitinase enzyme was studied over a range of temperature 20–80 °C. It showed no activity at 20 °C, while maximum activity was observed at 60 °C. Up to 20% enzyme activity was retained till 80 °C.
Fig. 2.
(a) The partially purified enzyme was checked for its activity at different incubation temperature with the substrate. Briefly, the active fractions of DEAE Sephacel and DEAE Sepharose were pooled together. 100 μl of this enzyme was mixed with 100 μl of appropriate pH buffer i.e 50 mM sodium acetate buffer, 50 mM citrate buffer, 50 mM Tris HCl buffer, 50 mM glycine NaOH buffer(followed by 200 μl of substrate (1% clloidal chitin).The reaction mixture was then incubated at 37 for 2 h. The rest process was carried out according to standard assay conditions i.e after incubation the reaction mixture was centrifuged, the supernatant was collected and 100 μl of potassium tetra borate was added to it followed by boiling for 3 min. To this DMAB reagent was added at RT the colour development was measured at 585 nm. (b) The partially purified enzyme was checked for its activity at different incubation temperature with the substrate. Briefly, the active fractions of DEAE Sephacel and DEAE Sepharose were pooled together. 100 μl of this enzyme was mixed with 100 μl of water followed by 200 μl of substrate (1% colloidal chitin).The reaction mixture was then incubated at 37 for 2 h. The rest process was carried out according to standard assay conditions i.e after incubation the reaction mixture was centrifuged, the supernatant was collected and 100 μl of potassium tetraborate was added to it followed by boiling for 3 min. To this DMAB reagent was added at RT the colour development was measured at 585 nm.
4.3. Effect of metal ion, surfactants and inhibitors on chitinase activity
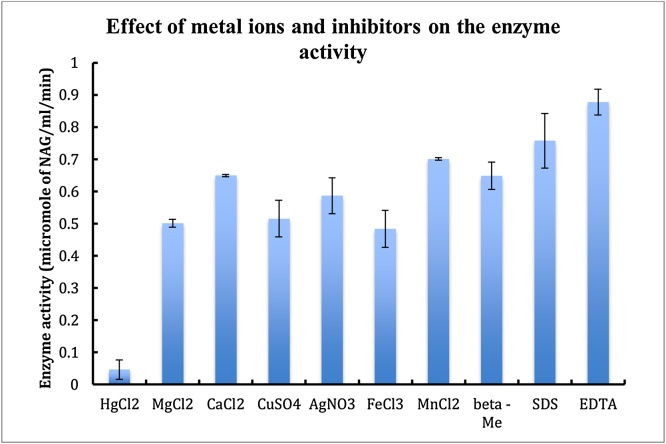
The enzyme was active in presence of inhibitors such as Mercuric chloride, Magnesium chloride, Manganese chloride, Calcium chloride, Copper sulphate, Ferric chloride, EDTA, SDS, AgNO3 and B-mercaptoethanol (Fig. 3).
Fig. 3.
The partially purified enzyme was checked for the its activity in presence of various metal ions and inhibitors. Briefly, the active fractions of DEAE Sephacel and DEAE Sepharose were pooled together. 100 μl of this enzyme was mixed with 100 μl of metal ion and inhibitor solution with a final concentration of 1 μmol, followed by 200 μl of substrate (1% clloidal chitin).The reaction mixture was then incubated at 37 for 2 h. The rest of the process was carried out according to standard assay conditions i.e after incubation the reaction mixture was centrifuged, the supernatant was collected and 100 μl of potassium tetraborate was added to it followed by boiling for 3 min. To this DMAB reagent was added at RT the colour development was measured at 585 nm.
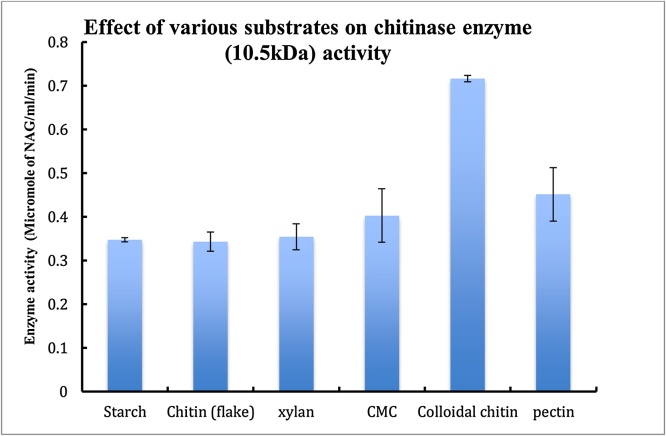
4.4. Effect of various substrates on enzyme activity
The effect of substrate glycol chitin concentration on enzyme activity was analyzed by hyperbolic regression analysis of the initial velocity and substrate concentration data. The apparent Vmax and Km for glycol chitin were 0.01 μM and 2.24U respectively at 37 °C (Fig. 4).
Fig. 4.
The partially purified enzyme was checked for the its activity in presence of various substrate. Briefly, the active fractions of DEAE Sephacel and DEAE Sepharose were pooled together. 100 μl of the enzyme the rest of the process was carried out according to standard assay conditions i.e after incubation the reaction mixture was cnterifuged, the supernatant was collected and 100 μl of potassium tetra borate was added to it followed by boiling for 3 min. To this DMAB reagent was added at RT the colour development was measured at 585 nm.
Substrate binding profile of the enzyme was studied using colloidal chitin, starch, carboxymethylcellulose (CMC) and xylan. Maximum activity was observed in case of colloidal chitin, residual activity of 15% was observed with starch. However, no enzymatic activity was observed in case of CMC and xylan.
4.5. N –terminal amino acid sequence analysis of purified 10.5 kDa chitinase
The amino acid sequence of the partially purified enzyme is GSPQLG.
4.6. Analysis of the hydrolytic products
The enzyme was also checked for formation of degradation product by reacting with chito-oligosaccharide substrates such as Diacetyl chitobiose (GlcNAc)2 (stock, 2.5 mM), N,N,N-Triacetyl chitotriose (GlcNAc)3 (stock, 2.5 mM), OF N,N,N,N,N-Pentacetyl chitopentaose (GlcNAc)5 (stock, 1 mM), and N,N,N,N,N,N-Hexacetyl chitohexaose (GlcNAc)6 (stock, 1 mM) (Sigma Aldrich, Germany). It was observed on the TLC plate that the enzyme was capable of degrading chitohexaose, chitopentaose and chitotriose. However with chitobiose the formation of degradation of degaradtion product were not evident. (Fig. 5)
Fig. 5.
Degradation products seen during reaction of partially purified chitinase (10.5 kD) with chito-oligosaccharides. 200ul of substrate (0.2 mM working conc.) and 200ul of enzyme solution (2 Units/mg protein) was mixed. The enzyme substrate mixture was incubated at 37 °C for 0, 5 s, 15 s, 30 s, 1 min, 5 min, 10 min, 15 min, 30 min, 1 h separately. Enzyme mixture with and without substrate served as negative control. The reaction was stopped by boiling the mixture at 100 °C and then concentrated using a vacuum evaporator (Eyela, Japan). The analysis was done on TLC plates (Silica gel 60, F 254 (20 x 20 cm) with butanol:methanol:ammonia:water (5:4:2:1) as mobile phase and aniline diphenylamine reagent as spraying reagent. The plate was developed by heating at 120 °C for 10–15 minutes. Dark brown-greenish spots were identified and Rf value was compared to that of standards.
Lane 1, mixture of all substrates; lane 2, individual substrate; lane 3–10, 15 s, 30 s, 1 min, 5 min, 10 min, 15 min, 30 min, 1 h; lane 11–12, negative control (enzyme with/without substrate); lane 13, mixture of all substrates.
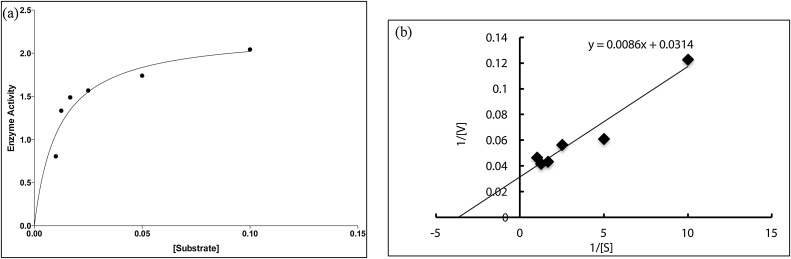
4.7. Kinetic characterization
The chitinase exhibited Km and Vmax values are o.02 mM and 3.184 mol/min respectively (Fig. 6)
Fig. 6.
(a) Determination of Km and Vmax by Michaelis-Menten equation. (b) Lineweaver Burk plot.
5. Discussion
Streptomyces species may harbor several chitinase genes which is known as multiplicity of the gene. Four chititnases were first time reported and purified from Streptomyces olivaceoviridis, showing multiplicity of chitinases in Streptomyces [6].
In the current work a novel isolate Streptomyces chlikensis RC1830 was studied for the presence of chitinase enzymes and the enzyme was partially purified by anion exchange chromatography. Presence of chitinase was observed in presence of colloidal chitin as a substrate at 48 h by the strain RC1830. This enzyme was thought to be inducible since enzyme activity was absent in the absence of chitin as a substrate. This was confirmed enzyme activity analysis by SDS- PAGE where pronounced bands were seen in culture supernatant with chitin. Hence, the RC1830 culture was grown in large quantities in the presence of chitin and used as the starting material for enzyme purification. The enzyme could not be precipitated using the usual ammonium sulfate precipitation process probably due to intolerance of the enzyme to the salt. Hence concentration of the enzyme was achieved using centrifugal device of different sizes (50, 30, 10, 5 and 3 kDa). Analysis of the concentrated upper part and diluted lower fraction of the 50 kD and 30 kD concentrator revealed that the protein with chitinase activity may have a low molecular weight. Thus, using the 3 kD concentrator, it was evident that maximum activity (121 units/mg protein) was retained in the upper part of the concentrator. It was clear that the molecular weight of the enzyme may be 10.5 kD or less. This was also confirmed by the SDS PAGE and measurement of the enzyme activity (by GlcNAc determination method) as well. The concentrated enzyme solution was then partially purified using anion exchange chromatography in DEAE Sepharose column since the enzyme was unable to bind to several other column resins (DEAE Sepahcel, SP Sephadex C-50, and gel filtration resins Sepahadex G-50, G-75 and G-100). Since the enzyme could not bind to other resins, the complete purification of the enzyme could not be accomplished.
The partial purification revealed the presence of a chitinases of molecular weights (MW) 10.5 kD. The chitinase activity was measured by GlcNAc release (chitin to GlcNAc) assay. Several other chitinolytic Streptomyces strains have reported been found to contain chitinases of MW ranging from 62-20 kD [6]. This reveals that probably the isolate RC 1830 contains novel chitinases not previously reported with MW less than equal to 10.5 kD. Enzyme activity of fractions containing MW 10.5 kD showed chitinase activity against glycol chitin in activity staining method (using SDS PAGE).
Using DEAE Sephacel™, protein with MW 10.5 kD (chitinase)) could be partially purified by 5.3-fold with 29% recovery. The specific activity of partially purified 10.5 kD chitinase enzyme, was enhanced from 11.21 units/mg protein to 60.53 units/mg protein after partial purification by anion exchange chromatography. Since, the purification was carried out using wild type bacterial enzyme concentrate, the amount of inducible protein was less.
Chitinase activity in the form of degradation spots on TLC and in GlcNAc release assay was observed for colloidal chitin (mixture of oligomers and monomers), chitotriose (GlcNAc)3, chitopentaose (GlcNAc)5 and chitohexaose (GlcNAc)6 except for chitobiose (GlcNAc)2. Chitinases with similar activities have been reported earlier in Streptomyces sp. IK, with MW 49 kD (Margino, 2010).
The partially purified 10.5 kD chitinase was characterized with respect to optimum temperature and pH as well as stability at different temperatures and pH. The 10.5 kD chitinase enzyme was found to be active over a pH range of pH 6–9, the optimum was 7. The activity decreased at pH10. The enzyme was active over a temperature range of 30 °C–60 °C, the optimum was 37 °C. However, the stability of the enzyme decreased above 60 °C. The temperature range for other similar chitinases from Streptomyces like S. aureofaciens was reported to be 30 °C−50 °C (Lumyong et al 2003), for S. venezuelae, 30 °C to 40 °C [10] and for Streptomyces sp M-20 [11,12], 30 °C −50 °C.The optimum pH and temperature for 10.5 kD chitinase were 7 and 37 °C, respectively. However optimum pH for other similar strains such as Streptomyces M-20 is 5–6 and the pH stability of the enzyme is 4–8 [11,12], for Streptomyces sp IK optimum activity of the chitinase enzyme was reported at 6–7 (Margino, 2010), and for S.venezuelae P10 the optimum pH is 6–8 [10].
Up to 40% of residual enzyme activity was retained till the pH 11 and 60 °C till 24 h for both enzymes. Substrate binding profile of 10.5 kDa chitinase revealed that colloidal chitin (0.5%, w/v) was the best substrate with 90% of enzyme binding to the substrate followed by starch (10%), but no binding was observed with CMC and xylan. This revealed that 10.5 kDa chitinase possesses a chitin binding domain either at N-terminal or C-terminal end. The end terminals of the protein still need to be characterized.
Funding
No funding was received.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors do not have any conflict of interest.
Acknowledgments
We are grateful to the Chilika Development Authority (CDA), Bhubaneswar. The staff of the CDA are acknowledged for their help and for providing the necessary facilities for the collection of sediment samples.
We are also thankful to English Language Expert Ms Chinmayee Nanda, Asst Prof (English) School of Law, (KIIT Deemed to be University) to help us in language correction and editing.
Contributor Information
Lopamudra Ray, Email: lopamudra.ray@kls.ac.in.
Vishakha Raina, Email: vishakha.raina@gmail.com.
References
- 1.Sakai K.A., Yokota H., Kurokawa M., Wakayama, Moriguchi M. Purification and characterization of three thermostable endochitinases of a novel Bacillus strain, MH-1, isolated from chitin containing compost. Appl. Environ. Microbiol. 1998;64:3397–3402. doi: 10.1128/aem.64.9.3397-3402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gooday G.W. The ecology of chitin degradation. Adv. Microb. Ecol. 1990;11:387–430. [Google Scholar]
- 3.Gopalakrishnan S., Kiran B.K., Humayun P., Vidya M.S., Deepthi K., Rupela O.P. Biocontrol of charcoal-rot of sorghum by actinomycetes isolated from herbal vermicompost. Afr. J. Biotechnol. 2011;10(79):18142–18152. [Google Scholar]
- 4.Al-Askar A.A., Abdulkhair W.M., Rashad Y.M., Hafez E.E., Ghoneem K.M., Baka Z.A. Streptomyces griseorubens E44G: a potent antagonist isolated from soil in Saudi Arabia. J. Pure Appl. Microbiol. 2014;8:221–230. (Spl. Edn. 2) [Google Scholar]
- 5.Arasu M.V., Duraipandiyan V., Ignacimuthu S. Antibacterial and antifungal activities of polyketide metabolite from marine Streptomyces sp. AP-123 and its cytotoxic effect. Chemosphere. 2013;90(2):479–487. doi: 10.1016/j.chemosphere.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Romaguera A., Menge U., Breves R., Diekmann H. Chitinases of Streptomyces olivaceoviridis and significance of processing for multiplicity. J. Bacteriol. 1992;174(11):3450–3454. doi: 10.1128/jb.174.11.3450-3454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reissig J.L., Strominger J.L., Lelor L.F. A modified colorimetric method for estimation of N-acetylamino sugars. J. Biol. Chem. 1955;217 959-956. [PubMed] [Google Scholar]
- 8.Liau C.Y., Lin C.S. A modified Coomassie Brilliant Blue G-250 staining method for the detection of chitinase activity and molecular weight after polyacrylamide gel electrophoresis. J. Biosci. Bioeng. 2008;106(1):111–11372. doi: 10.1263/jbb.106.111. [DOI] [PubMed] [Google Scholar]
- 9.Laemmli U.K. Cleavage of structural proteins during assembly of head of bacetriophages T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee G., Sen S.K. Purification, characterization and antifungal activity of chitinase from Streptomyces venezuelae P10. Curr. Microbiol. 2006;53:265–269. doi: 10.1007/s00284-005-0412-4. [DOI] [PubMed] [Google Scholar]
- 11.Kim K.J., Yang Y.J., Kim J.G. Purification and characterization of chitinase from Streptomyces species. M-20. J. Biochem. Mol. Biol. 2003;36(2):185–186. doi: 10.5483/bmbrep.2003.36.2.185. [DOI] [PubMed] [Google Scholar]
- 12.Kim Kyoung-Ja, Yang Yong-Joon, Kim Jong-Gi. Purification and characterization of chitinase from Streptomyces sp. M-20. J. Biochem. Mol. Biol. 2003;36(2):185–189. doi: 10.5483/bmbrep.2003.36.2.185. [DOI] [PubMed] [Google Scholar]