Abstract
Objectives
To investigate the contribution to virulence of the surface protein internalin B (InlB) in the Listeria monocytogenes lineage I strain F2365, which caused a deadly listeriosis outbreak in California in 1985.
Methods
The F2365 strain displays a point mutation that hampers expression of InlB. We rescued the expression of InlB in the L. monocytogenes lineage I strain F2365 by introducing a point mutation in the codon 34 (TAA to CAA). We investigated its importance for bacterial virulence using in vitro cell infection systems and a murine intravenous infection model.
Results
In HeLa and JEG-3 cells, the F2365 InlB+ strain expressing InlB was ≈9-fold and ≈1.5-fold more invasive than F2365, respectively. In livers and spleens of infected mice at 72 hours after infection, bacterial counts for F2365 InlB+ were significantly higher compared to the F2365 strain (≈1 log more), and histopathologic assessment showed that the F2365 strain displayed a reduced number of necrotic foci compared to the F2365 InlB+ strain (Mann-Whitney test).
Conclusions
InlB plays a critical role during infection of nonpregnant animals by a L. monocytogenes strain from lineage I. A spontaneous mutation in InlB could have prevented more severe human morbidity and mortality during the 1985 California listeriosis outbreak.
Keywords: Epidemic, Infection, Internalin B, Invasion, Listeria monocytogenes
Introduction
Listeria monocytogenes is a facultative intracellular bacterium that causes listeriosis [1]. After ingestion of contaminated food, L. monocytogenes disseminates to the liver, spleen, brain and/or placenta [1]. L. monocytogenes infections can be fatal, as exemplified by the 2017–2018 outbreak of listeriosis in South Africa affecting 1060 patients, 216 of whom died (http://www.nicd.ac.za/wp-content/uploads/2018/07/Listeriosis-outbreak-situation-report-_26July2018_fordistribution.pdf). Strains of L. monocytogenes are grouped into lineage I, lineage II and lineage III [1]. Major listeriosis epidemics have been associated with lineage I strains [2]. However, most reports investigating listeriosis pathophysiology have studied what are essentially strains from lineage II (e.g. EGD, EGD-e and 10403S) [3], [4].
The most important virulence factors of L. monocytogenes strains are encoded in the inlA-inlB locus and in the pathogenicity islands LIPI-1, LIPI-3 and LIPI-4 [2], [3]. The inlA-inlB locus encodes for internalin A (InlA) and internalin B (InlB), two bacterial surface proteins that bind the host cell receptors E-cadherin and Met, respectively, to induce bacterial uptake into nonphagocytic eukaryotic cells [1]. Expression of the inlA-inlB locus and LIPI-1 is regulated by the transcriptional regulator PrfA [1], [5]. Importantly, the strain EGD displays a PrfA mutation leading to constitutive production of InlA and InlB [5], [6]. However, one isolate carrying a PrfA mutation that leads to the constitutive production of InlA, InlB and LIPI-1 virulence factors has been found in a L. monocytogenes variant that diverged from a clinical isolate [2], [6].
All studies performed to understand the role of InlB in deep organ infection have used the EGD strain [7], [8], [9], [10]. While a clear contribution for InlB has been demonstrated for placental invasion [10], in spleen and liver infections it has been observed either as a contribution for InlB in conventional mice [7] or as no contribution for InlB in a transgenic humanized E-cadherin mouse model [10].
The genome of the lineage I strain F2365 responsible for the 1985 California outbreak, one of the deadliest bacterial foodborne outbreaks ever reported in the United States [11], shows that the F2365 isolate carries a nonsense mutation in inlB (codon number 34 is TAA) (http://genolist.pasteur.fr/) [12]. We thus decided to restore the expression of InlB in the F2365 strain and to examine the consequences of InlB expression during in vitro and in vivo infections.
Materials and methods
An isogenic mutant strain (F2365 InlB+, BUG3824) containing a functional InlB (a point mutation was introduced in the codon 34 (TAA to CAA)) was used. The InlB amino acid sequence of L. monocytogenes EGDe (lineage II) and F2365 (lineage I) strains has 94% amino acid sequence identity (Supplementary Fig. 1). Cell infection was performed as previously described [13] using multiplicity of infection values of 2 (phagocytic RAW 264.7), 5 (epithelial JEG-3 with InlA and InlB-dependent entry) or 25 (epithelial HeLa with only InlB-dependent entry). Luciferase reporter system experiments were performed by creating a transcriptional fusion by cloning 308 nucleotides upstream from the inlB initiation codon into SwaI- and SalI-digested pPL 2lux as described [3]. For in vivo bioluminescence experiments, mice were infected orally with 5 × 109 F2365 InlB+inlB::lux (BUG4155) as described elsewhere [3]. Mouse infections were performed intravenously with 104 CFU of the indicated strain as reported elsewhere [13]. Half of the organ was used to assess bacteria load, and the other half was used for histopathologic analysis at 72 and 96 hours after infection.
This study was carried out in accordance with the French and European laws (86/609/EEC) and was approved by the animal experiment committee of the Institut Pasteur (approval 03-49).
Results
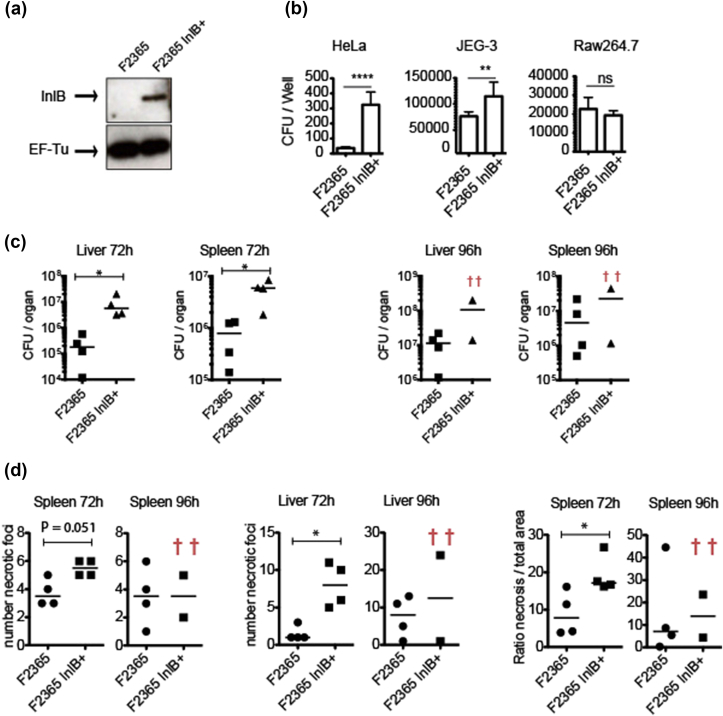
A point mutation in the inlB codon 34 (TAA to CAA) was performed to generate a F2365 strain carrying a functional inlB, termed F2365 InlB+ (Fig. 1(A)) [13]. Both F2365 and F2365 InlB+ strains were tested for entry into epithelial cells which express only the InlB receptor Met (HeLa), epithelial cells expressing both the InlB and the InlA receptors Met and E-cadherin, respectively (JEG-3), or RAW 264.7 macrophages. Quantification of the number of viable intracellular L. monocytogenes showed that in HeLa and JEG-3 cells, the F2365 InlB+ strain was ≈9-fold and ≈1.5-fold more invasive than F2365, respectively. We thus report for the first time that chromosomal restoration of InlB promotes a gain of entry associated to the presence of the InlB receptor Met (Fig. 1(B)). In macrophages, F2365 InlB+ and F2365 strains invaded similarly, showing that InlB does not play a role in entry into phagocytic cells.
Fig. 1.
(A) InlB protein levels detected by Western blot analysis. Levels of loading control protein, EF-Tu, are shown for comparison. (B) Role of InlB in epidemic Listeria monocytogenes entry. Eukaryotic cells were incubated with bacteria for 30 minutes (JEG-3 and RAW 264.7) or 1 hour (HeLa) at 37 °C. After incubation, extracellular bacteria were neutralized by adding complete fresh medium containing 40 μg/mL of gentamicin. At 2 hours after infection, cells were lysed to determine number of viable intracellular L. monocytogenes. Bars show numbers of viable intracellular bacteria. Mean and standard deviation are shown. Three independent experiments with 6 replicates in each experiment were performed. Data from one representative experiment are shown. ****p <0.0001, **p <0.01; ns indicates not statistically significant by Student's t test. Error bars represent standard deviation. (C) BALB/c mice were intravenously inoculated with 104L. monocytogenes F2365 and F2365 InlB+. Half of organ was used to assess bacteria load and other half was used for histopathologic analysis. CFUs in spleen and liver were assessed at 72 and 96 hours after infection. Each dot represents value for one mouse. Two mice infected with F2365 InlB+ strain died before 96 hours. Statistically significant differences were evaluated by Mann-Whitney test. *p <0.05. (D) Histopathologic analysis of number of necrotic foci and ratio of necrotic area to total area were recorded in spleens and livers from same infected mice (*p <0.05 by Mann-Whitney test).
In L. monocytogenes EGD, inlA and inlB are transcribed in vitro both individually and in an operon by PrfA-dependent and -independent mechanisms [4], [14]. Here, we investigated whether inlB is transcribed in vivo from its own promoter in the epidemic lineage I L. monocytogenes F2365. For this purpose, we fused the 308 nt located upstream from the inlB initiation codon to a Lux reporter plasmid and integrated it into the chromosome of the F2365 strain (F2365InlB+inlB:lux). Upon oral infection of 12 conventional BALB/c mice with 5 × 109 L. monocytogenes F2365InlB+inlB:lux, no bioluminescent signal was detected in organs of infected animals from 24 to 72 hours after infection (Supplementary Fig. 2(A)). To discard the possibility that the absence of bioluminescence in the liver and spleen could be due to a low number of CFUs in these organs, the two organs were dissected, homogenized, serially diluted and plated onto brain–heart infusion plates. Mice orally infected yielded ≈1 × 107.5 CFU in the liver or spleen at 48 hours after infection (Supplementary Fig. 2(B)).
To analyse the potential contribution of InlB to the F2365 InlB+ virulence, we performed intravenous inoculations of BALB/c mice with the F2365 and F2365 InlB+ strains. In all the organs tested at 72 hours, bacterial counts for F2365 InlB+ were significantly higher compared to the F2365 strain (≈1 log more) (Fig. 1(C)) (Mann-Whitney test was used for statistical significance).
Furthermore, histopathologic assessment showed that the F2365 strain displayed a reduced number of necrotic foci in the spleen and liver 72 hours after infection compared to the F2365 InlB+ strain (Fig. 1(D) and Supplementary Fig. 3). The ratio of necrotic area to total area was significantly higher in the spleen of mice infected with the F2365 InlB+ strain at 72 hours (Fig. 1(D)).
Discussion
In our study, chromosomal restoration of InlB promoted a gain of entry into eukaryotic cells associated with the presence of the InlB receptor Met.
Previous studies performed in our laboratory demonstrated that less than ≈107 L. monocytogenes CFUs distributed across the entire intestine were sufficient to produce a bioluminescent signal in this organ [3]. The present results therefore suggest that in vivo, inlA and inlB are transcribed in an operon from a promoter located upstream of inlA.
The present in vitro and in vivo results demonstrate that InlB expression increases the virulence of the F2365 InlB+ strain and show that InlB plays an essential role in spleen and liver infection by lineage I L. monocytogenes. InlB is highly conserved in the genome of L. monocytogenes, suggesting a critical role for this molecule during infections [2]. In conclusion, it could be speculated that a spontaneous mutation in InlB could have prevented a more severe L. monocytogenes disease during the 1985 California outbreak.
Transparency declaration
This work was supported by the Institut Pasteur, the Institut National de la Sante et de la Recherche Medicale (INSERM Unite 604), the Institut National de la Recherche Agronomique (INRA Unite Sous Contrat 2020), Universite Paris Diderot, grants from Region Ile-de-France, the Institut Pasteur ‘Programmes Transversaux de Recherche’ (PTR521 to JPC), Agence Nationale de la Recherche (ANR-15-CE15-0017 StopBugEntry to JPC), Fondation Le Roch Les Mousquetaires, European Research Council (advanced grant 670823 BacCellEpi to PC) and Region Ile-de-France (DIM-MALINF to JMT). PC is an international senior research scholar of the Howard Hughes Medical Institute. JGL is supported by a ‘Ramón y Cajal’ contract of the Spanish Ministry of Economy and Competitiveness (RYC-2014-16735). All authors report no conflicts of interest relevant to this article.
Editor: G. Lina
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2018.08.022.
Contributor Information
J.J. Quereda, Email: juan.quereda@uchceu.es.
J. Pizarro-Cerdá, Email: javier.pizarro-cerda@pasteur.fr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maury M.M., Tsai Y.H., Charlier C., Touchon M., Chenal-Francisque V., Leclercq A. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quereda J.J., Dussurget O., Nahori M.A., Ghozlane A., Volant S., Dillies M.A. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci U S A. 2016;113:5706–5711. doi: 10.1073/pnas.1523899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becavin C., Bouchier C., Lechat P., Archambaud C., Creno S., Gouin E. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio. 2014;5 doi: 10.1128/mBio.00969-14. e00969–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quereda J.J., Andersson C., Cossart P., Johansson J., Pizarro-Cerda J. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet Res. 2018;49:13. doi: 10.1186/s13567-017-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripio M.T., Dominguez-Bernal G., Lara M., Suarez M., Vazquez-Boland J.A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol. 1997;179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khelef N., Lecuit M., Bierne H., Cossart P. Species specificity of the Listeria monocytogenes InlB protein. Cell Microbiol. 2006;8:457–470. doi: 10.1111/j.1462-5822.2005.00634.x. [DOI] [PubMed] [Google Scholar]
- 8.Gregory S.H., Sagnimeni A.J., Wing E.J. Internalin B promotes the replication of Listeria monocytogenes in mouse hepatocytes. Infect Immun. 1997;65:5137–5141. doi: 10.1128/iai.65.12.5137-5141.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parida S.K., Domann E., Rohde M., Muller S., Darji A., Hain T. Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol Microbiol. 1998;28:81–93. doi: 10.1046/j.1365-2958.1998.00776.x. [DOI] [PubMed] [Google Scholar]
- 10.Disson O., Grayo S., Huillet E., Nikitas G., Langa-Vives F., Dussurget O. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 11.Linnan M.J., Mascola L., Lou X.D., Goulet V., May S., Salminen C. Epidemic listeriosis associated with Mexican-style cheese. N Engl J Med. 1988;319:823–828. doi: 10.1056/NEJM198809293191303. [DOI] [PubMed] [Google Scholar]
- 12.Nightingale K.K., Milillo S.R., Ivy R.A., Ho A.J., Oliver H.F., Wiedmann M. Listeria monocytogenes F2365 carries several authentic mutations potentially leading to truncated gene products, including inlB, and demonstrates atypical phenotypic characteristics. J Food Protect. 2007;70:482–488. doi: 10.4315/0362-028x-70.2.482. [DOI] [PubMed] [Google Scholar]
- 13.Quereda J.J., Nahori M.A., Meza-Torres J., Sachse M., Titos-Jimenez P., Gomez-Laguna J. Listeriolysin S is a streptolysin S–like virulence factor that targets exclusively prokaryotic cells in vivo. MBio. 2017;8 doi: 10.1128/mBio.00259-17. e00259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lingnau A., Domann E., Hudel M., Bock M., Nichterlein T., Wehland J. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect Immun. 1995;63:3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.