Abstract
The aberrant expression of miR107-5p is closely related to the development of several types of human cancers. However, the role of miR-107-5p in endometrial carcinoma (EC) has not been fully confirmed. In the present study, we aimed to explore the function of miR-107-5p in EC carcinogenesis. EC samples and normal endometrial tissues were obtained by laser capture microdissection. It was determined that the expression of miR-107-5p in EC was significantly higher than that in normal endometrium, and higher miR-107-5p expression was related to advanced FIGO stages, lymph node metastasis and myometrial invasion in EC patients. Blocking miR-107-5p significantly inhibited cell proliferation, migration and invasion of EC cells in vitro and in vivo. The results of bioinformatic algorithms and luciferase reporter assays revealed that estrogen receptor α (ERα) was a direct target of miR-107-5p. miR-107-5p downregulated the expression of ERα mRNA and protein. In conclusion, our results highlighted that miR-107-5p is a novel prognostic factor that targets ERα to promote tumor proliferation and invasion of EC.
Keywords: endometrial carcinoma, miR-107-5p, ERα, proliferation, invasion, prognosis
Introduction
Endometrial carcinoma (EC) is the most common gynecological malignancy worldwide. Diagnosis of 63,230 new cases in the USA, resulting in 11,350 disease-related deaths, were predicted in 2018 (1). According to its clinical and pathological characteristics, EC is traditionally classified into two types (2). Type I includes tumors of endometrioid histology graded as 1, 2 and 3 which are estrogen-dependent and usually preceded by endometrial hyperplasia with a favorable prognosis. Conversely, type II tumors are predominantly papillary serous carcinomas caused by atrophy of the endometrium. They are not consistently correlated with estrogen exposure and reveal a strong ability of aggression (3,4).
It is undeniable that estradiol (E2) plays a critical role in the development of EC (5). E2 and its analogs regulate the expression of selective estrogen-responsive genes by combining to the cytoplasmic estrogen receptor α (ERα), and subsequently, the complex migrates to the nucleus to activate the target genes (6). However, the therapeutic efficiency of hormones for treating EC is decreasing due to drug insensitivity. Thus, it is essential to raise our awareness of the molecular and cellular mechanisms underlying EC and to develop new therapeutic strategies to prevent the progression of EC.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (~22 nt). It is well known that miRNAs induces RNA interference by base-pairing with the 3′ untranslated region (UTR) of a complementary messenger RNA (mRNA), thus resulting with either mRNA translational repression or RNA degradation (7). In human cancers, specific miRNAs exist in different tissues. The aberrant expression of miRNAs is correlated with carcinogenesis (8), including EC carcinogenesis (9). Furthermore, miRNAs interact with certain transcription factors (TFs) to regulate mutual sets of target genes, thus realizing the coordinated modulation of gene transcriptional and post-transcriptional expression. We previously uncovered a regulatory loop involving TrkB/miR-204-5p that is critical in the tumorigenesis of EC (10), and these observations led us to hypothesize that specific miRNAs may control ERα expression post-transcriptionally during EC progression.
In the present study, the target gene of miR-107 containing ESR1 was predicted by bioinformatics analysis. Aberrant expression of miR-107 has been revealed to promote the tumorigenesis of glioma and lung cancer (11,12). However, the mechanism by which miR-107 participates in EC is poorly understood. Notably, our findings revealed that anti-miR-107-5p inhibited the proliferation and invasion of EC cells by targeting the 3′UTR of ESR1 mRNA directly. Our findings provide new ideas for further understanding the mechanism of cell proliferation mediated by miR-107-5p in EC.
Materials and methods
Clinical samples
Seventy-one EC patients who underwent hysterectomy with lymph node dissection at the Shanghai General Hospital Affiliated with Shanghai Jiao Tong University School of Medicine from August 2014 to May 2016 were included in our study (Table I). The stages and histological grades of these tumors were determined according to the criteria of the Federation International of Gynecology and Obstetrics (FIGO) surgical staging system (2009) (13). Twenty-six samples of normal endometrium collected from October 2014 to June 2015 were obtained from patients who underwent hysterectomy for other diseases, such as myoma or adenomyosis. None of the patients had received hormone therapy, radiotherapy, or chemotherapy prior to surgery. The resected specimens were stained with hematoxylin and eosin (H&E) for histological examination. The original tissues were collected from each patient immediately after resection and snap-frozen in liquid nitrogen until further use. The present study was approved by the Human Investigation Ethical Committee of Shanghai General Hospital Affiliated with Shanghai Jiao Tong University. The samples of EC and normal endometrial tissues were collected after informed consent was signed by the patients.
Table I.
Clinicopathological parameters of EC patients.
| No. patients (%) | ||
|---|---|---|
| Variable | n | % |
| Total | 71 | 100 |
| Age (years) | ||
| ≤50 | 15 | 21.1 |
| >50 | 56 | 78.9 |
| FIGO stage | ||
| I | 60 | 84.5 |
| II | 7 | 9.8 |
| III | 4 | 5.7 |
| IV | 0 | 0 |
| Grade (endometrioid, n=57) | ||
| G1 | 32 | 56.1 |
| G2 | 18 | 31.6 |
| G3 | 7 | 12.3 |
| Histological type | ||
| Endometrioid | 57 | 80.3 |
| Non-endometrioid | 14 | 19.7 |
| Myometrial invasion | ||
| <1/2 | 67 | 94.4 |
| ≥1/2 | 4 | 5.6 |
| Lymph node metastasis | ||
| Negative | 63 | 88.7 |
| Positive | 8 | 11.3 |
EC, endometrial carcinoma; FIGO, Federation International of Gynecology and Obstetrics.
Laser capture microdissection (LCM)
Each specimen was cut into 10–20 serial frozen sections of 8-µm thickness that were then mounted on polyethylene membrane glass slides (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and then stored at −80°C. The sections were fixed at −20°C for 2 min with 70% ethanol and then stained with 25 µl HistoGene (Applied Biosystems; Thermo Fisher Scientific) using a frozen section staining kit. Then, they were rinsed with ice-cold RNA nuclease-free water at −20°C, incubated in xylene at −20°C for 2 min and air-dried for 2 min. The targeted cells were microdissected using a UV cutting and laser capture procedure with an LCM system (LMD7000; Leica Microsystems GmbH, Wetzlar, Germany). The EC cells and normal epithelial cells (Fig. 1A) were cut and captured on CapSure Macro LCM Cap (Applied Biosystems; Thermo Fisher Scientific, Inc.). The caps containing the captured cells were placed into a microfuge tube (Applied Biosystems; Thermo Fisher Scientific, Inc.) that contained 100 µl of RLT lysis buffer (Qiagen, Inc., Valencia, CA, USA) and 1% of β-mercaptoethanol (Bio Basic, Inc., Markham, ON, Canada). For the polymerase chain reaction (PCR) experiments, each sample collected 10–20 serial sections of tissue with 10 caps.
Figure 1.
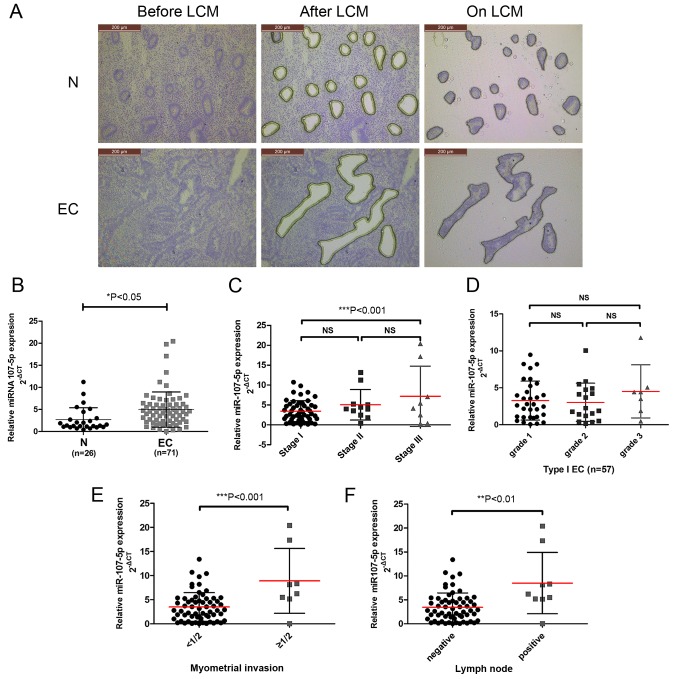
miR-107-5p expression in EC specimens and its association with clinical parameters. (A) LCM of normal endometrial cells and EC cells. Sections were stained with HistoGene solution. The upper panel reveals frozen sections from normal endometrium (normal epithelial cells before LCM, after LCM and on LCM caps) (magnification, ×100). The lower panel reveals frozen sections from endometrioid endometrial cancer stage IA (malignant epithelial cells before LCM, after LCM, and on LCM caps) (magnification, ×100). (B) miR-107-5p was differentially expressed between the EC specimens (n=71) and normal endometrium (N) (n=26) as assessed by qRT-PCR. ΔCt describes the expression of miR-107 relative to U6B expression (−ΔCt = CtU6 - CtmiR-107). *P<0.05, unpaired t-test. (C-F) miR-107 expression was significantly increased in (C) stage III EC patients compared to stage I patients, (E) myometrial invasion and (F) lymph node positive disease, but not in (C) stage I vs. stage II patients or (E) grades (NS, not significant; **P<0.01 and ***P<0.001). The bars represent the mean ± SD. EC, endometrial carcinoma; LCM, laser capture microdissection.
Cell culture
We obtained the human endometrial cancer cell lines (Ishikawa and HEC-1B) and 293 cells from the Chinese Academy of Sciences Committee Type Culture Collection (Shanghai, China). The cell lines were cultured in Dulbecco's modified Eagle's medium/F12 (11030; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS) (16000-44; Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2.
Cell transfections
The miR-107 mimics (miR-107m), miR-107 mimic negative control (miR-107m NC), miR-107 inhibitor (miR-107i) and miR-107 inhibitor negative control (miR-107i NC) are synthetic, chemically modified short single- or double-stranded RNA oligonucleotides, which were synthesized from Shanghai GenePharma Co., Ltd. (Shanghai, China) (Table II). Cells were seeded in 6-well plates at 70–80% confluence and grown overnight before transfection. Transfection of Ishikawa cells (or HEC-1B cells) with miR-107i (or miR-107m) or a counterpart negative control was performed via Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions.
Table II.
Oligo sequences used in the study.
| Identifier | Sense primer sequences | Antisense primer sequences |
|---|---|---|
| miR-107i | 5′-UGAUAGCCCUGUACAAUGCUGCU-3′ | 5′-TCATTGGCATGTACCATGCAGCT-3′ |
| miR-107i NC | 5′-CAGUACUUUUGUGUAGUACAA-3′ | |
| miR-107m | 5′-AGCAGCAUUGUACAGGGCUAUCA-3′ | 5′-AUAGCCCUGUACAAUGCUGCUUU-3′ |
| miR-107m NC | 5′-UUCUCCGAACGUGUCACGUTT-3′ | 5′-ACGUGACACGUUCGGAGAATT-3′ |
| ESR1-3′UTR-WT | 5′-TGCTCTATGCTGCAAAGACCTGAATACCAC-3′ | 5′-GCTCTATGCTGCTTGTTCCTCCCATAT-3′ |
| ESR1-3′UTR-MT | 5′-TATTACTACGACGATGAAACAAGCAATGC-3′ | 5′-TGAAAAACGATGAATTTCATAG-3′ |
Quantitative real-time reverse transcription (RT)-PCR
Total RNA was extracted from the cultured cells and tissues using TRI-reagent (Molecular Research Center, Inc., Cincinnati, OH, USA). For the miRNA analysis, TaqMan microRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.) was used to reverse transcribe mature miRNA from total RNA. According to the manufacturer's instructions, real-time PCR was performed using TaqMan MicroRNA Assay primers with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific Inc.) and analyzed with an ABI PRISM7000 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6B was used as an internal reference for the expression of miRNAs. The assay names were ‘hsa-miR-107-5p’ for miR-107-5p and ‘RNU6B’ for U6B miRNA (Table III). All reagents were purchased from Applied Biosystems; Thermo Fisher Scientific, Inc. For all the experiments, values on the y-axis were equal 2−ΔCq, where ΔCq was the difference between gene Cq and normalizer gene Cq (14). The thermocycling conditions were as follows: 95°C for 10 min, 45 cycles of 95°C for 15 sec and 60°C for 1 min, for both miRNA and mRNA PCR. The data were obtained in triplicate in 3 independent experiments.
Table III.
Primers used for quantitative real-time PCR analysis.
| mRNA/miRNA | Primer sequence |
|---|---|
| miR-107 | F: 5′-AGCAGCATTGTACAGGGCTATCA-3′ |
| R: 5′-GCGAGCACAGAATTAATACGAC-3′ | |
| U6 | F: 5′-AGAGCCTGTGGTGTCCG-3′ |
| R: 5′-CATCTTCAAAGCACTTCCCT-3′ | |
| ERα (ESR1) | F: 5′-CTCAACAGCGTGTCTCCG-3′ |
| R: 5′-GGCTCGTTCTCCAGGTAG-3′ | |
| β-actin | F: 5′-CAGCCATGTACGTTGCTATCCAGG-3′ |
| R: 5′-AGGTCCAGACGCAGGATGGCATG-3′ |
F, forward; R, reverse; ERα, estrogen receptor α.
For quantification of ERα, cDNA was generated using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), and real-time PCR was carried out on an ABI PRISM 7000 Sequence Detection System with SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.). The primer sequences are presented in Table II. For all experiments, values on the y-axis equal 2−ΔΔCq. The data were obtained in triplicate in 3 independent experiments.
CCK-8 proliferation assays
Infected cells at 70–80% confluence were serum-starved for 24 h and then cultured in 96-well plates at 5% CO2 and 37°C with a density of 1,000 cells/well. At selected time-points, a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay was applied to assess the rate of cell proliferation. CCK-8 reagent was added to the wells, and the absorbance at 450 nm was measured with a SpectraMax 190 microplate reader (Bio-Rad Model 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell migration and invasion assays
For wound-healing experiments, cells were seeded in 6-well plates, with 6×106 cells/well. Then, the cells were allowed to adhere for 24 h. Cell monolayers were scratched with a sterile micropipette tip and incubated in serum-free medium for 24 h. For each sample, 3 defined regions were monitored, and images were captured at 0, 24 and 48 h to assess the migration index, a value which was obtained by calculating the distance traveled by the cell monolayer relative to the gap made using the pipette tip at 0 h (15).
For invasion assays, cells were trypsinized, centrifuged at 1,000 × g and resuspended in serum-free medium. The cells were then plated at a density of 2×105 cells/well in invasion chambers (8-µm pore size; BD Biosciences, Franklin Lakes, NJ, USA) with Matrigel coating for the invasion assays. The medium containing 10% FBS was added to 24-well plates as a chemoattractant. After 6 h of incubation for the migration assay or after 24 h of incubation for the invasion assay, the cells were fixed with 4% paraformaldehyde for 1 h. The cells on the apical side of each insert were removed by mechanical scraping. The cells that migrated to the basal side of the membrane were stained with 0.1% crystal violet and visualized under a Leica DMI 3000B microscope (Leica Microsystems GmbH). The cells were counted at a 200-fold magnification.
Western blot analysis
The protein samples were washed with ice-cold PBS and then lysed in lysis buffer containing 10 mM Tris (pH 7.5), 150 mM NaCl, 10 mM ethylenediaminetetraacetic acid (EDTA), 1% sodium dodecyl sulfate (SDS), 1 mM sodium orthovanadate, and a mixture of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 µg/ml pepstatin A and 2 µg/ml aprotinin). After protein extraction (Beyotime Institute of Biotechnology, Shanghai, China), all the sample were detected with BCA protein determination method according to the protocol (Beyotime Institute of Biotechnology). Equal amounts (25 µg) of the cell lysates were separated via 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto a polyvinylidene difluoride (PVDF) membrane. Following blocking and washing 4 times for 15 min with Tris-buffered saline with Tween-20 (TBST) at room temperature, the PVDF membrane was incubated with rabbit polyclonal primary antibodies at 1:1,000 to detect ERα (cat. no. ab75635; Abcam, Cambridge, MA, USA). Following extensive washing, the membranes were incubated with secondary peroxidase-conjugated goat anti-rabbit IgG (dilution 1:5,000; cat. no. 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 1 h. The results were visualized using an enhanced chemiluminescence kit (ECL Kit; Pierce; Thermo Fisher Scientific, Inc.) using Kodak XAR-5 film (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). β-actin was used as the internal reference.
Construction of reporter plasmids and luciferase assays
A DNA fragment comprised ofa partial wild-type 3′UTR of ESR1 or a corresponding mutant 3′UTR of ESR1 was chemically synthesized and cloned into the pGL3-REPORT luciferase vector containing Renilla luciferase and firefly luciferase (Ambion; Thermo Fisher Scientific, Inc.) downstream of the luciferase gene (designated pGL3-ESR1-3′UTR-WT and pGL3-ESR1-3′UTR-MT, respectively). The nucleotide sequences of the plasmid constructs were confirmed by DNA sequencing. All steps of the luciferase reporter assay were performed in accordance with procedures previously described (16,17). Briefly, 293 cells (5×104) were seeded into 24-well plates and transfected with 0.2 µg of either pGL3-ESR1-3′UTR-WT or pGL3-ESR1-3′UTR-MT, with or without 50 nM miR-107m or miR-107m NC using Lipofectamine 2000 transfection reagent according to the manufacturer's protocol. After 48 h, we used the dual luciferase reporter assay system to assess the luciferase activity (Promega Corp., Madison, WI, USA) and the results were expressed as the relative luciferase activity (firefly LUC/Renilla LUC). Meanwhile, ERα was the predicted target of miR-107 using the miR target prediction algorithm TargetScan (http://targetscan.org/), Pictar and miRanda.
Xenograft tumor formation assays
Animal research was conducted in accordance with the recommendations in the Guidelines for the Care and Use of Laboratory Animals of China strictly. The protocol was approved by the Ethics Committee of Animal Experiments of Shanghai General Hospital Affiliated with Shanghai Jiaotong University School of Medicine (Permit no. SYXK (hu) 2009-0086). All efforts were made to minimize suffering. Ten female BALB/c nude mice (5 weeks of age, ~20 g weight) were obtained from the Chinese Academy of Sciences (Shanghai, China). The mice were housed in an environmentally controlled room (22±2°C; 40–60% humidity and a 12-h light/dark cycle). Ishikawa cells were harvested and resuspended at a density of 5×106 cells/200 µl in sterile saline. Five mice/group were subcutaneously injected with Ishikawa cells in the subdermal space on the medial side of the neck. One week after treatment, when the tumors reached an average volume of ~30 mm3, they were directly injected with a cocktail of antagomiRs (Dharmacon, Milan, Italy) targeting miR-107 or with a control antagomiR [40 ml of phosphate-buffered saline (PBS) containing 1 µg of each anti-miR-107 or control antagomiR] at days 0, 5 and 9, for a total of 3 injections/tumor (18). The tumor volume was assessed every 7 days until the end of the experiment, using the formula: largest diameter × smallest diameter2 ×0.5. The tumor weight was determined after the animals were sacrificed with cervical dislocation at the completion of the xenograft experiments.
Immunohistochemistry (IHC)
The 4-µm-thick sections of paraformaldehyde-fixed and paraffin-embedded xenograft tumor tissue were used for immunohistochemical examination of ERα expression. The standard avidin-biotin immunohistochemical techniques with the use of anti-Ki-67 (dilution 1:100; Wuhan Boster Biological Engineering Co., Ltd., Wuhan, China) was used to detect Ki-67 expression in the tumor tissues according to the manufacturer's instructions.
Statistical analyses
Each experiment was performed at least 3 times. Independent samples t-tests were used to compare the 2 groups and one-way analysis of variance (ANOVA) followed by post hoc Tukey's test were performed to compare multiple groups. Data are presented as the means ± SD. All P-values were two-sided, and P<0.05 was considered to indicate a statistically significant result. All statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA).
Results
Increased miR-107-5p expression in EC is associated with myometrial invasion and lymph node metastasis
To investigate the potential clinicopathological implications of altered miR-107-5p expression, the expression levels of miR-107-5p in 26 normal endometrium samples (N) and 71 EC tissue samples (EC) were compared by TaqMan PCR. While collecting the endometrial cells captured by LCM (Fig. 1A), it was concluded that the expression of the miR-107-5p in the EC tissues was significantly higher than that in the normal endometrium (P<0.05) (Fig. 1B).
To further examine the clinical implications of miR-107-5p in EC, the association between miR-107-5p expression levels and clinicopathological characteristics of EC were assessed. Higher miR-107-5p expression levels were observed in FIGO stage III tumors than in stage I tumors (P<0.05) (Fig. 1C). There were no statistical associations found with respect to tumor grade (P>0.05) (Fig. 1D). However, a statistically significant association was observed between miR-107-5p expression and EC myometrial invasion and lymph node metastasis (P<0.001 and P<0.01) (Fig. 1E and F). These results indicated that reduced miR-107-5p expression was closely related to advanced FIGO stage, myometrial invasion and lymph node metastasis in EC.
miR-107-5p promotes the proliferation of EC cells
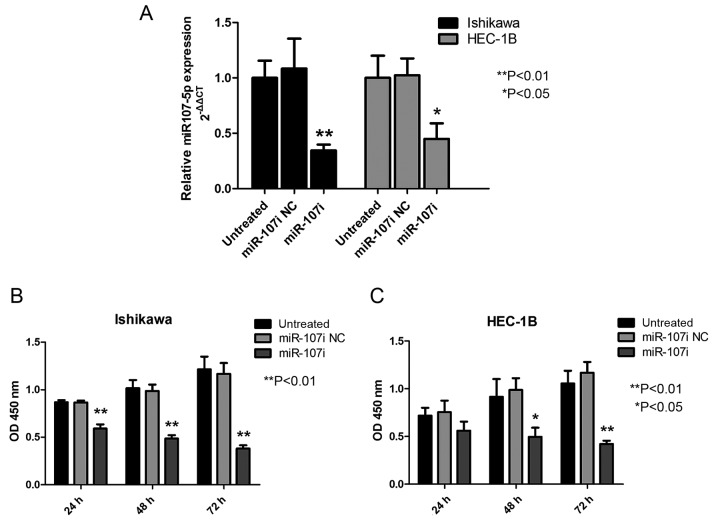
To determine the biological role of miR-107-5p, the effect of modulation of miR-107-5p expression on the proliferation of EC cells was assessed using miR-107 inhibitor in Ishikawa and HEC-1B cells. Transfection efficiency was verified at 24 h post-transfection (Fig. 2A). As revealed in Fig. 2B, compared with miR-107i NC and untreated cells, treatment with miR-107i significantly inhibited the growth of the Ishikawa cells as assessed by the CKK-8 assay (Fig. 2B; P<0.01). HEC-1B cells treated with miR-107i grew more slowly than untreated HEC-1B cells and miR-107i NC (Fig. 2B; P<0.05). All the aforementioned results revealed that miR-107-5p promoted EC cell proliferation.
Figure 2.
miR-107-5p promotes the proliferation of EC cells. (A) To verify transfection efficiency, the expression of miR-107-5p was assessed by TaqMan PCR at 24 h after transfection of Ishikawa and HEC-1B cells with a control (miR-107i NC) or miR-107 inhibitor (miR-107i). Values were calculated relative to U6B expression. *P<0.05, **P<0.01. (B and C) CCK-8 analysis of the growth of 3 groups of Ishikawa and HEC-1B cells. *P<0.05, **P<0.01. All values represent the means ± SD of 3 independent experiments performed in triplicate. EC, endometrial carcinoma; CCK-8, Cell Counting Kit-8.
miR-107-5p enhances the migration and invasion of EC cells
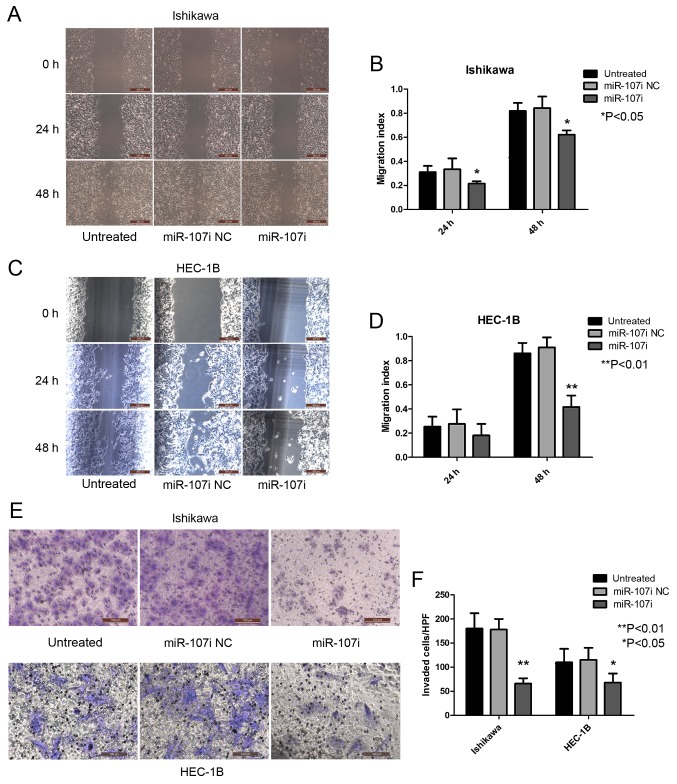
To explore the role of miR-107-5p in the regulation of metastatic function, we used wound-healing and Transwell invasion assays to examine the migration and invasion abilities of Ishikawa and HEC-1B cells after transfection with miR-107 inhibitor. Untreated Ishikawa and miR-107 NC cells displayed an enhanced rate of migration compared with Ishikawa cells transfected with miR-107i (Fig. 3A and B; P<0.05). HEC-1B cells transfected with miR-107i exhibited a low migration rate compared with untreated HEC-1B cells and miR-107i NC cells (Fig. 3C and D; P<0.01). These results indicated that miR-107-5p enhanced EC cell migration.
Figure 3.
miR-107-5p enhances the migration and invasion of EC cells. (A and B) Cell migration was continuously assessed by wound-healing assay. Ishikawa cells transfected with miR-107i revealed a significant decrease in the migration index compared with untreated cells or cells transfected with miR-107i NC at the indicated time-points (*P<0.05). (C and D) Wound healing assays revealed that HEC-1B cells transfected with miR-107i exhibited a significant decrease in the migration index compared to untreated cells or cells transfected with miR-107i NC at the indicated time-points (**P<0.01). (E) Ishikawa or HEC-1B cells (untreated or transfected with miR-107i NC or miR-107i) were seeded onto a Transwell filter. Invasive cells on the lower surface of the Transwell filter were stained after 24 h (magnification, ×200). (F) The mean ± SD number of invasive cells from panel E. *P<0.05, **P<0.01. All experiments were carried out in triplicate and repeated at least 3 times. EC, endometrial carcinoma; NC, negative control.
In accordance with the aforementioned results, the invasion rate of Ishikawa and HEC-1B cells transfected with miR-107i was significantly lower than that of untreated cells and miR-107i NC cells (Fig. 3E and F; P<0.01 and P<0.05). These results demonstrate a functional role for miR-107-5p in regulating migration and invasion in EC cells.
ERα is a functional target of miR-107-5p
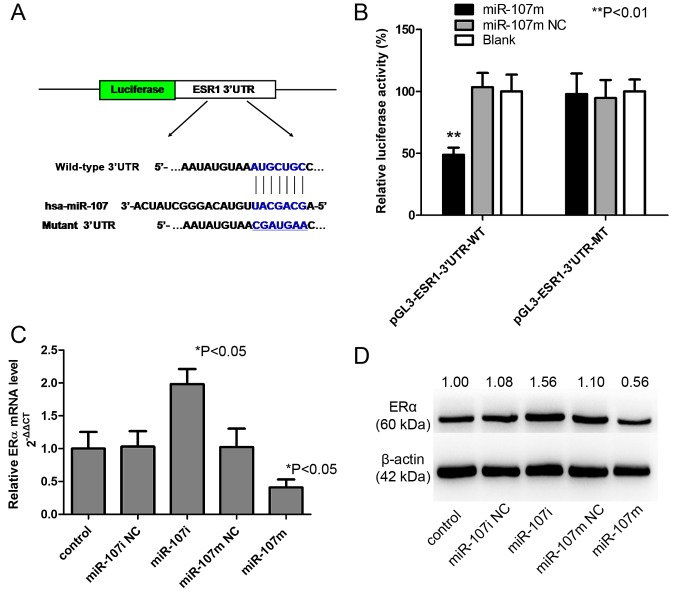
To further illuminate the underlying mechanism of miR-107-5p in EC carcinogenesis, the target of miR-107-5p must be determined. Notably, by searching for the potential targets of miR-107, we found the miR-107-5p targeting site within the 3′UTR of ESR1, as detected by 3 software algorithms (TargetScan, Pictar and miRanda) (data not shown). To directly examine whether this site was responsible for the regulation of ERα expression by miR-107-5p, a vector containing wild-type or mutant ESR1 3′UTR was constructed and directly fused downstream of firefly luciferase reporter gene (Fig. 4A). The wild-type or mutant vector was co-transfected into 293 cells with miR-107m or miR-107m NC. As revealed in Fig. 4B, miR-107 significantly decreased the relative luciferase activity of the wild-type ESR1 3′UTR (>50% inhibition, P<0.01), whereas the miR-107 NC had no significant effect. This inhibition was specific to the wild-type 3′UTR sequence, given that no reduction in luciferase activity was observed for the mutant ESR1 3′UTR. Collectively, these results indicated that ERα is a direct downstream target of miR-107-5p in 293 cells.
Figure 4.
miR-107-5p directly targets and post-transcriptionally regulates the 3′UTR of ESR1. (A) Putative miR-107 binding site in the ESR1 3′UTRas predicted by TargetScan. Schematic of the construction of pGL3-ESR1-3′UTR-WT and pGL3-ESR1-3′UTR-MT vectors is shown. The mutant binding site is underlined. (B) Luciferase assay of the pGL3-ESR1-3′UTR-WT and pGL3-ESR1-3′UTR-MT vectors transfected with miR-107m or 107m NC into 293 cells. miR-107m downregulated the activity of the wild-type ESR1 3′UTR but did not affect the activity of the mutant ESR13′UTR (**P<0.01). (C) Relative ERα mRNA expression in Ishikawa cells after transfection with a negative control mimic/inhibitor (miR-107m/107i NC) or with an miR-107 mimic/inhibitor (miR-107m/107i). Untransfected Ishikawa cells are shown for comparison. The expression levels (means ± SD of 3 independent experiments) were determined by RT-PCR and normalized to β-actin mRNA expression (*P<0.05). (D) Western blotting of ERα protein expression in Ishikawa cells after transfection with a negative control mimic/inhibitor (miR-107m/107i NC) or with an miR-107 mimic/inhibitor (miR-107m/107i). Untransfected cells are presented for comparison. β-actin was included as an internal control. All experiments were carried out in triplicate and repeated at least 3 times. 3′UTR, 3′ untranslated region; ERα, estrogen receptor α.
To further investigate the functional significance of the deregulated miR-107-5p observed in EC cell lines, Ishikawa cells were transfected with a control mimic/inhibitor (miR-107m NC/miR-107i NC) or miR-107 mimic/miR-107 inhibitor (miR-107m/miR-107i), and the effects on the expression of ERα were examined. Expression of miR-107m significantly decreased the expression of ERα at both the mRNA (Fig. 4C) and protein levels (Fig. 4D). In contrast, inhibition of miR-107-5p significantly increased the expression of ERα mRNA (Fig. 4C) and protein (Fig. 4D), further supporting its role as a functional suppressor of ERα.
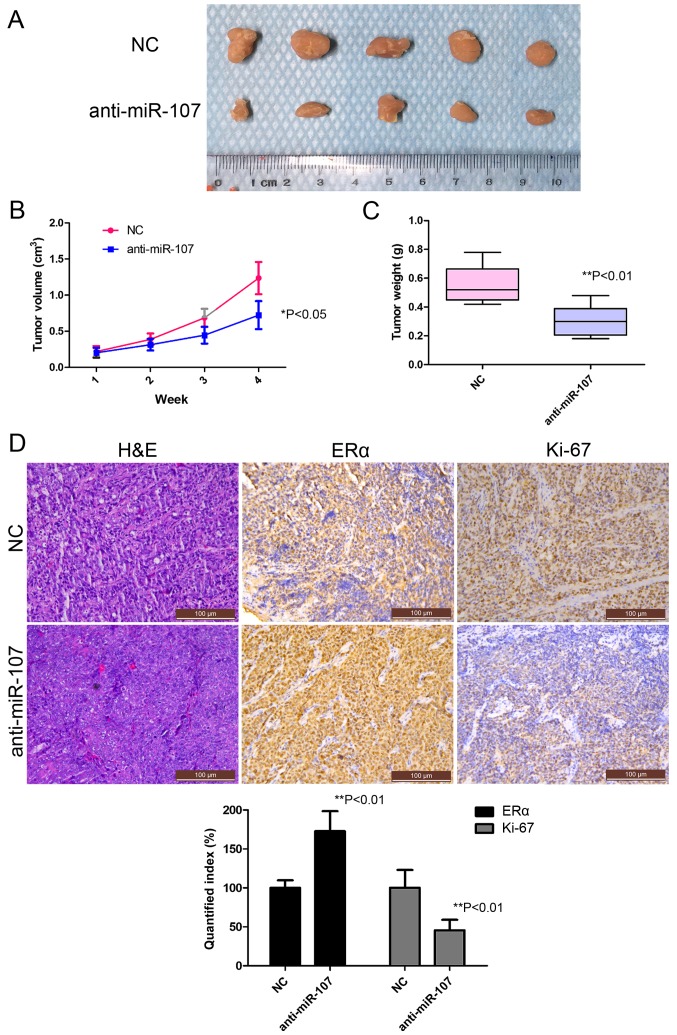
Blocking miR-107-5p inhibits tumorigenicity in vivo
To examine the potential role of miR-107-5p in tumorigenicity, we further assessed the feasibility of direct anti-miR-107 treatment in vivo. Ishikawa cells were subcutaneously injected into the flanks of nude mice in order to yield tumors that were then treated by direct intratumoral injection as soon as they became clearly palpable. For the treated group, the tumor on the flank was injected with a mixture of anti-miR-107 antagomiR, while for the control group, the tumor was injected with a control antagomiR. Tumor progression was monitored over a 28-day course. Inhibition of miR-107-5p caused a marked reduction in tumor size and tumor weight compared with the NC group over the time period tested (Fig. 5A-C). To ascertain the effects on protein expression, tumor tissue was embedded in paraffin and then stained with H&E for histological examination. As expected, Ki-67 levels were detectable in the control xenografts, while they were significantly decreased in the treated group (Fig. 5D), while ERα levels were significantly increased in the treated group (Fig. 5D, lower panel), thus confirming the role of miR-107-5p as a negative regulator of the miR-107/ERα pathway in vivo.
Figure 5.
Tumorigenicity assay in a mouse xenograft model. (A) Tumor cells were subcutaneously injected into mice in the subdermal space on the medial side of the neck. The short and long diameters of the tumors were assessed every 7 days and tumor volumes (cm3) were calculated. The results are presented as the mean ± SD of 5 mice. (A) Image of tumors derived from anti-miR-107 and anti-miR-107 NC Ishikawa cells in nude mice (n=5). (B and C) Mean ± SD of the tumor growth curve and the weights of tumors from the 2 groups. *P<0.05, **P<0.01. (D) Upper panel, representative H&E staining histopathology of anti-miR-107 NC and anti-miR-107 tumor tissues in mice (left panels). ERα and Ki-67 expression was detected by IHC (middle and right panels) (magnification, ×200). The results are representative of 3 independent experiments. Lower panel, quantified index of ERα and Ki-67 expression by IHC. **P<0.01. H&E, hematoxylin and eosin; IHC, immunohistochemistry; NC, negative control.
Discussion
In Western countries, endometrial carcinoma (EC) is the most common cancer of the female reproductive system. EC occurs mainly in peri- and postmenopausal women, although it may also be present in premenopausal women, particularly in the context of hyperestrogenism. From a clinical point of view, EC is classified into two different types: types I and II. Type I tumors are low-grade and estrogen-associated endometrioid ECs, which usually develop in perimenopausal women and coexists with or prior to endometrial hyperplasia (19). These tumors often occur in the environment of estrogen overexposure and early diagnosis has a good prognosis. However, the incidence rate of EC has risen over the last decade (20). In addition, due to obesity and physical inactivity, the onset age of patients is earlier. For early disease, surgical intervention with or without adjuvant whole pelvic radiation is the standard treatment, whereas the use of progestogen-based endocrine therapy is mainly restricted to patients with terminal or recurrent cancer. Non-invasive targeted therapies without side-effects or resistance have yet to be found (21).
ERα is an important mediator of estrogenic action in the female genital tract. E2 and its receptors are closely related to a variety of human diseases, including hormone-dependent tumors (22,23). Estrogens promote physiological actions after binding to their estrogen receptor (ER) subtypes (ERα and ERβ). The subtype ERα exists on the surface of the cell membrane and activates rapid E2-ER signaling. Amplification of the ERα gene is regarded as a common phenomenon in EC (24). Notably, presently, there is increasing coherent evidence supporting the view that ERα expression decreases in late stage and poorly differentiated ECs (25,26). Multiple hypotheses have been proposed to explain the ERα deletion, including ERα gene polymorphism (27), hypermethylation of CpG islands (28), and overexpression of estrogen receptor-associated receptors (29). However, the underlying regulatory mechanisms and the downstream biological effects of decreased expression of ERα on EC malignancy remain unclear.
Epigenetic effects are a key aspect of the relationship between miRNAs and carcinogenesis (30). Modulated expression of miRNAs has recently been related to the carcinogenesis of EC (31–33). Numerous studies have demonstrated the distinct miRNA expression profiles between EC and normal cells (32,34). Boren et al studied miRNA expression in EC, normal endometrium and complex atypical hyperplasia and found 13 miRNAs associated with EC (31). Then, Chung et al studied miRNA expression in endometrioid EC cells and normal cells of women from Hong Kong including endometrium in the proliferative and secretory phases as well as the postmenopausal atrophic endometrium. Thirty miRNAs were significantly upregulated in EC cells (33). This finding indicated that the miRNA distribution of cancer cells and normal cells is different. Notably, four types of miRNAs, namely, miR-103, miR-106a, miR-107 and miR-210, were found to be deregulated in both studies, highlighting their potential role in the pathogenesis of disease. In those four deregulated miRNAs, miR-107 piqued our interest since ERα was the predicted target of miR-107 using the miR target prediction algorithm TargetScan (http://targetscan.org/).
Recent studies indicate that the role of miR-107 in tumor development and progression is contradictory and differs in a context-dependent manner. miR-107 may play the role of a tumor suppressor gene by inducing cell cycle arrest in lung cancer and glioma (35,36) whereas it may be an oncogene that promotes tumor invasion and metastasis in breast and gastric cancer (37,38). However, the role of miR-107 in the progression and metastasis of EC remains unclear. In order to confirm the relationship between the expression of miR-107 and the progression and metastasis of EC, miR-107-5p expression with RT-PCR in 71 EC patients was examined and it was revealed that miR-107-5p was significantly upregulated in EC compared with normal endometrium. The upregulation of miR-107 was associated with an increased and more advanced histological grade and lymph node metastasis. Functionally, our data revealed that miR-107 promoted the proliferation and invasion of EC cells. Next, we found that miR-107 was able to directly bind to the 3′UTR of ESR1 mRNA in EC cells. Moreover, we observed that miR-107 could inhibit the expression of ERα at the mRNA and protein levels in the cells, indicating that miR-107 plays an important role in EC through ERα.
Some limitations in the present study should be addressed. Firstly, in a loss-of-function model, the function of miR-107 was evaluated by miR-107 gene knockout. Gain-of-function studies with overexpression of miR-107 in EC cell lines are required to verify our findings. Secondly, in order to validate the role of miR-107 in tumor development and progression, the mechanism warrants further study. It has been reported by Zhou et al that miR-107 activated the ATR/Chk1 pathway and suppressed cervical cancer invasion by targeting MCL1 (39).
In conclusion, our data demonstrated that miR-107-5p targeted ERα and that suppression of miR-107 decreased the proliferation, migration and invasion of EC in vitro and in vivo. It is thus proposed that miR-107 may serve as a useful therapeutic strategy for EC.
Acknowledgements
Not applicable.
Funding
The present study was sponsored by the National Natural Science Foundation of China (nos. 81402134 and 81502232), the Shanghai PuJiang Program (no. 17PJD032) and the Project Young Elite of the Shanghai Health System (no. 2017YQ063).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
WB, SW and YZhu conceived and designed the study. WB, YZhang, SL, QF, MQ, YW, YL, XJ and YeY performed the experiments. SL, QF, MQ, YW, YL, XJ, YeY, ZS, WX and YoY analyzed tha data. WB, YZhang, ZS, WX and YoY wrote the manuscript. WB, YZhang, SW and YZhu reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Human Investigation Ethical Committee of Shanghai General Hospital Affiliated with Shanghai Jiao Tong University. The samples of EC and normal endometrial tissues were collected after informed consent was signed by the patients. All experimental protocols were approved by the Ethics Committee of Animal Experiments of Shanghai General Hospital Affiliated with Shanghai Jiaotong University School of Medicine (Permit no. SYXK (hu) 2009-0086).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen HB, Haldorsen IS, Trovik J. Markers for individualised therapy in endometrial carcinoma. Lancet Oncol. 2012;13:e353–e361. doi: 10.1016/S1470-2045(12)70213-9. [DOI] [PubMed] [Google Scholar]
- 4.Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, Wolk A, Wentzensen N, Weiss NS, Webb PM, et al. Type I and II endometrial cancers: Have they different risk factors? J Clin Oncol. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuffa LG, Lupi-Júnior LA, Costa AB, Amorim JP, Seiva FR. The role of sex hormones and steroid receptors on female reproductive cancers. Steroids. 2017;118:93–108. doi: 10.1016/j.steroids.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Banno K, Yanokura M, Kisu I, Yamagami W, Susumu N, Aoki D. MicroRNAs in endometrial cancer. Int J Clin Oncol. 2013;18:186–192. doi: 10.1007/s10147-013-0526-9. [DOI] [PubMed] [Google Scholar]
- 10.Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, Zhang HJ, Wang LH, Wan XP. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155. doi: 10.1186/1476-4598-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Li ZY, Xu SY, Zhang XJ, Zhang Y, Luo K, Li WP. Upregulation of miR-107 inhibits glioma angiogenesis and VEGF expression. Cell Mol Neurobiol. 2016;36:113–120. doi: 10.1007/s10571-015-0225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, Xie Z, Peng Q. MiRNA-107 enhances chemosensitivity to paclitaxel by targeting antiapoptotic factor Bcl-w in non small cell lung cancer. Am J Cancer Res. 2017;7:1863–1873. [PMC free article] [PubMed] [Google Scholar]
- 13.Creasman W. Revised FIGO staging for carcinoma of the endometrium. Int J Gynaecol Obstet. 2009;105:109. doi: 10.1016/j.ijgo.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Bao W, Qiu H, Yang T, Luo X, Zhang H, Wan X. Upregulation of TrkB promotes epithelial-mesenchymal transition and anoikis resistance in endometrial carcinoma. PLoS One. 2013;8:e70616. doi: 10.1371/journal.pone.0070616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Zhou X, Xiao Q, Wang T, Shao G, Li Y, Zhang Z. MiR-107 suppresses cell proliferation and tube formation of Ewing sarcoma cells partly by targeting HIF-1β. Hum Cell. 2018;31:42–49. doi: 10.1007/s13577-017-0183-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Chen F, Zhao M, Yang Z, Zhang S, Ye L, Gao H, Zhang X. MiR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3′UTR. Biochem Biophys Res Commun. 2016;480:455–460. doi: 10.1016/j.bbrc.2016.10.070. [DOI] [PubMed] [Google Scholar]
- 18.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One. 2008;3:e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeramian A, Moreno-Bueno G, Dolcet X, Catasus L, Abal M, Colas E, Reventos J, Palacios J, Prat J, Matias-Guiu X. Endometrial carcinoma: Molecular alterations involved in tumor development and progression. Oncogene. 2013;32:403–413. doi: 10.1038/onc.2012.76. [DOI] [PubMed] [Google Scholar]
- 20.Schouten LJ, Goldbohm RA, van den Brandt PA. Anthropometry, physical activity, and endometrial cancer risk: Results from the Netherlands cohort study. Int J Gynecol Cancer. 2006;16(Suppl 2):S492. doi: 10.1111/j.1525-1438.2006.00676.x. [DOI] [PubMed] [Google Scholar]
- 21.Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8:261–271. doi: 10.1038/nrclinonc.2010.216. [DOI] [PubMed] [Google Scholar]
- 22.Chen WY. Exogenous and endogenous hormones and breast cancer. Best Pract Res Clin Endocrinol Metab. 2008;22:573–585. doi: 10.1016/j.beem.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuffa LG, Seiva FR, Fávaro WJ, Amorim JP, Teixeira GR, Mendes LO, Fioruci-Fontanelli BA, Pinheiro PF, Martinez M, Martinez FE. Melatonin and ethanol intake exert opposite effects on circulating estradiol and progesterone and differentially regulate sex steroid receptors in the ovaries, oviducts, and uteri of adult rats. Reprod Toxicol. 2013;39:40–49. doi: 10.1016/j.reprotox.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Lebeau A, Grob T, Holst F, Seyedi-Fazlollahi N, Moch H, Terracciano L, Turzynski A, Choschzick M, Sauter G, Simon R. Oestrogen receptor gene (ESR1) amplification is frequent in endometrial carcinoma and its precursor lesions. J Pathol. 2008;216:151–157. doi: 10.1002/path.2405. [DOI] [PubMed] [Google Scholar]
- 25.Jongen V, Briët J, de Jong R, ten Hoor K, Boezen M, van der Zee A, Nijman H, Hollema H. Expression of estrogen receptor-alpha and -beta and progesterone receptor-A and -B in a large cohort of patients with endometrioid endometrial cancer. Gynecol Oncol. 2009;112:537–542. doi: 10.1016/j.ygyno.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Rahman MT, Nakayama K, Rahman M, Ishikawa M, Katagiri H, Katagiri A, Ishibashi T, Sato E, Iida K, Ishikawa N, et al. ESR1 gene amplification in endometrial carcinomas: A clinicopathological analysis. Anticancer Res. 2013;33:3775–3781. [PubMed] [Google Scholar]
- 27.Ohshiro K, Mudvari P, Meng QC, Rayala SK, Sahin AA, Fuqua SA, Kumar R. Identification of a novel estrogen receptor-alpha variant and its upstream splicing regulator. Mol Endocrinol. 2010;24:914–922. doi: 10.1210/me.2009-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giacinti L, Claudio PP, Lopez M, Giordano A. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist. 2006;11:1–8. doi: 10.1634/theoncologist.11-1-1. [DOI] [PubMed] [Google Scholar]
- 29.Pelekanou V, Kampa M, Gallo D, Notas G, Troullinaki M, Duvillier H, Jacquot Y, Stathopoulos EN, Castanas E, Leclercq G. The estrogen receptor alpha-derived peptide ERα17p (P295-T311) exerts pro-apoptotic actions in breast cancer cells in vitro and in vivo, independently from their ERα status. Mol Oncol. 2011;5:36–47. doi: 10.1016/j.molonc.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boren T, Xiong Y, Hakam A, Wenham R, Apte S, Wei Z, Kamath S, Chen DT, Dressman H, Lancaster JM. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110:206–215. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 33.Chung TK, Cheung TH, Huen NY, Wong KW, Lo KW, Yim SF, Siu NS, Wong YM, Tsang PT, Pang MW, et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, Sasano H, Yaegashi N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Zhang R, Li P, Liu Y, Qin K, Fa ZQ, Liu YJ, Ke YQ, Jiang XD. P53-induced microRNA-107 inhibits proliferation of glioma cells and down-regulates the expression of CDK6 and Notch-2. Neurosci Lett. 2013;534:327–332. doi: 10.1016/j.neulet.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Zhang Y, Shi Y, Dong G, Liang J, Han Y, Wang X, Zhao Q, Ding J, Wu K, et al. MicroRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancer. J Cell Mol Med. 2011;15:1887–1895. doi: 10.1111/j.1582-4934.2010.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Zhou C, Li G, Zhou J, Han N, Liu Z, Yin J. miR-107 activates ATR/Chk1 pathway and suppress cervical cancer invasion by targeting MCL1. PLoS One. 2014;9:e111860. doi: 10.1371/journal.pone.0111860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.