Abstract
Pancreatic adenocarcinoma (PAC) is the most common type of pancreatic cancer, which commonly has an unfavorable prognosis. The present study aimed to develop a novel prognostic prediction strategy for PAC patients. mRNA sequencing data of PAC (the training dataset) were extracted from The Cancer Genome Atlas database, and the validation datasets (GSE62452 and GSE79668) were acquired from the Gene Expression Omnibus database. The differentially expressed genes (DEGs) between good and poor prognosis groups were analyzed by limma package, and then prognosis-associated genes were screened using Cox regression analysis. Subsequently, the risk score system was constructed and confirmed using Kaplan-Meier (KM) survival analysis. After the survival associated-clinical factors were screened using Cox regression analysis, they were performed with stratified analysis. Using DAVID tool, the DEGs correlated with risk scores were conducted with enrichment analysis. The results revealed that there were a total of 242 DEGs between the poor and good prognosis groups. Afterwards, a risk score system was constructed based on 6 prognosis-associated genes (CXCL11, FSTL4, SEZ6L, SPRR1B, SSTR2 and TINAG), which was confirmed in both the training and validation datasets. Cox regression analysis showed that risk score, targeted molecular therapy, and new tumor (the new tumor event days after the initial treatment according to the TCGA database) were significantly related to clinical prognosis. Under the same clinical condition, 6 clinical factors (age, history of chronic pancreatitis, alcohol consumption, radiation therapy, targeted molecular therapy and new tumor (event days) had significant associations with clinical prognosis. Under the same risk condition, only targeted molecular therapy was significantly correlated with clinical prognosis. In conclusion, the 6-gene risk score system may be a promising strategy for predicting the outcome of PAC patients.
Keywords: pancreatic adenocarcinoma, differentially expressed genes, risk score system, COX regression analysis, survival analysis
Introduction
Pancreatic cancer (PC) originates from the pancreas, and the cancerous cells have the ability to invade other parts of the body (1). PC patients in early stages often do not have obvious signs or symptoms that are specific enough to suggest pancreatic cancer, and most patients are diagnosed with late stage disease or metastasis to other organs (2). Most cases of PC occur in individuals over the age of 70 years, and PC can be induced by diabetes, tobacco smoking, obesity, and genetic conditions (3,4). PC usually has a poor prognosis, and was responsible for 411,600 deaths globally in 2015 (5). The most common type of PC is pancreatic adenocarcinoma (PAC), which consists of ~85% of all PC cases (6). Therefore, it is important to determine new biological or pathological indicators related to the prognosis of PAC in addition to conventional prognostic approaches such as clinicopathologic staging, tumor biology and molecular genetics, perioperative factors and the use of postoperative adjuvant therapy (7).
In the past decade, research has uncovered the genes affecting the survival of PC patients. For example, genetic alterations and accumulation of cyclin-dependent kinase inhibitor 2A (CDKN2A)/p16, tumor protein p53 (TP53), and SMAD family member 4 (SMAD4)/DPC4 are highly correlated with the malignant potential of PAC, and their expression levels may predict the prognosis of PAC patients (8). B-cell-specific Moloney murine leukemia virus insertion site 1 (BMI1) is reported to be significantly upregulated in PC, and its expression has a positive association with lymph node metastases and a negative correlation with the survival rates of PC patients (9,10). The expression levels of aldehyde dehydrogenase 1 family, member A1 (ALDH1A1) (11,12) and insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) could be used to predict the prognosis of PAC (13). Overexpression of homeo box B7 (HOXB7) contributes to the invasive behavior of PAC (14,15). Nevertheless, the prognostic mechanisms of PAC warrant further investigation.
Bioinformatic analysis is a new way for revealing the pathogenesis of diseases and identifying novel therapeutic targets (16). To screen the key genes correlated with the prognosis of PAC and develop novel prognostic prediction strategies, we downloaded and analyzed the public datasets of PAC. Through a series of bioinformatic analyses, a risk score system of PAC was constructed and assessed in the present study. The present study may provide a novel means for predicting the outcome of PAC patients and helping in selecting appropriate therapeutic methods.
Materials and methods
Data source
The mRNA sequencing data of PAC (the training dataset; platform: Illumina HiSeq 2000 RNA Sequencing; downloaded in March 30, 2017; including 178 PAC samples) and correlative clinical information were extracted from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) database. Meanwhile, ‘PAC’ was used as the search words for selecting relevant datasets from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. The inclusive criteria were as follows: i) the samples were human tissues (not cell lines); ii) the samples were provided with prognostic information. Finally, GSE79668 (17) [platform: GPL11154 Illumina HiSeq 2000 (Homo sapiens); 51 samples] and GSE62452 (18) [platform: GPL6244 (HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array (transcript (gene) version); 69 samples] were selected and considered as the validation datasets. The clinical information of the training dataset and the validation datasets are presented in Table I.
Table I.
Clinical information of The Cancer Genome Atlas (TCGA) dataset and the validation datasets (GSE79668 and GSE62452).
| Clinical factors | TCGA (n=178) | GSE79668 (n=51) | GSE62452 (n=69) |
|---|---|---|---|
| Age, years (mean ± SD) | 64.69±11.09 | 64.04±11.57 | – |
| Sex (male/female/-) | 91/74/13 | 32/19 | – |
| Chronic pancreatitis history (yes/no/-) | 13/117/48 | – | – |
| Diabetes history (yes/no/-) | 36/99/43 | 22/29 | – |
| Alcohol (yes/no/-) | 97/57/24 | – | – |
| Tobacco (never/reform/current/-) | 59/56/19/44 | – | – |
| New tumor (yes/no/-) | 55/101/22 | – | – |
| Pathologic_M (M0/M1/-) | 75/3/100 | 48/1/2 | – |
| Pathologic_N (N0/N1/-) | 44/117/17 | 14/37 | – |
| Pathologic_T (T1/T2/T3/T4/-) | 8/19/134/3/14 | 3/12/31/5 | – |
| Pathologic_stage (I/II/III/IV/-) | 20/137/4/3/ | – | 4/46/13/6 |
| Radiation therapy (yes/no/-) | 38/107/33 | – | |
| Targeted molecular therapy (yes/no/-) | 102/48/28 | – | |
| Deceased (death/alive/-) | 83/8213 | 45/6 | 49/16/4 |
| Overall survival months (mean ± SD) | 17.11±15.35 | 26.78±26.12 | 20.21±16.69 |
-, Indicates information unavailable. SD, standard deviation.
Differential expression analysis
Among the 178 PAC samples in the training dataset, 163 PAC samples had prognostic information. The 17 PAC samples with follow-up time <6 months whose status was still alive at the last follow-up were considered as ineligible samples since the actual survival time was unknown (data not available) due to loss of follow-up. Then, these 17 ineligible samples were removed for analysis in our study. Afterwards, the remained 146 PAC samples were divided into good prognosis and poor prognosis groups. The PAC samples obtained from living patients with a survival time >24 months were classified into a good prognosis group, and the PAC samples obtained from deceased patients with a survival time <6 months were classified into the poor prognosis group. Under the thresholds of false discovery rate (FDR) <0.05 and |logfold change (FC)| >0.585, the differentially expressed genes (DEGs) between the good and poor prognosis groups were analyzed using the R package limma (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (19).
Identification of prognosis-associated gene
The 146 PAC samples were applied for identifying prognosis-associated genes. Using univariate and multivariate Cox regression analyses in R package survival (20), prognosis-associated genes were selected from the DEGs. Then, significant P-values were obtained by log-rank test (21), and P-value <0.05 was taken as the threshold for screening prognosis-associated genes.
Construction and assessment of risk score system
Based on the prognosis-associated genes, a risk score system was constructed for the PAC patients. Firstly, the identified prognostic-associated genes were sorted by their individual P-value of the Cox regression analysis. Each gene was added one at a time in the risk score system, and the risk scores of the included gene were summed. This procedure was repeated until all the prognostic-associated genes were included. Finally, a set of minimum number of genes having the smallest P-value were selected for constructing the risk score system. Risk scores were obtained based on the linear combination of the gene expression values experiencing regression coefficient weighting. The risk score for each patient was calculated as the sum of each genes score, which was obtained by multiplying the expression level of a gene by its corresponding coefficient (β)s using the following formula:
Risk score = βgene1 × Exp gene1 + βgene2 × Exp gene2+ ··· + βgene(n) × Exp gene(n)
Subsequently, the risk of the PAC patients in the validation datasets were assessed using the β value acquired from the training dataset. Meanwhile, the differences in survival ratio were analyzed between high- and low-risk groups which were divided using the median cut-off of the risk scores as the threshold with log-rank test in Kaplan-Meier (KM) survival analysis. The differences between the low-risk and high-risk groups for expressions of the 6 genes were compared with t-test.
Correlation analysis between risk score system and clinical factors
Using the risk score system, risk scores were calculated for the samples in the training and validation datasets. According to the median of the risk scores, the samples were divided into high- and low-risk groups. Based on the clinical information corresponding to the samples, COX regression analysis (22) was used to perform correlation analysis for screening the survival associated-clinical factors.
Stratified analysis
Furthermore, stratified analysis was performed for the survival associated-clinical factors based on the following strategies: i) under the same clinical condition, the correlation between survival prognosis and high-/low-risk groups was analyzed; and ii) under the same risk condition, the correlation between survival prognosis and different clinical conditions was analyzed.
Enrichment analysis
According to the risk scores, the samples were classified into high- and low-risk groups. For the training dataset, the DEGs between high and low risk groups were identified using limma package (19). The DEGs were defined as genes with FDR <0.05. Afterwards, correlation analysis for the DEGs and risk scores were conducted. To screen significantly enriched biological processes and pathways, the DEGs positively and negatively related to risk scores were conducted with enrichment analysis using DAVID tool (https://david.ncifcrf.gov/) (23).
Results
Differential expression analysis
Among the 146 PAC samples, 18 and 19 PAC samples separately were divided into poor and good prognosis groups. Under the screening thresholds, 242 DEGs between the two groups were selected.
Construction and assessment of risk score system
Based on univariate Cox regression analysis, 165 prognosis-associated genes were selected. Moreover, the 165 prognosis-associated genes were conducted with multivariate Cox regression analysis and 8 prognosis-associated genes were further screened. Finally, 6 prognosis-associated genes [chemokine (C-X-C motif) ligand 11], CXCL11; follistatin-like 4, FSTL4; seizure related 6 homolog (mouse)-like, SEZ6L; small proline-rich protein 1B, SPRR1B; somatostatin receptor 2, SSTR2; and tubulointerstitial nephritis antigen, TINAG) were selected for constructing the risk score system (Table II). The formula was as follows:
Table II.
The 6 prognosis-associated genes to establish the risk score system.
| Genes | coef | HR | P-value |
|---|---|---|---|
| CXCL11 | 0.451453 | 0.6367 | 0.0031 |
| FSTL4 | 0.54981 | 0.5771 | 0.0025 |
| SEZ6L | −1.18976 | 3.2863 | <0.0001 |
| SPRR1B | 0.37643 | 0.6863 | 0.0004 |
| SSTR2 | 1.17541 | 0.3087 | 0.0035 |
| TINAG | 0.26515 | 0.7671 | 0.0163 |
HR, hazard ratio.
Risk score = 0.451 × Exp CXCL11 + 0.5498 × Exp FSTL4 + (−1.1897) × Exp SEZ6L + 0.376 × Exp SPRR1B + 1.175 × Exp SSTR2 + 0.265 × Exp TINAG
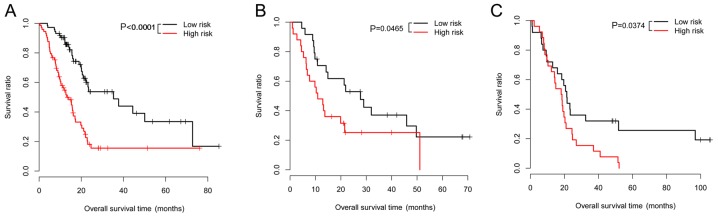
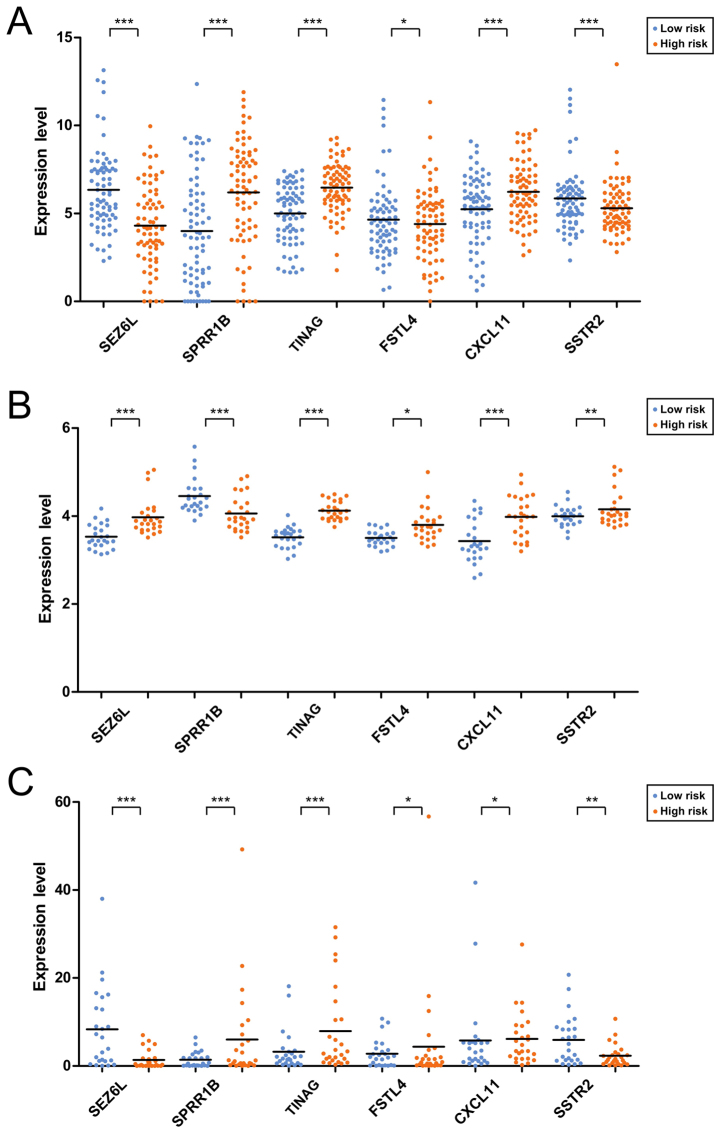
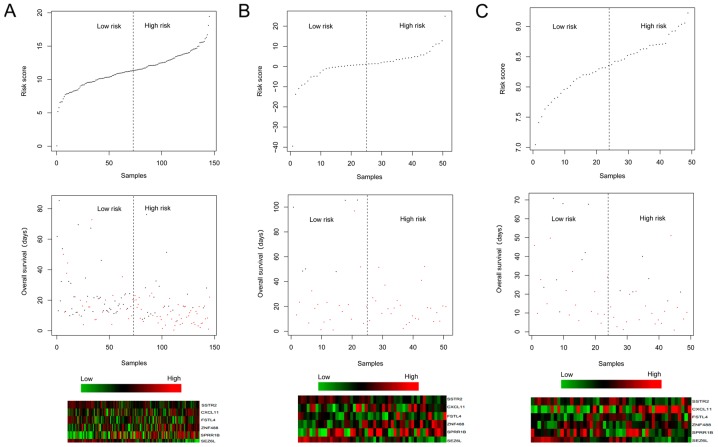
The risk scores were calculated for the samples using the risk score system. Afterwards, the 6 prognosis-associated genes were utilized for performing risk evaluation for the PAC patients. According to the median risk scores, the patients in the training dataset were classified into high-(83 patients) and low-(83 patients) risk groups. In relation to the high-risk group with the average overall survival (OS) time of 16.88±14.92 months, the low-risk group with the average OS time of 18.84±13.91 months had a higher survival ratio (P<0.0001; Fig. 1A). For the validation dataset GSE62452, the low-risk group (24 patients; average OS time=25.1±18.79 months) also had a higher survival ratio (P=0.0465) in comparison with the high-risk group (25 patients; average OS time=16.78±16.21 months) (Fig. 1B). For the validation dataset GSE79668, the low-risk group (25 patients; average OS time=37.07±32.15 months) had a higher survival ratio (P=0.0374) compared with the high-risk group (26 patients; average OS time=17.55±15.50 months) (Fig. 1C). The expression distributions of the 6 prognosis-associated genes in the high- and low-risk groups of the 3 datasets are exhibited in Fig. 2. The expression levels of SPRR1B, TINAG and CXCL11 were significantly lower, those of SEZ6L and SSTR2 were higher in the low-risk group of The Cancer Genome Atlas (TCGA) dataset (Fig. 2A). However, an obviously decreased expression level of SSTR2 was observed in the low-risk group of GSE62452 (Fig. 2B) which may be due to the fact that the gene expression model in the validation datasets could not be exactly the same as those in the training dataset.
Figure 1.
Overall survival of pancreatic adenocarcinoma (PAC) patients in low- and high-risk groups in The Cancer Genome Atlas (TCGA) dataset (A), GSE62452 (B), and GSE79668 (C). Red and black separately represent high- and low-risk groups.
Figure 2.
Expression distributions of the 6 prognosis-associated genes in the high- and low-risk groups of The Cancer Genome Atlas (TCGA) dataset (A), GSE62452 (B) and GSE79668 (C). *0.01≤P<0.05; **0.005≤P<0.01; ***P<0.005.
Correlation analysis between risk score system and clinical factors
The clinical factors significantly related to prognosis were selected by Cox regression analysis. Our results showed that risk score, targeted molecular therapy, and new tumor (event days) were significantly correlated with survival time (Table III). According to different clinical factors, the samples were divided into groups and then differential expression analysis was conducted (Table IV).
Table III.
Cox regression analysis for selecting the clinical factors significantly related to prognosis.
| Clinical characteristics | Univariable Cox P-value | Multivariable Cox P-value |
|---|---|---|
| Age in years (above/below median) | 0.0781 | |
| Sex (male/female) | 0.5630 | |
| Pathologic_M (M0/M1) | 0.3020 | |
| Alcohol (yes/no) | 0.8190 | |
| Tobacco (never/reform/current) | 0.1490 | |
| Chronic pancreatitis history (yes/no) | 0.6990 | |
| Diabetes history (yes/no) | 0.6830 | |
| Pathologic_N (N0/N1) | 0.0128 | 0.2683 |
| Pathologic_T (T1/T2/T3/T4) | 0.0338 | 0.2258 |
| Radiation therapy (yes/no) | 0.0371 | 0.5174 |
| New tumor (yes/no) | 0.0107 | 0.0269 |
| Targeted molecular therapy (yes/no) | 0.0010 | <0.0001 |
| Risk score | <0.0001 | <0.0001 |
P-values in bold print indicate significant correlations.
Table IV.
Results of differential expression analysis after dividing the samples into groups according to different clinical factors.
| Clinical factors | Downregulated genes | Upregulated genes |
|---|---|---|
| Age in years (above vs. below median) | WBSCR26, TRIM54, ARX, SERPINA4, MT1H, AQP5, PRSS21, MSLN, APOBEC1, CALHM3 | NTSR1, SPRR3, KLK10, SPRR1A, ALDH3A1, SERPINB3, CXCL17, SPRR1B, KLK1, SYCN, TRY6, CELA2B, PNLIPRP2, CLPS, CELA3A, REG1A, CELA3B, PNLIP, CELA2A |
| Sex (male vs. female) | NLRP2 | HOXA13, UPK1B |
| Chronic pancreatitis history | PNLIP, PNLIPRP1, CPA1, CELA2A, CLPS, CELA2B, TRY6, | PLEKHN1, POU2F3, CATSPER1, ABCA12, GPR110, WBSCR26, UGT1A6, |
| (yes vs. no) | REG3G, CELA3B, SYCN, TDRD9, ARX, KCNJ3, KIAA1409, ST18, TMEM132D, KCNMB2, SYT4 | HOXB9, MYEOV, S100P, GJB5, GJB4, GJB3, SFTPA2, NMU |
| Diabetes history (yes vs. no) | ABCA13 | NMUR2 |
| Alcohol (yes vs. no) | S100A2 | C5orf49 |
| Tobacco (never vs. reform) | – | – |
| Tobacco (never vs. current) | PPP1R1A, CRYBA2, SEZ6L, RIMBP2, LHFPL4, VWA5B2, PCSK1N, HMGCLL1, GRM4, ARX, TMEM63C, ASTN1, TCEAL2, LRRC10B, SSTR2, DUSP26, C1QL1, GCK, SNAP91, CACNA1A, JPH3, MSI1 | GPR110, DKK1, MUC4, SERPINB4, SERPINB3, MUC16 |
| Pathologic_M (M0 vs. M1) | – | – |
| Pathologic_T (T1+ T2 vs. T3+T4) | SYT4GPR98, SPTBN4, CELF4, ASTN1, CHD5, UNC13A, HAP1, HMGCLL1, FBLL1, PTPRT, LRRC24, ATP1A3, APOH, MSI1, PIPOX, LRRC4B, HPCA, | KPNA7, HOXA13, DSG3, UPK1B, AKR1B10 |
| Pathologic_N (N0 vs. N1) | LOC389332, GRM4, LRRC16B | MMP3, AIM2, HOXA13, ABCA13, CXCL11, EGF, GJB4, PIK3C2G, AKR1B10, ITGB6, C12orf36, KRT5, NMUR2, SERPINA4, UPK1B, GABRP, CXCL5, REG3G, CTRC, PNLIPRP1 |
| Radiation therapy (yes vs. no) | LOC554202, TNS4 | AIM2 |
| Targeted molecular therapy (yes vs. no) | ZNF683, KPNA7, RASEF, MT1H, GPR110, SERPINA4, EGF, UPK1B, KLK1, TRY6, CELA2B, SYCN | PNLIPRP2, PNLIPRP1, REG1A, CLPS, REG3G, CELA3B, CELA2A, REG1B, PNLIP, CTRC, CPA1, CELA3A, CTRB2 |
| New tumor (yes vs. no) | LOC389332, LRRC16B, FFAR2, PIPOX, RAB3C, JPH3 | CDSN, PLEKHN1, ABCA13, WBSCR26, TMEM105, GPR1, FAM83B, GJB4, CYP27C1, GJB5, LOC554202, FGFBP1, ABCA12 |
Tobacco: current, subjects who smoke at least once a month; reform, those who have tried smoking but have quit; never, those who have never tried tobacco). Pathologic_M: M0, no distant metastasis; M1, distant metastasis. Pathologic_T: T1, unilateral tumor 80 cm2 or less in area; T2, unilateral tumor more than 80 cm2 in area; T3, unilateral tumor rupture before treatment; T4, bilateral tumors. Pathologic_N: N0, no regional lymph node metastasis; N1, regional lymph node metastasis. New tumor, tumor metastasis or spread to other parts of the body.
Stratified analysis
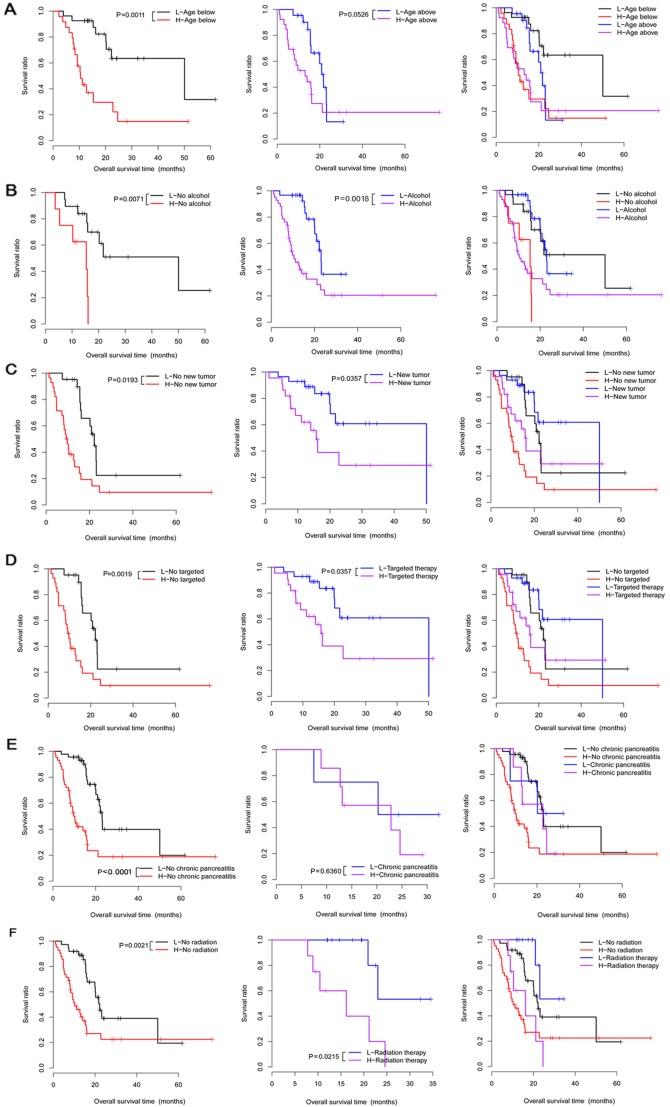
Correlation analysis under the same clinical condition showed that 6 clinical factors (age, chronic pancreatitis history, alcohol consumption, radiation therapy, targeted molecular therapy, and new tumor) under different groups were significantly correlated with survival time (Table V). Moreover, these 6 clinical factors were used to perform Kaplan-Meier (KM) survival analysis in the different groups (Fig. 3).
Table V.
Results of the stratified analysis under the same clinical condition.
| Clinical factors | P-value |
|---|---|
| Age (≥65 years, n=75) | 0.0526 |
| Age (<65 years, n=76) | 0.0011 |
| Sex (male, n=85) | 0.2520 |
| Sex (female, n=64) | 0.7210 |
| Chronic pancreatitis history (yes, n=13) | 0.6360 |
| Chronic pancreatitis history (no, n=109) | <0.0001 |
| Diabetes history (yes, n=32) | 0.1200 |
| Diabetes history (no, n=94) | 0.0936 |
| Alcohol (yes, n=90) | 0.0018 |
| Alcohol (no, n=51) | 0.0071 |
| Tobacco (never, n=54) | 0.3370 |
| Tobacco (reform, n=53) | 0.0502 |
| Tobacco (current, n=17) | 0.1180 |
| Pathologic_M (M0, N=68) | 0.6310 |
| Pathologic_M (M1, n=2) | – |
| Pathologic_N (N0, n=40) | 0.1930 |
| Pathologic_N (N1, n=105) | 0.0537 |
| Pathologic_T (T1+T2, n=24) | 0.1540 |
| Pathologic_T (T3+T4, n=124) | 0.0567 |
| Radiation therapy (yes, n=37) | 0.0215 |
| Radiation therapy (no, n=102) | 0.0021 |
| Targeted molecular therapy (yes, n=98) | 0.0357 |
| Targeted molecular therapy (no, n=45) | 0.0019 |
| New tumor (yes, n=54) | 0.0357 |
| New tumor (no, n=87) | 0.0193 |
Figure 3.
The Kaplan-Meier (KM) survival curves for the 6 clinical factors (age, alcohol use, new tumor, targeted molecular therapy, chronic pancreatitis history and radiation therapy) in high- and low-risk groups under the same clinical condition. (A) Survival curves for patients below the age of 65 (left), patients above the age of 65 (middle), and patients below or above the age of 65 years (right). (B) The survival curves for no-alcohol group (left), alcohol group (middle), and no-alcohol or alcohol groups (right). (C) Survival curves for no new tumor group (left), new tumor group (middle), and no new tumor or new tumor groups (right). (D) Survival curves for no targeted therapy group (left), targeted therapy group (middle), and no targeted therapy or targeted therapy groups (right). (E) Survival curves for no chronic pancreatitis group (left), chronic pancreatitis group (middle), and no chronic pancreatitis or chronic pancreatitis groups (right). (F) Survival curves for no radiation therapy group (left), radiation therapy group (middle), and no radiation therapy or radiation therapy groups (right). Red and black separately represent high- and low-risk groups.
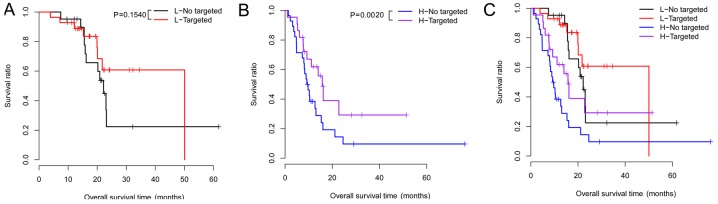
Under the same risk condition, the correlation analysis suggested that targeted molecular therapy had significant association with clinical prognosis (Table VI). KM survival analysis was also performed for targeted molecular therapy under different groups (Fig. 4). Meanwhile, the risk scores and survival time of the patients, and the expression heatmaps of the 6 prognosis-associated genes are presented in Fig. 5.
Table VI.
Results of the stratified analysis under the same risk condition.
| Clinical factors | High risk | Low risk |
|---|---|---|
| Age in years (above vs. below median) | 0.888 | 0.056 |
| Sex (male vs. female) | 0.622 | 0.939 |
| Pathologic_M (M0/M1) | 0.869 | 0.368 |
| Pathologic_N (N0 vs. N1) | 0.332 | 0.906 |
| Pathologic_T (T1 vs. T2 vs. T3) | 0.308 | 0.098 |
| Chronic pancreatitis history (yes vs. no) | 0.267 | 0.917 |
| Diabetes history (yes vs. no) | 0.643 | 0.997 |
| Alcohol (yes vs. no) | 0.803 | 0.977 |
| Tobacco (never vs. reform vs. current) | 0.210 | 0.534 |
| Radiation therapy (yes vs. no) | 0.668 | 0.173 |
| Targeted molecular therapy (yes vs. no) | 0.002 | 0.154 |
| New tumor (yes vs. no) | 0.389 | 0.997 |
Figure 4.
Kaplan-Meier (KM) survival curves for targeted molecular therapy in the high- and low-risk groups under the same risk condition. (A) Survival curve for no targeted therapy group. (B) Survival curve for targeted therapy group. (C) Survival curve for no targeted therapy or targeted therapy groups. Red and black separately represent high- and low-risk groups.
Figure 5.
Risk scores and survival time of the patients, as well as the expression heatmaps of the 6 prognosis-associated genes separately in The Cancer Genome Atlas (TCGA) dataset (A), GSE79668 (B) and GSE62452 (C).
Enrichment analysis
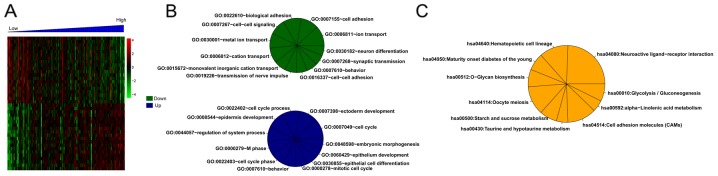
For the training set, there were 373 DEGs between the high- and low-risk groups. Correlation analysis showed that 179 and 194 DEGs separately were positively and negatively related to risk scores. Then, the top 20 DEGs were selected and conducted with clustering analysis (Fig. 6A). Additionally, multiple significantly enriched biological processes (Fig. 6B) and pathways (Fig. 6C) were obtained for these DEGs.
Figure 6.
Clustering heatmap for the top 20 differentially expressed genes (DEGs) positively or negatively related to risk scores (A), and the significantly enriched biological processes (B) and pathways (C) for the risk score-associated DEGs.
Discussion
In the present study, a total of 242 DEGs between the poor and good prognosis groups were selected. Then, 6 prognosis-associated genes (CXCL11, FSTL4, SEZ6L, SPRR1B, SSTR2 and TINAG) were selected for constructing a risk score system. The expression levels of SSTR2 were higher in the low-risk group of the TCGA dataset and GSE79668, while an obviously decreased expression level of SSTR2 was observed in the low-risk group of GSE62452. This discrepancy may be due to the fact that the gene expression model in the validation datasets could not be exactly the same as those in the training dataset. The patients in the TCGA training dataset and validation datasets (GSE62452 and GSE79668) were classified into high- and low-risk groups according to the median of risk scores which were calculated according to not only the expression levels of the 6 genes but also their regression coefficients. Moreover, the risk score system was confirmed in both the training and the two validation (GSE62452 and GSE79668) datasets, suggesting that the constructed 6-gene risk score system has prognostic prediction value. Therefore, it is necessary to select SSTR2 to build the 6-gene risk score system. Cox regression analysis showed that risk score and new tumor were significantly correlated with survival time. Under the same clinical condition, 6 clinical factors were significantly correlated with survival time. Although only targeted molecular therapy had a significant association with clinical prognosis under the same risk condition, the clinical impact was still unexplainable when various types of molecular-targeted agents were mixed. However, this association analysis was not performed since the specific method of targeted-therapy for each patient is unavailable from The Cancer Genome Atlas. In addition, multiple significantly enriched biological processes and pathways for the genes positively or negatively related to risk scores were obtained.
Angiogenesis is a typical feature of tumor cell growth, and the CXC chemokines have pleiotropic abilities in mediating tumor-correlated angiogenesis and tumor metastasis (24,25). Chemokine receptors chemokine (C-X-C motif) receptor 4 (CXCR4) and CXCR7 are co-expressed in PC samples (26). CXCL14 is highly expressed in PC tissues suggesting its correlation with the pathogenesis of PC (27). FSTL1 was found to have a low expression in PC, and inhibits the cell growth and proliferation in PC patients (28,29). The expression of SSTR2 is lost in the process of PAC development, which contributes to tumor cell growth via the activation of phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) signaling and the overexpression of CXCL16 (30). SSTR2 plays antitumor roles in PC, and its re-expression via gene transfer may be a promising gene therapy approach for the disease (31,32). Therefore, CXCL11, FSTL4 and SSTR2 may be related to the mechanisms of PAC.
However, little research has reported the involvement of SEZ6L, SPRR1B and TINAG in PAC. As a transmembrane protein with multiple domains, SEZ6L protein plays roles in signal transduction and protein-protein interaction (33). SEZ6L expression is elevated in lung cancer tissues, and SEZ6L variants are correlated with the progression of lung cancer and can increase the risk of the disease (34,35). The mRNA expression of SPRR1 is caused before the formation of Chinese hamster ovary (CHO) cells in G0 phase, and thus SPRR1 expression is responsive to growth-arresting signals (36). As a basement membrane glycoprotein, TINAG can be recognized by autoantibodies in some types of human tubulointerstitial nephritis (37). The TINAG-related protein (TINAG-RP) was found to have higher expression levels in a colorectal adenocarcinoma cell line (38). SEZ6L, SPRR1B and TINAG play roles in other types of malignant tumors, indicating that they may also function in the development and progression of PAC.
Furthermore, the following limitations should be mentioned in this study. On the one hand, the prognostic prediction model based on the expression levels of these 6 prognosis-associated genes should be validated in an independent patient cohort by clinical experiments. Whether our model is superior to conventional prognostic factors still needs to be explored based on more research. On the other hand, the prediction accuracy of the risk score system may be influenced by data heterogeneity, platform differences and sample size differences of the training and validation datasets. Thus, further experiments are still needed to confirm these results.
In conclusion, 242 DEGs between the poor and good prognosis groups were screened, and 6 prognosis-associated genes (CXCL11, FSTL4, SEZ6L, SPRR1B, SSTR2 and TINAG) were selected for constructing a risk score system. Moreover, the 6-gene risk score system may be utilized for predicting the clinical prognosis of PAC patients. However, further research is still needed to validate the prognostic prediction value based on the expression levels of these 6 prognosis-associated genes in an independent patient cohort with PAC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YL performed the data analyses and wrote the manuscript. DZ, HX and YH contributed significantly in data analyses and manuscript revision. YS conceived and designed the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
In the original article of the datasets, the trials were approved by the local institutional review boards of all participating centers, and informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Husain K. Pancreatic cancer treatment. Cancer Sci. 2014;5:1100–1100. [Google Scholar]
- 2.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.Maisonneuve P, Lowenfels AB. Risk factors for pancreatic cancer: A summary review of meta-analytical studies. Int J Epidemiol. 2015;44:186–198. doi: 10.1093/ije/dyu240. [DOI] [PubMed] [Google Scholar]
- 4.Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:197–209. doi: 10.1016/j.bpg.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 5.McGuire S. World cancer report 2014. Geneva, Switzerland: World health organization, international agency for research on cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond-Smith G, Banga N, Hammond TM, Imber CJ. Pancreatic adenocarcinoma. BMJ. 2012;344:e2476. doi: 10.1136/bmj.e2476. [DOI] [PubMed] [Google Scholar]
- 7.Yeo CJ, Cameron JL. Prognostic factors in ductal pancreatic cancer. Langenbecks Arch Surg. 1998;383:129–133. doi: 10.1007/s004230050104. [DOI] [PubMed] [Google Scholar]
- 8.Oshima M, Okano K, Muraki S, Haba R, Maeba T, Suzuki Y, Yachida S. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann Surg. 2013;258:336–346. doi: 10.1097/SLA.0b013e3182827a65. [DOI] [PubMed] [Google Scholar]
- 9.Song W, Tao K, Li H, Jin C, Song Z, Li J, Shi H, Li X, Dang Z, Dou K. Bmi-1 is related to proliferation, survival and poor prognosis in pancreatic cancer. Cancer Sci. 2010;101:1754–1760. doi: 10.1111/j.1349-7006.2010.01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proctor E, Waghray M, Lee CJ, Heidt DG, Yalamanchili M, Li C, Bednar F, Simeone DM. Bmi1 enhances tumorigenicity and cancer stem cell function in pancreatic adenocarcinoma. PLoS One. 2013;8:e55820. doi: 10.1371/journal.pone.0055820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlert C, Bergmann F, Beck J, Welsch T, Mogler C, Herpel E, Dutta S, Niemietz T, Koch M, Weitz J. Low expression of aldehyde deyhdrogenase 1A1 (ALDH1A1) is a prognostic marker for poor survival in pancreatic cancer. BMC Cancer. 2011;11:275. doi: 10.1186/1471-2407-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshino Y, Nishida J, Katsuno Y, Koinuma D, Aoki T, Kokudo N, Miyazono K, Ehata S. Smad4 decreases the population of pancreatic cancer-initiating cells through transcriptional repression of ALDH1A1. Am J Pathol. 2015;185:1457–1470. doi: 10.1016/j.ajpath.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Schaeffer DF, Owen DR, Lim HJ, Buczkowski AK, Chung SW, Scudamore CH, Huntsman DG, Ng SS, Owen DA. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer. 2010;10:59. doi: 10.1186/1471-2407-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen Kovochich A, Arensman M, Lay AR, Rao NP, Donahue T, Li X, French SW, Dawson DW. HOXB7 promotes invasion and predicts survival in pancreatic adenocarcinoma. Cancer. 2013;119:529–539. doi: 10.1002/cncr.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chile T, Fortes MA, Corrêa-Giannella ML, Brentani HP, Maria DA, Puga RD, de Paula Vde J, Kubrusly MS, Novak EM, Bacchella T, et al. HOXB7 mRNA is overexpressed in pancreatic ductal adenocarcinomas and its knockdown induces cell cycle arrest and apoptosis. BMC Cancer. 2013;13:451. doi: 10.1186/1471-2407-13-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Szustakowski J, Schinke M. Bioinformatics analysis of microarray data. Methods Mol Biol. 2009;573:259–284. doi: 10.1007/978-1-60761-247-6_15. [DOI] [PubMed] [Google Scholar]
- 17.Kirby MK, Ramaker RC, Gertz J, Davis NS, Johnston BE, Oliver PG, Sexton KC, Greeno EW, Christein JD, Heslin MJ, et al. RNA sequencing of pancreatic adenocarcinoma tumors yields novel expression patterns associated with long-term survival and reveals a role for ANGPTL4. Mol Oncol. 2016;10:1169–1182. doi: 10.1016/j.molonc.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, He P, Wang J, Schetter A, Tang W, Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al. A novel MIF signaling pathway drives the malignant character of pancreatic cancer by targeting NR3C2. Cancer Res. 2016;76:3838–3850. doi: 10.1158/0008-5472.CAN-15-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Limma: Linear models for microarray data. Bioinformatics Comput Biol Solutions Using R and Bioconductor. 2005:397–420. doi: 10.1007/0-387-29362-0_23. [DOI] [Google Scholar]
- 20.Therneau T. A package for survival analysis. R package. 2012:2.37–2. [Google Scholar]
- 21.Kleinbaum DG, Klein M. Kaplan-meier survival curves and the log-rank test. Statistics Biol Health. 2012:45–82. [Google Scholar]
- 22.Gui J, Li H. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005;21:3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeley EC, Mehrad B, Strieter RM. CXC chemokines in cancer angiogenesis and metastases. Adv Cancer Res. 2010;106:91–111. doi: 10.1016/S0065-230X(10)06003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeley EC, Mehrad B, Strieter RM. Chemokines as mediators of tumor angiogenesis and neovascularization. Exp Cell Res. 2011;317:685–690. doi: 10.1016/j.yexcr.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinrich EL, Lee W, Lu J, Lowy AM, Kim J. Chemokine CXCL12 activates dual CXCR4 and CXCR7-mediated signaling pathways in pancreatic cancer cells. J Transl Med. 2012;10:68. doi: 10.1186/1479-5876-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wente MN, Mayer C, Gaida MM, Michalski CW, Giese T, Bergmann F, Giese NA, Büchler MW, Friess H. CXCL14 expression and potential function in pancreatic cancer. Cancer Lett. 2008;259:209–217. doi: 10.1016/j.canlet.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Viloria K, Munasinghe A, Asher S, Bogyere R, Jones L, Hill NJ. A holistic approach to dissecting SPARC family protein complexity reveals FSTL-1 as an inhibitor of pancreatic cancer cell growth. Sci Rep. 2016;6:37839. doi: 10.1038/srep37839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trojan L, Schaaf A, Steidler A, Haak M, Thalmann G, Knoll T, Gretz N, Alken P, Michel MS. Identification of metastasis-associated genes in prostate cancer by genetic profiling of human prostate cancer cell lines. Anticancer Res. 2005;25:183–191. [PubMed] [Google Scholar]
- 30.Chalabidchar M, Cassantsourdy S, Duluc C, Fanjul M, Lulka H, Samain R, Roche C, Breibach F, Delisle MB, Poupot M, et al. Loss of somatostatin receptor subtype 2 promotes growth of KRAS-induced pancreatic tumors in mice by activating PI3K signaling and overexpression of CXCL16. Gastroenterology. 2015;148:1452–1465. doi: 10.1053/j.gastro.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Du ZY, Qin RY, Xia W, Tian R, Kumar M. Gene transfer of somatostatin receptor type 2 by intratumoral injection inhibits established pancreatic carcinoma xenografts. World J Gastroenterol. 2005;11:516–520. doi: 10.3748/wjg.v11.i4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrere N, Vernejoul F, Souque A, Asnacios A, Vaysse N, Pradayrol L, Susini C, Buscail L, Cordelier P. Characterization of the bystander effect of somatostatin receptor sst2 after in vivo gene transfer into human pancreatic cancer cells. Human Gene Ther. 2005;16:1175–1193. doi: 10.1089/hum.2005.16.1175. [DOI] [PubMed] [Google Scholar]
- 33.Pigoni M, Wanngren J, Kuhn PH, Munro KM, Gunnersen JM, Takeshima H, Feederle R, Voytyuk I, De Strooper B, Levasseur MD, et al. Seizure protein 6 and its homolog seizure 6-like protein are physiological substrates of BACE1 in neurons. Mol Neurodegener. 2016;11:67. doi: 10.1186/s13024-016-0134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishioka M, Kohno T, Takahashi M, Niki T, Yamada T, Sone S, Yokota J. Identification of a 428-kb homozygously deleted region disrupting the SEZ6L gene at 22q12.1 in a lung cancer cell line. Oncogene. 2000;19:6251–6260. doi: 10.1038/sj.onc.1204031. [DOI] [PubMed] [Google Scholar]
- 35.Gorlov IP, Meyer P, Liloglou T, Myles J, Boettger MB, Cassidy A, Girard L, Minna JD, Fischer R, Duffy S, et al. Seizure 6-like, SEZ6L) gene and risk for lung cancer. Cancer Res. 2007;67:8406–8411. doi: 10.1158/0008-5472.CAN-06-4784. [DOI] [PubMed] [Google Scholar]
- 36.Tesfaigzi Y, Wright PS, Belinsky SA. SPRR1B overexpression enhances entry of cells into the G0 phase of the cell cycle. Am J Physiol Lung Cell Mol Physiol. 2003;285:L889–L898. doi: 10.1152/ajplung.00065.2003. [DOI] [PubMed] [Google Scholar]
- 37.Yoshioka K, Takemura T, Hattori S. Tubulointerstitial nephritis antigen: Primary structure, expression and role in health and disease. Nephron. 2002;90:1–7. doi: 10.1159/000046307. [DOI] [PubMed] [Google Scholar]
- 38.Wex T, Lipyansky A, Brömme NC, Wex H, Guan XQ, Brömme D. TIN-ag-RP, a novel catalytically inactive cathepsin B-related protein with EGF domains, is predominantly expressed in vascular smooth muscle cells. Biochemistry. 2001;40:1350–1357. doi: 10.1021/bi002266o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.