Abstract
The microbiota and bacterial metabolites in the colon are regarded as alternative targets for colon cancer prevention and therapy. Among these metabolites, short-chain fatty acids (SCFAs) exhibit anticancer effects and suppress inflammation in the colon. However, the molecular mechanisms and target development of SCFAs require additional study. In the present study, using RNA-seq results from colon cancer samples derived from the Cancer Genome Atlas (TCGA) portal, overexpressed epigenetic modifiers were identified and RT-PCR and qRT-PCR analysis was performed to select target genes that responded to treatment with propionate in HCT116 cells. Downregulation of protein arginine methyltransferase 1 (PRMT1), a histone arginine methyltransferase, was observed after sodium propionate (SP) treatment. Moreover, phospho-array analysis demonstrated that the mTOR pathway was involved in propionate and siPRMT1 treatment, and regulation of this pathway was associated with apoptosis in HCT116 cells. The present study, to the best of our knowledge, was the first to demonstrate that PRMT1 levels were reduced by propionate treatment in HCT116 cells and that downregulation of PRMT1 induced cell apoptosis. Thus, this novel mechanism of sodium propionate treatment for colon cancer therapy may indicate more effective approaches, such as dietary therapy, for CRC patients.
Keywords: colon cancer, PRMT1, propionate, apoptosis
Introduction
Colorectal cancer (CRC; colon and rectal cancer) is the most common cancer worldwide. To overcome the challenges of CRC treatment, recent studies of microbiota and metabolites derived from the microbiome in the colon, have investigated alternative methods for colon cancer prevention and treatment (1). Among these metabolites, short-chain fatty acids (SCFAs; butyrate, acetate and propionate) are produced from dietary fiber in the colon via bacterial fermentation to maintain host health (2). Moreover, in a disease context, SCFAs are related to colonic inflammation, insulin resistance and high-fat-diet-induced obesity (3). The signaling components activated by SCFAs have been divided into two main pathways: a histone deacetylase (HDAC) inhibitor pathway and a G-protein coupled receptor (GPCR) activation pathway (2). In colon cancer, treatment with SCFAs was revealed to induce hyperacetylation and suppress colon cancer proliferation and metastasis (4–7). Specifically, butyrate induced apoptosis in HCT116 cells by regulating p38 MAPK. Additionally, in an in vivo study, SCFA mixtures suppressed azoxymethane/dextran sodium sulfate (AOM/DSS)-induced tumors by inhibiting COX-2 expression (7–10). In addition, propionate treatment was revealed to reduce free fatty acid receptor 2 (FFAR2; also known as GPR43) expression in leukemic cell lines and to reduce leukemic cell proliferation (11). However, despite studies on the roles of SCFAs in the colon, their molecular mechanisms and target development require further investigation. Moreover, since SCFA studies have focused mainly on butyrate treatment, the relationship between propionate and colon cancer was examined in this study.
SCFA treatment in colon cancer has been revealed to induce hyper-acetylation by inhibiting HDAC activity, suggesting that SCFAs can act as an HDAC inhibitor. Although major studies of the anticancer effects of SCFAs (mainly butyrate) have focused on inhibiting HDAC activity, the molecular mechanism of propionate in colon cancer remains unclear. Epigenetic modifiers, such as histone methyltransferase, acetylase and DNA methyltransferase, have become important target genes for developing anticancer drugs for colon cancer (12,13). Among these genes, protein arginine methyltransferase 1 (PRMT1) is a histone arginine methyltransferase that mainly mono- and dimethylates histone H4 arginine 3, which is an activation site for gene expression (14,15). In colon cancer, PRMT1 was revealed to be involved in epidermal growth factor (EGF) receptor methylation during resistance to cetuximab treatment, and methylation of methyl-DNA binding domain 2 (MBD2) by PRMT1 inhibited binding to methyl-CG DNA in colon cancer (16,17). Moreover, PRMT1 overexpression was revealed to be clearly associated with poor prognosis in colon cancer cohorts. Although the relationship between colon cancer and PRMT1 has been studied, regulation of PRMT1 expression in colon cancer is unclear.
Therefore, in this study, the HCT116 cell line was treated with sodium propionate (SP) and cellular apoptosis was observed upon downregulation of PRMT1 expression. This is the first study, to the best of our knowledge, revealing that downregulation of PRMT1 expression by propionate, induced apoptosis by inhibiting p70 S6 kinase phosphorylation. Thus, PRMT1 inhibitors and propionate treatment are expected to reveal synergistic anticancer effects which may be used in the treatment of CRC patients.
Materials and methods
Cell culture and reagents
The human colon cancer cell lines HCT116 and SW480 were purchased from the Korean Cell Line Bank (Seoul, South Korea) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humidified atmosphere with 5% CO2 at 37°C. Sodium propionate (product no. P5436) was purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
CRC patients in TCGA data
The mRNA expression (RNA-Seq) data of 572 colon-related samples (51 normal samples and 521 tumor samples) was obtained from TCGA data portal (http://cancergenome.nih.gov). The RNA-Seq quantification data (HTSeq-FPKM) was downloaded and the mean value for the expression levels of each gene across samples was calculated. These mean values represented the expression each gene in normal and tumor samples.
Cell viability assay
Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Rockville, MD, USA) was used to conduct cell viability assays. Cells were seeded in 6-well plates starting at 4×105 cells/well and incubated for 24 h. After sodium propionate treatment and siRNA transfection for 2 days, CCK-8 solution and RPMI-1640 medium with 10% FBS mixture were added into each well and incubated with 5% CO2 at 37°C for 5 min or 10 min. The absorbance was assessed using a microplate reader at 450 nm. For crystal violet staining, the cells were fixed with methanol for 5 min and stained with 0.1% crystal violet after sodium propionate treatment and siRNA infection for 48 h.
Human phospho-kinase array
Human phospho-kinase array was performed using Proteome Profiler Human Phospho-Kinase Array kit (cat. no. ARY003B) purchased from R&D Systems, Inc. (Minneapolis, MN, USA). HCT116 cells, treated with sodium propionate and PRMT1 siRNA, were lysed in lysis buffer. Cell lysates were centrifuged at 14,000 × g for 5 min at 4°C and diluted to 400 µg per array set. Cell lysates were incubated with membranes overnight at 4°C on a rocking platform shaker. Then, the membranes were incubated with detection antibody cocktail for 2 h and Streptavidin-HRP for another 30 min at room temperature. The signal was detected by incubating the membranes with Chemi Reagent Mix and exposing to film.
FACS analysis
After treatment with SP or knockdown of PRMT1 for 2 days, the cells were collected and incubated with the MUSE Annexin V and Dead Cell Assay kit (cat. no. MCH100105; Merck KGaA) for 20 min at room temperature. After incubation, ~5×104 cells were analyzed using a Muse Cell analyzer (Merck KGaA) (18). The FACS result was analyzed using Muse 1.5 Analysis (Merck KGaA).
siRNA transfection
siRNA duplexes against PRMT1 (siPRMT1; 5′-GAGUUCACACGCUGCCACATT-3′, 5′-UGUGGCAGCGUGUGAACUCTT-3′) were purchased from ST Pharm Co., Ltd. (Seoul, South Korea). Negative control siRNA (siCont; 5′-AUGAACGUGAAUUGCUCAATT-3′, 5′-UUG AGCAAUUCACGUUCACTT-3′) was used as a control (19). The siRNAs (100 nM) were transfected into cancer cell lines using RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 48 h.
Semi-quantitative reverse transcription PCR and quantitative real-time PCR
Total RNA was isolated from the indicated cell lines using a Qiagen RNeasy Mini kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturers instructions. RNA aliquots of 1 µg were then reverse-transcribed using the iScript™ cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA), according to the standard protocols provided by the manufacturer. For semi-quantitative RT-PCR, cDNA was used as a template for PCR using AccuPower® HotStart PCR PreMix (Bioneer, Daejeon, South Korea). For quantitative RT-PCR, PCR reactions (PRMT1: annealing temperature 55°C, 30 cycles; ACTB: annealing temperature 58°C, 28 cycles) were performed using the SimpliAmp Thermal Cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.) following the manufacturers instructions. Quantitative real-time PCR was performed on cDNA samples using Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix and the signal was detected using AriaMx Real-Time PCR System (both from Agilent Technologies, Inc., Santa Clara, CA, USA). The fluorescence threshold value was calculated using Agilent Aria 1.6 software (Agilent Technologies, Inc.) (20,21). The PCR primers were as follows: PRMT1 forward, 5′-GGGCTACTGCCTCTTCTACGAGTC-3′ and reverse, 5′-GTCTTTGTACTGCCGGTCCTCGTAG-3′; and ACTB forward, 5′-ACTCTTCCAGCCTTCCTTCC-3′ and reverse, 5′-CAATGCCAGGGTACATGGTG-3′.
Western blot analysis
The cells were washed once with phosphate-buffered saline (PBS) and then lysed in cold lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF and 1X protease inhibitor cocktail). Cell lysates were centrifuged at 14,000 × g for 15 min at 4°C and then boiled in 5X sample buffer following protein determination (BSA, cat. no. 23208; Thermo Fisher Scientific, Inc.). The protein samples were subjected to western blot analysis. For western blot analysis, nitrocellulose membranes (cat. no. 1620145; Bio-Rad Laboratories) and blocking reagent (5% skim milk, 1 h at room temperature), 4–20% precasting gel (cat. no. 456-1094; Bio-Rad Laboratories) were used with the indicated antibodies at a 1:1,000 dilution ratio (22). The samples were stained with the following antibodies: anti-PRMT1 (cat. no. 2449), PARP (cat. no. 9542), caspase-3 (cat. no. 9662), phospho-p70 S6 kinase (Thr389) (cat. no. 9234), p70 S6 kinase (cat. no. 9202) all from Cell Signaling Technology (Danvers, MA, USA) and PRMT1 (cat. no. sc-166963) and ACTB (cat. no. sc-47778) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Secondary antibodies (rabbit; cat. no. SC-2357, mouse; cat. no. SC-2031; Santa Cruz Biotechnology) were incubated at room temperature for 1 h, and ECL solution (cat. no. 170-5060; Bio-Rad Laboratories) was used for visualization. Chemiluminescence imaging system (Mini HD9; UVitec, Cambridge, UK) was used for imaging.
Statistical analysis
Results were expressed as the mean ± SD (error bars) of three independent experiments. One-way analysis of variance (ANOVA) with Bonferroni post hoc test was performed using Graphpad Prism 5.0 (Graphpad Software, Inc., La Jolla, CA, USA). The cut-off for significance was P<0.05.
Results
SP suppresses the proliferation of the HCT116 and SW480 cell lines
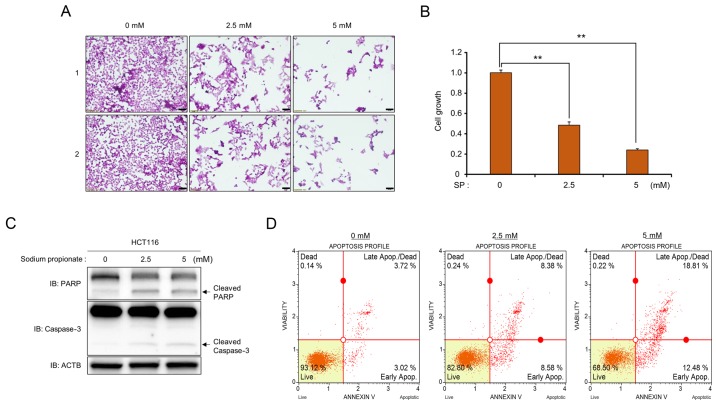
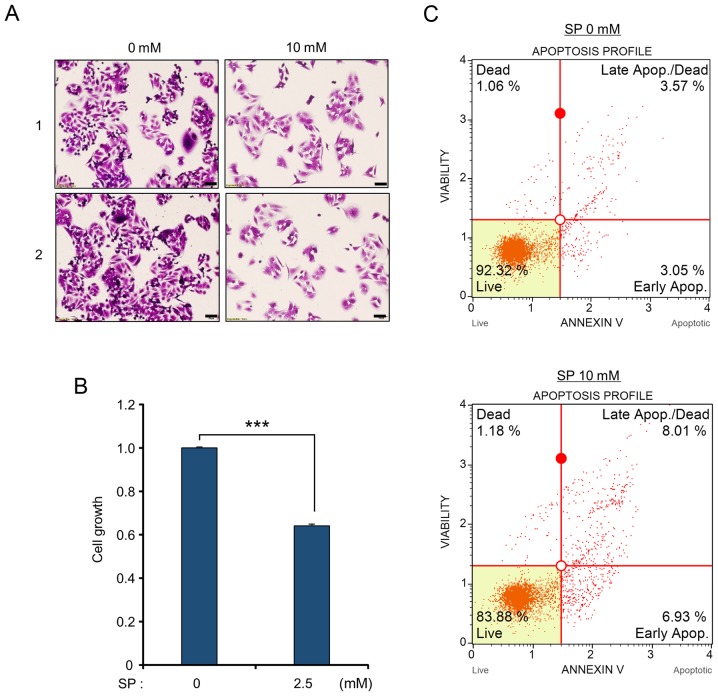
First, to verify the anticancer activity of propionate in colon cancer, a cell growth assay and CCK-8 analysis were performed. Following treatment with SP, a significant SP dose-dependent reduction of the number of cells and growth rates was observed compared to those of the control (Figs. 1A and B and 2A and B). In addition, in western blot analysis, cleaved PARP and cleaved caspase-3 were clearly increased after SP treatment, implying that propionate suppressed cell proliferation in CRC cell lines via the apoptosis pathway (Fig. 1C). Moreover, FACS analysis using Annexin V staining after treatment with SP revealed that the proportion of apoptotic cells (early and late apoptotic cells) increased compared to that of the control group in a dose-dependent manner in HCT116 and SW480 cell lines (Figs. 1D and 2C). Thus, treatment of CRC cell lines with propionate revealed anticancer effects similar to those of butyrate treatment.
Figure 1.
SP induces cell apoptosis in the HCT116 cell line. (A and B) Cell growth assay (crystal violet staining and CCK-8 analysis). (A) After treatment with SP (0, 2.5 and 5 mM), cell fixation with 100% methanol and cell staining with crystal violet were performed. ‘1’ and ‘2’ are results of two independent experiments after treatment with SP. The cells were imaged under a microscope. Scale bar, 50 µm. (B) In addition, CCK-8 solution was added to the culture medium, and an incubation step was performed for 10 min at 37°C. The intensity of cell growth was assessed by a microplate reader (450 nm). P-values were calculated using one-way ANOVA (**P<0.01). (C) Western blot analysis after treatment with SP using anti-PARP and anti-caspase-3 antibodies. ACTB was used as the internal control. (D) FACS analysis using Annexin V staining was performed after treatment with SP. The lower right and upper right quadrants indicate early apoptosis and late apoptosis, respectively. SP, sodium propionate, ACTB, β-actin.
Figure 2.
Induction of cell apoptosis of SW480 cells by SP treatment. (A and B) Cell growth assay. (A) After treatment with SP (0 and 10 mM), cell fixation with 100% methanol and cell staining with crystal violet were performed. ‘1’ and ‘2’ are results of two independent experiments after treatment with SP. The cells were imaged under a microscope. Scale bar, 50 µm. (B) CCK-8 solution was added to the culture medium, and an incubation step was performed for 5 min at 37°C. The intensity of cell growth was assessed by a microplate reader (450 nm). P-values were calculated using Students t-tests (***P<0.001). (C) FACS analysis using Annexin V staining was performed after treatment with SP. The lower right and upper right quadrants indicate early apoptosis and late apoptosis, respectively. SP, sodium propionate.
Treatment with propionate reduces PRMT1 expression in the HCT116 cell line
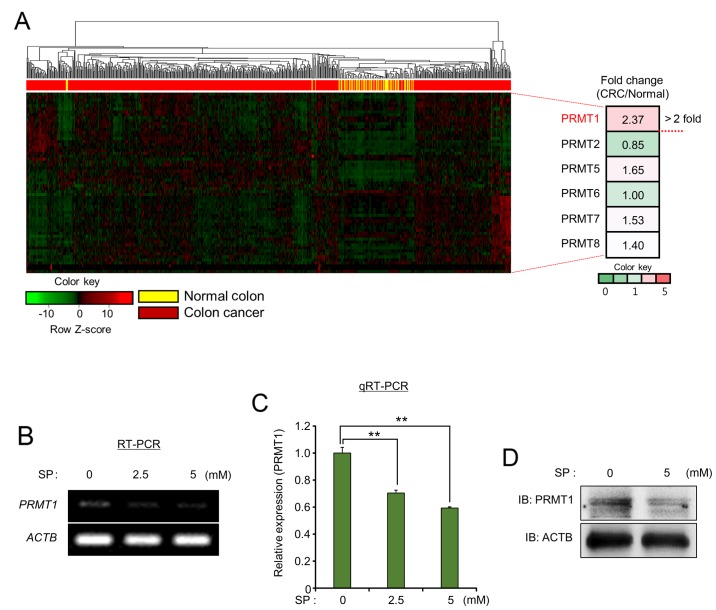
Recently, epigenetic modifiers related to DNA methylation and histone modification have been strongly associated with cell proliferation and metastasis in several types of cancer (23). Therefore, in the present study, the identification of the novel epigenetic modifiers controlled by propionate treatment in colon cancer, was attempted. To investigate the relationship between epigenetic modifiers and propionate treatment in colon cancer, an in silico epigenetic panel (61 histone methyltransferases/demethylases, 28 histone acetylases/deacetylases and 7 DNA methyltransferases/demethylases) was established to select epigenetic modifiers overexpressed in colon cancer (data not shown). To examine the expression levels of epigenetic modifiers, the RNA-seq results for colon cancer (n=521) and normal samples (n=51) derived from the TCGA portal were evaluated, and PRMT1, which is a histone arginine methyltransferase 1 (>2-fold change), was selected from the PRMTs (Fig. 3A). Since the knockdown of PRMT1 was revealed to suppress cell proliferation in hepatocellular carcinoma (HCC), lung cancer and bladder cancer (15,24), RT-PCR and qRT-PCR analysis after treatment with SP in the HCT116 cell line was performed to assess the relationship between propionate treatment and PRMT1 expression. As observed in Fig. 3B and C, the expression level of PRMT1 was decreased by SP treatment in a dose-dependent manner. Concurrently, the reduction of PRMT1 expression by SP treatment using western blot analysis was confirmed (Fig. 3D). Collectively, these data revealed the negative regulation of PRMT1 expression by propionate treatment in colon cancer, indicating that PRMT1 downregulation induced by SP treatment may lead to anticancer effects via the induction of cell apoptosis.
Figure 3.
PRMT1 expression is downregulated by SP treatment. (A) Heat map of gene expression related to PRMT genes, sorted by fold change of the CRC/normal FPKM value. (B and C) RT-PCR and qRT-PCR analysis of PRMT1. ACTB was used as an internal control. P-values were calculated using one-way ANOVA (**P<0.01). (D) Western blot analysis of PRMT1 after treatment with SP. ACTB was used as the internal control. PRMT1, protein arginine methyltransferase 1; SP, sodium propionate; CRC, colorectal cancer; ACTB, β-actin.
Overexpression of PRMT1 in colon cancer
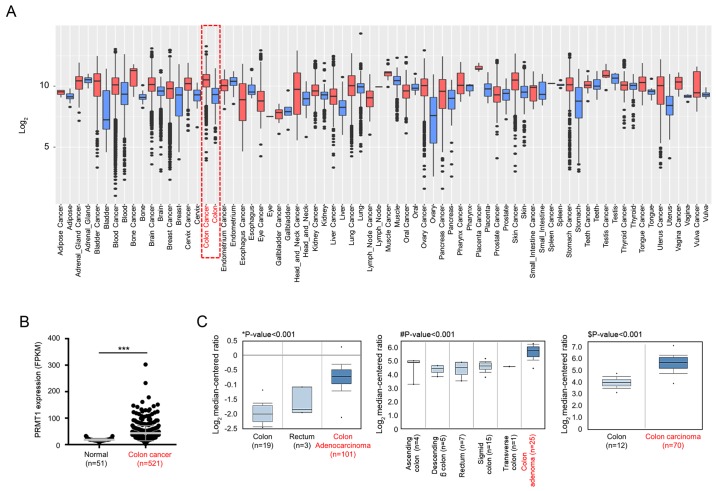
GENT2 analysis (http://gent2.appex.kr/gent2/) revealed that PRMT1 was overexpressed in several types of cancer, including colon cancer (Fig. 4A), and dot blot graph revealed that the expression levels of PRMT1 were significantly increased in colon cancer, with data derived from the TCGA portal (Fig. 4B). Additionally, in Oncomine data (https://www.oncomine.org), the expression level of PRMT1 in colon cancer samples was higher than that in normal colon and rectum samples (Fig. 4C). Thus, in colon cancer, PRMT1 was overexpressed, and the functions of PRMT1 in colon cancer may be involved in colon cancer proliferation. These results indicated that overexpressed PRMT1 may become a useful molecular target for propionate treatment.
Figure 4.
PRMT1 is overexpressed in colon cancer. (A) Tissue-wide expression profile of PRMT1 in normal/cancer tissues using GENT2 analysis (http://gent2.appex.kr/gent2/). (B) Expression levels of PRMT1 using normal and CRC samples (***P<0.001). (C) Overexpression of PRMT1 in Oncomine data (www.oncomine.org). *P<0.001 vs. colon adenocarcinoma; #P<0.001 vs. colon adenoma; $P<0.001 vs. colon carcinoma. The cut-off for significance was P<0.05. PRMT1, protein arginine methyltransferase 1; CRC, colorectal cancer; FPKM, fragments per kilobase million.
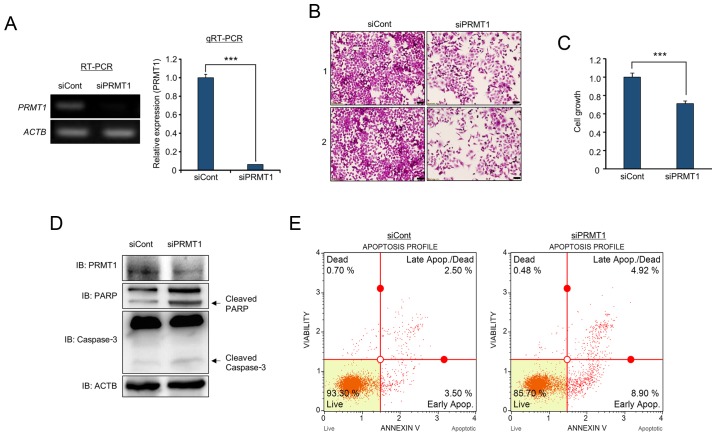
Knockdown of PRMT1 suppresses cell proliferation via the apoptosis pathway
To determine whether overexpressed PRMT1 is related to colon cancer proliferation, a PRMT1-specific siRNA was designed and PRMT1 knockdown analysis was performed. In RT-PCR and qRT-PCR analysis, PRMT1 expression was significantly decreased by treatment with siPRMT1 compared to that in cells treated with siCont (Fig. 5A). In cell growth analysis, cell staining with crystal violet revealed that the number of stained cells was reduced by PRMT1 knockdown (Fig. 5B). In addition, the proliferation of HCT116 cells was clearly suppressed after the knockdown of PRMT1 using CCK-8 analysis (Fig. 5C). Additionally, as revealed in Fig. 1D, PRMT1 knockdown induced cleaved PARP and caspase-3 compared to the levels in cells treated with siCont; moreover, in FACS analysis, the proportion of apoptosis was increased in PRMT1 knockdown samples (Fig. 5D and E). Thus, it was demonstrated that PRMT1 expression was associated with colon cancer proliferation, and cell apoptosis induced by propionate treatment was linked to the PRMT1-related apoptosis pathway.
Figure 5.
PRMT1 knockdown induces cell apoptosis in the HCT116 cell line. (A) RT-PCR and qPCR analysis of PRMT1 after treatment with siCont (negative control) and siPRMT1. ACTB was used as an internal control. P-values were calculated using Students t-tests (***P<0.001). (B and C) Cell growth assay (crystal violet staining and CCK-8 analysis). (B) After treatment with siPRMT1, cell fixation with 100% methanol and cell staining with crystal violet were performed. ‘1’ and ‘2’ are results of two independent experiments after treatment with siRNAs. The cells were imaged under a microscope. Scale bar, 50 µm. (C) In addition, CCK-8 solution was added to culture medium, and an incubation step was performed for 5 min at 37°C. The intensity of cell growth was assessed by a microplate reader (450 nm). P-values were calculated using Students t-tests (***P<0.001). (D) Western blot analysis after PRMT1 knockdown using anti-PRMT1, anti-PARP and anti-caspase-3 antibodies. ACTB was used as the internal control. (E) FACS analysis using Annexin V staining was performed after treatment with siCont and siPRMT1. The lower right and upper right quadrants indicate early apoptosis and late apoptosis, respectively. PRMT1, protein arginine methyltransferase 1.
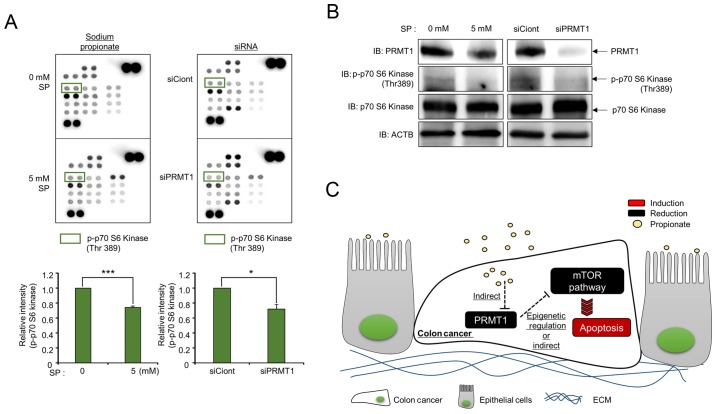
p70 S6 kinase phosphorylation status is regulated by SP treatment and PRMT1 knockdown
To assess the pathway for propionate/PRMT1-induced apoptosis, a phospho-array was purchased from R&D Systems. Fig. 6A revealed that the intensity of positive spots after treatment with 0 mM propionate was similar to that of the positive spots after treatment with 5 mM propionate; however, the p70 S6 kinase (Thr389) phosphorylation status was reduced by 5 mM SP treatment compared to that of the control. Tang et al reported that the phosphorylation of mTOR, 4E-BP1 and p70 S6 kinase was reduced depending on the concentration of propionate and the incubation time, and the mTOR pathway was involved in the suppression of colon cancer proliferation and the stimulation of cell apoptosis (9,25,26). Notably, the results of the phospho-array using cell lysates from cells subjected to PRMT1 knockdown also demonstrated that the phosphorylation of p70 S6 kinase was reduced compared to that of cells subjected to SP treatment (Fig. 6A). To confirm the phospho-array results, western blot analysis was carried out using a phospho-antibody. After treatment of HCT116 cells with SP, the downregulation of PRMT1 expression was clearly detected, and the phosphorylation of p70 S6 kinase was reduced after both PRMT1 knockdown and 5 mM SP treatment compared to that of cells treated with siCont and 0 mM SP (Fig. 6B). Collectively, these results indicated that treatment with SP induced cell apoptosis via the inhibition of the mTOR signaling pathway by PRMT1 downregulation in colon cancer (Fig. 6C). Therefore, this novel PRMT1-related mechanism induced by propionate treatment in colon cancer may help to increase our understanding of the use of propionate treatment for colon cancer patients.
Figure 6.
Phosphorylation of p70 S6 kinase is regulated by SP and PRMT1 knockdown. (A) Phospho-array with lysates from SP-treated and PRMT1-knockdown cells. The phospho-array was purchased from R&D Systems. Approximately 400 µg of cell lysate was used. Green boxes revealed blots of p-p70 S6 kinase (Thr389). P-values were calculated using Students t-tests (*P<0.05, ***P<0.001). (B) Western blot analysis after treatment with SP and siPRMT1 using anti-PRMT1 and anti-phospho-p70 S6 kinase antibodies. ACTB was used as the internal control. (C) Schematic summary of the effects of SP and PRMT1 on colon cancer. PRMT1, protein arginine methyltransferase 1; SP, sodium propionate; ACTB, β-actin.
Discussion
Epigenetic modifiers [histone methyltransferase (HMT)/demethylase (HDM]), histone acetyltransferase (HAT)/deacetylase (HDAC) and DNA methyltransferase/demethylase] have been investigated as therapeutic targets for developing anticancer drugs in several cancers. In colon cancer, overexpression of epigenetic modifiers plays an important role in proliferation and metastasis. For example, EZH2, SMYD3, EHMT2 and LSD1 have been linked with colon cancer proliferation. In addition, p300 and HDAC are important factors in colon cancer (27–30). Thus, studies on reducing epigenetic modifier expression are required to treat colon cancer.
In the present study, it was demonstrated that PRMT1 is overexpressed in several cancers, including colon cancer, using GENT2, the TCGA portal and Oncomine data (Fig. 4). Generally, benign and malignant tumors are the main tumor types. In malignant tumors, carcinoma (carcinoma, squamous cell carcinoma and adenocarcinoma), sarcoma and undifferentiated carcinoma are divided. In colon cancer progression, several steps occur (Polyp >> precancer [adenoma] >> Stage I–IV). Thus, using Oncomine data, we focused on the high PRMT1 expression in several tumor types including malignant tumors and adenoma compared with the normal colon and rectum. In Fig. 4C, PRMT1 was overexpressed in colon adenoma, carcinoma and adenocarcinoma according to Oncomine data. Thus, it was indicated that PRMT1 was overexpressed in early colon cancer stages, and its overexpression may help to acquire malignant characteristics for colon cancer progression. In addition, it was clearly demonstrated that propionate treatment downregulated PRMT1 in the HCT116 cell line (Fig. 3). Subsequently, PRMT1 downregulation induced apoptosis by inhibiting phospho-p70 S6 kinase as revealed by siPRMT1 treatment (Figs. 5 and 6), however, to confirm the anticancer effect of SP, cell growth assays with crystal violet were performed after SP treatment (0 and 5 mM) in normal enterocytes and the HCT116 cell line. Notably, the effect of SP treatment could not be detected in normal enterocytes (data not shown). Therefore, it is suggested that SP may play a role as an anticancer agent for colon cancer treatment.
Recently, our group reported that PRMT1 could serve as a prognostic and therapeutic marker in hepatocellular carcinoma (HCC), and PRMT1 overexpression downregulated CDKN1A to promote HCC proliferation (15). Additionally, Mathioudaki et al reported that high PRMT1 expression was associated with poor survival rates in patients with colon cancer (31). Therefore, PRMT1 should be recognized as a useful therapeutic marker for CRC treatment, and developing novel methods for downregulating PRMT1 expression or activity is important for CRC therapy. AMI-1 is a pan-PRMT inhibitor, and treatment with this agent suppressed cell proliferation by reducing arginine methylation. However, problems have been reported regarding the specificity and adverse effects of PRMT inhibitors (32). Thus, downregulation of PRMT1 by propionate treatment should be recognized as a safe procedure for CRC treatment since propionate is a bacterial fermentative product in the colonic epithelium and a safe compound authorized in the EU for food use. Therefore, developing PRMT1 inhibitors and establishing guidelines for co-treatment with propionate and PRMT1 are required to increase the curative value of this approach in CRC treatment.
As revealed in Fig. 3, although propionate treatment decreased PRMT1 expression, the definite mechanisms that link propionate and PRMT1 regulation remain unknown. Similar to butyrate, propionate was also revealed to inhibit HDAC (5). First, it was hypothesized that the reduction in HDAC activity by propionate would decrease PRMT1 expression. However, as hyperacetylation by propionate is associated with reconstructing the euchromatin structure from heterochromatin to induce gene expression, PRMT1 downregulation by propionate may not be involved in the hyperacetylation mechanism. However, upregulation of unknown regulators by propionate-induced hyperacetylation may affect the suppression of PRMT1 expression. Thus, the relationship between PRMT1 and propionate should be investigated, and the status of H4R3me2 after SP treatment should be validated in further studies.
SCFA treatment induced apoptosis in colon cancer via mitochondrial defects and mTOR pathway inhibition (25,26,33). Additionally, the cAMP/PKA/CREB pathway is controlled by SCFAs for dysregulating inflammatory suppressors in the colonic epithelium (34). In the present study, to ascertain the common signaling pathway for cellular apoptosis used by propionate and PRMT1 knockdown, phospho-array analysis was performed after treatment with propionate and siPRMT1 and a reduction in p70 S6 kinase phosphorylation after separate treatments with propionate and siPRMT1 was observed (Fig. 6A). This is the first study, to the best of our knowledge, which revealed PRMT1-related mTOR signaling pathway effects in colon cancer. Liao et al demonstrated that overexpressed PRMT1 methylated the EGF receptor to increase EGFR signaling activation. Subsequently, the ERK and AKT pathways were activated for survival during cetuximab treatment (16). Thus, it is suggested that: i) the AKT pathway, which is regulated by PRMT1, may affect the mTOR pathway; or ii) the epigenetic mechanism induced by PRMT1 may regulate the mTOR pathway. Therefore, this study was the first to report PRMT1 downregulation by propionate treatment in the HCT116 cell line. Suppressed cell growth via mTOR signaling pathway inhibition was also observed. Thus, this mode-of-action (MOA) study of propionate treatment in colon cancer may provide a basis for new guidelines, such as a dietary therapy, for CRC patients.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- SP
sodium propionate
- SCFA
short-chain fatty acid
- TCGA
The Cancer Genome Atlas
- MBD2
methyl-DNA binding domain 2
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- siCont
siRNA control
- HDAC
histone deacetylase
- CCK-8
Cell Counting Kit-8
- HCC
hepatocellular carcinoma
- EU
European union
- MOA
mode of action
Funding
The present study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2018M3A9H3023077) and the KRIBB Research Initiative Program. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors contributions
DSK and HSC were involved in the conception and design of the study. TYR, TSH and KK were involved in the development of the methodology. DSK, MYS and JKM were involved in the analysis and interpretation of the data (e.g., statistical analysis, biostatistics and computational analysis). KK, TYR, JK, DSK, MYS, JKM and HSC were involved in the writing, reviewing and/or revision of the manuscript. JK and JKM were responsible for administrative, technical or material support. DSK and HSC supervised the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rea D, Coppola G, Palma G, Barbieri A, Luciano A, Del Prete P, Rossetti S, Berretta M, Facchini G, Perdonà S, et al. Microbiota effects on cancer: From risks to therapies. Oncotarget. 2018;9:17915–17927. doi: 10.18632/oncotarget.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 3.McNabney SM, Henagan TM. Short chain fatty acids in the colon and peripheral tissues: A focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients. 2017;9:1348. doi: 10.3390/nu9121348. doi: 10.3390/nu9121348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(Suppl):2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 5.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 6.Ooi CC, Good NM, Williams DB, Lewanowitsch T, Cosgrove LJ, Lockett TJ, Head RJ. Structure-activity relationship of butyrate analogues on apoptosis, proliferation and histone deacetylase activity in HCT-116 human colorectal cancer cells. Clin Exp Pharmacol Physiol. 2010;37:905–911. doi: 10.1111/j.1440-1681.2010.05403.x. [DOI] [PubMed] [Google Scholar]
- 7.Tian Y, Xu Q, Sun L, Ye Y, Ji G. Short-chain fatty acids administration is protective in colitis-associated colorectal cancer development. J Nutr Biochem. 2018;57:103–109. doi: 10.1016/j.jnutbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Fung KY, Brierley GV, Henderson S, Hoffmann P, McColl SR, Lockett T, Head R, Cosgrove L. Butyrate-induced apoptosis in HCT116 colorectal cancer cells includes induction of a cell stress response. J Proteome Res. 2011;10:1860–1869. doi: 10.1021/pr1011125. [DOI] [PubMed] [Google Scholar]
- 9.Tang Y, Chen Y, Jiang H, Nie D. The role of short-chain fatty acids in orchestrating two types of programmed cell death in colon cancer. Autophagy. 2011;7:235–237. doi: 10.4161/auto.7.2.14277. [DOI] [PubMed] [Google Scholar]
- 10.Yu DC, Bury JP, Tiernan J, Waby JS, Staton CA, Corfe BM. Short-chain fatty acid level and field cancerization show opposing associations with enteroendocrine cell number and neuropilin expression in patients with colorectal adenoma. Mol Cancer. 2011;10:27. doi: 10.1186/1476-4598-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bindels LB, Porporato PE, Ducastel S, Sboarina M, Neyrinck AM, Dewulf EM, Feron O, Lestavel S, Cani PD, Staels B, et al. Ffar2 expression regulates leukaemic cell growth in vivo. Br J Cancer. 2017;117:1336–1340. doi: 10.1038/bjc.2017.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/A:1025806911782. [DOI] [PubMed] [Google Scholar]
- 13.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e1212. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Ryu JW, Kim SK, Son MY, Jeon SJ, Oh JH, Lim JH, Cho S, Jung CR, Hamamoto R, Kim DS, et al. Novel prognostic marker PRMT1 regulates cell growth via downregulation of CDKN1A in HCC. Oncotarget. 2017;8:115444–115455. doi: 10.18632/oncotarget.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, Arold ST, Chou CK, Tsou PH, Yamaguchi H, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125:4529–4543. doi: 10.1172/JCI82826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan CP, Nakielny S. Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol Cell Biol. 2006;26:7224–7235. doi: 10.1128/MCB.00473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SK, Kim K, Ryu JW, Ryu TY, Lim JH, Oh JH, Min JK, Jung CR, Hamamoto R, Son MY, et al. The novel prognostic marker, EHMT2, is involved in cell proliferation via HSPD1 regulation in breast cancer. Int J Oncol. 2019;54:65–76. doi: 10.3892/ijo.2018.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K, Son MY, Jung CR, Kim DS, Cho HS. EHMT2 is a metastasis regulator in breast cancer. Biochem Biophys Res Commun. 2018;496:758–762. doi: 10.1016/j.bbrc.2018.01.074. [DOI] [PubMed] [Google Scholar]
- 20.Kim DS, Ryu JW, Son MY, Oh JH, Chung KS, Lee S, Lee JJ, Ahn JH, Min JS, Ahn J, et al. A liver-specific gene expression panel predicts the differentiation status of in vitro hepatocyte models. Hepatology. 2017;66:1662–1674. doi: 10.1002/hep.29324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son MY, Jung CR, Kim DS, Cho HS. Comparative in silico profiling of epigenetic modifiers in human tissues. Mol Biol Rep. 2018;45:309–314. doi: 10.1007/s11033-018-4163-2. [DOI] [PubMed] [Google Scholar]
- 22.Jung KB, Lee H, Son YS, Lee MO, Kim YD, Oh SJ, Kwon O, Cho S, Cho HS, Kim DS, et al. Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat Commun. 2018;9:3039. doi: 10.1038/s41467-018-05450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebbioso A, Tambaro FP, Dell'Aversana C, Altucci L. Cancer epigenetics: Moving forward. PLoS Genet. 2018;14:e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimatsu M, Toyokawa G, Hayami S, Unoki M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, et al. Dysregulation of PRMT1 and PRMT6, type I arginine methyltransferases, is involved in various types of human cancers. Int J Cancer. 2011;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 25.Kapral M, Wawszczyk J, Jesse K, Paul-Samojedny M, Kusmierz D, Weglarz L. Inositol hexaphosphate inhibits proliferation and induces apoptosis of colon cancer cells by suppressing the AKT/mTOR signaling pathway. Molecules. 2017;22:1657. doi: 10.3390/molecules22101657. doi: 10.3390/molecules22101657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, Chen Y, Jiang H, Nie D. Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differ. 2011;18:602–618. doi: 10.1038/cdd.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Ding J, Wang Z, Zhu J, Wang X, Du J. Identification of downstream metastasis-associated target genes regulated by LSD1 in colon cancer cells. Oncotarget. 2017;8:19609–19630. doi: 10.18632/oncotarget.14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo CW, Wang JY, Hung WC, Peng G, Tsai YL, Chang TM, Chai CY, Lin CH, Pan MR. G9a governs colon cancer stem cell phenotype and chemoradioresistance through PP2A-RPA axis-mediated DNA damage response. Radiother Oncol. 2017;124:395–402. doi: 10.1016/j.radonc.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Sarris ME, Moulos P, Haroniti A, Giakountis A, Talianidis I. Smyd3 is a transcriptional potentiator of multiple cancer-promoting genes and required for liver and colon cancer development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto I, Nosho K, Kanno S, Igarashi H, Kurihara H, Ishigami K, Ishiguro K, Mitsuhashi K, Maruyama R, Koide H, et al. EZH2 expression is a prognostic biomarker in patients with colorectal cancer treated with anti-EGFR therapeutics. Oncotarget. 2017;8:17810–17818. doi: 10.18632/oncotarget.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathioudaki K, Papadokostopoulou A, Scorilas A, Xynopoulos D, Agnanti N, Talieri M. The PRMT1 gene expression pattern in colon cancer. Br J Cancer. 2008;99:2094–2099. doi: 10.1038/sj.bjc.6604807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H, Qian K, Ho MC, Zheng YG. Small molecule inhibitors of rotein arginine methyltransferases. Expert Opin Investig Drugs. 2016;25:335–358. doi: 10.1517/13543784.2016.1144747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan P, Oshima K, Huang YW, Agle KA, Drobyski WR, Chen X, Zhang J, Yearsley MM, Yu J, Wang LS. Loss of FFAR2 promotes colon cancer by epigenetic dysregulation of inflammation suppressors. Int J Cancer. 2018;143:886–896. doi: 10.1002/ijc.31366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.