Abstract
It has been revealed by our previous proteomic study that the expression profile is different between well-differentiated and poorly differentiated hepatocellular carcinoma (HCC). Among those differently expressed proteins, peroxiredoxin2 (PRDX2) was our protein of interest. The present study aimed to further investigate the value of PRDX2 as a prognostic factor in HCC. Tissue microarrays were used to investigate the expression difference between HCC tissues and their adjacent normal liver tissues. The expression of PRDX2 at both mRNA and protein levels was examined by q-RT-PCR, western blotting and immunohistochemical assessment in HCC tissues and cell line HCCLM3. Silencing of PRDX2 in HCCLM3 was achieved usingpGMLV-SC1 lentiviral vectors. Cell Counting Kit-8 (CCK-8) and Transwell migration assays were used to assess cell proliferation and migration, respectively. Categorical variables were assessed using the Chi-square test, and ordinal variables were examined using the Mann-Whitney U test. The difference of continuous variables between groups were compared with t-tests. The Kaplan-Meier method was used to calculate the overall survival (OS) and disease-free survival (DFS) of patients, and the log-rank test was used to analyze the differences between groups. The results revealed that the expression of PRDX2 was decreased at both the mRNA and protein levels in an HCC cell line compared to that of a normal human liver cell line. PRDX2 protein expression levels were significantly downregulated in HCC tissues and were positively linked to overall survival (OS) and disease-free survival (DFS) of HCC patients. Patients with high PRDX2 expression levels had longer OS and DFS times than those with lower PRDX2 expression. Silencing of PRDX2 in the HCC cell line HCCLM3 promoted cancer cell proliferation and migration. Our findings indicated that PRDX2 may play an important role in HCC development; PRDX2 may serve as a useful prognostic factor and a therapeutic target.
Keywords: peroxiredoxin2, hepatocellular carcinoma, prognosis, proliferation, migration
Introduction
Hepatocellular carcinoma (HCC) is now the third leading cause of cancer-related deaths worldwide. Almost half a million people are diagnosed with this disease every year (1), and it is quite prevalent among developing countries (2,3). Although the standard protocols and treatment methods have been well accepted and used for clinical HCC management, the prognosis for patients who are candidates to receive curative treatments such as surgical resection and ablation, is mainly unfavorable (4). Therefore, early diagnosis appears to be critical. HCC can be diagnosed by non-invasive imaging such as contrast-enhanced computerized tomography (CT) or magnetic resonance imaging (MRI), and by several reported biomarkers, such as α-fetoprotein (AFP), glypican-3 and des-gamma-carboxy prothrombin (5). Although these markers are currently reported to have been accepted for HCC screening, their sensitivity and specificity are somehow unsatisfactory (6). Furthermore, in the updated practice guidelines by the American Association for the Study of Liver Disease (AASLD) for HCC management, obtaining the AFP serum level was not recommended as a screening test due to its inadequate sensitivity (7). Hence, there is still urgent need to find a sensitive biomarker for HCC prognosis and potential therapeutic targeting.
Peroxiredoxins (PRDXs), which are widely expressed in human and animal cells, refer to a thiol-specific anti-oxidant enzyme family (8,9). It has been indicated that prolonged oxidative stress leading to the activation of multiple signaling pathways and alterations of transcription factors is a driving mechanism of currently known carcinogenesis (10,11). As a family member of PRDXs, PRDX2 is capable of regulating intracellular redox status byscavenging peroxides and reactive oxygen species (ROS), which are believed to be the most vital stimulito trigger several signaling pathways related to oxidative stress and cancer progression (12,13). Emerging studies have demonstrated that PRDX2 is frequently overexpressed in several types of cancers, such as lung, colorectal and ovarian cancer (14–16). It can be acknowledged from these studies that a high level of PRDX2 may generate a protective effect to cancer cells, and PRDX2-induced ROS reduction is beneficial for cancer cell survival. A recent study led by Zhou et al reported that PRDX2 silencing promoted H2O2-induced ROS production and led to increased cell death in HCC SMMC-7721 cells; while PRDX2 upregulation inhibited cancer cell death (17). According to our recently published research study led by Zhao et al, using 2-dimensional gel electrophoresis and matrix-assisted laser desorption ionization time-of-flight mass spectrometry, 11 different proteins were identified from 10 poorly differentiated HCC tissues and 10 well differentiated HCC tissues, among which PRDX2 was found to be upregulated in poorly differentiated HCC tissues compared to the well differentiated HCC tissues. Using bioinformatics tools, it was revealed that PRDX2 may be correlated with tumor invasion, metastasis, and poor prognosis, thus serving as a potentially useful diagnostic and therapeutic biomarker (18). However, whether the expression level of PRDX2 has clinical significance in HCC patients has yet to be reported. Thus, our study aimed to investigate the prognostic value of PRDX2 in HCC patients.
Materials and methods
Cell lines and antibodies
The human HCC cell line HCCLM3 and normal liver cell line L-02 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were routinely incubated (37°C, 5% CO2) in high-glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS). The rabbit monoclonal anti-PRDX2 antibody (EPR5154) (cat. no. ab109367) was purchased from Abcam (Cambridge, MA, USA). The HRP-conjugated anti-rabbit IgG (cat. no. A9169) was purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
Pathological tissues and follow-up of patients
Paraffin-embedded HCC and corresponding adjacent normal liver tissue samples were obtained from 180 cases of patients who underwent radical surgery at The First Affiliated Hospital of Lanzhou University (Gansu, China) from January 2009 to April 2010. A total of 176 cases of HCC patients were enrolled in tissue microarray analysis. The follow-up period was defined as the time interval from the date of surgery to the date of death or last follow-up date. The latest follow-up information was updated in June 2016. All patients were regularly followed-up at an outpatient clinic every 3 months during the first year, every 6 months until the 5th year, and then annually. All included patients had complete follow-up information until death or the latest follow-up date. At follow-up visits, all medical data regarding preoperative diagnosis, surgery, recurrence, clinical staging, adjuvant treatment, clinical follow-up, and cause of death were re-assessed and recorded to our database by a surgery specialist. The current vital status of each patient was reviewed by confirming deaths from the patient registry of the hospital or, if uncertain, from the Gansu Branch of China Population Register Centre (Lanzhou City, China). Overall survival (OS) was defined as the interval from the date of surgery to the time that the patient succumbed to HCC. Patients alive at the end of follow-up were censored. Disease-free survival (DFS) was defined as the interval between the day that surgery was performed and the day that recurrence was first detected. If recurrence was not diagnosed, the date of death due to HCC or the last follow-up was used. The tumor-node-metastasis (TNM) staging was classified according to the criteria proposed by the Standard American Joint Committee on Cancer (AJCC). The present study was approved by the Ethics Committee on human research at The First Affiliated Hospital of Lanzhou University (approval no. LDYYLL-2010-56). Informed and written consents were obtained from the patients or their relatives for the use of these clinical materials for research, which were performed in accordance with the Declaration of Helsinki of the World Medical Association.
Tissue microarrays and immunohistochemistry
Hematoxylin and eosin (H&E) staining was applied on sections obtained from a total of 180 paraffin-embedded HCC tissue samples and their matched adjacent normal liver tissues samples; 1-mm core samples for tissue microarray construction (Beecher Instruments, Inc., Sun Prairie, WI, USA) were obtained from typical regions from each section of 176 enrolled tumor samples and their matched normal liver tissue samples (total 352 cores). Regions of benign liver and HCC were determined by an experienced pathologist for each patient. Tumor staging and pathological grading were determined by a qualified pathologist before scoring any immunostaining. Finally, 2 sets of tissue microarrays entirely carrying 176 paired cores were successfully constructed to further implement PRDX2 immunostaining.
An immunohistochemical kit (GeneTech, Co., Ltd., Shanghai, China) was used to perform immunostaining of PRDX2 on tissue microarrays, the applied concentration of rabbit monoclonal antibodies to PRDX2 was: 1.0%. All steps were performed according to the manufacturer's protocols. The PRDX2 immunostaining score was defined as follows: Score 0, negative; score 1, <30% positive cancer cells; score 2, 30–50% positive cancer cells; score 3, 51–70% positive cancer cells; and score 4, >70% positive cancer cells. The PRDX2 staining intensity score was defined as: 0, no color or extremely weak; 1, light yellow; 2, light brown; or 3, brown (19,20). Microarrays were cross-assessed by 2 experienced pathologists.
Quantitative real-time reverse-transcription PCR analysis
To assess PRDX2 expression at transcript levels, RT-PCR was performed to amplify PRDX2 cDNA fragments from total RNAs purified from isolated cells and frozen specimens using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (1 µg) was reverse-transcribed with the PrimeScript™ RT reagent Kit and gDNA Eraser (Takara Bio, Inc., Kusatsu, Japan). All the reactions were performed in triplicate, and the GAPDH gene was used as the internal control. Primer sequences used for the amplification of human genes were as follows: PRDX2 forward, 5′-GGACTCTCAGTTCACCCACCT-3′ and reverse, 5′-GCCCTCATCTGTTTTCAGCA-3′; and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. Primers were designed to span different exons of PRDX2 genes to ensure the amplification of cDNAs instead of contaminated genomic DNA, and RNAs without reverse transcription were used as negative controls. The relative expression levels of mRNAs were calculated using the 2 - (ΔΔCq sample - ΔΔCq control) method (21).
Western blot analysis
To assess PRDX2 expression at protein levels, the tumor tissues and cells were rinsed with PBS, and lysates were prepared using RIPA buffer (Sigma-Aldrich; Merck KGaA). Total protein concentrations were determined using a BCA protein concentration assay kit (Beyotime Institute of Biotechnology, Haimen, China). Before blotting, 10% separating gel (Beyotime Institute of Biotechnology) and 5% stacking gel (Beyotime Institute of Biotechnology) were prepared. In both cell line and tissue western blot experiments, a quantity of 50 µg protein sample was loaded to each lane before electrophoresis began. Then the polyvinylidene fluoride (Millipore; Merck KGaA) was used to perform protein transfer. Blotted membranes were firstly moved to blocking buffer containing 5% bovine serum albumin (BSA; cat. no. 180549; MP Biomedicals, LLC., Solon, OH, USA), shaking at room temperature for 1 h, then incubated with 0.879 mg/ml rabbit monoclonal anti-PRDX2 (diluted 1:1,000; cat. no. ab109367; Abcam, Cambridge, MA, USA) and polyclonal anti-actin (diluted 1:1,000; cat. no. ab8227; Abcam) primary antibodies at 4°C overnight, and then incubated with 2 mg/ml HRP-conjugated anti-rabbit IgG (diluted 1:5,000; cat. no. A9169; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. After washing, ECL Western Blotting reagent (Millipore; Merck KGaA) was applied for the detection. The Glyco Bandscan software (version 5.0; Glyco, Inc., Madison, WI, USA) was used for densitometry.
PRDX2 knockdown by lentivirus-expressed short hairpin RNAs
Lentivirus-expressing short hairpin RNAs (shRNAs) designed to target different sites of the PRDX2 transcript were obtained from Shanghai GeneChem Co., Ltd. (Shanghai, China). To knock down PRDX2 gene expression in HCC cell line HCCLM3, a total of 4 shRNA-expressing lentiviruses (PRDX2-shRNA1-PRDX2-shRNA4) and corresponding control empty viruses were used to transfect the HCC cell line HCCLM3 following previously described protocols (22). Finally, 2 lentivirus-transfected HCCLM3 clones, HCCLM3-sh-PRDX2-1 and HCCLM3-sh-PRDX2-3 were used in further experiments. In brief, HCC cells were co-cultured with 8 mg/ml hexadimethrine bromide (Sigma-Aldrich; Merck KGaA) in the presence of 1 ml lentiviral particles containing medium with a multiplicity of infection of 0.5–1. Cells were incubated at 37°C in a humidified incubator in an atmosphere of 5% CO2 for 20 h, and then washed and cultured in fresh media. The cells were cultured for another 4 days, and then the levels of PRDX2 protein were assessed by western blotting. Stably transfected clones were selected and validated for PRDX2 expression by q-RT-PCR and western blotting.
Cell proliferation assay
Cells (2,000/well) were dispensed with culture media (100 µl) into 96-well plates. At specified time-points (0, 24, 48, 72 and 96 h), 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well. The absorbance at 450 nm was assessedat the end of the incubation (4 h). All experiments were performed in triplicate.
Cell migration assay
Cell migration assays were performed using a 24-cluster Transwell plate (8-µm pore size; EMD Millipore, Billerica, MA, USA). A total of 1×105 cells were dispensed to upper chambers, suspended in DMEM (upper, 100 µl with 1% FBS; lower, 600 µl with 20% FBS). Folowing incubation (48 h), the cells remaining in the upper chamber were retrieved via cotton swabs. Cells on the lower membrane surface were fixed in 4% paraformaldehyde and stained (Giemsa), and 5 microscopic fields (magnification, ×200) were counted. All experiments were performed in triplicate.
Statistical analysis
Categorical variables were assessed using the Chi-square test, and ordinal variables were assessed using the Mann-Whitney U test. The difference of continuous variables between groups were compared with t-tests. The comparisons of paired ordinal data were performed using the Wilcoxon signed-rank test. The Kaplan-Meier method was used to calculate the disease-free and overall survival of patients, and the log-rank test was used to analyze the differences between groups. The Cox proportional hazards regression model was applied for univariate and multivariate analysis, and P<0.05 was considered to indicate a statistically significant difference. All the statistical analyses were conducted using IBM SPSS Statistics 20 (IBM Corp., Armonk, NY, USA).
Results
PRDX2 is downregulated in HCC
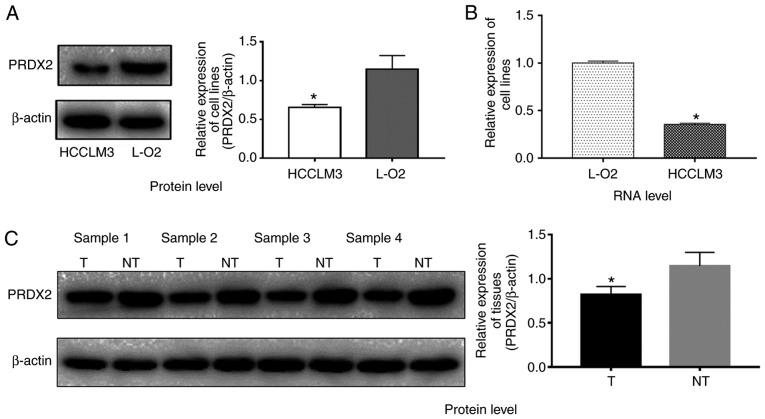
Due to the expression differences of PRDX2 at mRNA and protein levels between HCC cell lines and normal liver cell lines are not studied according to recently published studies (17,23), to clarify this issue, western blot and q-RT-PCR analyses were employed to investigate the PRDX2 expression levels in HCC cell line HCCLM3 and normal liver cell line L-02. The results revealed that HCCLM3 cells expressed relatively less PRDX2 at the protein level when compared to normal liver cell line L-02 (Fig. 1A; P<0.05). The PRDX2 mRNA level in HCCLM3 cells was much lower than that of L-02 as well (Fig. 1B; P<0.05).
Figure 1.
Low PRDX2 expression level in HCC cell line and HCC tissue samples. HCC cell line HCCLM3 expressed relatively less PRDX2 at both (A) protein (y-axis represents the PRDX2 density/β-actin density ratio; a t-test was used to compare the difference between groups; *P<0.05 compared to the normal liver cell line L-02) and (B) transcriptional levels (a t-test was used to compare the difference between groups, *P<0.05 compared to the normal liver cell line L-02). (C) PRDX2 was downregulated in all 4 HCC tissue samples when compared to their adjacent counterparts. The y-axis represents the PRDX2 density/β-actin density ratio; each column represents one sample; a t-test was used to compare the difference between groups; *P<0.05. PRDX2, peroxiredoxin2; HCC, hepatocellular carcinoma.
PRDX2 protein expression level is downregulated in HCC tissues
Four paired HCC tissue samples and their matched adjacent normal liver tissue samples were used to compare PRDX2 expression at the protein level by western blotting. As shown in Fig. 1C, PRDX2 expression was downregulated in all 4 HCC tissue samples compared to that of their matched normal liver tissue samples (Fig. 1C; P<0.05), indicating downregulation of PRDX2 may be a universal phenomenon in HCCs. To validate this hypothesis, immunohistochemitry was applied on 2 sets of tissue microarrays carrying a total of 176 paired samples. In these paired cancer samples, 80.7% (142/176) of the normal tissues exhibited strong staining (score 3); 10.8% (19/176) of the normal tissues exhibited moderate staining (score 2); 8.5% (15/176) of the normal liver tissues exhibited weak staining (score 1); none of the normal liver tissues exhibited negative staining (score 0); conversely, 5.7% (10/176) of the HCC tissues exhibited negative staining (score 0); 35.2% (62/176) of the HCC tissues exhibited weak staining (score 1); 47.7% (84/176) of the tumor tissues exhibited moderate staining (score 2), and 11.4% (20/176) exhibited strong staining (score 3) (Table I). The Wilcoxon signed-rank test revealed that the PRDX2 staining index between HCC tissues and normal liver tissues was significantly different (Table I; P<0.05). Collectively, these results indicated that PRDX2 protein expression was frequently downregulated in HCC.
Table I.
Difference of PRDX2 staining intensity index between HCC tissues and normal liver tissues.
| Normal | |||||
|---|---|---|---|---|---|
| HCC | Score 0 (N=0) | Score 1 (N=15) | Score 2 (N=19) | Score 3 (N=142) | P-value |
| Score 0 | |||||
| (N=10) | 0 | 3 | 5 | 2 | |
| Score 1 | |||||
| (N=62) | 0 | 3 | 6 | 53 | |
| Score 2 | <0.001 | ||||
| (N=84) | 0 | 6 | 4 | 74 | |
| Score 3 | |||||
| (N=20) | 0 | 3 | 4 | 13 | |
For analysis of PRDX2 staining intensity index between HCC tissues and normal liver tissues, the Wilcoxon signed-rank test was used. P<0.05 was considered statistically significant. The PRDX2 staining intensity score was defined as: 0, no color or extremely weak; 1, light yellow; 2, light brown; or 3, brown. PRDX2, peroxiredoxin2; HCC, hepatocellular carcinoma.
Association between PRDX2 expression and the clinicopathological features of HCC patients
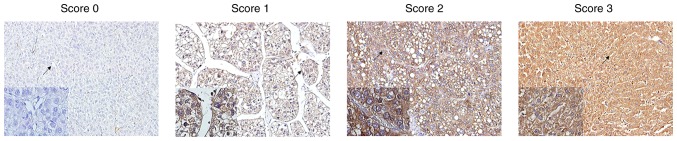
To investigate if the expression of PRDX2 was associated with the clinicopathological features of HCC patients, HCC patients were divided into 2 subgroups depending on their relatively low or high PRDX2 expression levels. High PRDX2 expression was defined as a staining index score ≥4, and low PRDX2 expression was defined as a staining index <4 (Staining index = staining intensity + tumor cell staining grade) (19,20) (Fig. 2). In addition, as observed in Fig. 2, it was also clear that PRDX2 expression was cytoplasmic. It was revealed that high PRDX2 expression was negatively associated with tumor size (P<0.001), microvascular invasion (P<0.001), tumor encapsulation (P=0.047), tumor differentiation (P=0.016), and TNM stage of HCC (P=0.015) (Table II). However, no significant association was observed between the expression level of PRDX2 and the age of patients (P=0.865), sex (P=0.718), HBsAg level (P=0.556), serum AFP level (P=0.487), liver cirrhosis (P=0.417) or tumor number (P=0.665) (Table II).
Figure 2.
PRDX2 immunostaining and staining intensity index in HCC tissue spots illustrated by immunohistochemistry. PRDX2 immunostaining score was defined as follows: Score 0, negative; score 1, <30% positive cancer cells; score 2, 30–50% positive cancer cells; score 3, 51–70% positive cancer cells; and score 4, >70% positive cancer cells. The staining intensity index was defined as 0, no color or extremely weak; 1, light yellow; 2, light brown; or 3, brown. PRDX2, peroxiredoxin2; HCC, hepatocellular carcinoma.
Table II.
Association between PRDX2 expression and clinicopathologic characteristics of HCC patients.
| PRDX2 expression | ||||
|---|---|---|---|---|
| Variables | No. | Low | High | P-value |
| No. | 176 | 72 | 104 | |
| Age (years) | 0.865 | |||
| ≤50 | 72 | 30 | 42 | |
| >50 | 104 | 42 | 62 | |
| Sex | 0.718 | |||
| Male | 144 | 58 | 86 | |
| Female | 32 | 14 | 18 | |
| HBsAg | 0.556 | |||
| Negative | 26 | 12 | 14 | |
| Positive | 150 | 60 | 90 | |
| AFP (ng/ml) | 0.487 | |||
| ≤20 | 54 | 20 | 34 | |
| >20 | 122 | 52 | 70 | |
| Liver cirrhosis | 0.417 | |||
| No | 24 | 8 | 16 | |
| Yes | 152 | 64 | 88 | |
| Tumor size (cm) | <0.001a | |||
| ≤5 | 111 | 35 | 76 | |
| >5 | 65 | 37 | 28 | |
| Tumor no. | 0.665 | |||
| Single | 144 | 60 | 84 | |
| Multiple | 32 | 12 | 20 | |
| Microvascular invasion | <0.001a | |||
| No | 72 | 16 | 56 | |
| Yes | 104 | 56 | 48 | |
| Tumor encapsulation | 0.047a | |||
| None | 82 | 40 | 42 | |
| Complete | 94 | 32 | 62 | |
| Tumor differentiation | 0.016a | |||
| Good | 116 | 40 | 76 | |
| Poor | 60 | 32 | 28 | |
| TNM stage | 0.015a | |||
| T1 | 54 | 18 | 36 | |
| T2 | 60 | 20 | 40 | |
| T3 | 62 | 34 | 28 | |
For analysis of the association between PRDX2 expression and clinical characteristics of HCC patients, all category variables were compared using the Chi-square test; when comparing TNM stages, the Mann-Whitney U test was used.
P<0.05, indicates statistical significance. AFP, α-fetoprotein; PRDX2, peroxiredoxin2; TNM, tumor-node-metastasis.
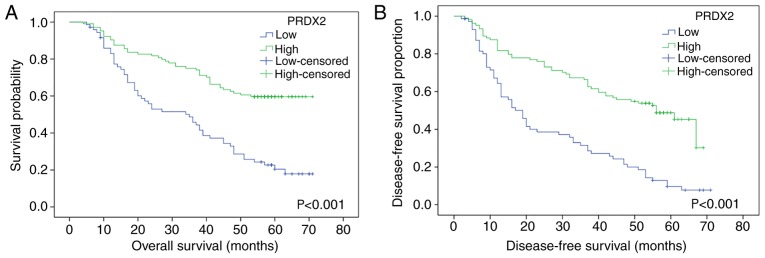
PRDX2 expression level is positively associated with overall survival (OS) and disease-free survival (DFS) of HCC patients
As revealed in Fig. 3, high expression of PRDX2 was associated with longer OS and DFS times, while low PRDX2 expression revealed less OS and DFS times. The log-rank test revealed the difference between the high PRDX2 group and the low PRDX2 group in terms of the survival rate of patients, and a statistical difference was revealed between these 2 groups (for OS, P<0.001; for DFS, P<0.001, respectively).
Figure 3.
The Kaplan-Meier method was used to calculate the DFS and OS between the group of patients with high PRDX2 expression and the group of patients with low PRDX2 expression, and the log-rank test was used to analyze the difference between groups. It was revealed that among HCC patients, high expression of PRDX2 was associated with longer (A) OS (P<0.001) and (B) DFS (P<0.001), while low PRDX2 expression revealed shorter (A) OS (P<0.001) and (B) DFS (P<0.001). OS, overall survival; DFS, disease-free survival; PRDX2, peroxiredoxin2; HCC, hepatocellular carcinoma.
Univariate and multivariate analyses were performed to further investigate whether PRDX2 was an independent prognostic factor for HCC patients. When univariate analysis was applied, the results indicated that the expression level of PRDX2 was associated with both OS and DFS [hazard ratio (HR)=0.714, 95% CI, 0.624–0.817, P<0.001; HR=0.711, 95% CI, 0.629–0.804, P<0.001, respectively] (Tables III and IV). The multivariate analysis was then conducted to further evaluate all the significant variables. The results revealed that the expression level of PRDX2 was an independent prognostic factor for OS (HR=0.785, 95% CI, 0.680–0.905, P=0.001), as well as DFS (HR=0.809, 95% CI, 0.709–0.922, P=0.002), in addition to other variables (Tables III and IV).
Table III.
Univariate and multivariate analyses of prognostic factors for overall survival of HCC patients.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (year, >50 vs. ≤50) | 1.000 (0.666–1.502) | 0.998 | ||
| Sex (female vs. male) | 0.629 (0.357–1.108) | 0.109 | ||
| HBsAg (positive vs. negative) | 0.771 (0.451–1.319) | 0.343 | ||
| AFP, ng/ml (>20 vs. ≤20) | 1.167 (0.753–1.810) | 0.489 | ||
| Liver cirrhosis (yes vs. no) | 0.847 (0.480–1.493) | 0.565 | ||
| Tumor size (cm) (>5 vs. ≤5) | 4.939 (3.262–7.478) | <0.00 | 2.719 (1.710–4.323) | <0.001 |
| Tumor no. (multiple vs. single) | 1.459 (0.907–2.347) | 0.120 | ||
| Microvascular invasion (with vs. without) | 7.409 (4.295–12.779) | <0.001 | 4.125 (2.238–7.602) | <0.001 |
| Tumor encapsulation (complete vs. none) | 0.982 (0.660–1.460) | 0.927 | ||
| Tumor differentiation (poor vs. good) | 0.929 (0.609–1.418) | 0.734 | ||
| TNM stage | ||||
| T1 | 1.000 | 1.000 | ||
| T2 | 1.257 (0.667–2.368) | 0.479 | 1.452 (0.754–2.795) | 0.264 |
| T3 | 7.570 (4.265–13.439) | <0.001 | 4.266 (2.284–7.970) | <0.001 |
| PRDX2 (high vs. low) | 0.714 (0.624–0.817) | <0.001 | 0.785 (0.680–0.905) | 0.001 |
HCC, hepatocellular carcinoma; AFP, α-fetoprotein; CI, confidence interval; HR, hazard ratio; PRDX2, peroxiredoxin2; TNM, tumor-node-metastasis.
Table IV.
Univariate and multivariate analyses of prognostic factors for disease-free survival of HCC patients.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (year, >50 vs. ≤50) | 0.991 (0.685–1.436) | 0.963 | ||
| Sex (female vs. male) | 0.569 (0.336–0.965) | 0.036 | 0.478 (0.280–0.816) | 0.007 |
| HBsAg (positive vs. negative) | 1.029 (0.607–1.746) | 0.914 | ||
| AFP, ng/ml (>20 vs. ≤20) | 1.354 (0.902–2.033) | 0.143 | ||
| Liver cirrhosis (yes vs. no) | 0.948 (0.559–1.608) | 0.844 | ||
| Tumor size (cm) (>5 vs. ≤5) | 3.463 (2.379–5.042) | <0.001 | 2.337 (1.540–3.547) | <0.001 |
| Tumor no. (multiple vs. single) | 1.620 (1.059–2.479) | 0.026 | 0.990 (0.635–1.544) | 0.965 |
| Microvascular invasion (yes vs. no) | 5.384 (3.450–8.404) | <0.001 | 3.146 (1.896–5.221) | <0.001 |
| Tumor encapsulation (complete vs. none) | 0.831 (0.579–1.193) | 0.316 | ||
| Tumor differentiation (poor vs. good) | 1.281 (0.881–1.863) | 0.194 | ||
| TNM stage | ||||
| T1 | 1.000 | |||
| T2 | 0.799 (0.472–1.352) | 0.403 | 0.945 (0.547–1.634) | 0.840 |
| T3 | 4.969 (3.091–7.990) | <0.001 | 3.226 (1.933–5.384) | <0.001 |
| PRDX2 (high vs. low) | 0.711 (0.629–0.804) | <0.001 | 0.809 (0.709–0.922) | 0.002 |
HCC, hepatocellular carcinoma; AFP, α-fetoprotein; CI, confidence interval; HR, hazard ratio; PRDX2, preoxiredoxin2; TNM, tumor-node-metastasis.
Collectively, these data indicated that the expression level of PRDX2 could be used as an independent factor for the prognosis of HCC patients.
Silencing of PRDX2 in HCCLM3 promotes cancer cell proliferation and migration
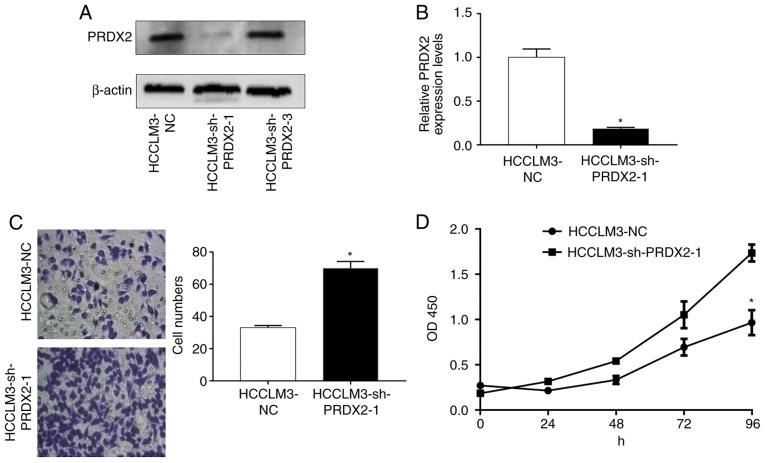
Since PRDX2 expression is usually downregulated in HCC and is negatively associatedwith HCC progression, it was hypothesized that PRDX2 inhibited HCC malignancies. To assessthis hypothesis, stable cell lines whose PRDX2 expression was permanently inhibited in HCCLM3 were established usinga lentiviral system (HCCLM3-shPRDX2) (Fig. 4A and B; P<0.05).
Figure 4.
PRDX2 silencing in HCCLM3 cell line. (A and B) PRDX2 silencing in HCCLM3 cells. A t-test was used to compare the difference between groups, *P<0.05 compared to HCCLM3-NC. (C) PRDX2 silencing in HCCLM3 cells promoted cancer cell migration ability. A t-test was used to compare the difference between groups; *P<0.05 compared to HCCLM3-NC. (D) PRDX2 silencing in HCCLM3 cells promoted cancer cell proliferation ability. At-test was used to compare the difference between groups; *P<0.05 compared to HCCLM3-NC. PRDX2, peroxiredoxin2.
A Transwell assay was used to assess cell migration ability after PRDX2 silencing. As revealed in Fig. 4C, silencing of PRDX2 in HCCLM3 significantly promoted cancer cell migration ability compared with that of lentiviral green fluorescent protein (GFP) infected control cell (HCCLM3) (Fig. 4C; P<0.05).
Additionally, cell proliferation was also assessed by counting the number of cells at the specific time-points indicated, and as revealed in Fig. 4D (P<0.05) after PRDX2 was inhibited, HCCLM3 presented enhanced cell proliferation abilities.
Discussion
The roles thatPRDX2 has been revealed to play in carcinogenesis have been described in emerging studies as a ‘dual-effect’ manner and its clinical importance as a prognostic factor has started drawing increased attention from researchers in recent years (24). The role of PRDX2 as a tumor promoter is supported by mounting evidence in recent years (25), particularly, it is currently well established in colorectal cancers. According to Lu et al, it was determinedthat PRDX2 had higher expression levels in colorectal cancer tissues when compared with their matched adjacent normal mucosa tissues, and PRDX2 expression was positively linked to cancer metastasis and TNM stage by downregulating oxidation-induced cancer cell apoptosis (26). In another related study, it was indicated that PRDX2 silencing in colorectal cancer cell lines resulted in elevated cancer cell apoptosis and increased endogenous ROS production, further leading to Wnt signaling pathway-associated protein alterations (27). It has also been reported that a high PRDX2 expression level was associated with tumor progression and poor survival in colorectal cancer patients (28). Silencing of PRDX2 was revealed to increase sensitivity of colorectal cancer cells to 5-FU treatment by suppressing the PI3K/AKT signaling pathway (29). Cruz et al also demonstrated that PRDX2 was involved in cancer cell resistance to bortezomib, paclitaxel, and carboplatin in ovarian cancer (30). According to Diao et al, PRDX2 may serve as a downstream target of microRNA-122a, which was found to be significantly downregulated in HCC and may possess a tumor-inhibiting effect (23); Zhou et al determined that PRDX2 downregulation increased H2O2-induced HCC cell death, thus concluding that PRDX2 is a tumor promoter in HCC (17). In contrast, evidence to support the notion that PRDX2 possesses tumor-inhibiting ability is emerging. In a study published in 2013, Lee et al revealed that in metastatic melanoma, PRDX2 downregulation resulted in increased proliferative and migratory activities of tumor cells, and this tumor-inhibiting effect was mediated by PRDX2 by promoting ERK-dependent E-cadherin expression and the Src-dependent retention of β-catenin in the adherens junctions (31). Moreover, another study revealed that in colon cancer cells, PRDX2 inhibited TGFβ1-induced epithelial-mesenchymal transition (EMT) and reduced the invasive phenotype by modulating downstream transcription factors, such as Twist1, Snail, ZEB1 and ZEB2 (32). Thus, as revealed by all this evidence, the function of PRDX2 in cancer progression remains controversial.
The reason for thismay not only be partially attributed to different experimental methods and materials adopted but also, much more importantly, to the underlying mechanism of how PRDX2 affects redox balancing which is complicated and has not been fully elucidated by currently known studies (33). Not to mention the multiple roles that ROS plays during cancer development (11,34), which have been proposed to both accelerate and delay cancer initiation and progression (35), orthe clinical trials clouded by the failure of oral antioxidant administration in the treatment of several types of cancer, such as in human skin cancer and mice lung cancer models (36–39), which was substantiated by the results from a significant vitamin E randomized clinical trial, namely the Selenium and Vitamin E Cancer Prevention Trial (SELECT), by revealingthat α-tocopherol supplementation increased prostate cancer incidence (40). In addition, the decreased PRDX2 level in HCC cells may weaken ROS scavenging, thus resulting in intensified oxidative stress, which was believed to play a pivotal role in non-oncogene addiction (41).
However, the clinical importance and function of PRDX2 in HCC has not been comprehensively studied. A previous study conducted in our laboratory illustrated upregulation of PRDX2 in poorly differentiated HCC tissues by proteomic approaches and indicated the involvement of PRDX2 in HCC progression (18). Notably, in our present study, our results revealed the protein expression levels of PRDX2 in HCC tissues were relatively lower than that of their adjacent normal liver tissues, not only in all 4 matched tissue specimens, but also in the protein microarrays which were comprised of 176 paired tissue spots. These results were consistent with our results obtained from western blotting and q-RT-PCR experiments applied in an HCC cell line. Furthermore, there were statistically significant differences between HCC patients who had high PRDX2 expression levels and those who had low expression levels in terms of OS and DFS of patients. Moreover, the hypothesis that PRDX2 possesses prognostic value was further supported by the result that PRDX2 was an independent prognostic factor in HCC patients. Data from experiments in cell lines favor the concept that PRDX2 could serve not only as a prognostic marker but also as a therapeutic target for treatment since silencing of PRDX2 promoted cancer cell proliferation and migration. Further experiments are required to reveal the underlying signaling mechanism to support the potential pharmaceutical value of PRDX2.
Collectively, our findings revealed that the expression level of PRDX2 was positively linked to the OS and DFS of HCC patients. It was also revealed that PRDX2 was an independent prognostic indicator. PRDX2 could serve as a useful biomarker for the prognosis of HCC patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 31570509, 81702326 and 81872036), the Science and Technology Major Project of Gansu Province (grant no. 1602FKDA001), the Natural Science Foundation of Gansu Province (grant no. 17JR5RA187), the Gansu Provincial Administration of Traditional Chinese Medicine (grant no. GZK-2015-69), the Gansu Province Science Foundation for Youths (grant no. 17JR5RA259), the Provincial Youth Science and Technology Foundation of Gansu (grant nos. 1606RJYA276 and17JR5RA259), the Talent Innovation and Entrepreneurship Program of Lanzhou City (grant no. 2016-RC-57), the Science and Technology Project of Chenguan District (grant no. 2017SHFZ0014) and the Foundation of the First Hospital of Lanzhou University (grant nos. LDYYYN 2017-04, LDYYYN 2017-02 and LDYYYN 2015-01).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XL was the designer and the principal investigator (PI) of the present study. BB performed the major clinical and experimental data collection, the statistical analysis, the immunohistochemistry experiments and the cell function experiments, and also writing of the present study (in English). YL also performed the data collection, the statistical analysis, the immunohistochemistry experiments and the cell function experiments. JH performed the Transwell experiments. HW performed the statistical analysis. LL performed the CCK-8 experiments. SZ analyzed the correlation between the patients' clinicopathological variables and PRDX2 expression. JZ performed the western blot experiments and the image processing. WM performed the lentivirus transfection experiments. PY the performed cell culture and the mRNA and protein extraction. ZB performed the PCR experiments. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study does not contain any studies with animals performed by any of the authors. The present study was approved by the Ethics Committee on human research of the First Affiliated Hospital of Lanzhou University (Approval no. LDYYLL-2010-56). Informed and written consents were obtained from the patients or their relatives for the use of these clinical materials for research, which were performed in accordance with the Declaration of Helsinki of the World Medical Association.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: Current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med. 2012;4:19–37. doi: 10.2147/HMER.S16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda M, Morizane C, Ueno M, Okusaka T, Ishii H, Furuse J. Chemotherapy for hepatocellular carcinoma: Current status and future perspectives. J Clin Oncol. 2018;48:103–114. doi: 10.1093/jjco/hyx180. [DOI] [PubMed] [Google Scholar]
- 5.Behne T, Copur MS. Biomarkers for hepatocellular carcinoma. Int J Hepatol 2012. 2012:859076. doi: 10.1155/2012/859076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Gao Q, Pei L, Wang C, Jin L, Liao F. Current status and future prospects of biomarkers in the diagnosis of hepatocellular carcinoma. Int J Biol Markers. 2017;32:e361–e369. doi: 10.5301/ijbm.5000299. [DOI] [PubMed] [Google Scholar]
- 7.Heimbach JK. Overview of the updated AASLD guidelines for the management of HCC. Gastroenterol Hepatol. 2017;13:751–753. [PMC free article] [PubMed] [Google Scholar]
- 8.Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 9.Hillas PJ, del Alba FS, Oyarzabal J, Wilks A, Ortiz De Montellano PR. The AhpC and AhpD antioxidant defense system of mycobacterium tuberculosis. J Biol Chem. 2000;275:18801–18809. doi: 10.1074/jbc.M001001200. [DOI] [PubMed] [Google Scholar]
- 10.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 11.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Fu Z, Wei J, Guo J, Liu M, Du K. Peroxiredoxin 2 is involved in vasculogenic mimicry formation by targeting VEGFR2 activation in colorectal cancer. Med Oncol. 2015;32:414. doi: 10.1007/s12032-014-0414-9. [DOI] [PubMed] [Google Scholar]
- 13.Duan T, Fan K, Chen S, Yao Q, Zeng R, Hong Z, Peng L, Shao Y, Yao B. Role of peroxiredoxin 2 in H2O2-induced oxidative stress of primary leydig cells. Mol Med Rep. 2016;13:4807–4813. doi: 10.3892/mmr.2016.5147. [DOI] [PubMed] [Google Scholar]
- 14.Stresing V, Baltziskueta E, Rubio N, Blanco J, Arriba MC, Valls J, Janier M, Clézardin P, Sanz-Pamplona R, Nieva C, et al. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32:724–735. doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Wei J, Zhang S, Wu X, Guo J, Liu M, Du K, Xu J, Peng L, Lv Z, et al. Peroxiredoxin 2 is essential for maintaining cancer stem cell-like phenotype through activation of hedgehog signaling pathway in colon cancer. Oncotarget. 2016;7:86816–86828. doi: 10.18632/oncotarget.13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinina EV, Berezov TT, Shtil' AA, Chernov NN, Glazunova VA, Novichkova MD, Nurmuradov NK. Expression of peroxiredoxin 1, 2, 3, and 6 genes in cancer cells during drug resistance formation. Bull Exp Biol Med. 2012;153:878–881. doi: 10.1007/s10517-012-1849-7. (In English, Russian) [DOI] [PubMed] [Google Scholar]
- 17.Zhou S, Han Q, Wang R, Li X, Wang Q, Wang H, Wang J, Ma Y. PRDX2 protects hepatocellular carcinoma SMMC-7721 cells from oxidative stress. Oncol Lett. 2016;12:2217–2221. doi: 10.3892/ol.2016.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao S, Su G, Yang W, Yue P, Bai B, Lin Y, Zhang J, Ba Y, Luo Z, Liu X, et al. Identification and comparison of differentiation-related proteins in hepatocellular carcinoma tissues by proteomics. Technol Cancer Res Treat. 2017;16:1092–1101. doi: 10.1177/1533034617732426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z, Cao Y, Fan J, Huang XW, Zhou J. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63:1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Lu Y, Qin A, Qiao Z, Jiang X. Overexpression of RAB34 correlates with poor prognosis and tumor progression in hepatocellular carcinoma. Oncol Rep. 2017;38:2967–2974. doi: 10.3892/or.2017.5957. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, Zhu G, Tamada K, Flies DB, van Deursen JM, Chen L. B7-H1 determines accumulation and deletion of intrahepatic CD8+ T lymphocytes. Immunity. 2004;20:327–336. doi: 10.1016/S1074-7613(04)00050-0. [DOI] [PubMed] [Google Scholar]
- 23.Diao S, Zhang JF, Wang H, He ML, Lin MC, Chen Y, Kung HF. Proteomic identification of microRNA-122a target proteins in hepatocellular carcinoma. Proteomics. 2010;10:3723–3731. doi: 10.1002/pmic.201000050. [DOI] [PubMed] [Google Scholar]
- 24.Nicolussi A, D'Inzeo S, Capalbo C, Giannini G, Coppa A. The role of peroxiredoxins in cancer. Mol Clin Oncol. 2017;6:139–153. doi: 10.3892/mco.2017.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park MH, Jo M, Kim YR, Lee CK, Hong JT. Roles of peroxiredoxins in cancer, neurodegenerative diseases and inflammatory diseases. Pharmacol Ther. 2016;163:1–23. doi: 10.1016/j.pharmthera.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu W, Fu Z, Wang H, Feng J, Wei J, Guo J. Peroxiredoxin 2 is upregulated in colorectal cancer and contributes to colorectal cancer cells' survival by protecting cells from oxidative stress. Mol Cell Biochem. 2014;387:261–270. doi: 10.1007/s11010-013-1891-4. [DOI] [PubMed] [Google Scholar]
- 27.Lu W, Fu Z, Wang H, Feng J, Wei J, Guo J. Peroxiredoxin 2 knockdown by RNA interference inhibits the growth of colorectal cancer cells by downregulating Wnt/β-catenin signaling. Cancer Lett. 2014;343:190–199. doi: 10.1016/j.canlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Peng L, Wang R, Shang J, Xiong Y, Fu Z. Peroxiredoxin 2 is associated with colorectal cancer progression and poor survival of patients. Oncotarget. 2017;8:15057–15070. doi: 10.18632/oncotarget.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Zhang S, Wang R, Wu X, Zeng L, Fu Z. Knockdown of PRDX2 sensitizes colon cancer cells to 5-FU by suppressing the PI3K/AKT signaling pathway. Biosci Rep. 2017;37:BSR20160447. doi: 10.1042/BSR20160447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz G, Fernandois D, Paredes AH. Ovarian function and reproductive senescence in the rat: Role of ovarian sympathetic innervation. Reproduction. 2017;153:R59–R68. doi: 10.1530/REP-16-0117. [DOI] [PubMed] [Google Scholar]
- 31.Lee DJ, Kang DH, Choi M, Choi YJ, Lee JY, Park JH, Park YJ, Lee KW, Kang SW. Peroxiredoxin-2 represses melanoma metastasis by increasing E-Cadherin/β-Catenin complexes in adherens junctions. Cancer Res. 2013;73:4744–4757. doi: 10.1158/0008-5472.CAN-12-4226. [DOI] [PubMed] [Google Scholar]
- 32.Feng J, Fu Z, Guo J, Lu W, Wen K, Chen W, Wang H, Wei J, Zhang S. Overexpression of peroxiredoxin 2 inhibits TGF-β1-induced epithelial-mesenchymal transition and cell migration in colorectal cancer. Mol Med Rep. 2014;10:867–873. doi: 10.3892/mmr.2014.2316. [DOI] [PubMed] [Google Scholar]
- 33.Neumann CA, Fang Q. Are peroxiredoxins tumor suppressors? Curr Opin Pharmacol. 2007;7:375–380. doi: 10.1016/j.coph.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Poprac P, Jomova K, Simunkova M, Kollar V, Rhodes CJ, Valko M. Targeting free radicals in oxidative stress-related human diseases. Trends Pharmacol Sci. 2017;38:592–607. doi: 10.1016/j.tips.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Chandel NS, Tuveson DA. The promise and perils of antioxidants for cancer patients. N Engl J Med. 2014;371:177–178. doi: 10.1056/NEJMcibr1405701. [DOI] [PubMed] [Google Scholar]
- 36.Miller ER, III, Pastor-Barriuso R, Dalal D, Riemersma RA, Appel LJ, Guallar E. Meta-analysis: High-dosage vitamin E supplementation may increase all-cause mortality. Ann Intern Med. 2005;142:37–46. doi: 10.7326/0003-4819-142-1-200501040-00110. [DOI] [PubMed] [Google Scholar]
- 37.Chen CS, Wells PG. Enhanced tumorigenesis in p53 knockout mice exposed in utero to high-dose vitamin E. Carcinogenesis. 2006;27:1358–1368. doi: 10.1093/carcin/bgi325. [DOI] [PubMed] [Google Scholar]
- 38.Hercberg S, Ezzedine K, Guinot C, Preziosi P, Galan P, Bertrais S, Estaquio C, Briançon S, Favier A, Latreille J, et al. Antioxidant supplementation increases the risk of skin cancers in women but not in men. J Nutr. 2007;137:2098–2105. doi: 10.1093/jn/137.9.2098. [DOI] [PubMed] [Google Scholar]
- 39.Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med. 2014;6:221ra15. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 40.Klein EA, Thompson IM, Jr, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, Minasian LM, Ford LG, Parnes HL, Gaziano JM, et al. Vitamin E and the risk of prostate cancer: The selenium and vitamin E cancer prevention trial (SELECT) JAMA. 2011;306:1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.