Abstract
The present study aimed to further explore the molecular mechanisms of miRNA-223 in non-small cell lung cancer (NSCLC). Data prospectively collected from NSCLC patients and volunteers from March 2016 to May 2016 at Tsinghua Changgung Hospital were analyzed. Cell proliferation was measured using MTT assay, while cell apoptosis and caspase-3/9 activity were measured using flow cytometry and caspase-3/9 activity kit. Bax, EGFR, PI3K and p-Akt protein were also investigated using western blotting. The results revealed that the serum levels of miRNA-223 in NSCLC patients were downregulated. In an in vitro model, overexpression of miRNA-223 induced apoptosis while reducing cell proliferation. In contrast, downregulation of the expression of miRNA-223 inhibited apoptosis whereas it increased cell proliferation. Meanwhile, overexpression of miRNA-223 suppressed the protein expression of EGFR, PI3K and p-Akt in NSCLC cells. An EGFR inhibitor promoted the anticancer effects of miRNA-223 in NSCLC cells through the EGFR/PI3K/AKT pathway. Meanwhile, a PI3K inhibitor increased the anticancer effects of miRNA-223 in NSCLC cells through the PI3K/AKT pathway. Thus, a new pathway has been identified in the present study, and application of miRNA-223 may induce the apoptosis of NSCLC through the PI3K/AKT pathway by EGFR.
Keywords: miRNA-223, non-small cell lung cancer, EGFR, PI3K/AKT
Introduction
Lung cancer is a malignant tumor with the highest morbidity and mortality in the world. It is a major disease threatening human life and health (1). The world is undergoing an increasing cancer burden (1). This was suggested in World Cancer Reports from the International Agency for Research on Cancer (IARC) of WTO released in 2014. Lung cancer cases ranked first among all newly diagnosed cancers in 2012 (2). This figure is approximately 1.8 million. It accounts for 13% of the total number of common cancer cases. In addition, lung cancer is also the leading cause of cancer-related mortality. This number was approximately 1.6 million in 2015, which accounted for 19.4% of the total number of deaths (2). Of them, over 1/3 were Chinese cases (2).
It was discovered in recent years that circulatory miRNAs can serve as disease markers (3). This has aroused the interests of numerous scientific researchers. In particular, close attention has been paid to miRNAs as tumor diagnostic markers. miRNAs are a class of endogenous non-coding small molecule RNAs. They are highly conserved in evolution. They consist of approximately 18–23 basic groups in length (4). miRNAs have been verified in reports to play a certain role in cancer pathogenesis, genesis and development. In addition, they exert functions that are similar to oncogenes or tumor-suppressor genes (5).
Epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase receptor. It is approximately 170 kb in molecular weight (6). It consists of three parts, namely, the extracellular domain, the transmembrane domain and the intracellular domain. The intracellular domain is also the EGFR-tyrosine kinase domain (7). It is an important component for regulating tumor cell proliferation, invasion, angiogenesis, adhesion, metastasis and apoptosis. EGFR binds with its ligand in the extracellular domain (8). This can produce an EGFR dimer and induce intracellular phosphorylation (8). Thus, it can activate a series of downstream signaling pathways (9). Of these, the phosphoinositide 3-kinase (PI3K) pathway is a canonical pathway. PI3K is the major downstream effector of receptor tyrosine kinase and G-protein coupled receptor (9). It can produce activated serine/threonine protein kinase AKT and its downstream effector phospholipid. Consequently, it can transduce signals from all growth factors and cytokines to intracellular messenger (9). This is closely associated with tumor cell proliferation and survival. EGFR is notably correlated with NSCLC. The downstream PI3K/AKT pathway that it activates is abnormally expressed in numerous tumors including breast cancer, prostate cancer and gastric cancer (10). It was demonstrated that miRNA-223 suppressed cervical cancer cell growth via targeting the EGFR/AKT2/CCND1 pathway (11). Herein, our study further explored the molecular mechanisms of miRNA-223 in NSCLC.
Materials and methods
Patients
We analyzed the prospectively collected data of NSCLC patients and healthy volunteers between March 2016 and May 2016 at the Tsinghua Changgung Hospital. The study was approved by Jining First People's Hospital. Basic data of all patients with NSCLC were collected and are documented in Table I. Ten milliliters of peripheral blood was centrifuged at 1,000 × g for 10 min at 4°C, and serum was collected and saved at −80°C.
Table I.
Characteristics of the NSCLC patients.
| Variables | NSCLC cases (n=6) | Healthy subjects (n=6) |
|---|---|---|
| Age (years) | ||
| ≤55 | 4 | 3 |
| >55 | 2 | 3 |
| Sex | ||
| Female | 0 | 0 |
| Male | 6 | 6 |
| Tumor size (cm) | ||
| ≤3.0 | 2 | 0 |
| >3.0 | 4 | 0 |
| Edmondson grade | ||
| I | 0 | 0 |
| II | 2 | 0 |
| III | 4 | 0 |
NSCLC, non-small cell lung cancer.
Cell culture and transfection
Human NSCLC cell line A549 was purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA) and was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a 5% CO2 incubator. Anti-miRNA-223, miRNA-223 and control negative mimics were designed and obtained from GenePharma (Shanghai, Beijing, China). A549 cells were transfected with anti-miRNA-223, miRNA-223 and the control negative mimics using Invitrogen™ Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). AG1478, an EGFR special inhibitor and LY294002, a PI3K special inhibitor were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany).
qRT-PCR analysis
Total RNA was extracted from prepared cells or serum using TRizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized with the PrimiScript RT reagent kit (Takara, Shiga, Japan). qRT-PCR analysis was performed using SYBR Green PCR Master Mix (PE Applied Biosystems; Thermo Fisher Scientific, Inc.) in the Applied Biosystems 7900HT real-time PCR machine (Applied Biosystems; Thermo Fisher Scientific, Inc.). The probes for miRNA-223 were: 5′-GCGTGTATTTGACAAGCTGAGTT-3′ and 5′-GTGTCAGTTTGTCAAATACCCCA-3′ and U6: 5′-CCGCCCGCCGCCAGGCCCC-3′ and 5′-ATATGGAACGCTTCACGAATT-3′. The reaction conditions were as follows: hot start at 94°C for 5 min; 40 cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec at 72°C, and 10 min at 72°C. The relative expression of miRNAs was calculated with the 2−∆∆Ct method.
Microarray analysis
RNA (200 ng) was amplified using Cy3 using the Low Input Quick-Amp Labeling kit (both from Agilent Technologies, Santa Clara, CA, USA). The cRNAs were subjected to hybridization into Agilent SurePrint G3 Mouse GE 8X 60 K Microarray Chips (Agilent Technologies). Data were quantified by Feature Extraction 10.5.1.1 image analysis software (Agilent Technologies).
Cell proliferation assay and lactate dehydrogenase (LDH)
At 24, 48, 72 h after transfection, cell proliferation was measured using the MTT assay and incubation was carried out for 4 h at 37 °C. DMSO was added into the cells and then incubated for 20 min. Optical density (OD) value was read using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) at 490 nm.
LDH activity was measured using LDH activity kits (Beyotime Biotechnology, Co., Ltd., Shanghai, China). Optical density (OD) value was read using a microplate reader (Bio-Rad Laboratories) at 450 nm.
Cell apoptosis assay and caspase-3/9 activity assay
Seventy-two hours after transfection, apoptotic cells were stained with an Annexin V-FITC/PI double staining apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, USA). Apoptotic cells were assessed by flow cytometry (BD FACSCalibur, BD Biosciences).
At 72 h after transfection, cells were lysed in RIPA buffer (Beyotime Biotechnology, Co., Ltd.) and protein content was measured using BCA assay (Beyotime Biotechnology, Co., Ltd.). Equal amounts of protein were used to determine caspase-3/9 activity using a caspase-3/9 activity kit (Beyotime Biotechnology, Co., Ltd.). Optical density (OD) value was read using a microplate reader (Bio-Rad Laboratories) at 405 nm.
Dual luciferase reporter gene assay
EGFR and miRNA-223 plasmids were co-transfected into A549 cells using Lipofectamine 2000. Reporter gene assays were performed using the Dual Luciferase Reporter Assay kit (Promega, Madison, WI, USA) for 24 h.
Cell migration assay
A549 cells (5×105 cell/ml) were seeded on 24-well plates and were added to the upper chamber of each migration well, and 500 µl of DMEM with 10% FBS was added to the lower part for 16 h. Cells that migrated to the lower side were fixed with 75% ice-alcohol for 30 min and stained with 1% crystal violet solution for 1 h. A549 cells were fixed and counted under a fluorescence microscope (Axio version II, Carl Zeiss Inc., Oberkochen, Germany).
Western blot analysis
Seventy-two hours after transfection, cells were lysed in RIPA buffer (Beyotime Biotechnology, Co., Ltd.) and protein content was measured using the BCA assay (Beyotime Biotechnology, Co., Ltd.). Equal amounts of protein (40 µg) were subjected to 8–12% SDS gel electrophoresis and transferred to a Amersham polyvinylidene difluoride membrane (GE Healthcare Life Sciences, Little Chalfont, UK). Western blot analysis was performed with anti-Bax (sc-6236, 1:500), anti-EGFR (sc-365829, 1:500), anti-PI3K (sc-7174, 1:500), anti-p-Akt (sc-135651, 1:500) and anti-GAPDH (sc-25778, 1:2000, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) antibodies at 4°C overnight. The membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (1:2000, Santa Cruz Biotechnology, Inc.) for 1 h at room temperature and visualized with the chemiluminescent detection method using the SuperSignal West Pico Substrate (Pierce; Thermo Fisher Scientific, Inc.).
Statistical analyses
Data are expressed as mean ± SD. Data among groups were analyzed using one-way analysis of variance followed by a Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant result.
Results
Serum levels of miRNA-223 in NSCLC patients
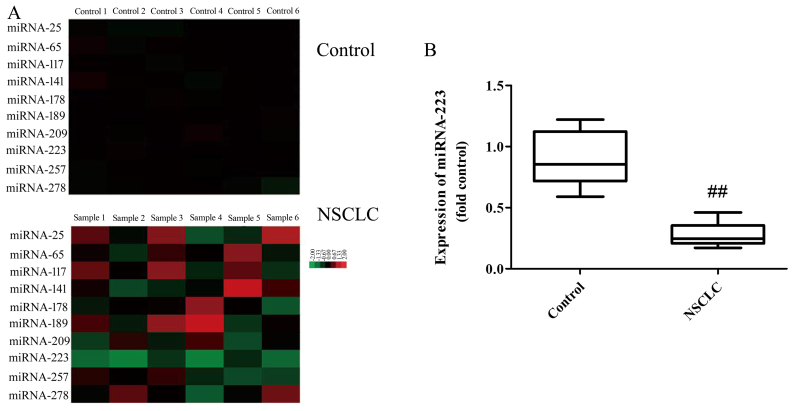
Gene chip shows that the serum levels of miRNA-223 in NSCLC patients were downregulated compared with the normal group (Fig. 1A). Fig. 1B shows that the serum levels of miRNA-223 in NSCLC patients were downregulated compared with the normal group. Taken together, miRNA-223 may be an important strategy for the anticancer effect in NSCLC.
Figure 1.
Serum levels of miRNA-223 in NSCLC patients. (A) Gene chip for miRNA-223 expression, and (B) qPCR of miRNA-223 expression in the serum of non-small cell lung cancer (NSCLC) cases and controls. Control, volunteer group; NSCLC, NSCLC patient group. ##P<0.01 compared with the control group.
miRNA-223 affects the proliferation and migration of A549 cells
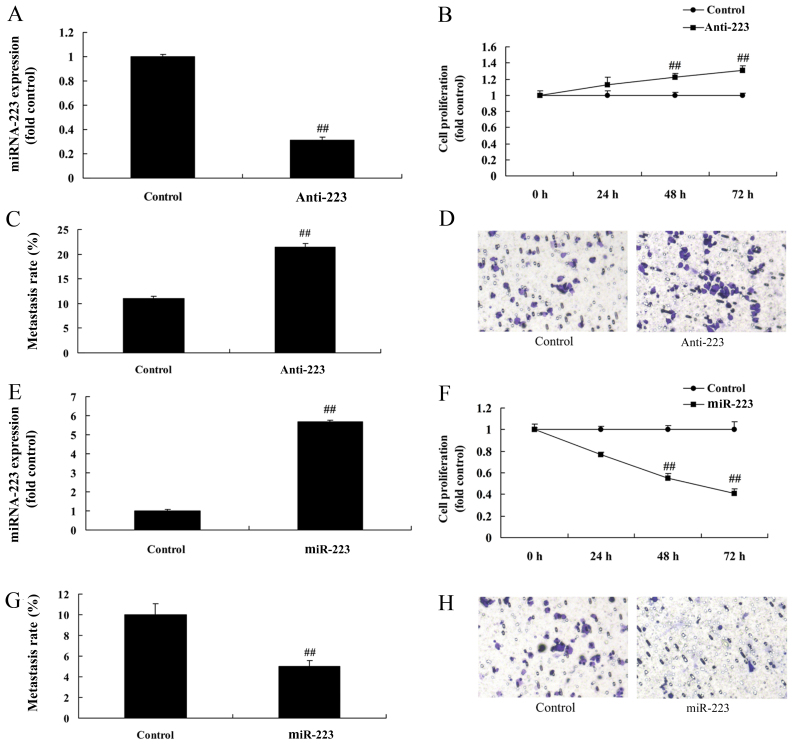
To confirm the function of miRNA-223, miRNA-223 expression was downregulated or upregulated using anti-miRNA-223 mimics or miRNA-223 mimics. As shown in Fig. 2A, miRNA-223 expression in A549 cells was significantly inhibited following transfection with the anti-miRNA-223 mimics compared with the control group. Downregulation of miRNA-223 expression significantly promoted the proliferation and migration of A549 cells compared with the negative group (Fig. 2B-D). In contrast, miRNA-223 mimics significantly upregulated miRNA-223 expression in A549 cells, which significantly reduced cell proliferation and migration when compared with the negative group (Fig. 2E-H).
Figure 2.
miRNA-223 affects the proliferation and migration of A549 cells. (A and E) miRNA-223 expression, (B and F), cell proliferation and (C, D, G and H) migration rate. Control, control group; Anti-223, downregulated miRNA-223 expression group; miR-223, upregulated miRNA-223 expression group. ##P<0.01 compared with the control group.
miRNA-223 affects the apoptosis and LDH activity in A549 cells
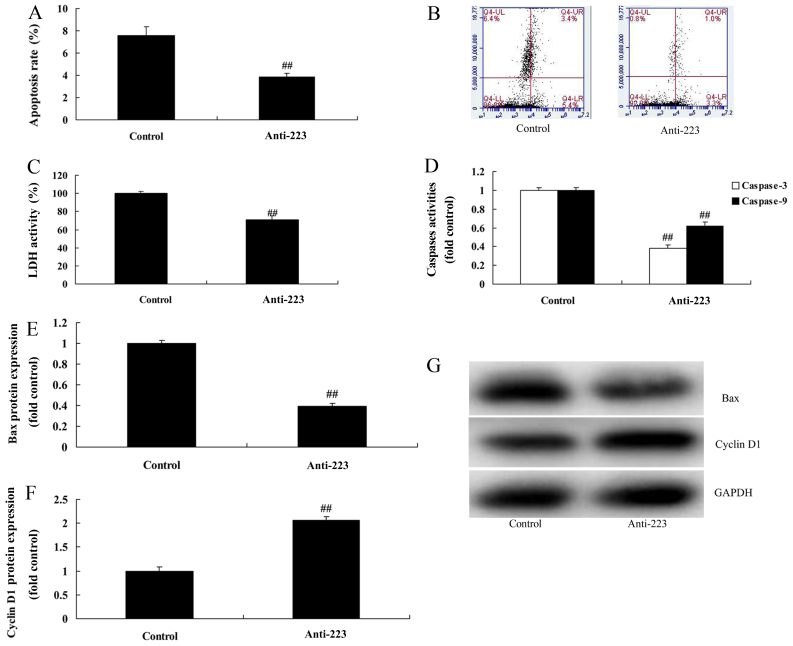
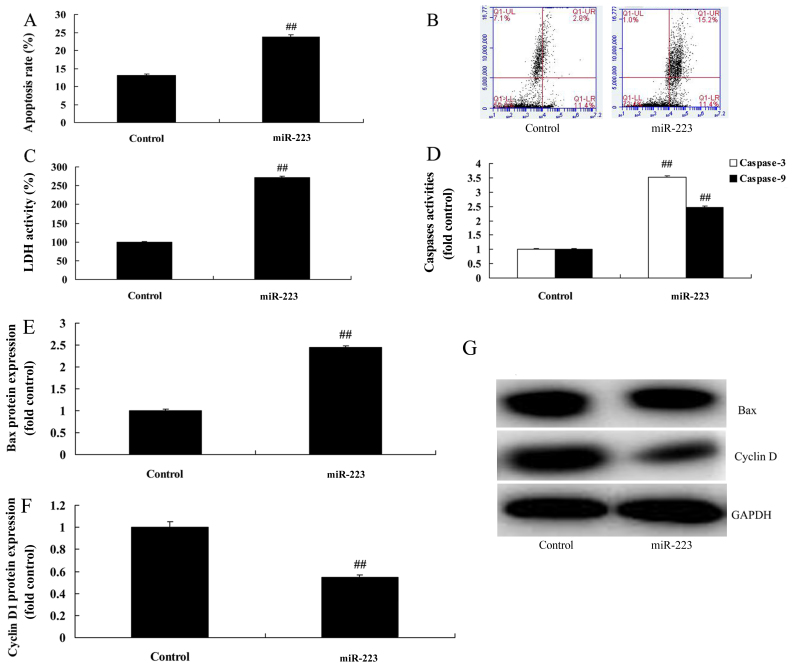
We subsequently found that miRNA-223 downregulation evidently reduced the apoptosis rate, LDH activity and caspase-3/9 activity in A549 cells compared with the control group (Fig. 3A-D). Compared with the control group, the protein expression of Bax and cyclin D1 was markedly suppressed and induced by downregulation of miRNA-223 expression in A549 cells, respectively (Fig. 3E-G). However, upregulation of miRNA-223 evidently increased the apoptosis rate, LDH activity and caspase-3/9 activity in the A549 cells compared with the control group (Fig. 4A-D). Compared with the control group, miRNA-223 upregulation significantly induced Bax protein expression and suppressed cyclin D1 protein expression in the A549 cells, respectively (Fig. 4E-G).
Figure 3.
Anti-miRNA-223 affects the apoptosis and LDH activity of A549 cells. (A and B) Apoptosis rate, (C) LDH activity, (D) caspase-3/9 activity, (E and F) Bax and cyclin D1 protein expression by statistical analysis and (G) western blot assays. Control, control group; Anti-223, downregulated miRNA-223 expression group. ##P<0.01 compared with the control group.
Figure 4.
miRNA-223 affects the apoptosis and LDH activity of A549 cells. (A and B) Apoptosis rate, (C) LDH activity, (D) caspase-3/9 activity, (E and F) Bax and cyclin D1 protein expression by statistical analysis and (G) western blot assays. Control, control group; miR-223, upregulated miRNA-223 expression group. ##P<0.01 compared with the control group.
miRNA-223 affects EGFR, PI3K and p-Akt protein expression in A549 cells
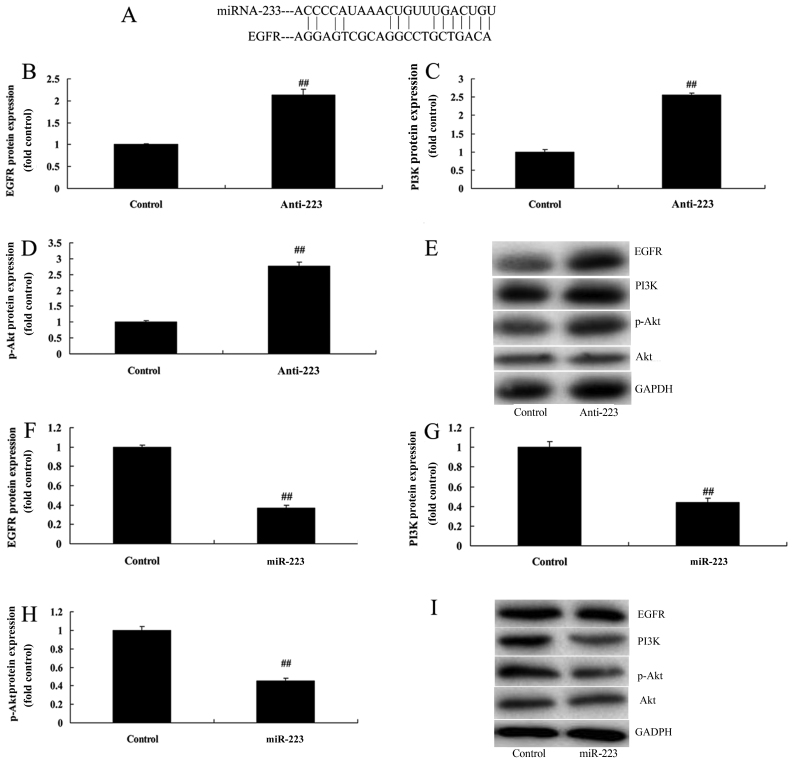
We determined whether miRNA-223 achieved its effects on NSCLC through the EGFR/PI3K/AKT pathway. Schematic representation of putative miRNA-223-binding sites in EGFR mRNA was found (Fig. 5A). However, downregulation of the expression of miRNA-223 dramatically induced EGFR, PI3K and p-Akt protein expression in A549 cells compared with the control group (Fig. 5B-E). In comparison, upregulation of miRNA-223 significantly suppressed EGFR, PI3K and p-Akt protein expression in A549 cells compared with the control group (Fig. 5F-I). There results indicate that miRNA-223 may be suppressed by the EGFR/PI3K/AKT pathway in NSCLC for its anticancer effects.
Figure 5.
miRNA-223 affects EGFR, PI3K and p-Akt protein expression of A549 cells. (A) miRNA-223 binding site with EGFR. (B-D) EGFR, PI3K and p-Akt protein expression by statistical analysis and (E) western blot assays following downregulation of miRNA-223 expression. (F-H) EGFR, PI3K and p-Akt protein expression by statistical analysis and (I) western blot assays following upregulation of miRNA-223 expression. Control, control group; Anti-223, downregulated miRNA-223 expression group; miR-223, upregulated miRNA-223 expression group. ##P<0.01 compared with the control group.
EGFR inhibition increases the anticancer effects of miRNA-223 on EGFR protein expression in A549 cells
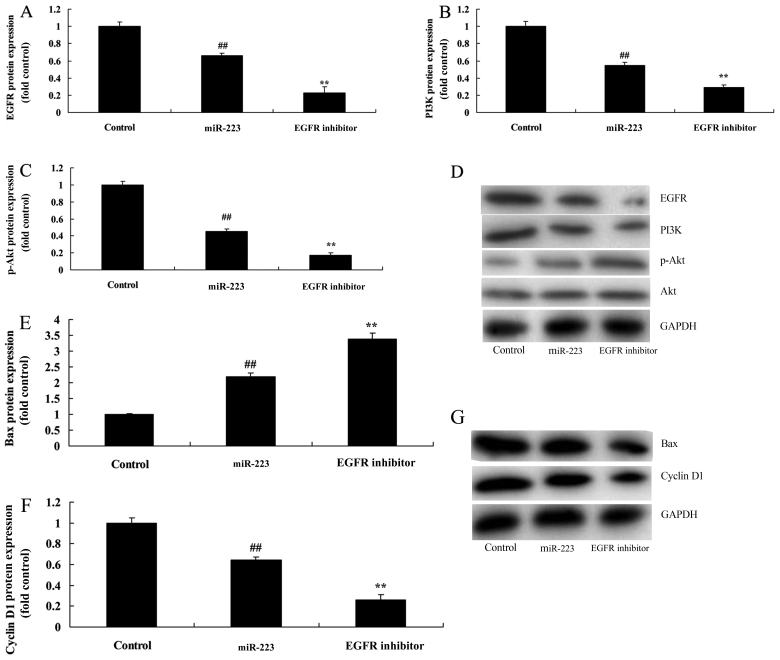
We next examined the impact of EGFR on the anticancer effects of miRNA-223 on the apoptosis of A549 cells. Treatment with 300 nM of AG1478 (EGFR inhibitor) for 48 h notably suppressed EGFR, PI3K and p-Akt protein expression in the A549 cells following miRNA-223 transfection, compared with the miRNA-223 group (Fig. 6A-D). Compared with the miRNA-223 group, EGFR inhibition evidently increased the anticancer effects of miRNA-223 on the induction of Bax protein expression and the suppression of cyclin D1 protein expression in A549 cells (Fig. 6E-G).
Figure 6.
Inhibition of EGFR increases the anticancer effects of miRNA-223 on EGFR protein expression in A549 cells. (A-C) EGFR, PI3K and p-Akt protein expression by statistical analysis and (D) western blot assays. (E and F) Bax and cyclin D1 protein expression by statistical analysis and (G) western blot assays. Control, control group; miR-223, upregulated miRNA-223 expression group; EGFR inhibitor, upregulated miRNA-223 expression and EGFR inhibitor group. ##P<0.01 compared with the control group; **P<0.01 compared with the upregulated miRNA-223 expression group.
EGFR inhibition increases the anticancer effects of miRNA-223 in A549 cells
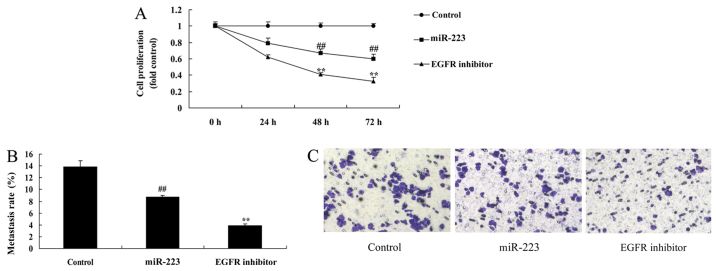
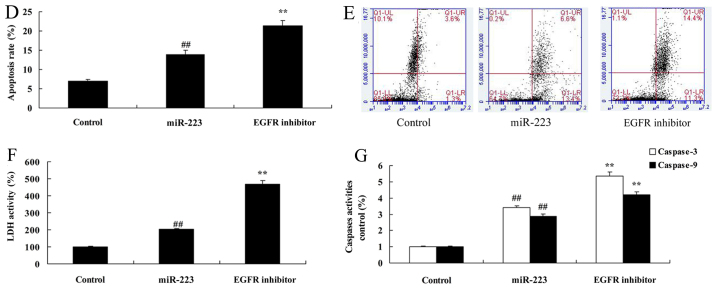
It was found that EGFR inhibition dramatically enhanced the anticancer effects of miRNA-223 in regards to inhibition of cell proliferation and migration (Fig. 7A-C), while promoting apoptosis, LDH activity and caspase-3/9 activity in A549 cells (Fig. 7D-G), compared with the miRNA-223 group.
Figure 7.
Inhibition of EGFR increases the anticancer effects of miRNA-223 in A549 cells. (A) Cell proliferation, (B and C) migration rate. Control, control group; miR-223, upregulated miRNA-223 expression group; EGFR inhibitor, upregulated miRNA-223 expression and EGFR inhibitor group. ##P<0.01 compared with the control group; **P<0.01 compared with the upregulated miRNA-223 expression group. Inhibition of EGFR increases the anticancer effects of miRNA-223 in A549 cells. (D and E) Apoptosis rate, (F) LDH activity, and (G) caspase-3/9 activity. Control, control group; miR-223, upregulated miRNA-223 expression group; EGFR inhibitor, upregulated miRNA-223 expression and EGFR inhibitor group. ##P<0.01 compared with the control group; **P<0.01 compared with the upregulated miRNA-223 expression group.
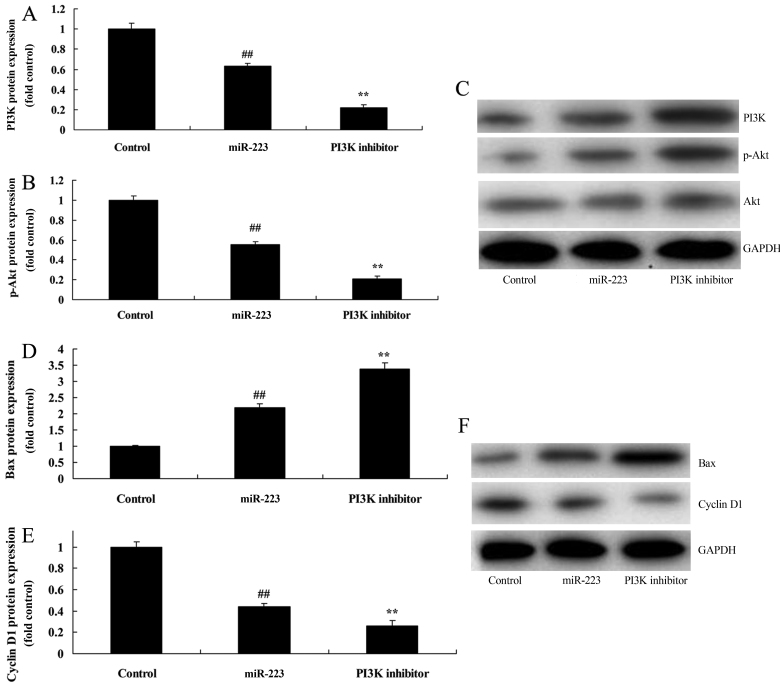
PI3K inhibition increases the anticancer effects of miRNA-223 on PI3K protein expression in A549 cells
To further identify the potential role of PI3K in the anticancer effects of miRNA-223 on the apoptosis of NSCLC, we comparatively analyzed PI3K protein expression in A549 cells. As shown in Fig. 8A-C, treatment of 200 nM of LY294002 (PI3K inhibitor) for 48 h evidently suppressed PI3K and p-Akt protein expression in A549 cells following miRNA-223 transfection, compared with the miRNA-223 group. Compared with the miR-223 group, PI3K inhibition significantly promoted the anticancer effects of miRNA-223 on the induction of Bax protein expression and the suppression of cyclin D1 protein expression in A549 cells (Fig. 8D-F).
Figure 8.
Inhibition of PI3K increases the anticancer effects of miRNA-223 on PI3K protein expression in A549 cells. (A and B) PI3K and p-Akt protein expression by statistical analysis and (C) western blot assays. (D and E) Bax and cyclin D1 protein expression by statistical analysis and (F) western blot assays. Control, control group; miR-223, upregulated miRNA-223 expression group; PI3K inhibitor, upregulated miRNA-223 expression and PI3K inhibitor group. ##P<0.01 compared with the control group; **P<0.01 compared with the upregulated miRNA-223 expression group.
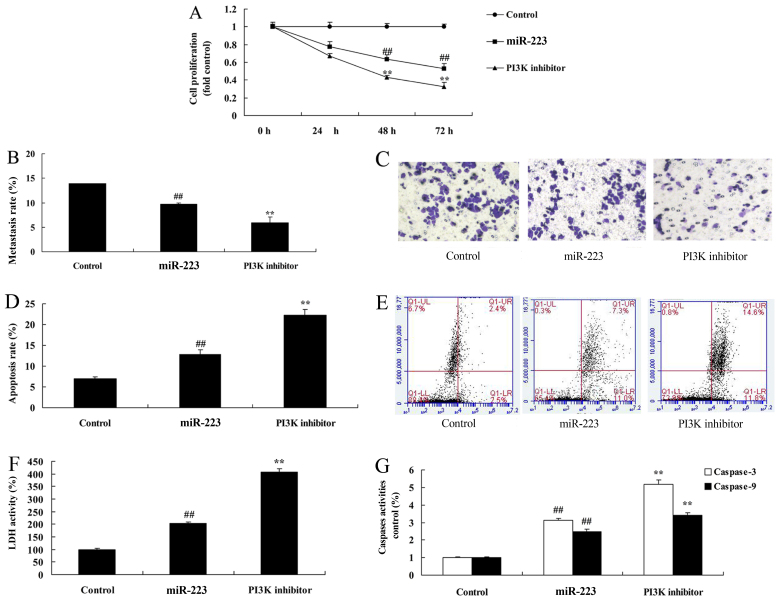
PI3K inhibition enhances the anticancer effects of miRNA-223 in A549 cells
Compared with the miRNA-223 group, PI3K inhibition significantly increased the anticancer effects of miRNA-223 in regards to the inhibition of cell proliferation and migration (Fig. 9A-C) while promoting apoptosis, LDH and caspase-3/9 activities (Fig. 9D-G) in A549 cells.
Figure 9.
Inhibition of PI3K increases the anticancer effects of miRNA-223 in A549 cells. (A) Cell proliferation, (B and C) migration rate, (D and E) apoptosis rate, (F) LDH activity, and (G) caspase-3/9 activity. Control, control group; miR-223, upregulated miRNA-223 expression group; PI3K inhibitor, upregulated miRNA-223 expression and PI3K inhibitor group. ##P<0.01 compared with the control group; **P<0.01 compared with the upregulated miRNA-223 expression group.
Discussion
Lung cancer is associated with high morbidity and poor patient prognosis (12). It has become the leading major malignant tumor threatening the life and health of individuals worldwide (12). Treatments for lung cancer include surgery, radiotherapy, chemotherapy and molecular-targeted therapy. Of these, surgical treatment exhibits the best efficacy on lung cancer (12). Moreover, the overall postoperative 5-year survival rate has reached 46.4% (13). The 5-year survival rates for stage I, II, III and IV patients are 58.6, 25.9, 16.8 and 3.9%, respectively. Surgical treatment has provided the greatest chance of healing for stage I and II patients (13). However, 80% of Chinese patients with lung cancer are in the advanced stage at diagnosis. As a result, they are not suitable for surgical treatment. At present, chemotherapy-oriented comprehensive treatment has become an important therapeutic means for advanced NSCLC (14).
In recent years, it has been discovered that miRNAs are factors involved in cancer (15). They have biological regulatory function. They are involved in biological processes such as cell division, proliferation, differentiation and development. Moreover, they exhibit functions that are similar to oncogenes and tumor-suppressor genes (16). Thus, they play a vital role in tumor genesis and development (16). Approximately 1/3 of human genes are regulated by miRNAs (16). miRNAs display complicated biological functions during the genesis and development of lung cancer (17). We first demonstrated that serum levels of miRNA-223 in NSCLC patients were downregulated; the downregulation of the expression of miRNA-223 increased cell proliferation and migration of A549 cells.
EGFR plays a regulatory role in tumor cell proliferation, differentiation and anti-apoptosis (6). Its abnormal expression can promote tumor cell proliferation, adhesion, invasion and metastasis (6). In addition, it induces tumor angiogenesis. Research indicates that expression of phosphorylated EGFR in NSCLC tissue is higher than that in para-carcinoma tissue (7). This has revealed that high EGFR expression is closely related to the genesis and development of NSCLC (18). Binding of EGFR with related ligands can activate the tyrosine signaling pathway and PI3K/AKT signaling pathway. Meanwhile, it can upregulate the expression of vascular endothelial growth factor (VEGF) and epidermal growth factor (EGF). Consequently, excessive activation of such pathways may enhance tumor cell proliferation, invasion and metastasis. Meanwhile, it inhibits cell apoptosis (18). EGFR may activate the PI3K/AKT signaling pathway (19). Our study reported that overexpression of miRNA-223 suppressed EGFR protein expression in A549 cells. It was demonstrated that miRNA-223 suppressed cervical cancer cell growth by targeting the EGFR/AKT2/CCND1 pathway (11). These results were in keeping with our results, and miRNA-223 suppressed EGFR protein expression to regulate the cell apoptosis of NSCLC.
Theoretically, any inhibitor for blocking expression of EGFR and PI3K/AKT signal pathway-related proteins can be used to treat NSCLC. The application of EGFR tyrosine inhibitor in NSCLC has ushered in a new horizon of targeted drug therapy aimed at the activated signaling pathway (20). However, the expected efficacy cannot be achieved in NSCLC patients after applying EGFR tyrosine kinase inhibitor. PI3K inhibitor at the current stage mainly targets catalytic subunit P110 (20). As reported in the literature, abnormality in the PI3K/AKT signaling pathway is related to tumor growth, maintenance and chemoresistance (20). Therefore, inhibiting such a signaling pathway can reverse chemoresistance and poor prognosis induced by activation of this pathway (21). At the same time, the combined application of a PI3K/AKT signaling pathway inhibitor and a traditional chemotherapeutic agent can enhance the chemotherapeutic effect and enhance radiotherapy sensitivity (22). Our study showed that overexpression of miRNA-223 suppressed PI3K and p-Akt protein expression in A549 cells. It was previously found that miRNA-223 suppressed cervical cancer cell growth via targeting the EGFR/AKT2/CCND1 pathway (11). Downregulation of miR-223 was also found to promote degranulation via the PI3K/Akt pathway by targeting IGF-1R in mast cells (23), which demonstrated that the miRNA-223/EGFR/PI3K/Akt pathway regulates the cell growth of NSCLC.
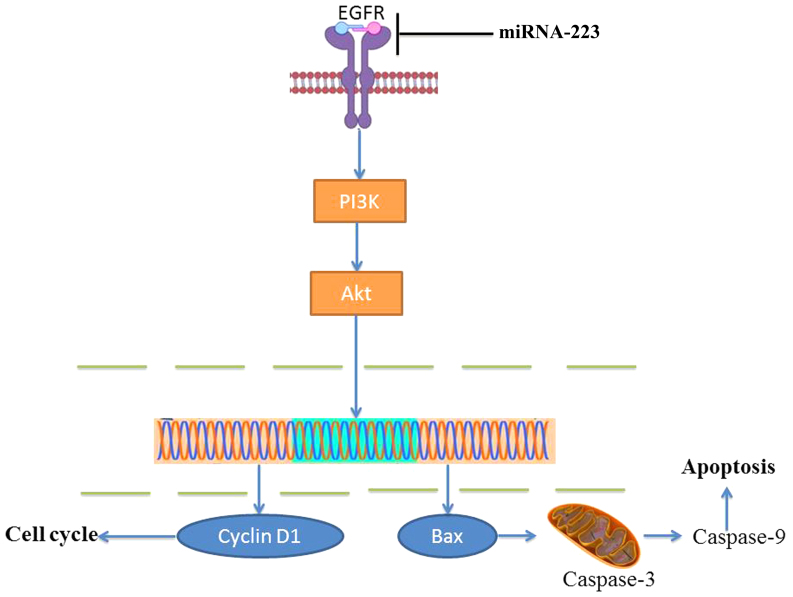
In conclusion, for the first time, we revealed that overexpression of miRNA-223 increased the apoptosis of non-small cell lung cancer cells through the PI3K/AKT pathway by targeting EGFR (Fig. 10). This study provides promising results supporting miRNA-223 as a novel antitumor candidate target for non-small cell lung cancer and has potential for further development as a single method for anticancer therapy.
Figure 10.
miRNA-223 may be an anticancer gene in NSCLC through the PI3K/AKT pathway by targeting EGFR. NSCLC, non-small cell lung cancer; EGFR, epidermal growth factor receptor.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.
Authors' contributions
XYL designed the experiment. HPM, WXK, WL, YZ and YW performed the experiment. HPM and XYL analyzed the data. XYL wrote the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The study was approved by Jining First People's Hospital, Jining, China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Price TJ, Peeters M, Kim TW, Li J, Cascinu S, Ruff P, Suresh AS, Thomas A, Tjulandin S, Zhang K, et al. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): A randomised, multicentre, open-label, non-inferiority phase 3 study. Lancet Oncol. 2014;15:569–579. doi: 10.1016/S1470-2045(14)70118-4. [DOI] [PubMed] [Google Scholar]
- 2.Denis F, Lethrosne C, Pourel N, Molinier O, Pointreau Y, Domont J, Bourgeois H, Senellart H, Trémolières P, Lizée T, et al. Randomized Trial Comparing a Web-Mediated Follow-up With Routine Surveillance in Lung Cancer Patients. J Natl Cancer Inst. 2017;109:109. doi: 10.1093/jnci/djx029. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, Sun W, Gong T, Chai Y, Wang J, Hui B, Li Y, Song L, Gao Y. miR-30a radiosensitizes non-small cell lung cancer by targeting ATF1 that is involved in the phosphorylation of ATM. Oncol Rep. 2017;37:1980–1988. doi: 10.3892/or.2017.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Zhou H, Luo J, Huang J. MicroRNA-17-5p inhibits proliferation and triggers apoptosis in non-small cell lung cancer by targeting transforming growth factor β receptor 2. Exp Ther Med. 2017;13:2715–2722. doi: 10.3892/etm.2017.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju L, Han M, Li X, Zhao C. MicroRNA signature of lung adenocarcinoma with EGFR exon 19 deletion. J Cancer. 2017;8:1311–1318. doi: 10.7150/jca.17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Liu Y, Shi W, Xu H, Hu H, Dong Z, Zhu G, Sun Y, Liu B, Gao H, et al. Droplet digital PCR improved the EGFR mutation diagnosis with pleural fluid samples in non-small-cell lung cancer patients. Clin Chim Acta. 2017;471:177–184. doi: 10.1016/j.cca.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Ni J, Weng L, Liu Y, Sun Z, Bai C, Wang Y. Dynamic monitoring of EGFR mutations in circulating cell-free DNA for EGFR-mutant metastatic patients with lung cancer: Early detection of drug resistance and prognostic significance. Oncol Lett. 2017;13:4549–4557. doi: 10.3892/ol.2017.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao JW, Zhan P, Qiu XY, Jin JJ, Lv TF, Song Y. Erlotinib-based doublet targeted therapy versus erlotinib alone in previously treated advanced non-small-cell lung cancer: A meta-analysis from 24 randomized controlled trials. Oncotarget. 2017;8:73258–73270. doi: 10.18632/oncotarget.18319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou G, Zhang F, Guo Y, Huang J, Xie Y, Yue S, Chen M, Jiang H, Li M. miR-200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3K/Akt signaling pathway and inhibites cell migration via targeting ZEB1. Biomed Pharmacother. 2017;85:113–119. doi: 10.1016/j.biopha.2016.11.100. [DOI] [PubMed] [Google Scholar]
- 10.Liu XL, Zhang XT, Meng J, Zhang HF, Zhao Y, Li C, Sun Y, Mei QB, Zhang F, Zhang T. ING5 knockdown enhances migration and invasion of lung cancer cells by inducing EMT via EGFR/PI3K/Akt and IL-6/STAT3 signaling pathways. Oncotarget. 2017;8:54265–54276. doi: 10.18632/oncotarget.17346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Wang Y, Chen Q, Qiu N, Zhao Y, You X. MiR-223 inhibited cell metastasis of human cervical cancer by modulating epithelial-mesenchymal transition. Int J Clin Exp Pathol. 2015;8:11224–11229. [PMC free article] [PubMed] [Google Scholar]
- 12.Soria JC, Adjei AA, Bahleda R, Besse B, Ferte C, Planchard D, Zhou J, Ware J, Morrissey K, Shankar G, et al. A phase IB dose-escalation study of the safety and pharmacokinetics of pictilisib in combination with either paclitaxel and carboplatin (with or without bevacizumab) or pemetrexed and cisplatin (with or without bevacizumab) in patients with advanced non-small cell lung cancer. Eur J Cancer. 2017;86:186–196. doi: 10.1016/j.ejca.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Wakelee HA, Dahlberg SE, Keller SM, Tester WJ, Gandara DR, Graziano SL, Adjei AA, Leighl NB, Aisner SC, Rothman JM, et al. ECOG-ACRIN: Adjuvant chemotherapy with or without bevacizumab in patients with resected non-small-cell lung cancer (E1505): An open-label, multicentre, randomised, phase 3 trial. Lancet Oncol. 2017;18:1610–1623. doi: 10.1016/S1470-2045(17)30691-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melichar B, Adenis A, Lockhart AC, Bennouna J, Dees EC, Kayaleh O, Obermannova R, DeMichele A, Zatloukal P, Zhang B, et al. Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: A five-arm phase 2 study. Lancet Oncol. 2015;16:395–405. doi: 10.1016/S1470-2045(15)70051-3. [DOI] [PubMed] [Google Scholar]
- 15.Schofield P, Ugalde A, Gough K, Reece J, Krishnasamy M, Carey M, Ball D, Aranda S. A tailored, supportive care intervention using systematic assessment designed for people with inoperable lung cancer: A randomised controlled trial. Psychooncology. 2013;22:2445–2453. doi: 10.1002/pon.3306. [DOI] [PubMed] [Google Scholar]
- 16.Schuette W, Behringer D, Stoehlmacher J, Kollmeier J, Schmager S, Fischer von Weikersthal L, Schumann C, Buchmann J. CHAMP: A phase II study of panitumumab with pemetrexed and cisplatin versus pemetrexed and cisplatin in the treatment of patients with advanced-stage primary nonsquamous non-small-cell lung cancer with particular regard to the KRAS status. Clin Lung Cancer. 2015;16:447–456. doi: 10.1016/j.cllc.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, Guan GH, Liu Q, Qian YH, Xie D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:10. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 18.Daly C, Castanaro C, Zhang W, Zhang Q, Wei Y, Ni M, Young TM, Zhang L, Burova E, Thurston G. FGFR3-TACC3 fusion proteins act as naturally occurring drivers of tumor resistance by functionally substituting for EGFR/ERK signaling. Oncogene. 2017;36:471–481. doi: 10.1038/onc.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JY, Welsh EA, Fang B, Bai Y, Kinose F, Eschrich SA, Koomen JM, Haura EB. Phosphoproteomics reveals MAPK inhibitors enhance MET- and EGFR-driven AKT signaling in KRAS-mutant lung cancer. Mol Cancer Res. 2016;14:1019–1029. doi: 10.1158/1541-7786.MCR-15-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li X, Zhao C, Zhang L, Cai W, Zhou C. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7:49948–49960. doi: 10.18632/oncotarget.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres AF, Nogueira C, Magalhaes J, Costa IS, Aragao A, Gomes Neto A, Martins F, Tavora F. Expression of EGFR and molecules downstream to PI3K/Akt, Raf-1-MEK-1-MAP (Erk1/2), and JAK (STAT3) pathways in invasive lung adenocarcinomas resected at a single institution. Anal Cell Pathol (Amst) 2014. 2014:352925. doi: 10.1155/2014/352925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Schmid-Bindert G, Wang D, Zhao Y, Yang X, Su B, Zhou C. Blocking the PI3K/AKT and MEK/ERK signaling pathways can overcome gefitinib-resistance in non-small cell lung cancer cell lines. Adv Med Sci. 2011;56:275–284. doi: 10.2478/v10039-011-0043-x. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Zhao DY, Xu H, Zhou H, Yang QY, Liu F, Zhou GP. Down-regulation of microRNA-223 promotes degranulation via the PI3K/Akt pathway by targeting IGF-1R in mast cells. PLoS One. 2015;10:e0123575. doi: 10.1371/journal.pone.0123575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.