Abstract
Background: Colorectal cancer is an increasing cause of death. Circulating microRNAs (miRs) could be great diagnostic and prognostic biomarkers of colorectal cancer, but further continuation of their utility is needed for their comprehensive application. Materials and Methods: Twenty-seven patients with colonic cancer, 16 with rectal cancer and 12 healthy volunteers as controls, were involved in this study. Expression of miR-155, miR-21, miR-221, miR-30a, miR-34a and miR-29a were determined by reverse transcription polymerase chain reaction (RT-PCR) from sera of patients. Results: Expression of miR-155, miR-21 and miR-221 was significantly higher in rectal cancer than in colonic cancer. There was no difference found between those with TNM1 cancer and controls for both cancer types. miR-155, miR-34a and miR-29a were down-regulated in all patients with cancer compared to controls. We did not find any statistically significant up-regulation of miR-221 in patients with colonic cancer compared to controls. In contrast, in patients with rectal cancer, miR-221 expression was higher than in controls. Advanced stage was also linked to higher miR-221 expression compared to early stage. Slight, but statistically significant increase was observed in miR-30a expression in patients with colon cancer compared to control individuals. Conclusion: Our results partly support previous findings. Here we report on differences in the expression of circulating microRNA between colonic and rectal tumours for the first time.
Keywords: Circulating microRNA, colon, rectum, biomarker, human
Colorectal tumours are becoming more and more significant in cancer mortality. In 2015, they ranked globally in second place. Lifestyle changes would have primarily played a role in this trend. In addition, aging of the population is also an important risk factor. Since aging of the world population will continue, diagnostics and therapy need to be improved in order to reduce mortality (1).
An understanding of the microRNA (miR) profile of patients with cancer can lead to an accurate diagnosis and effective therapy (2). Most published research on microRNA has compared normal and tumour tissue. Fortunately, some articles have appeared in recent years analysing the levels of circulating microRNA in peripheral blood. These data have great diagnostic and prognostic potential. Circulating microRNAs may indicate the presence of the tumour before the appearance of clinical symptoms and may lead to longer survival (3). Furthermore, taking samples from the tumour itself is unnecessary. However, the databank of circulating microRNA expression profiles of tumours has only just begun to develop. Moreover, for accurate diagnosis and therapy, differentiating colonic and rectal cancer is also essential (4).
Altered expression of miR-155 (5), miR-21 (6), miR-221 (7), miR-30a (8), miR-34a (9) and miR-29a (10) has been measured as promising diagnostic and prognostic biomarkers in the blood of patients with colorectal cancer, but whether their expression differs between colonic or rectal cancer has not been reported, as far as we are aware.
The purpose of this study was to compare the expression of these microRNAs in the serum of patients with colonic or rectal tumours and healthy controls.
Materials and Methods
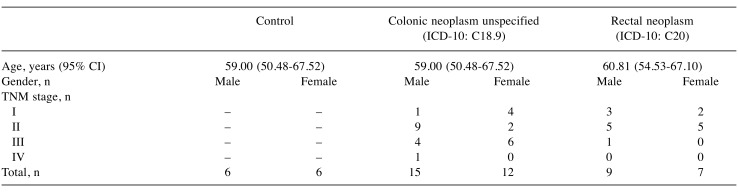
Study participants. A total of 43 patients with histopathologically confirmed colorectal cancer (27 with colonic cancer, 16 with rectal cancer) (Table I) and 12 controls participated at the National Oncology Institute between 2012 and 2015. Those who did not show evidence of disease (including cancer, precancerous lesion, and chronic illnesses) were selected as controls. Participants gave their written informed consent to use of study materials (Scientific and Research Ethics Commission No. 52088-11/2015/EKU).
Table I. Selected characteristics of the study population.
RNA isolation. The miRNAs were isolated from serum with TRI Reagent (Sigma-Aldrich Inc., Budapest, Hungary) according to the manufacturer’s instruction. RNA quality was checked by nano-drop absorption photometry and only RNA fractions with A>2.0 at 260/280 nm were used for reverse transcription.
cDNA synthesis and reverse transcription polymerase chain reaction (PCR). RNA templates of 5 μg/μl were used for cDNA synthesis by Universal cDNA Synthesis Kit (Quiagen, Woburn, MA, USA) and random hexamer priming. miRNA expression was determined in a Roche LC480 system (Roche, Budaörs, Hungary). PCR primers for miR-155, miR-21, miR-221, miR-30a, miR-34a and miR-29a were designed using a primer finder database (www.applied-science.roche.com) and were synthesised at TIB Molbiol, ADR Logistics (Roche Warehouse, Budapest, Hungary). PCR master mix contained 2 μl of specific primers, 8 μl DNA template and 10 μl LC480 SYBR Green I Master mix in a total volume of 20 μl. PCR was performed on a LightCycler 2.0 carousel-based PCR system (Roche, Berlin, Germany). The 20 μl PCR reaction mixture contained 7.2 μl of LightCycler RNA master SYBR Green I fluorescently labelled dye, 1.3 μl Mn(OAc)2 stock solution, 2 μl specific primer at a final concentration of 0.5 μM, 1 μl template miRNA and 8.2 μl of water. The reaction mixture was incubated for 30 s at 95˚C, followed by 45 three-step amplification cycles (95˚C for 5 s, 50˚C for 15 s, and 72˚C for 5 s). All runs, including template controls (nuclease-free water), were performed in three copies. Inter-run calibrators were used to calculate correction factors to avoid differences between runs. Data were normalized using 5S rRNA and U6sn RNA as endogenous references. Relative miRNA expression levels were compared using the 2−ΔΔCT method.
Statistical analyses were performed using IBM SPSS Version 21 (IBM Corp., Armonk, NY, USA). Mann–Whitney U-test was used to compare continuous and categorical variables based on data analysis.
Results
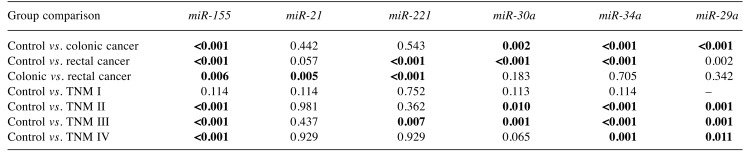
miR expression in controls vs. patients, and in colonic vs. rectal cancer. The expression of miR-155 was significantly higher in sera of control individuals than that of patients with colonic (p<0.001) or rectal (p<0.001) cancer. Furthermore, miR-155 expression was significantly higher in rectal cancer (p=0.006) than in colonic cancer.
In the case of miR-21, expression was significantly (p=0.005) elevated in serum obtained from patients with rectal, compared to colonic cancer. miR-221 was increased in serum from patients with rectal cancer compared to controls (p<0.001) and those with colonic cancer (p<0.001). The expression of miR-30a was similarly different, although expression was lowest in controls.
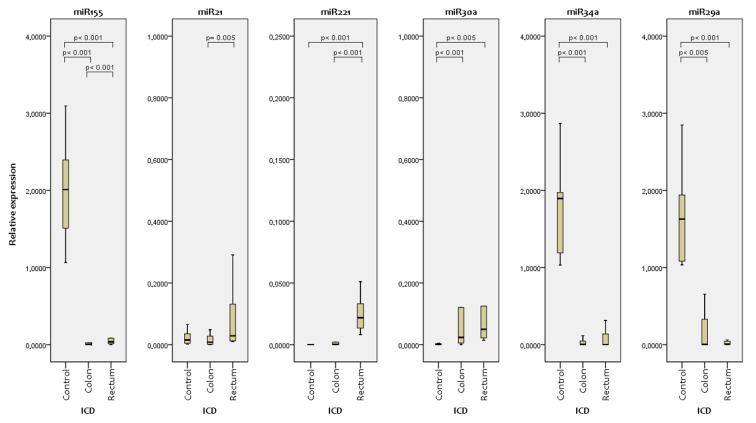
For miR-29a and miR-34, levels in controls were significantly higher than in patients with cancer, and differed in colonic compared with rectal cancer (Table II, Figure 1).
Table II. Statistical analysis of microRNA expression: p-Values for Man–Whitney U-test. Significant differences are shown in bold.
Figure 1. Comparison of relative microRNA (miR) expression in control, colonic cancer and rectal cancer.
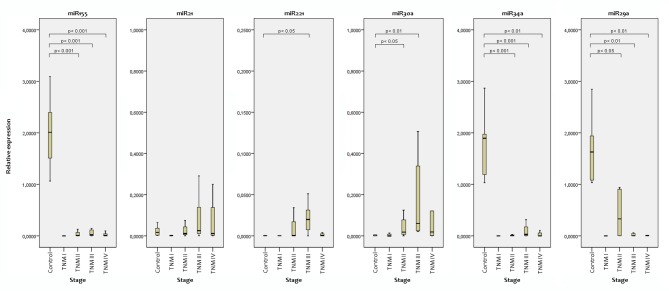
Expression by TNM stage. Expression in patients with TNM I cancer was not significantly different from that of the controls for any of the studied miRNAs. Expression of miR-155, miR-34a and miR-29a in TNM II, III and IV were significantly lower than those of the controls. Expression of miR-221 was elevated only in TNM III compared to the controls (p=0.007). mir-30a expression was significantly increased in TNM II (p=0.01) and TNM III (p=0.001) in comparison with the controls (Figure 2).
Figure 2. Comparison of relative microRNA (miR) expression in control and cancer according to cancer stage.
Discussion
We found microRNAs whose expression significantly differed from control in colonic and rectal cancer. In some cases, we measured altered expression levels in the serum of patients with colon cancer and rectal cancer. Furthermore, in those with advanced tumour, expression of some microRNAs was higher than for the controls.
According to the limited published data, high miR-155 expression, measured in blood, is a characteristic of normal healthy tissue (characterizes controls) and high expression results in better survival (11). We confirmed this with the results of our study, and distinguished colorectal cancer from rectal cancer since miR-155 expression was lower in colonic than in rectal cancer.
Toiyama and colleagues reported elevated expression of miR-21 in the sera of patients with colonic adenoma and colorectal cancer (12). In our study, we found the expression of miR-21 in patients with rectal cancer was statistically significantly higher than in those with colonic cancer, and differed from that of controls with only borderline significance (p=0.057).
Increased expression of miR-21 and miR-221 was previously found in peripheral blood samples in patients with colorectal cancer (10,13). We also found that miR-221 had higher expression in rectal than in colonic cancer and than in healthy controls. Moreover, expression of miR-21 and miR-221 was more pronounced at an advanced tumour stage (11-13).
Our results showed that the expression of miR-30a was slightly, but statistically significantly higher in the blood of patients with colonic or rectal cancer and in stage II and III compared to controls. This is contradictory to the literature since many publications mention that miR-30a has an anti-metastatic effect and expression was lower in patients with colorectal tumours (14-17).
Acherne et al. mentioned that the level of miR-34a was significantly lower in the blood of patients with colorectal cancer than in healthy individuals (18). Our results are consistent with this because significantly diminished expression was measured in sera of patients with colonic and rectal cancer and at stages II-IV compared to the controls.
The tendencies for miR-29a expression were practically the same as those for miR-34a. Our results confirm the data described in the literature (9,19), namely, the expression of miR-29a in the blood was significantly lower in colorectal cancer and in stages greater than I.
Overall, it can be stated that with the exception of miR-30a, we obtained similar results to those previously found. Furthermore, we found that the expression of miR-155, miR-21 and miR-221 in rectal cancer was significantly higher than in colonic cancer. Our results may contribute to the better application of studied microRNAs in the diagnosis and prognosis of colorectal cancer.
References
- 1.Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Raju GS, Chang DW, Lin SH, Chen Z, Wu X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer. 2018;124:785–796. doi: 10.1002/cncr.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Q, Ge B, Hu M, Zhou J, Bai X. Circulating microRNAs in colorectal cancer. J Mol Genet Med. 2017;11:274. [Google Scholar]
- 4.Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–713. doi: 10.1016/j.biopha.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 5.Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901–913. doi: 10.1002/jcp.25801. [DOI] [PubMed] [Google Scholar]
- 6.Ulivi P, Canale M, Passardi A, Marisi G, Valgiusti M, Frassineti GL, Calistri D, Amadori D, Scarpi E. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int J Mol Sci. 2018;19:307. doi: 10.3390/ijms19010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroen Hepatol. 2010;25:1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Bioph Res Co. 2017;493:1322–1328. doi: 10.1016/j.bbrc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106:947–952. doi: 10.1002/jso.23174. [DOI] [PubMed] [Google Scholar]
- 10.Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E, Pisa A, Casalots À, Serra-Aracil X, Oliva JC, Ruiz A, Saigí E. MicroRNA expression profile in stage III colorectal cancer: Circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep. 2013;30:320–326. doi: 10.3892/or.2013.2475. [DOI] [PubMed] [Google Scholar]
- 11.Lv ZC, Fan YS, Chen HB, Zhao DW. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumor Biol. 2015;36:1619–1625. doi: 10.1007/s13277-014-2760-9. [DOI] [PubMed] [Google Scholar]
- 12.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer I. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarlinova M, Halasa M, Mistuna D, Musak L, Iliev R, Slaby O, Mazuchova J, Valentova V, Plank L, Halasova E. miR-21, miR-221 and miR-150 are deregulated in peripheral blood of patients with colorectal cancer. Anticancer Res. 2016;36:5449–5454. doi: 10.21873/anticanres.11124. [DOI] [PubMed] [Google Scholar]
- 14.Yang LH, Yin SY, He RQ, Mo WJ, Pang YY, Wu YZ, Peng ZG, Gan TQ. Prospective target genes and pathways of miR-30a-5p in colorectal cancer: an investigation using TCGA and bioinformatics analysis. Int J Clin Exp Med. 2017;10:4373–4385. [Google Scholar]
- 15.Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, Zöllner H, Munding J, Klein-Scory S, Reinacher-Schick A, Schwarte-Waldhoff I, Schmiegel W, Hahn SA. miR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis. 2012;33:732–739. doi: 10.1093/carcin/bgs020. [DOI] [PubMed] [Google Scholar]
- 16.Zhong M, Bian Z, Wu Z. miR-30a suppresses cell migration and invasion through down-regulation of PIK3CD in colorectal carcinoma. Cell Physiol Biochem. 2013;31:209–218. doi: 10.1159/000343362. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Shi X, Zhao Y, Peng M, Kong Y, Qin D, Lv X. MicroRNA-30a mediates cell migration and invasion by targeting metadherin in colorectal cancer. Technol Cancer Res T. 2018;17:1533033818758108. doi: 10.1177/1533033818758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aherne ST, Madden SF, Hughes DJ, Pardini B, Naccarati A, Levy M, Vodicka P, Neary P, Dowling P, Clynes M. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer. 2015;15:329. doi: 10.1186/s12885-015-1327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen WY, Zhao XJ, Yu ZF, Hu FL, Liu YP, Cui BB, Dong XS, Zhao YS. The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int J Clin Exp Patho. 2015;8:7092–7101. [PMC free article] [PubMed] [Google Scholar]