Abstract
MicroRNAs (miRNAs) play critical roles in the growth, metastasis and therapeutic resistance of liver cancer. Accumulating evidence suggests that miR-498 is aberrantly expressed in several human malignancies. However, the role and underlying mechanism of miR-498 in liver cancer remain unclear. In the present study, we investigated the potential roles and clinical value of miR-498 in liver cancer. We found that the miR-498 expression level was significantly lower in liver cancer patient tissues than that in healthy control tissues. The expression of miR-498 was also decreased in liver cancer cell lines compared to that noted in a normal human normal liver cell line. miR-498 overexpression markedly inhibited liver cancer cell proliferation, migration and invasion. miR-498 overexpression induced cell cycle arrest and apoptosis while it suppressed epithelial-mesenchymal transition (EMT) in liver cancer cells. Bioinformatic analysis and luciferase reporter assay further identified zinc finger E-box binding homeobox 2 (ZEB2) as a novel target of miR-498. Furthermore, ZEB2 knockdown recapitulated the inhibitory effects of miR-498 overexpression in liver cancer cells. ZEB2 overexpression rescued the inhibition of liver cancer cell proliferation, migration, and invasion by miR-498, indicating that ZEB2 acts as a downstream effector of miR-498 in liver cancer cells. Thus, we demonstrated that miR-498 suppresses the growth and metastasis of liver cancer cells, partly at least, by directly targeting ZEB2, suggesting that miR-498 may serve as a potential biomarker for the diagnosis and therapy of liver cancer.
Keywords: liver cancer, miR-498, ZEB2, proliferation, migration
Introduction
Liver cancer is the fifth most common cancer and the third leading cause of cancer-related mortality worldwide (1). Despite recent advances in disease diagnosis and treatment, the long-term prognosis of liver cancer patients remains poor. For advanced-stage liver cancer patients, the overall 5-year survival rate is <5%. The main challenges for liver cancer treatment include intrahepatic recurrence and metastasis, which leads to the poor outcome for liver cancer patients (2). The molecular mechanisms underlying liver cancer recurrence and metastasis have not been fully elucidated.
The important roles of microRNAs (miRNAs) in health and diseases have been revealed in the past decade. miRNAs are important non-coding RNAs that negatively regulate gene expression at the post-transcriptional or translational levels. miRNAs participate in the regulation of many cellular processes, including cell proliferation, apoptosis, migration and invasion. Increasing evidence suggests that a variety of miRNAs are aberrantly expressed in human malignancies, including liver cancer (3,4). The deregulated expression of miRNAs is critically involved in the pathogenesis of cancer. miRNAs may function as tumor-suppressor genes or oncogenes, contributing to the development and progression of liver cancer (5–7). Therefore, further study of the expression pattern and roles of miRNAs may provide novel diagnostic and therapeutic targets for liver cancer.
miR-498 has been previously shown to be abnormally expressed in several human cancers. For instance, miR-498 is downregulated in non-small cell lung (8), ovarian (9), esophageal (10) and colorectal cancer (11). Low expression level of miR-498 is associated with disease progression and poor clinical outcome in cancer patients, indicating that miR-498 may act as a potential tumor-suppressor gene. miR-498 has also been shown to inhibit ovarian cancer cell growth by targeting human telomerase reverse transcriptase (hTERT) (12) and FOXO3 (13). However, it remains unknown whether miR-498 is involved in liver cancer growth and metastasis.
In the present study, we reported that miR-498 expression was low in human liver cancer and miR-498 overexpression suppressed liver cancer cell growth in vitro and in vivo. miR-498 overexpression induced cell cycle arrest and cell apoptosis in liver cancer cells. Furthermore, miR-498 inhibited liver cancer cell migration and invasion by reversing EMT. Mechanistically, miR-498 targets zinc finger E-box binding homeobox 2 (ZEB2), which consequently inactivates the TGF-β/SMAD and Wnt/β-catenin pathways. These findings provide a basis for the better understanding of the roles of miRNAs in liver cancer and a new target for the diagnosis and therapy of liver cancer.
Materials and methods
Microarray data analysis
Human microarray datasets GSE59856 (14) and GSE26323 (15) were obtained from the Gene Expression Omnibus (GEO database, http://www.ncbi.nlm.nih.gov/geo/). The GSE59856 dataset contained the transcriptome data of 202 serum samples from 52 liver cancer patients and 150 healthy controls. The GSE26323 dataset contained 6 samples of paired primary liver cancer tissues and lung metastatic tissues. The data were normalized by the Robust Multichip Average (RMA) algorithm (16). The z-score of log2 format of normalized data was used for further analysis.
Cell culture
Human normal liver cell line (HL-7702), human liver cancer cell lines [HepG2 (hepatoblastoma) and HCC-LM3 (hepatocellular carcinoma)] and human 293T cells were purchased from the Institutes for Biological Sciences at the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All the cells were cultured in a humidified incubator with 5% CO2 at 37°C.
Gene transfection
Cells were seeded in 6-well plates at a density of 2×105/well and cultured overnight. The overexpressing plasmid and knockdown shRNA (Hanbio, Shanghai, China) were transfected into the cells using LipoFiter transfection reagent (Hanbio) in serum-free medium. Cells were added to complete medium at 6 h after transfection and cultured for another 36 h.
Luciferase reporter assay
293T cells were co-transfected with miR-498 mimics and the luciferase reporter vector containing wild-type (WT) or mutant (MUT) 3′-UTR of ZEB2 as indicated. At 36 h after transfection, the cells were lysed and the luciferase activity was detected using the Dual-Luciferase Assay Kit (Promega Corporation, Madison, WI, USA).
Cell cycle analysis
Cell cycle analysis was conducted with a cell cycle detection kit (Fcmacs, Jiangsu, China). The transfected cells were collected and fixed in 95% ethanol overnight. Afterwards, the cells were stained with 50 µg/ml propidium iodide (PI) for 30 min in the dark. The cell cycle distribution was analyzed on a flow cytometer (BD FACSCalibur; BD Biosciences, San Jose, CA, USA) using CellQuest software (BD Biosciences).
Cell apoptosis assay
Cell apoptosis was determined using the Annexin V-Alexa Fluor 647/PI apoptosis detection kit (Fcmacs). The transfected cells were digested with collagenase, collected, and stained with Annexin V-Alexa Fluor 647 and PI for 15 min at room temperature. The cell apoptosis rate was analyzed using flow cytometry.
Cell counting and cell colony formation assays
For the cell counting assay, the transfected cells were seeded in 24-well plates (1×104/well) and were counted for 6 days. For the cell colony formation assay, the transfected cells were seeded in 6-well plates (1×103/well) and cultured for 10 days at 37°C. The medium was changed every 3 days. At the end of the experiments, the cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet. The number of colonies was calculated under a Nikon Eclipse Ti-S inverted microscope (Nikon Corp., Tokyo, Japan).
Transwell migration assay
The transfected cells were collected and seeded into the upper Transwell chamber (8 µm) (Corning Inc., Corning, NY, USA) at a density of 1×105 cells/well. The lower chamber was filled with 500 µl culture medium supplemented with 10% FBS. The cells were let to migrate for 12 h at 37°C. Then, the cells on the upper surface of the membrane were removed with a cotton swab. Then, the lower cells were fixed with formaldehyde and stained with 1% crystal violet for 30 min at room temperature. The number of migrated cells was counted under a Nikon Eclipse Ti-S inverted microscope (Nikon Corp.).
Cell invasion assay
The diluted basement Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added into each chamber and let to polymerize at 37°C for 30 min. The transfected cells were seeded into the upper chamber at a density of 2×105 cells/well. The lower chamber was filled with 500 µl culture medium supplemented with 10% FBS. The cells were allowed to invade to the lower membrane for 24 h. Subsequently, the cells on the upper surface of the membrane were removed with a cotton swab. The lower cells were then fixed with formaldehyde and stained with 1% crystal violet for 30 min at room temperature. The number of invaded cells was counted under a Nikon Eclipse Ti-S inverted microscope (Nikon Corp.).
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA using miScript reverse transcription kit (Bio-Rad Laboratories, Hercules, CA, USA). The relative expression of target genes was detected on a Bio-Rad CFX96 quantitative real-time PCR system with the SYBR-Green method (Qiagen, Inc., Valencia, CA, USA). PCR was conducted at 95°C for 15 min, followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec, and 63°C for 30 sec. U6 and β-actin served as internal controls. The expression of miR-498 was analyzed by 2−ΔΔCq method. The sequences of the primers are listed in Table I.
Table I.
Primers used for qRT-PCR.
| Gene | Sequence (5′-3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|
| E-cadherin | F: 5′-CGCATTGCCACATACACTCT-3′ | 252 | 55 |
| R: 5′-TTGGCTGAGGATGGTGTAAG-3′ | |||
| N-cadherin | F: 5′-AGTCAACTGCAACCGTGTCT-3′ | 337 | 55 |
| R: 5′-AGCGTTCCTGTTCCACTCAT-3′ | |||
| Vimentin | F: 5′-GAGCTGCAGGAGCTGAATG-3′ | 344 | 55 |
| R: 5′- AGGTCAAGACGTGCCAGAG-3′ | |||
| Slug | F: 5′-CCTGGTTGCTTCAAGGACAC-3′ | 395 | 55 |
| R: 5′-TCCATGCTCTTGCAGCTCTC-3′ | |||
| Twist | F: 5′-ACGAGCTGGACTCCAAGATG-3′ | 484 | 55 |
| R: 5′-GGCACGACCTCTTGAGAATG-3′ | |||
| Snail | F: 5′-GCGAGCTGCAGGACTCTAAT-3′ | 310 | 55 |
| R: 5′-GCCTCCAAGGAAGAGACTGA-3′ | |||
| Bcl-2 | F: 5′-GGATCCAGGATAACGGAGGC-3′ | 150 | 55 |
| R: 5′-CCAGATAGGCACCCAGGGT-3′ | |||
| Cyclin D1 | F: 5′-CCGAGAAGCTGTGCATCTAC-3′ | 221 | 55 |
| R: 5′-CTTCACATCTGTGGCACAGAG-3′ |
F, forward; R, reverse.
Western blot analysis
The cells were collected and lysed with RIPA buffer containing 1% protease inhibitors. Equal amounts of proteins were separated on 12% SDS-polyacrylamide gels and transferred onto polyvinylidene fluoride (PVDF) membranes, followed by blocking with 5% non-fat milk for 1 h. The membranes were incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: Anti-E-cadherin (cat. no. 4695S; Cell Signaling Technology, Inc., Inc., Beverly, MA, USA), anti-N-cadherin (cat. no. 4370S; Cell Signaling Technology, Inc.), anti-Bcl2 (cat. no. H-108; Santa Cruz Biotechnology, Dallas, TX, USA), anti-cyclin D1 (cat. no. H-108; Santa Cruz Biotechnology), anti-Slug (cat. no. 9585S; Cell Signaling Technology, Inc., Inc.), anti-Snail (cat. no. 3879S; Cell Signaling Technology, Inc.), anti-vimentin (cat. no. 5741S; Cell Signaling Technology, Inc.), anti-β-catenin (cat. no. 8480S; Cell Signaling Technology, Inc.), anti-c-Myc (cat. no. 5605S; Cell Signaling Technology, Inc.), anti-TGF-β1 (3711S; Cell Signaling Technology, Inc.), anti-SMAD2/3 (cat. no. 8685S; Cell Signaling Technology, Inc.), anti-p-SMAD2 (cat. no. 3108S; Cell Signaling Technology, Inc.), anti-p-SMAD3 (cat. no. 9520S; Cell Signaling Technology, Inc.) and anti-GAPDH (cat. no. MB001; Bioworld Technology, St. Louis Park, MN, USA). After incubation with the secondary antibodies (anti-rabbit secondary antibody, cat. no. 31460; and anti-mouse secondary antibody, cat. no. 31430; both from Bioworld Technology) at 37°C for 1 h, specific protein bands were visualized using an ECL chemiluminescent detection system (Millipore, Shanghai, China).
Clinical samples
The serum samples from liver cancer (n=20; 12 male and 8 female patients; age range, 56–78 years old) and sex- and age-matched healthy controls (n=20, 10 male and 10 female patients; age range, 53–72 years old) were obtained from the Affiliated People's Hospital of Jiangsu University between January 2018 and April 2018. Written informed consent was obtained from all the patients and this study was approved by the Institutional Ethics Committee of Jiangsu University (ref. no. 2017003). The patients included in the present study had not received any preoperative therapy.
Animal study
Ten male BALB/c nude mice (weight, 21–25 g; aged 4–6 weeks) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, Jiangsu, China) and maintained in specific pathogen-free (SPF) conditions in accordance with the institutional policies. The mice received sterile rodent chow and water ad libitum and were housed in sterile filter-top cages with 12-h light/dark cycles. Control or miR-498-transfected HepG2 cells were collected in PBS and subcutaneously injected into the mice (2×106 cells/mice, n=5). The mice were regularly fed and the tumors were measured twice a week. The tumor volume was calculated using the following formula: V (cm3) = 1/2 × length × width2. The protocol was approved by the Laboratory Animal Management Committee of Jiangsu University.
Statistical analysis
All the results are expressed as the mean ± SD. Differences between experimental groups were assessed by the Student's t-test or one-way analysis of variance (ANOVA) with the least significant difference (LSD) t-test using GraphPad Prism version 5.0 software (GraphPad Software, La Jolla, CA, USA). P<0.05 was considered as statistically significant.
Results
miR-498 is downregulated in human liver cancer
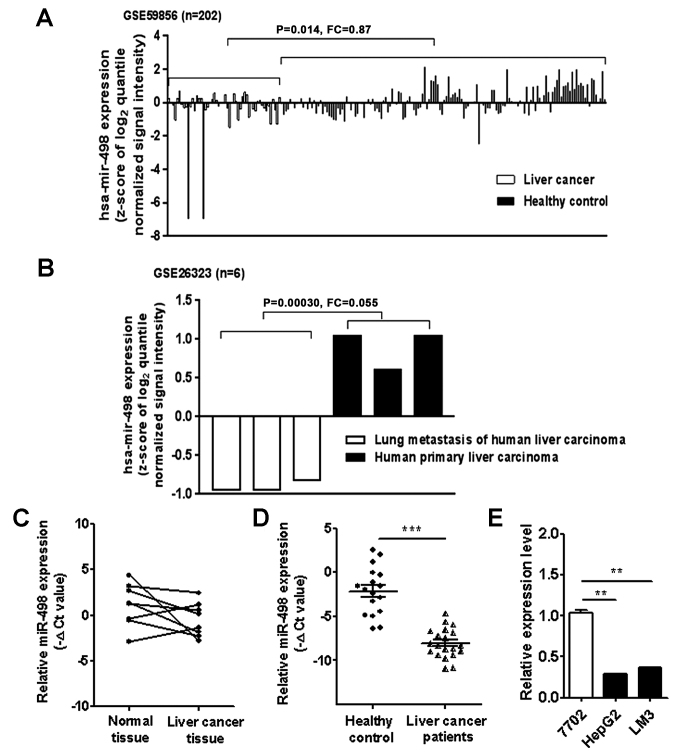
We first analyzed the expression levels of miR-498 in liver cancer patients using the microarray data downloaded from GEO (GSE59856 and GSE26323). The results showed that miR-498 expression level was downregulated in the serum of liver cancer patients compared to that from healthy controls (Fig. 1A). miR-498 expression level was also lower in the metastatic tumor tissues than that in the primary tumor tissues (Fig. 1B). To validate the findings of the GEO data analysis, we detected the expression of miR-498 in 8 pairs of liver cancer tissues and adjacent normal tissues using qRT-PCR. As shown in Fig. 1C, the expression of miR-498 was downregulated in 6 liver cancer tissues compared to that noted in the adjacent normal tissues. We further examined the expression of miR-498 in serum samples from liver cancer patients and healthy controls. The results showed that the expression levels of serum miR-498 were significantly lower in liver cancer patients than that in healthy controls (Fig. 1D). Moreover, miR-498 expression levels were detected in the normal liver cell line (HL-7702) and liver cancer cell lines [HepG2 (hepatoma) and HCC-LM3 (hepatocellular carcinoma)]. The expression levels of miR-498 in HepG2 and HCC-LM3 cells were significantly lower than that in the HL-7702 cells (Fig. 1E). Taken together, these findings suggest that miR-498 is downregulated in liver cancer.
Figure 1.
miR-498 is downregulated in human liver cancer tissues, serum samples and cell lines. (A) Analysis of GEO dataset GSE59856 (n=52 for liver cancer group; n=150 for healthy control group) showed decreased expression of miR-498 expression level in the serum samples of liver cancer patients. (B) Analysis of GEO dataset GSE26323 showed decreased expression levels of miR-498 in lung metastasis tissues compared to paired primary tumor tissues (n=3). (C) qRT-PCR analyses of miR-498 expression levels in paired liver cancer tissues and adjacent normal tissues (n=8). (D) qRT-PCR analyses of serum miR-498 expression levels in liver cancer patients (n=20) and healthy controls (n=20). (E) qRT-PCR analyses of miR-498 expression in HepG2, HCC-LM3 and HL-7702 cells. **P<0.01, ***P<0.001.
miR-498 overexpression inhibits the growth of liver cancer cells
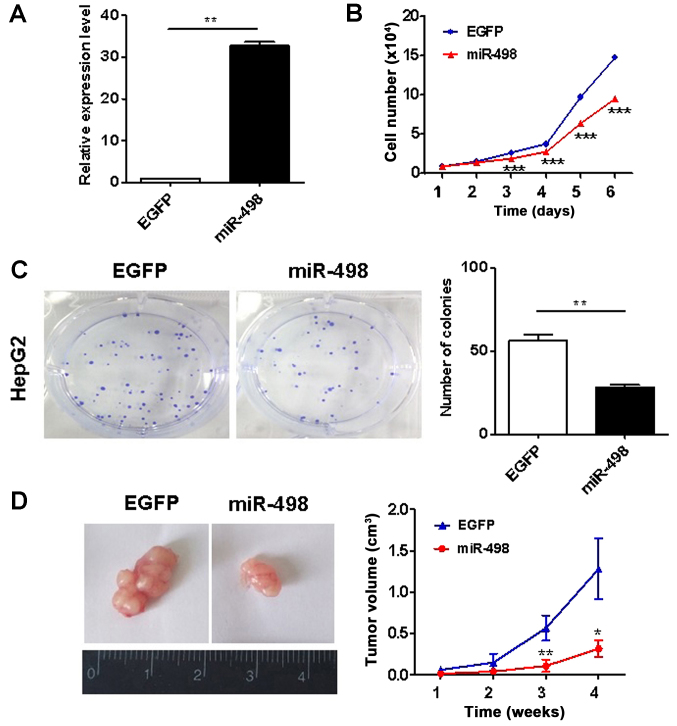
To investigate the roles of miR-498 in liver cancer, we overexpressed miR-498 in HepG2 cells using gene transfection. The efficacy of gene overexpression was validated (Fig. 2A). We then determined the proliferation abilities of HepG2 cells using cell counting and colony formation assays. The ectopic expression of miR-498 significantly inhibited the proliferation rate of HepG2 cells (Fig. 2B). The results of colony formation assay showed that HepG2 cells with miR-498 overexpression formed significantly less colonies than the control cells (P<0.01; Fig. 2C). Thus, these findings indicate that miR-498 suppresses liver cancer cell proliferation in vitro. We further determined the effects of miR-498 on liver cancer growth in vivo. As shown in Fig. 2D, the growth rate of xenograft tumors in the miR-498 overexpression group were significantly slower than that of xenograft tumors in in the control group.
Figure 2.
miR-498 overexpression inhibits the growth of liver cancer cells. (A) qRT-PCR analyses of miR-498 expression in control and transfected HepG2 cells. (B) Cell growth curves for HepG2 cells transfected with or without miR-498. (C) Colony formation assays for HepG2 cells transfected with or without miR-498. (D) The growth curves of xenograft tumors in control and miR-498 overexpression groups. *P<0.05, **P<0.01, ***P<0.001.
miR-498 overexpression induces cell cycle arrest and apoptosis in liver cancer cells
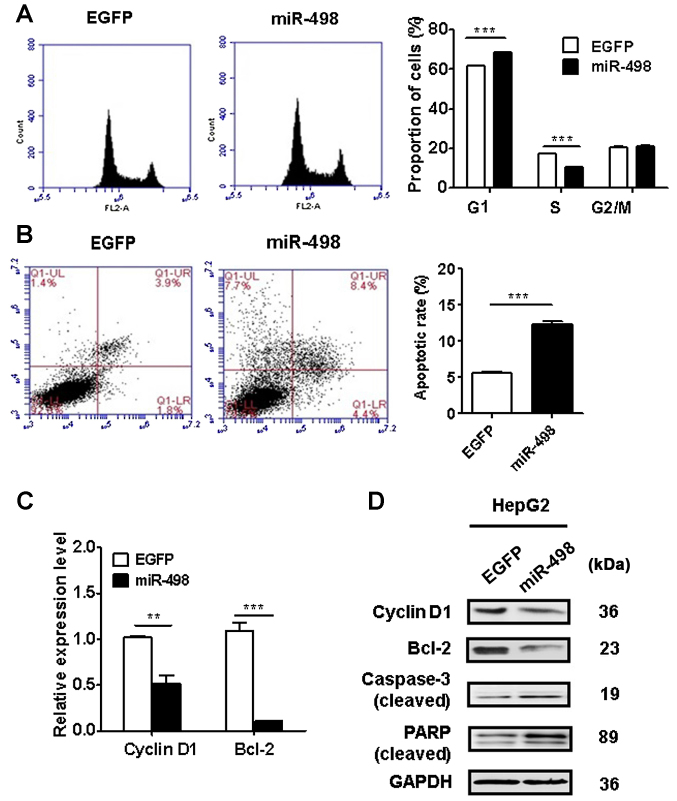
We performed flow cytometric analyses to determine the cell cycle distribution and apoptosis of HepG2 cells with or without miR-498 overexpression. The results of cell cycle analysis revealed that miR-498 overexpression in HepG2 cells significantly increased the proportion of cells in the G1 phase (P<0.01) while decreased that in the S phase (P<0.01) (Fig. 3A). We then evaluated the effect of miR-498 overexpression on cell apoptosis. As shown in Fig. 3B, miR-498 overexpression in HepG2 cells significantly increased the percentage of apoptotic cells. We determined the expression of cell cycle and apoptosis-related genes and proteins using qRT-PCR and western blot analysis. As shown in Fig. 3C and D, miR-498 overexpression led to a decrease in the expression of cyclin D1 and Bcl-2 in HepG2 cells. Moreover, western blot results showed that miR-498 overexpression promoted the cleavage of PARP and caspase-3 in HepG2 cells (Fig. 3D). Taken together, these results indicate that miR-498 overexpression inhibited liver cancer cell proliferation by inducing cell cycle arrest and apoptosis.
Figure 3.
miR-498 overexpression induces cell cycle arrest and apoptosis in liver cancer cells. (A) Flow cytometric analyses of cell cycles showed decreased G1 phase in miR-498-transfected HepG2 cells compared to control cells. (B) Flow cytometric analyses of cell apoptosis showed more apoptotic cells in the miR-498-transfected group than those in the control group. (C) qRT-PCR analyses of cyclin D1 and Bcl-2 gene expression in control and miR-498-overexpressing HepG2 cells. (D) Western blot analyses of cyclin D1, Bcl-2, poly(ADP-ribose) polymerase (PARP), and caspase-3 in control and miR-498-overexpressing HepG2 cells. **P<0.01, ***P<0.001.
miR-498 overexpression suppresses the metastatic abilities of liver cancer cells
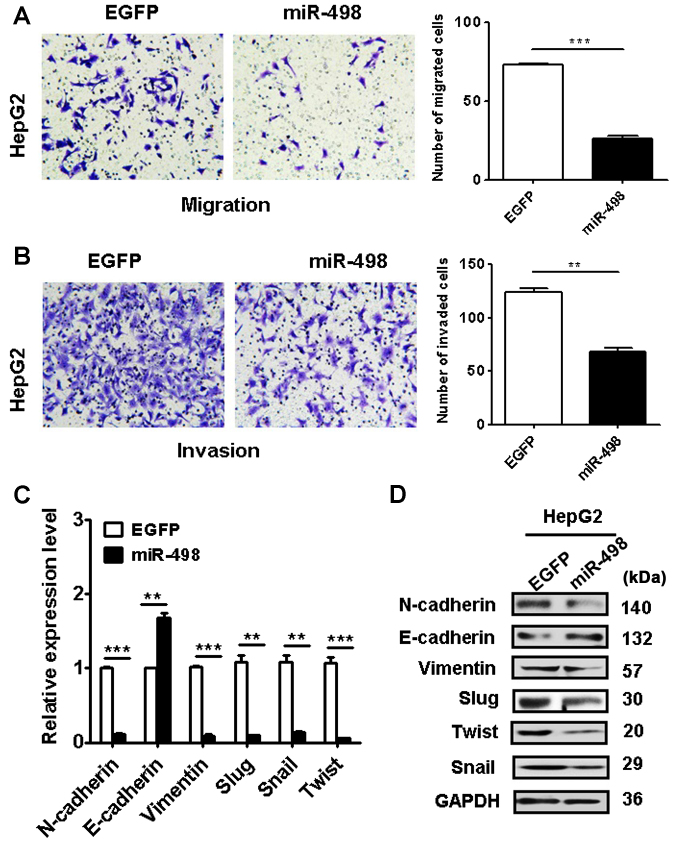
We next determined the effects of miR-498 overexpression on the metastatic potential of liver cancer cells. Our Transwell migration assay results showed that the miR-498 overexpression group had less migrated cells than that of the control group (Fig. 4A). In addition, the results of Matrigel invasion assays showed that miR-498 overexpression also suppressed the invasion abilities of HepG2 cells (Fig. 4B). Together, these data suggest that miR-498 has an inhibitory role in liver cancer cell migration and invasion.
Figure 4.
miR-498 overexpression inhibits the metastatic potential of liver cancer cells. (A) Transwell migration assays showed impaired migratory abilities in miR-498-overexpressing HepG2 cells. Magnification, ×200. (B) Matrigel invasion assays showed decreased invasive abilities of HepG2 cells in the miR-498 overexpression group. Magnification, ×200. (C) qRT-PCR and (D) western blot analyses of EMT-specific markers in HepG2 cells with miR-498 overexpression. **P<0.01, ***P<0.001.
To further understand the mechanisms by which miR-498 overexpression inhibits liver cancer cell migration and invasion, we determined the expression of EMT-related factors in HepG2 cells with or without miR-498 overexpression. We found that miR-498 overexpression upregulated the expression of E-cadherin gene while downregulated that of N-cadherin, vimentin, Slug, Snail and Twist genes in HepG2 cells (Fig. 4C). Western blot results also confirmed the same changes (Fig. 4D). These data indicate that miR-498 overexpression may inhibit EMT in liver cancer cells.
miR-498 targets ZEB2 in liver cancer cells
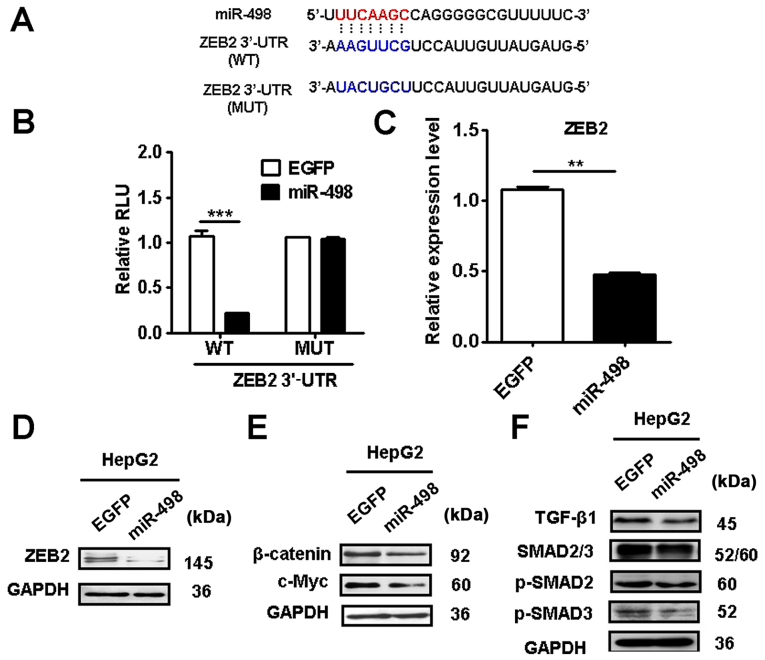
We searched for potential target genes of miR-498 using the bioinformatic algorithm TargetScan (http://www.targetscan.org) and miRDB (http://www.miRDB.org) and identified ZEB2 as a putative target of miR-498 (Fig. 5A). We further validated the prediction using luciferase reporter assay. The 3′-UTR of ZEB2 mRNA containing the wild-type (WT) or mutant (MT) miR-498 potential binding site were cloned and inserted into the luciferase reporter vector. The luciferase reporter assay results showed that miR-498 overexpression significantly decreased the activity of luciferase reporter gene in wild-type but not the mutant group (Fig. 5B). In addition, miR-498 overexpression significantly reduced the expression of ZEB2 gene and protein in HepG2 cells (Fig. 5C and D). Moreover, we found that miR-498 overexpression inhibited the expression of β-catenin and its downstream target c-Myc in HepG2 cells (Fig. 5E). The expression levels of TGF-β1, p-SMAD2 and p-SMAD3 were also suppressed by miR-498 overexpression (Fig. 5F). These results indicate that miR-498 targets ZEB2 to inactivate the Wnt/β-catenin and TGF-β/SMAD pathways in liver cancer cells.
Figure 5.
miR-498 inhibits ZEB2 expression in liver cancer cells. (A) TargetScan software prediction of miR-498 downstream genes. The seed sequences of miR-498 in the wild type (WT) or mutant type (MUT) of ZEB2 3′-UTR are indicated. (B) The WT or MUT ZEB2 3′-UTR were cloned into luciferase reporter vector. The luciferase activity in 293T cells transfected with the WT (or MUT) ZEB2 vector and miR-498. (C) qRT-PCR and (D) western blot analyses of ZEB2 expression levels in HepG2 cells transfected with miR-498. (E) Western blot analyses showed decreased expression of β-catenin and c-Myc in miR-498-transfected HepG2 cells. (F) Western blot analyses showed decreased expression of TGF-β, p-SMAD2 and p-SMAD3 in miR-498-transfected HepG2 cells. **P<0.01, ***P<0.001. ZEB2, zinc finger E-box binding homeobox 2.
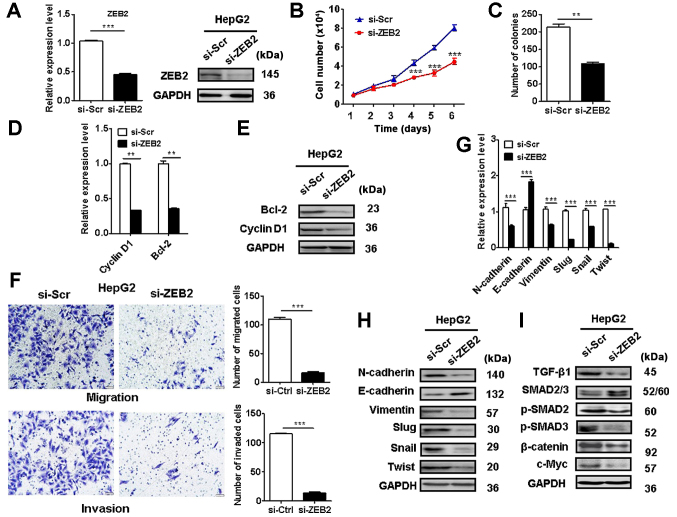
ZEB2 knockdown re-capitulates the inhibitory roles of miR-498 in liver cancer cell proliferation, migration and invasion
We further determined the role of ZEB2 in liver cancer progression. The efficacy of ZEB2 knockdown in liver cancer cells was validated (Fig. 6A). Cell counting and colony formation assay results showed that ZEB2 knockdown suppressed HepG2 cell proliferation (Fig. 6B and C). In addition, ZEB2 knockdown in HepG2 cells decreased the expression of cyclin D1 and Bcl-2 (Fig. 6D and E). ZEB2 knockdown also suppressed the migration and invasion of HepG2 cells (Fig. 6F). ZEB2 knockdown in liver cancer cells increased the expression of E-cadherin while decreased that of N-cadherin, vimentin, Slug, Snail, and Twist (Fig. 6G and H). Moreover, ZEB2 knockdown inactivated Wnt/β-catenin and TGF-β/SMAD pathways in HepG2 cells (Fig. 6I). These findings suggest that ZEB2 knockdown recapitulates the inhibitory roles of miR-498 in liver cancer cells.
Figure 6.
ZEB2 knockdown re-capitulates the inhibitory roles of miR-498 in liver cancer cells. HepG2 cells were transfected with scramble or ZEB2 siRNA. (A) qRT-PCR and western blot analyses of ZEB2 protein levels in control and ZEB2-knockdown HepG2 cells. (B) Cell counting and (C) colony formation assays showed decreased proliferation abilities of ZEB2-knockdown HepG2 cells compared to the control cells. (D) qRT-PCR and (E) western blot analyses of cyclin D1 and Bcl-2 levels in the control and ZEB2-knockdown HepG2 cells. (F) Transwell migration and Matrigel invasion assays showed decreased migratory and invasive abilities of HepG2 cells in the ZEB2-knockdown group compared to the control group. Magnification, ×200. (G) qRT-PCR and (H and I) western blot analyses of EMT-specific markers, and Wnt/β-catenin and TGF-β/SMAD pathway proteins in control and ZEB2-knockdown HepG2 cells. **P<0.01, ***P<0.001. ZEB2, zinc finger E-box binding homeobox 2.
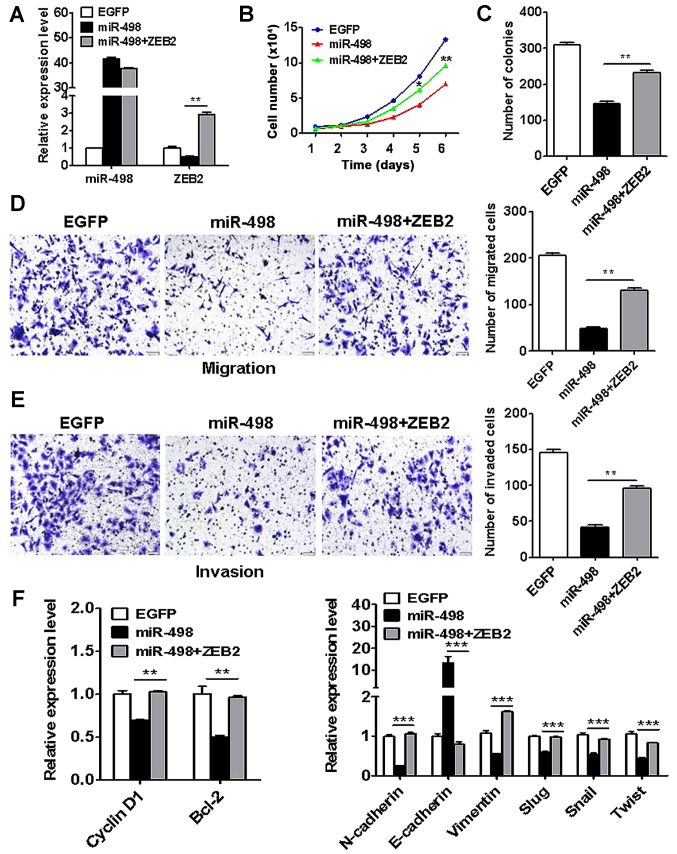
ZEB2 overexpression reverses the suppressive effects of miR-498 on liver cancer cells
To further demonstrate that ZEB2 is a key target of miR-498 in liver cancer, we co-transfected ZEB2 and miR-498 into HepG2 cells (Fig. 7A). The results of cell counting and colony formation assays showed that ZEB2 overexpresvicesion reversed the inhibition of proliferation by miR-498 in liver cancer cells (Fig. 7B and C). In addition, ZEB2 overexpression restored the migratory and invasive abilities of HepG2 cells that were inhibited by miR-498 (Fig. 7D and E). Moreover, ZEB2 overexpression reversed the cyclin D1, Bcl-2, N-cadherin, vimentin, Slug, Snail and Twist expression in the miR-498-transfected HepG2 cells (Fig. 7F). These results suggest that ZEB2 is key target of miR-498 in liver cancer.
Figure 7.
ZEB2 overexpression reverses the inhibition of liver cancer cell proliferation, migration and invasion by miR-498. HepG2 cells were co-transfected with miR-498 and ZEB2. (A) qRT-PCR analyses of miR-498 and ZEB2 expression in HepG2 cells. (B) Cell counting and (C) colony formation assays showed increased proliferation abilities of HepG2 cells in the ZEB2 co-transfection group compared to the miR-498 group. (D) Transwell migration and (E) Matrigel invasion assays showed increased migratory and invasive abilities of HepG2 cells in the ZEB2 co-transfection group compared to the miR-498 group. (F) qRT-PCR analyses of cell proliferation-related and EMT-specific markers. **P<0.01, ***P<0.001. ZEB2, zinc finger E-box binding homeobox 2.
Discussion
In the past decade, various miRNAs have been identified to play critical roles in the pathogenesis of liver cancer. For example, miR-146a suppresses liver cancer metastasis by inhibiting the nuclear accumulation of β-catenin and downregulating NF-κB p65 (17). Wu et al demonstrated that miR-655-3p targets ADAM10 to inhibit liver cancer growth (18). Yu et al found that miR-195 targets YAP to inhibit EMT in liver cancer cells (19). In the present study, we showed that miR-498 was frequently downregulated in liver cancer tissues and serum samples. Moreover, we found that decreased miR-498 expression was associated with liver cancer progression. We further investigated the function of miR-498 in liver cancer and found that miR-498 overexpression significantly inhibited the growth and metastasis of liver cancer cells in vitro and in vivo, indicating that miR-498 acts as a tumor suppressor in liver cancer. Although our preliminary results is in support of this notion, the detection of miR-498 expression in a large cohort of liver cancer tissue and serum samples need to be performed in future studies.
Our data suggest that the downregulation of miR-498 may contribute to the advanced progression of human liver cancer. Similar findings have been reported in other types of human cancers. For instance, Wang et al found that miR-498 inhibited the proliferation of non-small cell lung cancer (NSCLC) cells (8). Gopalan et al found that miR-498 suppressed cell proliferation in colorectal cancer (11). However, miR-498 is found to act as an oncogenic miRNA that targets cell cycle progression 1 (CCPG1) in retinoblastoma (20). miR-498 was found to be consistently upregulated in triple-negative breast cancer (TNBC) cells and miR-498 knockdown led to reduced proliferation in TNBC cells (21,22), suggesting that the functional roles of miR-498 is tumor type-specific and cell context-dependent.
Recurrence or metastasis is the major cause of the poor prognosis of liver cancer patients. miRNAs have been previously suggested to play important roles in cancer metastasis. We found that miR-498 overexpression decreased the metastatic ability of liver cancer cells. EMT is a key mechanism for cancer metastasis. During the process of EMT, the cancer cells lose the characteristics of epithelial cells and acquire the properties of mesenchymal cells. The previous studies demonstrated that miRNAs can regulate the occurrence of EMT in liver cancer. We found that miR-498 overexpression increased E-cadherin expression and decreased N-cadherin and vimentin expression. Furthermore, we confirmed that liver cancer cells with miR-498 overexpression had significantly lower level of several important EMT-related transcription factors. These findings suggest that miR-498 could suppress liver cancer metastasis by inhibiting EMT.
ZEB2 is highly expressed in human cancers and plays oncogenic roles (23,24). Several other studies have shown that ZEB2 is regulated by miRNAs. miRNA-30a-5p targets ZEB2 to promote the aggressiveness of clear cell renal cell carcinoma (25). In papillary thyroid cancer, miR-335 expression is decreased and its re-expression suppresses cancer cell growth, migration and invasion by directly targeting ZEB2 (26). Pang et al demonstrated that miR-154 targets ZEB2 to inhibit liver cancer cell proliferation, migration and invasion (27). Hu et al demonstrated that miR-204 targets ZEB2 to suppress liver cancer cell growth and metastasis (28). Our results showed that miR-498 overexpression markedly decreased the expression of ZEB2 in liver cancer cells. We identified ZEB2 as a novel target gene of miR-498. We further showed that ZEB2 knockdown could re-capitulate the suppressive roles of miR-498 in the proliferation, migration, and invasion of liver cancer cells. In glioma cells, ZEB2 knockdown was found to decrease β-catenin expression (29). Furthermore, Zhou et al reported that miRNA-145 targets ZEB2 to inactivate the Wnt/β-catenin pathway, inhibiting hepatic stellate cell activation and proliferation (30). In the present study, we found that miR-498 overexpression inactivated Wnt/β-catenin and TGF-β/SMAD pathways in liver cancer cells. miRNAs have been suggested to regulate multiple molecular pathways that are involved in the development and progression of tumorigenesis (31). Whether miR-498 could regulate other targets and pathways in liver cancer warrants further investigation in future studies.
In conclusion, we demonstrated in the present study that miR-498 acts as a novel tumor suppressor in liver cancer. miR-498 inhibits the proliferation, migration, and invasion of liver cancer cells, partly at least, via targeting ZEB2. The present study not only provides novel insights into the mechanism underlying the development and progression of liver cancer, but also suggests a promising diagnostic and therapeutic target for liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (nos. 81672416 and 81572075), the Key Research and Development Project of Zhenjiang (no. SH2015034), the Major Research and Development Project of Jiangsu Province (no. BE2015667), the Project of Jiangsu Provincial Commission of Health and Family Planning (no. LGY2017024), the Starting Foundation for Senior Talents of Jiangsu University (no. 13JDG086), the Qing Lan Project of Jiangsu Province, the ‘333’ project of Jiangsu Province and the Foundation for Young Academic Leader of Jiangsu University.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XZh conceived the idea of this study; XZh, XX, GG, XZa, MS, SZ, YZ, ZM, JZ and FM performed the experiments and acquired the data; XZh, HQ and WX analyzed and interpreted the data; XZh, XX, HQ and WX drafted and revised the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Written informed consent was obtained from all the patients and this study was approved by the Institutional Ethics Committee of Jiangsu University (ref no. 2017003). The protocol for the animal study was approved by the Laboratory Animal Management Committee of Jiangsu University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: Translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuguchi Y, Takizawa T, Yoshida H, Uchida E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol Res. 2016;46:391–406. doi: 10.1111/hepr.12606. [DOI] [PubMed] [Google Scholar]
- 4.Morishita A, Masaki T. miRNA in hepatocellular carcinoma. Hepatol Res. 2015;45:128–141. doi: 10.1111/hepr.12386. [DOI] [PubMed] [Google Scholar]
- 5.Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2017;23:2593–2604. doi: 10.1158/1078-0432.CCR-16-0851. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich P, Koch A, Fritz V, Hartmann A, Bosserhoff AK, Hellerbrand C. Wild type Kirsten rat sarcoma is a novel microRNA-622-regulated therapeutic target for hepatocellular carcinoma and contributes to sorafenib resistance. Gut. 2018;67:1328–1341. doi: 10.1136/gutjnl-2017-315402. [DOI] [PubMed] [Google Scholar]
- 7.Kabir TD, Ganda C, Brown RM, Beveridge DJ, Richardson KL, Chaturvedi V, Candy P, Epis M, Wintle L, Kalinowski F, et al. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67:216–231. doi: 10.1002/hep.29478. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Zhang Q, Wang J, Zhai Y. MicroRNA-498 is downregulated in non-small cell lung cancer and correlates with tumor progression. J Cancer Res Ther. 2015;11(Suppl 1):C107–C111. doi: 10.4103/0973-1482.163859. [DOI] [PubMed] [Google Scholar]
- 9.Cong J, Liu R, Wang X, Wang J, Wang H, Hou J. Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2015;19:4762–4765. [PubMed] [Google Scholar]
- 10.Islam F, Gopalan V, Law S, Tang JC, Chan KW, Lam AK. MiR-498 in esophageal squamous cell carcinoma: Clinicopathological impacts and functional interactions. Hum Pathol. 2017;62:141–151. doi: 10.1016/j.humpath.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Gopalan V, Smith RA, Lam AK. Downregulation of microRNA-498 in colorectal cancers and its cellular effects. Exp Cell Res. 2015;330:423–428. doi: 10.1016/j.yexcr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Kasiappan R, Shen Z, Tse AK, Jinwal U, Tang J, Lungchukiet P, Sun Y, Kruk P, Nicosia SV, Zhang X, et al. 1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J Biol Chem. 2012;287:41297–41309. doi: 10.1074/jbc.M112.407189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu R, Liu F, Li L, Sun M, Chen K. MiR-498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed Pharmacother. 2015;72:52–57. doi: 10.1016/j.biopha.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Kojima M, Sudo H, Kawauchi J, Takizawa S, Kondou S, Nobumasa H, Ochiai A. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS One. 2015;10:e0118220. doi: 10.1371/journal.pone.0118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, Wang L, Zhou J, Ren ZG, Li YX, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu M, Huang Z, Zang X, Pan L, Liang W, Chen J, Qian H, Xu W, Jiang P, Zhang X. Long noncoding RNA LINC00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 2017;5 doi: 10.1111/cpr.12425. doi: 10.1111/cpr.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer. 2015;14:5. doi: 10.1186/1476-4598-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G, Zheng K, Xia S, Wang Y, Meng X, Qin X, Cheng Y. MicroRNA-655-3p functions as a tumor suppressor by regulating ADAM10 and β-catenin pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:89. doi: 10.1186/s13046-016-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu S, Jing L, Yin XR, Wang MC, Chen YM, Guo Y, Nan KJ, Han LL. MiR-195 suppresses the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by inhibiting YAP. Oncotarget. 2017;8:99757–99771. doi: 10.18632/oncotarget.20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Wei N, Wang L, Wang X, Liu QH. miR-498 promotes cell proliferation and inhibits cell apoptosis in retinoblastoma by directly targeting CCPG1. Childs Nerv Syst. 2018;34:417–422. doi: 10.1007/s00381-017-3622-8. [DOI] [PubMed] [Google Scholar]
- 21.Matamala N, Vargas MT, González-Cámpora R, Arias JI, Menéndez P, Andrés-León E, Yanowsky K, Llaneza-Folgueras A, Miñambres R, Martínez-Delgado B, et al. MicroRNA deregulation in triple negative breast cancer reveals a role of miR-498 in regulating BRCA1 expression. Oncotarget. 2016;7:20068–20079. doi: 10.18632/oncotarget.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai C, Wu H, Wang B, Eisenstat DD, Leng RP. MicroRNA- 498 promotes proliferation and migration by targeting the tumor suppressor PTEN in breast cancer cells. Carcinogenesis. 2018;39:1185–1196. doi: 10.1093/carcin/bgy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai YH, Tang YP, Zhu HY, Lv L, Chu Y, Zhou YQ, Huo JR. ZEB2 promotes the metastasis of gastric cancer and modulates epithelial mesenchymal transition of gastric cancer cells. Dig Dis Sci. 2012;57:1253–1260. doi: 10.1007/s10620-012-2042-6. [DOI] [PubMed] [Google Scholar]
- 24.Li MZ, Wang JJ, Yang SB, Li WF, Xiao LB, He YL, Song XM. ZEB2 promotes tumor metastasis and correlates with poor prognosis of human colorectal cancer. Am J Transl Res. 2017;9:2838–2851. [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Zhang J, Zhang Z, Feng Z, Wei J, Lu J, Fang Y, Liang Y, Cen J, Pan Y, et al. The putative tumor suppressor microRNA-30a-5p modulates clear cell renal cell carcinoma aggressiveness through repression of ZEB2. Cell Death Dis. 2017;8:e2859. doi: 10.1038/cddis.2017.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kan Q, Su Y, Yang H. MicroRNA-335 is downregulated in papillary thyroid cancer and suppresses cancer cell growth, migration and invasion by directly targeting ZEB2. Oncol Lett. 2017;14:7622–7628. doi: 10.3892/ol.2017.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272–3279. doi: 10.3892/or.2015.4321. [DOI] [PubMed] [Google Scholar]
- 28.Hu B, Sun M, Liu J, Hong G, Lin Q. MicroRNA-204 suppressed proliferation and motility capacity of human hepatocellular carcinoma via directly targeting zinc finger E-box binding homeobox 2. Oncol Lett. 2017;13:3823–3830. doi: 10.3892/ol.2017.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi S, Song Y, Peng Y, Wang H, Long H, Yu X, Li Z, Fang L, Wu A, Luo W, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS One. 2012;7:e38842. doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou DD, Wang X, Wang Y, Xiang XJ, Liang ZC, Zhou Y, Xu A, Bi CH, Zhang L. MicroRNA-145 inhibits hepatic stellate cell activation and proliferation by targeting ZEB2 through Wnt/β-catenin pathway. Mol Immunol. 2016;75:151–160. doi: 10.1016/j.molimm.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Vasuri F, Visani M, Acquaviva G, Brand T, Fiorentino M, Pession A, Tallini G, D'Errico A, de Biase D. Role of microRNAs in the main molecular pathways of hepatocellular carcinoma. World J Gastroenterol. 2018;24:2647–2660. doi: 10.3748/wjg.v24.i25.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.