Environmental perturbations are central to the ability of pathobionts, such as Porphyromonas gingivalis, to promote the development of diseased sites. In the case of periodontal disease, increased local pH, a shift to anaerobic surroundings, and the accumulation of Gram-negative anaerobes at the expense of Gram-positive cocci are known ecological fluctuations prominently associated with progression toward disease. Importantly, in contrast, the alterations to subgingival food webs in disease sites remain poorly characterized. We hypothesized that given the dramatic shift in community structure during disease, it is possible that free carbohydrates, which would typically be readily metabolized by Gram-positive cocci after cleavage from glycoproteins, may increase in concentration locally and thereby affect the physiological state of the subgingival microbiota. In this study, we explored the impact of free monosaccharides on P. gingivalis to gain deeper insight into the effect of dysbiotic conditions on the growth and physiology of this periodontal pathogen.
KEYWORDS: microbial physiology, stress response
ABSTRACT
The asaccharolytic anaerobe Porphyromonas gingivalis metabolizes proteins it encounters in the periodontal pocket, including host-derived glycoproteins such as mucins and immunoglobulins. Often, these proteins are protected by a diverse array of carbohydrates tethered to the polypeptide chain via glycolytic bonds, and P. gingivalis produces enzymes capable of liberating these carbohydrates, exposing the proteinaceous core. In this study, we investigated the effect of individual monosaccharides, including galactose, l-fucose, mannose, and glucose, on the growth and physiology of P. gingivalis. Of the carbohydrates tested, only galactose noticeably altered the density of the bacterial culture, and we observed that cultures grown with galactose reached significantly higher densities during stationary phase. Importantly, electron micrographs and plating of P. gingivalis in stationary phase demonstrated that the presence of galactose did not increase cell numbers; instead, the higher densities resulted from the expansion of individual cells which contained large intracellular granules. Initial attempts to characterize these granules revealed only a subtle increase in soluble carbohydrates, suggesting they are likely not composed of stored carbohydrate. Also, an analysis of major surface polysaccharides via an enzyme-linked immunosorbent assay (ELISA) did not reveal significant differences between cells grown with or without galactose. Finally, an initial investigation of the transcriptional changes elicited by galactose in late exponential phase suggested that genes important for cell shape and for the general stress response may play roles in this phenomenon. Overall, galactose, a monosaccharide commonly present on the surfaces of host proteins, substantially alters the physiology of P. gingivalis via the production of large, currently undefined, intracellular granules.
IMPORTANCE Environmental perturbations are central to the ability of pathobionts, such as Porphyromonas gingivalis, to promote the development of diseased sites. In the case of periodontal disease, increased local pH, a shift to anaerobic surroundings, and the accumulation of Gram-negative anaerobes at the expense of Gram-positive cocci are known ecological fluctuations prominently associated with progression toward disease. Importantly, in contrast, the alterations to subgingival food webs in disease sites remain poorly characterized. We hypothesized that given the dramatic shift in community structure during disease, it is possible that free carbohydrates, which would typically be readily metabolized by Gram-positive cocci after cleavage from glycoproteins, may increase in concentration locally and thereby affect the physiological state of the subgingival microbiota. In this study, we explored the impact of free monosaccharides on P. gingivalis to gain deeper insight into the effect of dysbiotic conditions on the growth and physiology of this periodontal pathogen.
INTRODUCTION
Porphyromonas gingivalis, an asaccharolytic, highly proteolytic Gram-negative anaerobe, is a major etiological agent in the progression of chronic periodontal disease (1, 2). In large part due to the application of new genomic techniques, it has become increasingly apparent that P. gingivalis can be found in the periodontal pockets of healthy as well as diseased individuals, and these discoveries have shifted our understanding of the principle causes of periodontal disease. P. gingivalis is best classified as a pathobiont, i.e., a naturally present, host-associated microorganism that contributes to or exacerbates the disease state under certain environmental conditions (3, 4). Though our understanding of the precise mechanisms of periodontal disease development continues to evolve, it is broadly understood that alterations in the environment of the periodontal pocket due to inflammation and an increased flow of the gingival crevicular fluid results in a shift of the bacterial community from largely Gram-positive streptococci to primarily Gram-negative anaerobes, including P. gingivalis (5). Therefore, a better understanding of how P. gingivalis and closely associated bacteria react to environmental fluctuations is critical to defining the progression toward periodontal disease and to the development of new therapeutic interventions for preventing this disease.
To generate cellular energy, P. gingivalis relies primarily on the breakdown of proteins and subsequent fermentation of amino acids. In many cases, especially for host-derived proteins such as immunoglobulins and mucins, these proteins are decorated with an array of carbohydrates which can play functional roles as well as protect the underlying polypeptide chain from cleavage (6–8), and these carbohydrate residues must be removed by scavenging oral bacteria before the proteinaceous core can be hydrolyzed into short peptides and subsequently metabolized. While many of the genes required for the fermentation of carbohydrates are present in the genome of P. gingivalis, this organism, unlike many of the other black-pigmented Gram-negative anaerobes inhabiting the periodontal pocket, lacks key enzymes for catabolizing sugars and thus does not utilize carbohydrates as an energy source (9–12). A growth study of P. gingivalis on a complete panel of dietary carbohydrates is not available in the literature; however, it has been observed that the concentration of metabolic end products and pH remained essentially unchanged when cultures of P. gingivalis were grown in medium supplemented with glucose, arguably one of the simplest sugars to metabolize (13). While sugars may not be catabolized by P. gingivalis to generate energy, it has been observed that P. gingivalis W83 produces a neuraminidase capable of liberating sialic acid residues; it has been suggested that these free sugar residues could potentially be directly incorporated into the organism’s capsule (14). Importantly, P. gingivalis does not encode the enzymes required to synthesize sialic acid de novo (9, 14), but if this mechanism of inserting liberated sugar residues into surface structures is observed more broadly, it might represent an interesting evolutionary adaption with P. gingivalis taking advantage of liberated carbohydrates without directly internalizing and metabolizing these residues.
Though carbohydrate catabolism appears highly unlikely for P. gingivalis, the organism is nonetheless prolific in the synthesis of complex carbohydrates structures, many of which are exported across the cellular membrane and decorate the surface of the bacterium or bacterially derived surface proteins. Among these structures are key virulence determinants of P. gingivalis, including the K-antigen capsule (15), at least two distinct lipopolysaccharide (LPS) molecules (16–18), and gingipain proteases (17, 19), among others, and in some cases, these carbohydrate residues serve as important mediators of cell-to-cell associations and biofilm formation (20–23). Each of these cellular structures comprises complex, and in many cases, poorly characterized, carbohydrate chains. The disruption of genes encoding enzymes along the pathways for generating these complex carbohydrates can be highly detrimental to the physiology of the organism and often impedes its virulence, demonstrating the importance of these carbohydrates for survival. For example, deletion of the gene galE, which encodes a UDP-galactose 4-epimerase, in P. gingivalis 33277 produced a mutant strain unable to grow in high concentrations of galactose and which synthesized shorter O antigen chains in its LPS (24). This mutant also produced high levels of intracellular carbohydrates when grown with 0.1% galactose, generated more biomass than the parental strain in a standard biofilm assay, and appeared as long filamentous bacilli when viewed by electron microscopy (24). Overall, the highly regulated synthesis of complex carbohydrate chains and their proper attachment to the cell surface is an important factor in the persistence of P. gingivalis in the oral cavity and a key driver of the organism’s virulence.
In this study, we investigated the impact of several carbohydrates commonly found decorating the surfaces of host-derived extracellular proteins on the growth and physiology of P. gingivalis. After screening bacterial cultures using these sugars, we found that galactose significantly increased the optical density of bacterial cultures, but the number of CFU we recovered from cultures grown with this carbohydrate was essentially the same as from the control. Surprisingly, our study revealed that the inclusion of galactose in the growth medium caused cells of P. gingivalis to expand and develop large intracellular vacuoles. Though the precise mechanism of this phenomenon remains elusive, we found that known surface polysaccharides of P. gingivalis remained largely unaltered. The initial transcriptional changes occurring just hours after the addition of galactose have provided the first clues to understanding this profound physiological effect.
RESULTS
The optical density of P. gingivalis cultures was altered when galactose, but not other sugars, was included in the growth medium.
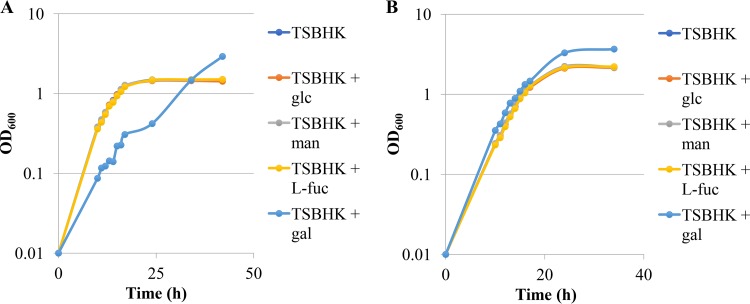
At least one previous study has investigated the effect of carbohydrates on the growth and metabolism of P. gingivalis (13). The authors included glucose in the growth medium at a concentration of 1% (approximately 55.5 mM) and found that supplementing the medium with this carbohydrate did not appreciably alter the growth or metabolism of P. gingivalis. To begin our experiments, we chose several carbohydrates commonly found decorating the surfaces of host-derived proteins, and these included mannose, l-fucose, and galactose as well as glucose, which was included primarily to confirm the results of Saito and colleagues (13). We chose two representative strains of P. gingivalis, i.e., the encapsulated strain P. gingivalis W83 and the nonencapsulated strain P. gingivalis 381. The two strains were subcultured in prereduced tryptic soy broth supplemented with 5 μg/ml hemin and 1 μg/ml menadione (TSBHK; we purchased the version not supplemented with glucose) or prereduced TSBHK supplemented with 0.1% (approximately 5.55 mM) of glucose, mannose, l-fucose, or galactose. As expected, we did not observe any effect on the growth of either strain of P. gingivalis when glucose was included in the medium, and similar results were seen for cultures where l-fucose or mannose was added (Fig. 1). Surprisingly, galactose initially inhibited the growth of P. gingivalis W83, and the cultures grown in medium supplemented with galactose reached a higher optical density than cultures grown in TSBHK alone (Fig. 1). Cultures of P. gingivalis 381 were not inhibited by the inclusion of galactose in the medium, but these cultures also consistently reached a higher optical density in stationary phase than the cultures grown with TSBHK alone (Fig. 1).
FIG 1.
Growth curves of P. gingivalis W83 (A) and 381 (B) on TSBHK alone or TSBHK supplemented with 0.1% of glucose (glc), mannose (man), l-fucose (l-fuc), or galactose (gal). The data are representative of at least three replications showing similar results.
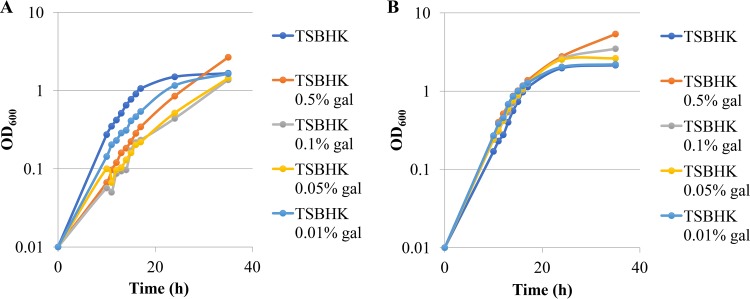
We next investigated whether there was a dose-dependent effect of galactose on the inhibition of P. gingivalis W83 during the initial phase of growth or on the optical density P. gingivalis W83 or P. gingivalis 381 achieved during stationary phase. Cultures of P. gingivalis W83 or P. gingivalis 381 were diluted in prereduced TSBHK or prereduced TSBHK supplemented with 0.01%, 0.05%, 0.1%, or 0.5% galactose, and the optical densities at 600 nm (OD600s) of the cultures were recorded. For P. gingivalis W83, we observed that the two intermediate concentrations (0.05% and 0.1%) of galactose produced the strongest inhibitory effect (Fig. 2). The cultures grown in TSBHK supplemented with the lowest concentration of galactose had only slightly inhibited growth of P. gingivalis (Fig. 2). The highest concentration (0.5%) of galactose produced a stronger inhibition than 0.01% galactose, but most strikingly, the cultures grown in 0.5% reached a much higher optical density once the cultures reached stationary phase (Fig. 2). As for P. gingivalis 381, the inclusion of galactose in the growth medium produced a dose-dependent response, i.e., increasing concentrations of galactose resulted in correspondingly higher optical densities of the cultures in stationary phase (Fig. 2). Our consistent observation of higher optical densities in cultures grown in medium supplemented with galactose led us to wonder if these cultures also contained higher numbers of cells. At the conclusion of this growth study, we serially diluted the bacterial cultures and spotted 10-μl dilutions of P. gingivalis W83 and P. gingivalis 381 on blood agar plates with hemin and menadione (BAPHK). We observed very little difference between the number of CFU recovered for cultures grown in TSBHK medium alone and the number in TSBHK supplemented with galactose (see Fig. S1 in the supplemental material). In fact, the highest concentration of galactose (0.5%) included in the study resulted in the lowest observed number of CFU recovered, suggesting either that galactose had an inhibitory effect on the growth of P. gingivalis W83 on blood agar plates or that galactose altered the cell size, which would impact the accuracy of pipetting and diluting the cultures. Importantly, these observations provide evidence that the high optical density resulting from growth with galactose is likely not the result of a larger population of bacterial cells.
FIG 2.
Growth curves of P. gingivalis W83 (A) and 381 (B) on TSBHK alone or TSBHK supplemented with 0.5%, 0.1%, 0.05%, or 0.01% galactose (gal). The data are representative of at least three replications showing similar results.
We consistently observed that TSBHK supplemented with higher concentrations of galactose inhibited the growth of P. gingivalis W83 during the initial phase of growth; however, we found that if cultures of the bacteria were grown in TSBHK medium to early exponential phase (OD600 of 0.5) before the addition of galactose, the inhibitory effect of galactose was eliminated. Furthermore, the optical densities of cultures of P. gingivalis W83 that received galactose increased at a higher rate than those of the culture that received equal volumes of sterile water, with a final optical density approximately twice that of the control cultures after 24 h (Table 1). This galactose-induced phenomenon was not unique to strains W83 and 381. We found that another encapsulated strain, P. gingivalis W50, reached a similar optical density to that of strain W83 after a 24-h incubation with 0.1% galactose (Table 1). Interestingly, the nonencapsulated (K-antigen minus) strains P. gingivalis 33277 and 381 as well as the reference strain AJW4 (capsule status not determined) displayed much weaker reactions to the presence of galactose in the medium; the increase in optical density was only approximately 1.75 for these strains after 24 h of growth (Table 1).
TABLE 1.
Optical density of P. gingivalis strains after 24 h of incubation with galactose
| Description | Galactose (%) | Strain | Average OD600 after 24 h | Fold change (with gal/no gal) | P valuea |
|---|---|---|---|---|---|
| Encapsulated strains | W50 | 1.35 | |||
| 0.1 | W50 | 3.43 | 2.54 | 6.4E−05 | |
| W83 | 1.62 | ||||
| 0.1 | W83 | 3.92 | 2.42 | 7.2E−05 | |
| Nonencapsulated strains | 33277 | 2.00 | |||
| 0.1 | 33277 | 3.52 | 1.76 | 6.4E−04 | |
| 381 | 2.08 | ||||
| 0.1 | 381 | 3.65 | 1.75 | 1.1E−04 | |
| Capsule status not determined | AJW4 | 2.22 | |||
| 0.1 | AJW4 | 3.95 | 1.78 | 2.9E−03 | |
| Capsule null W83 | W83 | 1.65 | |||
| 0.1 | W83 | 3.97 | 2.40 | 3.4E−04 | |
| W83 ΔPG0106 | 1.71 | ||||
| 0.1 | W83 ΔPG0106 | 3.75 | 2.20 | 2.2E−07 | |
| W83 fucP | W83 | 1.88 | |||
| 0.1 | W83 | 3.96 | 2.11 | 2.0E−03 | |
| W83 ΔPG2170 | 1.84 | ||||
| 0.1 | W83 ΔPG2170 | 3.25 | 1.77 | 2.1E−04 | |
| 381 rel | 381 | 2.00 | |||
| 0.1 | 381 | 3.48 | 1.74 | 4.0E−05 | |
| 381 Δrel | 2.08 | ||||
| 0.1 | 381 Δrel | 2.92 | 1.40 | 5.7E−03 | |
| 381 gingipain mutant | 381 | NDb | |||
| 0.1 | 381 | 3.83 | ND | ND | |
| 381 ΔrpgA | 2.43 | ||||
| 0.1 | 381 ΔrpgA | 4.13 | 1.70 | 2.1E−04 |
Determined by the Student's t test.
ND, not determined.
To determine the mechanism of this galactose-induced phenomenon, we selected a panel of P. gingivalis strains bearing mutations in genes we suspected to be related to carbohydrate utilization or to the sensing of carbohydrates in the environment, and these mutants were grown in TSBHK supplemented with galactose in an attempt to identify mutants that did not display an increase in culture density when this monosaccharide was introduced. The putative sugar permease encoded by fucP (PG2170 in P. gingivalis W83) is annotated as a transporter of sugars in P. gingivalis (9). To determine if the product of this gene contributes to the effect of galactose on P. gingivalis, a deletion replacement mutant of PG2170 was generated. We found that the deletion mutant achieved approximately the same optical density as the parent strain after a 24-h incubation in TSBHK containing 0.1% galactose (Table 1). We consistently observed that the optical density of encapsulated strains of P. gingivalis was higher than that of the other strains tested after incubation with galactose, and because of this, we also tested the effect of galactose on the capsule null mutant PG0106 (Table 1) (23). Similarly to the putative sugar permease, we observed a slightly lower increase in optical density when the PG0106 mutant was compared to the parental strain, but the overall effect on culture density remained. Due to the fact that the proteolytic gingipains of P. gingivalis are decorated with sugar residues, we also examined the possibility that their absence might alter the effect of galactose; but, we found that the rgpA mutant of P. gingivalis 381 (generously provided by Margaret Duncan, Forsyth Institute, Boston, MA) (25) retained the effect, though slightly less pronounced (Table 1). Finally, we hypothesized that the increase in optical density might be the result of a generalized stress response of P. gingivalis mediated by the stress response protein Rel. As with the previous deletions, we found that the P. gingivalis 381 Δrel mutant (381ΔPGN_0465; graciously provided by Hey-Min Kim, University of Florida, Gainesville, FL) consistently reached approximately the same optical density after a 24-h incubation in the presence of galactose as the parental strain (Table 1).
The increased optical density resulting from incubation with galactose is associated with an increase in cell size and the presence of large inclusion bodies.
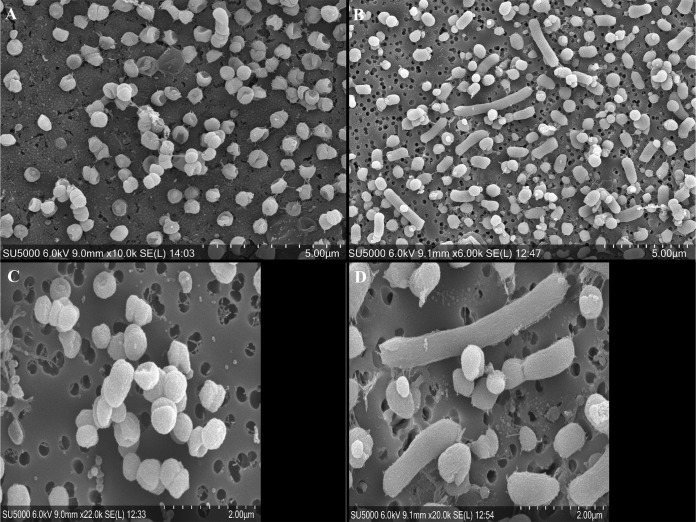
We consistently recovered approximately the same numbers, if not fewer, of CFU from stationary phase cultures of P. gingivalis grown in TSBHK supplemented with galactose as those from the TSBHK control (Fig. S1). Therefore, we speculated that the increase in culture density we observed after growth with galactose could not be attributed to an increase in the number of bacterial cells. We next asked if these differences could be explained by the size or morphology of the bacterial cells. During our initial growth studies with galactose, we viewed the stationary-phase cultures of P. gingivalis W83 grown in TSBHK supplemented with galactose under a simple light microscope and noted that the cells were approximately twice as large as cells from cultures grown in TSBHK alone (data not shown). To investigate these findings further, we grew P. gingivalis W83 to an OD600 of 0.5 and added either 0.1% galactose or sterile water as a control. We grew the cultures for 24 h and then observed them using a scanning electron microscope (SEM). After incubation with galactose, we observed that cells had become longer and distended and were easily distinguishable from the typical coccobacillus morphology of the cells grown in TSBHK (Fig. 3). In many cases, the cells grown with galactose lost their traditional coccus morphology and instead resembled long bacilli. Importantly, we noted some cells in the cultures treated with galactose that remained approximately the same size as cells from the control culture.
FIG 3.
The presence of galactose alters the size of P. gingivalis W83 cells. Cultures of P. gingivalis W83 were grown in TSBHK alone (A and C) or containing 0.1% galactose (B and D). Samples were prepared and viewed using scanning electron microscopy as described in Materials and Methods.
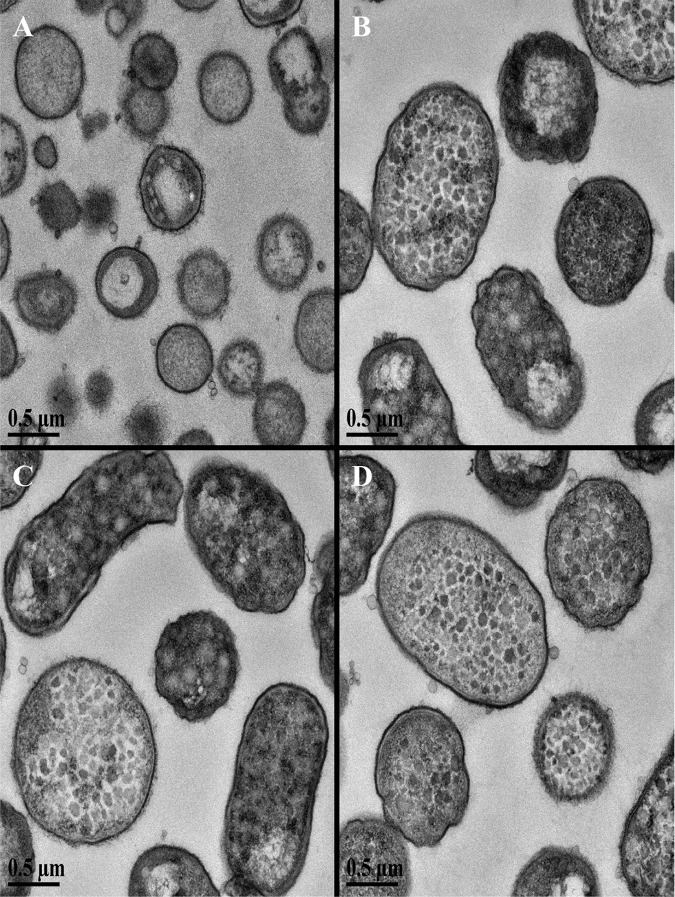
These images confirmed that the increase in culture density resulted from a change in the morphology of the cells and was not due to higher numbers of bacterial cells in the culture. We next asked if the incubation with galactose altered the internal cellular structures or the envelope of the cell. Again, we grew P. gingivalis W83 to an OD600 of 0.5 in prereduced TSBHK, added 0.1% galactose or water, and incubated the cultures for 24 h. These cultures were processed, observed using a transmission electron microscope (TEM), and treated with ruthenium red in order to reveal the internal cellular structures of the cell and to stain the surface polysaccharides. The micrographs of cultures grown in TSBHK supplemented with galactose revealed large elongated cells (Fig. 4), which were similar in appearance to the cells we observed with an SEM (Fig. 3). However, surprisingly, the TEM images revealed that the galactose-grown cells were filled with large vacuoles or inclusions, with some cells showing the shape of the inclusions across their surface, appearing as though the cell envelope was essentially stretched over a superstructure of bulbous inclusions (Fig. 4). Furthermore, we did not observe substantial differences in the density of the ruthenium red stain when the images of cells grown in the presence of galactose were compared to those of the corresponding control, providing initial evidence that galactose does not alter the presentation of carbohydrates on the cell surface.
FIG 4.
Cells of P. gingivalis W83 grown in the presence of galactose contain large intracellular inclusions. Cultures of P. gingivalis W83 were grown in TSBHK alone (A) or containing 0.1% galactose (B to D), and samples were stained with ruthenium red and viewed using scanning electron microscopy as described in Materials and Methods. We consistently observed large rod-shaped cells in cultures grown with galactose compared to the control cultures. Furthermore, the larger cells were packed with intracellular inclusions (presently undefined). We did not observe consistent differences in ruthenium red staining between cells grown with or without galactose, which suggests the cultures produce similar levels of surface polysaccharides.
We pursued a number of avenues in an attempt to determine the composition of the inclusion bodies we observed after the growth of P. gingivalis in the presence of galactose. As discussed in the introduction, previous authors have observed similar elongated cells when galE was deleted (24). In that study, the authors determined the total amount of carbohydrate in cells and concluded that the mutant cells became elongated due to a buildup of carbohydrates in the cell as the galE mutation hindered the construction of surface polysaccharides. We performed a similar assay comparing P. gingivalis W83 grown in TSBHK with or without 0.1% galactose. We consistently observed a larger amount of carbohydrate in cultures grown in the presence of galactose, but the numbers were lower than those observed in the previous study (24). Overall, though it might explain some of the difference in optical densities between the cells grown in the presence or absence of galactose, we did not think this slight increase in total carbohydrate fully accounted for such a dramatic difference in culture density (see Fig. S2).
One difficulty we consistently encountered was that the lyophilized cultures of P. gingivalis W83 grown in the presence of galactose were much harder to fully resuspend than the cultures grown in TSBHK alone. When left at the benchtop, the reconstituted suspension from cultures grown in the presence of galactose formed a large precipitate, even after repeated sonication. We found that treatment with proteinase K cleared this precipitate, and we began to suspect that the inclusion bodies might be stored proteins or short polypeptides. However, silver staining of the lysates revealed no obvious banding differences between the cultures grown in galactose and the control. In these experiments, a standard SDS-PAGE loading buffer and a buffer supplemented with urea (to fully denature the proteins) were compared. In all trials, no clear differences in banding were observed between samples grown with or without galactose (data not shown).
Finally, we explored several staining procedures in an attempt to identify the components of these intracellular structures; specifically, we utilized various staining techniques to look for signs of metabolic stress or nutrient storage. This included a Nile blue stain for inclusions containing polyhydroxybutyrate (26) and 4′,6-diamidino-2-phenylindole (DAPI) staining for the detection of inorganic polyphosphate (27), but we were unable to identify any obvious differences between the cultures grown with galactose and the control cultures (data not shown). In addition, we performed a live/dead stain on cultures of P. gingivalis W83 grown for 24 h in TSBHK alone or TSBHK supplemented with galactose. As expected, the cells grown with galactose were much larger than those in the control cultures, but we did not observe obvious differences in the numbers of dead cells. Overall, the staining of the live cells was similar between the two populations.
Galactose does not alter the surface polysaccharides produced by P. gingivalis.
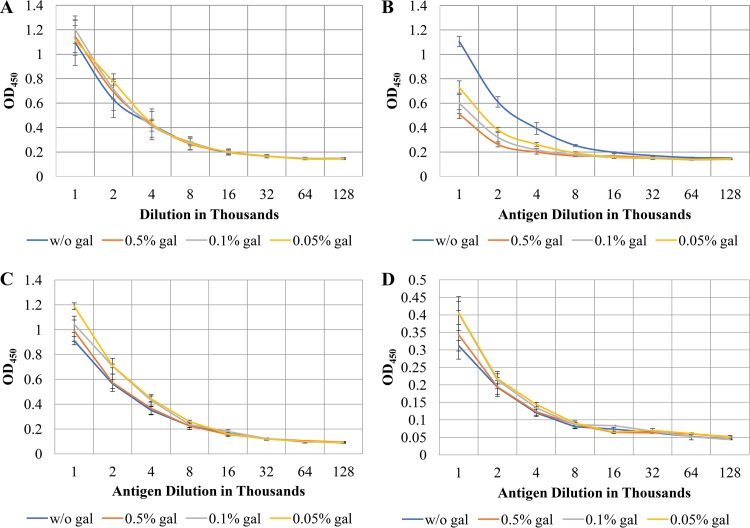
While P. gingivalis is incapable of metabolizing carbohydrates, it was suggested previously that the organism can scavenge carbohydrates, such as sialic acid, from the environment and incorporate these liberated residues into cell surface glycans (14). Based on these observations, we investigated whether the extracellular monosaccharide galactose could modify the presentation of surface polysaccharides by P. gingivalis. The encapsulated strain P. gingivalis W83 was grown to an OD600 of 0.5 and equal volumes of 0.5%, 0.1%, or 0.05% galactose or water were added to the cultures. The bacteria were grown for 24 h, at which point equal volumes of each culture were used to generate autoclaved cell extracts. We consistently found that the capsular polysaccharide, A-LPS, and O-LPS did not substantially differ between cultures grown in galactose and the controls (Fig. 5B to D).
FIG 5.
The presentation of major surface polysaccharides by P. gingivalis W83 is not altered by growth in the presence of galactose. Cultures of P. gingivalis W83 were grown in TSBHK alone or TSBHK supplemented with 0.5%, 0.1%, 0.05%, or 0.01% galactose (gal) for 24 h. Before generating autoclaved extracts, a fixed volume of the cultures was removed (A) or the cultures were normalized to an OD600 of 1.0 (B), and ELISAs were performed to detect capsular polysaccharides (A and B). The extracts generated using a fixed volume of culture as in panel A were used in subsequent ELISAs to detect the O-LPS (C) and A-LPS (D).
One challenge we faced with this experiment was that the samples for our enzyme-linked immunosorbent assays (ELISAs) were typically normalized by adjusting the cell density of the cultures prior to autoclaving. However, cultures of P. gingivalis grown in galactose increase in density faster than the control cultures while the numbers of cells remain roughly the same. For the ELISAs described above, we generated the autoclaved cell extracts using a set volume of bacterial culture. We reasoned that the total number of bacteria was the same and only the size was different. To further illustrate this point, we grew cultures of P. gingivalis W83 in TSBHK to an OD600 of 0.5 and added equal volumes of 0.5%, 0.1%, or 0.05% galactose or water. Before generating the autoclaved extracts, we normalized the cultures to an OD600 of 1.0. When the extracts were probed for capsular polysaccharide, we found that the highest concentration of galactose corresponded to the weakest staining of the antibody and observed a dose-dependent response for the ELISAs of cell extracts grown with the remaining concentrations of galactose. These results indicated to us that the galactose-grown cultures contained the same number of cells, but these cells were much larger than the cells in the control cultures (Fig. 5A).
The transcriptome of P. gingivalis was altered within one doubling period after galactose was added to the culture medium.
Finally, we extracted RNA from cultures of P. gingivalis W83 grown in medium supplemented with galactose and compared the transcriptome of the cultures with that of bacteria grown in TSBHK alone. We routinely observed that the most pronounced effect of galactose on cell morphology was after the cultures were grown with the carbohydrate for approximately 24 h. However, we wanted to ensure that each population was heterogeneous in order to generate comparable transcriptional data. To accomplish this, we grew P. gingivalis W83 to an OD600 of 0.5 and supplemented the medium with 0.1% galactose or water for the control. We extracted RNA from the cultures once the control cultured reached an OD600 of 1.0, which we have routinely observed is enough time for cultures of P. gingivalis W83 (as well as P. gingivalis 381) grown with galactose to begin achieving a higher optical density than the control cultures (see Fig. S3).
We performed RNA sequencing and compared samples of P. gingivalis W83 grown with or without 0.1% galactose (Table 2). We observed changes in a number of transporters, including a formate nitrate transporter and an iron ABC transporter, as well as a Na/H antiporter. Two transcripts related to CRISPRs and four encoding histone-like DNA-binding proteins were significantly altered. We observed a subset of genes related to stress response, which included relA, a heat shock protein, and two alkyl hydroperoxide reductase subunits. There were also a number of transcripts related to putative electron transport functions or fatty acid metabolism. Finally, another set of genes encoded proteins that function at the membrane but were not transporters, one of which is a peptidoglycan metabolic protein. Though only a modest set of genes was identified, these transcripts provide early indications of the structural changes taking place at the cell membrane when galactose is included in the growth medium and are our first clues to uncovering the mechanism behind the dramatic change in cell morphology after exposure to galactose.
TABLE 2.
Differential gene expression of P. gingivalis strain W83 grown for one doubling with or without 0.1% galactose (RNA-seq analysis)
| Annotation in W83 | Common name | Predicted product | P value | q value (<0.01) | Fold changea (with gal/no gal) |
|---|---|---|---|---|---|
| PG0129 | Mannosyltransferase | 6.8E−13 | 4.1E−11 | 1.51 | |
| PG0164 | Hypothetical protein | 4.4E−10 | 1.7E−08 | 1.52 | |
| PG0165 | hslR | Heat shock protein 15 | 2.4E−11 | 1.1E−09 | 1.52 |
| PG0209 | Formate/nitrite transporter | 9.6E−10 | 3.4E−08 | 1.70 | |
| PG0222 | Histone-like family DNA-binding protein | 4.5E−08 | 1.1E−06 | 0.67 | |
| PG0254 | nusA | Transcription elongation factor NusA | 1.2E−06 | 1.8E−05 | 1.78 |
| PG0282 | ABC transporter ATP-binding protein | 2.0E−03 | 8.0E−03 | 0.67 | |
| PG0306 | RnfABCDGE type electron transport complex subunit G | 2.6E−04 | 1.5E−03 | 1.51 | |
| PG0333 | Hypothetical protein | 6.3E−18 | 7.0E−16 | 1.67 | |
| PG0365 | 3′-to-5′ exonuclease | 5.8E−10 | 2.1E−08 | 1.50 | |
| PG0366 | Hypothetical protein | 1.2E−23 | 2.9E−21 | 1.82 | |
| PG0442 | Hypothetical protein | 1.5E−17 | 1.6E−15 | 0.50 | |
| PG0443 | Hemagglutinin-like protein | 1.4E−04 | 9.3E−04 | 0.65 | |
| PG0471 | Hypothetical protein | 7.6E−08 | 1.7E−06 | 1.59 | |
| PG0554 | Hypothetical protein | 8.2E−29 | 2.3E−26 | 0.36 | |
| PG0555 | Histone-like family DNA-binding protein | 2.9E−13 | 1.8E−11 | 0.42 | |
| PG0574 | Hypothetical protein | 1.2E−14 | 9.4E−13 | 1.73 | |
| PG0618 | Alkyl hydroperoxide reductase | 3.0E−14 | 2.1E−12 | 2.27 | |
| PG0619 | Alkyl hydroperoxide reductase | 2.1E−15 | 1.6E−13 | 1.58 | |
| PG0648 | Iron ABC transporter substrate-binding protein | 3.1E−09 | 1.0E−07 | 1.64 | |
| PG0681 | Hypothetical protein | 1.4E−09 | 4.6E−08 | 0.58 | |
| PG0684 | ABC transporter permease | 1.1E−09 | 4.0E−08 | 1.50 | |
| PG0717 | Putative lipoprotein | 1.9E−12 | 1.1E−10 | 0.54 | |
| PG0718 | Hypothetical protein | 1.1E−15 | 9.2E−14 | 0.47 | |
| PG0775 | Acyl-CoAb dehydrogenase | 1.5E−04 | 9.9E−04 | 1.57 | |
| PG0776 | etfA-1 | Electron transfer flavoprotein subunit alpha | 1.2E−08 | 3.6E−07 | 1.66 |
| PG0777 | etfB-1 | Electron transfer flavoprotein subunit beta | 9.4E−10 | 3.4E−08 | 1.65 |
| PG0778 | Hypothetical protein | 2.5E−10 | 1.0E−08 | 1.53 | |
| PG0835 | Hypothetical protein | 4.1E−12 | 2.1E−10 | 0.67 | |
| PG0901 | Hypothetical protein | 6.3E−16 | 5.5E−14 | 1.61 | |
| PG0987 | Hypothetical protein | 1.2E−03 | 5.3E−03 | 0.66 | |
| PG1020 | Hypothetical protein | 1.0E−04 | 7.2E−04 | 2.00 | |
| PG1110 | Hypothetical protein | 1.5E−04 | 9.9E−04 | 0.59 | |
| PG1148 | Hypothetical protein | 1.1E−04 | 7.7E−04 | 1.50 | |
| PG1167 | Hypothetical protein | 2.0E−11 | 9.5E−10 | 0.64 | |
| PG1205 | Histone-like family DNA-binding protein | 1.7E−08 | 5.0E−07 | 0.58 | |
| PG1240 | TetR family transcriptional regulator | 1.8E−05 | 1.7E−04 | 1.51 | |
| PG1306 | Metallo-beta-lactamase | 3.7E−11 | 1.7E−09 | 1.56 | |
| PG1357 | Hypothetical protein | 3.9E−04 | 2.0E−03 | 0.65 | |
| PG1374 | Hypothetical protein | 2.2E−04 | 1.3E−03 | 1.53 | |
| PG1392 | Rod shape-determining protein RodA | 3.0E−14 | 2.1E−12 | 1.64 | |
| PG1524 | menD | 2-Succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | 1.5E−13 | 9.9E−12 | 1.57 |
| PG1530 | Hypothetical protein | 1.1E−03 | 4.7E−03 | 0.66 | |
| PG1537 | Hypothetical protein | 2.6E−10 | 1.0E−08 | 1.55 | |
| PG1765 | acpP | Acyl carrier protein | 4.5E−05 | 3.4E−04 | 1.63 |
| PG1829 | Long-chain-fatty-acid-CoA ligase | 2.0E−23 | 4.4E−21 | 2.14 | |
| PG1877 | nhaA | Na+/H+ antiporter | 2.5E−05 | 2.2E−04 | 1.52 |
| PG1893 | Hypothetical protein | 2.6E−07 | 4.8E−06 | 0.62 | |
| PG1894 | Hypothetical protein | 2.5E−04 | 1.5E−03 | 0.65 | |
| PG1963 | Sua5/YciO/YrdC/YwlC family protein | 3.7E−12 | 2.0E−10 | 1.51 | |
| PG1980 | Hypothetical protein | 1.5E−03 | 6.5E−03 | 0.61 | |
| PG1987 | CRISPR-associated Csm1 family protein | 5.5E−04 | 2.8E−03 | 0.63 | |
| PG1988 | Hypothetical protein | 1.6E−04 | 1.0E−03 | 0.61 | |
| PG1989 | Hypothetical protein | 1.3E−04 | 8.4E−04 | 0.61 | |
| PG2019 | Hypothetical protein | 1.2E−04 | 8.3E−04 | 0.63 | |
| PG2036 | Ion transporter | 1.1E−10 | 4.8E−09 | 1.54 | |
| PG2038 | N-Acetylmuramoyl-l-alanine amidase | 1.9E−14 | 1.4E−12 | 1.56 | |
| PG2040 | Histone-like family DNA-binding protein | 4.9E−11 | 2.2E−09 | 1.74 | |
| PG2181 | nqrB | Na+-translocating NADH-quinone reductase subunit B | 7.2E−08 | 1.7E−06 | 1.91 |
Difference of >1.5-fold.
Acyl-CoA, acyl coenzyme A.
DISCUSSION
In this study, we demonstrated that the addition of a single monosaccharide routinely found in the diet of humans and on the surface of human extracellular proteins, as well as a constituent of the capsules of a variety of bacteria, significantly altered the physiology, especially the cell shape and intracellular composition, of the oral pathobiont P. gingivalis. In our review of the literature, we found that the most striking analog to our observations was a study conducted by Nakao and colleagues (24). These researchers found that a galE deletion mutant of P. gingivalis 33277, which is deficient for the gene that encodes a UDP-galactose 4-epimerase, could not grow on an agar plate containing 1% galactose, contained approximately twice the amount of carbohydrate as the parental strain after growth in the highest concentration of galactose tested, and increased in size compared to the parental strain when viewed with an SEM (24). The authors concluded that the GalE enzyme is important for the synthesis of the LPS as well as biofilm formation in P. gingivalis 33277 (24). When the results of that study are considered alongside the data we present in this study, it is tempting to draw the conclusion that the findings presented here can be explained by a similar mechanism. However, there are some striking differences, which cast doubt on this hypothesis. First, though two different strains (33277 and W83) were examined, we did not observe the same clear increase in the total carbohydrates present in cells grown in the presence of galactose. In fact, we routinely saw an increase in total carbohydrates of only approximately 1.3-fold (see Fig. S2 in the supplemental material), which is far lower than the ∼3-fold increase observed in strain 33277 for the same concentration of galactose (24). Related to the media, we routinely cultured P. gingivalis W83 in TSBHK without glucose, while in the previous studies with P. gingivalis 33277, the strain was grown in brain heart infusion (BHI) broth, which is supplemented with glucose. Second, the SEM images presented by Nakao et al. were taken of P. gingivalis 33277 and the galE mutant grown in medium without galactose on the surfaces of plastic sheets. Overall, the image of the mutant strain reveals long chained cells resembling thin noodles in contrast to those of the parental strain, which resemble the typical coccobacilli and elongated rods of biofilm-grown P. gingivalis. In contrast, the SEM images we present of P. gingivalis W83 cells cultured planktonically in medium supplemented with galactose appear very different. Truly, many of the cells grown in medium containing galactose were elongated, but they did not form long, continuous chained structures. Rather, the cells in our studies were, in many cases, swollen around the middle and were often enlarged cocci rather than resembling the multichained structures presented by Nakao et al. (Fig. 3) (24). These contrasts are more striking when the TEM images we present are considered (Fig. 4). In these images, the cells often appear as a thin layer of cell membrane stretched over numerous, bloated intracellular vacuoles. Importantly, many of the cells we observed appeared as swollen cocci and not multichain links stretched across the microscope field. Finally, and perhaps most strikingly, we did not observe substantial changes in the capsular polysaccharide, A-LPS, or O-LPS between cultures of P. gingivalis W83 grown in medium with or without the addition of galactose (Fig. 5). Though a comparison of the amounts of LPS by ELISA was not performed by Nakao and colleagues, these authors did observe differences in the lengths of the O-LPS when the galE mutant and parental strain were compared (24). Overall, though the work presented by Nakao et al. appears at first glance to bear some similarity to the results we present here, a more refined analysis argues that these observations may overlap to some degree but are largely distinct phenomena.
In this study, it was demonstrated that each strain of P. gingivalis we tested responded similarly to the addition of galactose to the medium, i.e., the culture typically doubled in optical density after 24 h in the presence of galactose, yet the numbers of CFU we recovered were essentially the same with or without the addition of galactose. These findings indicate that while the cells appeared to be stressed, the bacteria were viable and culturable. In fact, we recovered fewer CFU from the cultures grown in the highest concentrations of galactose than from the control (Fig. S1), suggesting that galactose might be stressing the cells or that the presence of larger cells impacts the accuracy of pipetting. Interestingly, when the encapsulated strain P. gingivalis W83 and the nonencapsulated strain P. gingivalis 381 were subcultured in TSBHK containing galactose, we found that only P. gingivalis W83 was inhibited during the initial growth phase. We also observed that the encapsulated strains P. gingivalis W83 and W50 reached higher optical densities after growth in the presence of galactose than the nonencapsulated or weakly encapsulated strains we tested (Table 1). Though these two observations may not be connected, it is possible that the presence of a capsule renders P. gingivalis more sensitive to galactose. This suggestion is in direct contrast to the observation that the capsule null mutant (ΔPG0106) grew as poorly and often more poorly (data not shown) than the parental strain in the presence of galactose during the initial growth phases, suggesting that this sensitivity may be attributable to a genetic difference between the two clades and may be unrelated to the synthesis of capsular polysaccharide.
However, though some striking variations in response to galactose were observed among the strains of P. gingivalis, we have not, thus far, found a wild-type or mutant strain that failed to achieve an OD600 of at least ∼1.40 times higher after growth in the presence of 0.1% galactose than cultures grown in galactose-free medium. In this initial study, we have attempted to characterize the intracellular vesicles we observed in our transmission electron micrographs, but the composition of these vacuoles remains a mystery. Our data indicate that the vacuoles are likely not storage granules of polyphosphate or carbon (polyhydroxybutyrate), yet galactose-exposed cells were altered in cell lysate solubility, which was eliminated after proteinase K treatment. This suggests that some lipid membrane compartmentalizing proteins mediate these galactose-induced effects. Overall, it is clear that further studies are required to more clearly elucidate the precise nature of the galactose-induced vacuoles.
Although the phenotype of the cells exposed to galactose is dramatic, transcriptome sequencing (RNA-seq) detected few and subtle changes in gene expression, suggesting that the impact of galactose may be predominantly posttranscriptional. No clear changes in the expression of operons related to carbohydrate metabolism were identified; yet, stress response genes and genes encoding proteins involved in DNA-binding were differentially expressed, as were various genes indicating a change in metabolism, in particular, genes predicted to be involved in electron transport. Of particular interest were the two hydroperoxide reductase genes, encoded by ahpCF, which were upregulated approximately 2-fold during growth with galactose. These enzymes reduce alkyl hydroperoxides to nontoxic alcohols, and it has been shown that P. gingivalis cells that adhered to epithelial cell monolayers have increased expression of these genes (28). While this upregulation is thought to be due to oxidative stress, this may not be the whole story. Perhaps this differential expression results after the release of galactose from the sialic acid residues on the epithelial cell surface. Lastly, it is interesting to note that galactose toxicity, in particular, the accumulation of toxic intermediates, is a well-documented phenotype in many eukaryotes (29, 30) as well as in the cariogenic bacterium Streptococcus mutans (31). However, the mechanism has not been fully characterized. In addition, galactose has recently been shown to disrupt the Fusobacterium nucleatum AI-2-initiated biofilm formation of periodontal pathogens, including P. gingivalis (32). Though these results appear to be more directly related to the disruption of AI-2 signaling by galactose, they may indicate a more general role of galactose as a signal in the formation/dispersal of oral biofilms. In conclusion, the results of this study demonstrate that galactose alters the cell shape and internal structure of P. gingivalis, but further work is required to determine the molecular mechanisms that drive the dramatic impact of galactose on this asaccharolytic bacterium.
MATERIALS AND METHODS
Bacterial strains and mutant construction.
The following P. gingivalis strains were used in this study: strain 381 (Howard Kuramitsu, State University of New York at Buffalo, Buffalo, NY, USA), P. gingivalis strain W83 (Christian Mouton, Laval University, Quebec City, QC, Canada), strain W50 (ATCC), strain 33277 (ATCC), and strain AJW4 (Ann Progulske-Fox, University of Florida, Gainesville, FL, USA). Strains were stored at −80°C and subcultured on blood agar plates (BAPHK) comprising Trypticase soy broth (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), 5 μg · ml−1 hemin, 1 μg · ml−1 menadione, and 5% defibrinated sheep blood (Northeast Laboratory Services, Winslow, ME, USA) with incubation at 37°C in an anaerobic chamber (Coy Lab Products, Grass Lake, MI, USA) with an atmosphere containing 5% hydrogen, 10% carbon dioxide, and 85% nitrogen. These cultures were used to verify purity and were subcultured to fresh BAPHK plates only once (minimal passaging). For all the experiments in this study, Trypticase soy broth without glucose was used as described below.
Mutant strains 381ΔPGN_0465 (381 Δrel) and W83 ΔPG2170 were generated using the NEBuilder HiFi DNA assembly cloning kit (New England BioLabs, Ipswich, MA, USA) according to the instructions provided by the manufacturer and as previously described (33). Briefly, primers (Table 3) were designed using the online NEBuilder Assembly Tool. These oligonucleotides were used to prime PCRs using genomic DNA (gDNA) from P. gingivalis and Phusion high-fidelity PCR master mix with HF buffer according to the manufacturer’s instructions. The products were purified and combined using the NEBuilder HiFi DNA Assembly master mix using the protocol provided by NEB. The final product was run on an agarose gel; bands of the appropriate size were excised and purified. Previously frozen cells of P. gingivalis were mixed with the purified DNA and transformed by electroporation as previously described (34). All mutants were confirmed by PCR using primers at least 100 bp outside the site of the lesion.
TABLE 3.
Primers used in this study
| Name | Sequence | Purpose |
|---|---|---|
| PGN0465_1100bpUP_FW | CCGCCACCATGTACATATTCAG | Sequencing of rel (PGF00004550) in 381 |
| PGN0465_1100bpDW_RV | TACGATCCCCTACCTTTTCGCTG | Sequencing of rel (PGF00004550) in 381 |
| pUC19_RV | GATGCGGTATTTTCTCCTTAC | Sequencing of rel (PGF00004550) on plasmid |
| pUC19_FW | GCGGGCCTCTTCGCTATTAC | Sequencing of rel (PGF00004550) on plasmid |
| relA_up_FW | ATGCGTAAGGAGAAAATACCGCATCGTGAAGGTGAGGGTCATATTC | Deletion of rel (PGF00004550) in 381 |
| relA_up_RV | CGGAAGCTATCGGGAGGGATACAGGATGGGATAATG | Deletion of rel (PGF00004550) in 381 |
| Pro_ermF_FW | TCCTGTATCCCTCCCGATAGCTTCCGCTATTG | Deletion of rel (PGF00004550) in 381 |
| Pro_ermF_RV | TTGCAAGGGGCTGTCATCTTGACAACCACCC | Deletion of rel (PGF00004550) in 381 |
| relA_down_FW | GTTGTCAAGATGACAGCCCCTTGCAAGTCTTTAG | Deletion of rel (PGF00004550) in 381 |
| relA_down_RV | CTGGCGTAATAGCGAAGAGGCCCGCTGAGGTCGCTGTCTCATG | Deletion of rel (PGF00004550) in 381 |
| PG2170seqF | ACGGCTATGTCGTCTTGG | Sequencing of fucP (PG2170) in W83 |
| PG2170seqR | ATGTATTGTCGAATAAGGACGTTC | Sequencing of fucP (PG2170) in W83 |
| W83_2170_SeqF | GTGCCGACATTTGTCATCCG | Sequencing of fucP (PG2170) in W83 |
| W83_2170_SeqR | TCGTTCCTACCTGCAAACC | Sequencing of fucP (PG2170) in W83 |
| PG2170_Seq_UPplus | GCCTCGGAGTTCGACATAGAC | Sequencing of fucP (PG2170) in W83 |
| PG2170A-1_fwd | GCTTTTGGCATGAATGTTC | Deletion of fucP (PG2170) in W83 |
| PG2170A-1_rev | CTTTTTTGTCATTATTATTCTGTATGGATTATTTGTTATTTC | Deletion of fucP (PG2170) in W83 |
| PG2170ermF_fwd | CAGAATAATAATGACAAAAAAGAAATTGCC | Deletion of fucP (PG2170) in W83 |
| PG2170ermF_rev | CAATTTACGACTACGAAGGATGAAATTTTTC | Deletion of fucP (PG2170) in W83 |
| PG2170A + 1_fwd | CTTCGTAGTCGTAAATTGCAAGTTTTCAGATAAAAG | Deletion of fucP (PG2170) in W83 |
| PG2170A + 1_rev | GGATGAAGATCCACCGCTG | Deletion of fucP (PG2170) in W83 |
| PG2170A_fwdnew | TGGCATTTAACACCGTGAGCG | Deletion of fucP (PG2170) in W83 |
| PG2170C_revnew | GGCATTGACCGAGGAAGAGAAGCAAGGA | Deletion of fucP (PG2170) in W83 |
| PG2170nest_fwd | GGCTTACCGCTCGTCACG | Deletion of fucP (PG2170) in W83 |
| PG2170nest_rev | AGCACCATCGAACGCACC | Deletion of fucP (PG2170) in W83 |
Growth studies.
For growth experiments, P. gingivalis parental and mutant strains were grown on BAPHK for 4 days under anaerobic conditions. Colonies of selected strains were inoculated in prereduced tryptic soy broth (TSB) without glucose (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) supplemented with 5 μg/ml hemin and 1 μg/ml menadione (TSBHK), grown overnight to stationary phase, and subcultured in TSBHK supplemented with various carbohydrates, including glucose, mannose, l-fucose, and galactose. We specifically used TSB without glucose to limit the amount of monosaccharides in the base medium. The optical density at 600 nm (OD600) of P. gingivalis cultures was monitored using a spectrophotometer. In cases where the OD600 was above 1.0, a sample of the culture was diluted with TSBHK and remeasured. Once it was established that some strains of P. gingivalis were inhibited by galactose when the carbohydrate was included in the growth medium of cultures with a low optical density, we modified our protocol by inoculating colonies from BAPHK in TSBHK, growing cultures to stationary phase, subculturing the cultures in TSBHK, and finally supplementing the medium with the desired carbohydrates (or sterile water as a control) after the cultures reached an OD600 of 0.5. We initially prereduced the carbohydrate solutions and water by placing them in the anaerobic chamber for 24 h before adding them to bacterial cultures. However, after confirming that the addition of 25 μl unreduced water did not appreciably affect the growth of a 5-ml culture of P. gingivalis at an OD600 of 0.5, we routinely moved the carbohydrate solutions and water into the anaerobic chamber approximately 1 h before their addition to the bacterial cultures. In some circumstances, the number of CFU was enumerated by plating P. gingivalis cultures on blood agar plates. Briefly, cultures were serially diluted in the wells of a microtiter plate using TSBH (without glucose). For each dilution, 10 μl was spotted on the surface of BAPHK in duplicates. The spots were permitted to dry for approximately 10 to 20 min, and CFU were observed after 5 days of incubation.
Electron microscopy.
P. gingivalis strain W83 was inoculated in prereduced TSBHK, grown anaerobically to stationary phase, and subcultured using TSBHK. Cultures were grown to an OD600 of 0.5, 0.1% galactose (final concentration) or an equal volume of sterile water was added to the cultures as described in “Growth studies,” and they were grown an additional 24 h. At this point, the samples were immediately removed from the anaerobic chamber, chilled on ice, and delivered to the electron microscopy core at the Interdisciplinary Center for Biotechnology Research (ICBR) located at the University of Florida for scanning and transmission electron microscope analyses (SEM and TEM, respectively). For imaging with an SEM, the samples were fixed with 2.5% glutaraldehyde and 4% paraformaldehyde in 1× phosphate-buffered saline (PBS; pH 7.24). The cells were washed with 1× PBS and deionized water, dehydrated in a graded ethanol series (25%, 50%, 75%, 95%, 100%), and then treated with hexamethyldisilazane (HMDS). Dried samples were mounted onto aluminum stubs with carbon adhesive tabs, sputter coated with Au/Pd (Denton Desk V), and imaged with a Hitachi SU5000 FE-SEM (Hitachi High Technologies America, Inc.). For TEM analysis, the cell pellets were washed with PBS, and the cell suspensions were fixed at room temperature for 2 h with 3.6% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), followed by secondary fixation at room temperature in 2% osmium tetroxide in 0.1 M phosphate buffer (pH 7.2) for 1 h. Ruthenium red (0.075%) and lysine (55 mM) were added to the glutaraldehyde fixative to preserve polysaccharide-containing material. The grids were viewed with an FEI Tecnai G2 Spirit Twin TEM (FEI Corp.), and digital images were acquired with a Gatan UltraScan 2k x 2k camera and Digital Micrograph software (Gatan Inc.).
Carbohydrate detection assay.
The carbohydrate detection assay was performed as previously described with minor modification (24). Briefly, colonies of P. gingivalis W83 were inoculated in prereduced TSBHK and grown to stationary phase, and a subculture of the bacteria was diluted in 500 ml of prereduced TSBHK. The bacteria were grown to an OD600 of 0.5. Next, the cultures were supplemented with 0.1% galactose (final concentration) or an equal volume of water as a control, and the cultures were grown an additional 24 h. The cultures were removed from the anaerobic chamber and washed 3 times with 1× PBS (pH 7.4) (with centrifugation at 5,000 × g for 20 min at 4°C) to reduce the concentration of extracellular sugars and medium components, and the cell suspensions were immediately lyophilized. To perform the colorimetric carbohydrate detection assay, 40-mg samples of lyophilized cells were resuspended in 5 ml of 0.1 M phosphate buffer (pH 7.2) supplemented with 1% Triton X-100, and each sample was sonicated on ice 5 times in 30-s intervals. The samples were briefly centrifuged (10,000 × g for 2 min at 4°C) to remove bulk cellular fragments, and a subsequent round of ultracentrifugation (100,000 × g for 60 min at 4°C) was performed to remove cell membrane components. After centrifugation, the supernate was carefully removed and used to perform a phenol-sulfuric acid carbohydrate detection assay as detailed previously by Nielsen (35). Briefly, a standard curve was prepared using increasing concentrations of glucose. Samples were diluted as appropriate, and 2 ml of each sample was combined with 50 μl of 80% phenol in a well-ventilated fume hood. The reaction was initiated by quickly adding 5 ml of sulfuric acid, and samples were firmly capped and vigorously vortexed to ensure sufficient mixing. After a 10-min incubation at 25°C, the samples were briefly vortexed, and the OD490 was recorded. Only samples falling within the standard curve were considered valid. The Student’s t test was performed to determine the significance of the results.
Enzyme linked immunosorbent assays.
ELISAs were performed essentially as previously detailed (36) with some modifications. Briefly, colonies of P. gingivalis W83 were inoculated in TSBHK and grown to stationary phase, and a small volume of the bacterial suspension was diluted in TSBHK. The cultures were grown to an OD600 of 0.5, and the medium was supplemented with various concentrations of galactose or an equal volume of sterile water (for the controls). After a 24-h incubation, equal volumes of cultures grown in each concentration of galactose were centrifuged and resuspended in one-half their initial volume with sterile water. Alternatively, the cultures were normalized to an OD600 of 1.0, centrifuged, and resuspended to one-half of their volume. All the samples were then autoclaved on the liquid cycle at 121°C for 30 min. Once the samples cooled, they were centrifuged, and the supernates were removed to generate the autoclaved cell extracts. To perform the ELISAs, the extracts were diluted in 50 mM carbonate-bicarbonate buffer (pH 9.6) and serially diluted across the wells of a 96-well microtiter plate. These plates were incubated overnight at 4°C. The buffer was removed, and the plates were blocked for at least 30 min using 1× PBS containing 5% nonfat milk. The plates were washed and incubated with the primary antibody targeting total capsular polysaccharide (a primary antibody generated against whole cells of P. gingivalis W83 [34]), the anionic polysaccharide of A-LPS (antibody 1B5 was a generous gift from Michael Curtis, Queen Mary University of London, England), or the O-LPS (antibody 7F12 was a generous gift from Richard Darveau, University of Washington, Seattle, WA) and incubated at 37°C for 1 h. These plates were washed with PBS-Tween (PBS with 0.05% Tween 20) and treated with an appropriate concentration of the secondary antibody for 1 h at 37°C. After an extensive final wash with PBS-Tween, 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich, St. Louis, MO, USA) was added to each well of the plate. Once the desired color change was achieved, 1 M HCl was added to stop the reaction, and the OD450 was recorded using a microtiter plate reader.
RNA extraction and sequencing.
Colonies of P. gingivalis W83 were inoculated in prereduced TSBHK and grown overnight to late stationary phase. The cultures were subcultured in prereduced TSBHK (with no added glucose) and grown to an OD600 of 0.5. Each culture received either 0.1% galactose (final concentration) or sterile water as a control. The cultures were grown for 3 h (to permit one complete doubling), and the OD600 was recorded. To avoid aerobic stress, the RNA extraction was performed in the anaerobic chamber using the Direct-zol RNA Miniprep kit (Zymo Research) with a slight modification. Briefly, cells were lysed using 600 μl of TRI reagent (Zymo Research). The samples were centrifuged at 14,000 × g for 10 min to remove cell debris, followed by mixing with equal volumes of 100% ethanol. The mixtures were transferred to a Zymo-Spin IIC column, centrifuged for 30 s at 14,000 × g, and washed with 400 μl of RNA wash buffer. The samples were incubated with 80 μl of DNase I (added directly to the column) at 30°C for 1 h, washed with 400 μl of RNA wash buffer, and incubated with 80 μl of DNase I a second time under the same conditions to digest the bacterial gDNA. Finally, the columns were washed with 400 μl of Direct-zol RNA Prewash buffer followed by 700 μl of RNA wash buffer, and the RNA samples were eluted by briefly incubating the column with nuclease free water, centrifuging into a clean microcentrifuge tube, and repeating this elution a second time to concentrate the sample.
RNA samples were delivered to the gene expression and genotyping core of the Interdisciplinary Center for Biotechnology Research (ICBR) at University of Florida for sample quality determination and sequencing. Quality control of RNA samples was performed using a Qubit 2.0 fluorometer (Thermo Fisher, Invitrogen, Grand Island, NY) to quantify the RNA, and an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) was utilized to assess the quality. Only samples with an RNA integrity number (RIN) of 7.0 or greater were retained for further analysis. To begin library construction, 600 ng of each sample was treated with the Illumina Ribo-Zero magnetic kit for bacterial RNA according to the manufacturer’s instructions to reduce the amount of ribosomal RNAs (rRNAs). Five microliters of each of the samples was fragmented, used for first strand cDNA synthesis with random primers, treated to create the second strand, and ligated with NEBNext adaptors after repairing the ends of DNA strands using the NEBNext Ultra RNA library prep kit for Illumina (New England Biolabs) according to the manufacturer’s instructions. Enrichment of the libraries was accomplished by PCR amplification, with subsequent purification of the samples performed using the Agencourt AMPure XP system (Beckman Coulter). For quality control of the library and pooling, barcoded libraries were sized on the bioanalyzer and then quantitated by Qubit assay kits (Invitrogen). Typically, a 200 to 1,000 broad library peak was observed. Quantitative PCR was used to validate the library’s functionality, using the KAPA library quantification kits for Illumina platforms (catalog no. KK4824; Kapa Biosystems). All samples were equimolar pooled for one lane of HiSeq 3000 with a 2 × 100-cycle run. Sequencing was performed on the Illumina HiSeq 3000 system instrument using the clustering and sequencing reagents provided by Illumina. Paired-end 2 × 100-cycle runs required adding together the reagents from the 150-cycle and 50-cycle kits (catalog no. FC-410-1002, FC-410-1001, and PE-410-1001). Sequencing reactions were set up using 5 μl of the library (2.5 nM).
RNA sequencing data analysis.
The cleaned data files were downloaded from the Illumina website and analyzed using the bioinformatics tool Rockhopper (37, 38), which was run using the default settings. The sequences annotated as tRNA and rRNA messages were eliminated, and of the remaining transcripts, we eliminated any with a P value of >0.01 and a fold change of less than 1.5.
Accession number(s).
The Illumina data files can be accessed in the NCBI database under GEO accession number GSE124206.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The National Institute of Dental and Craniofacial Research (NIDCR) of the National Institutes of Health (NIH) under awards R01DE019117 and R01DE024580 to M.E.D. Z.D.M. was supported by T90DE021990.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02268-18.
REFERENCES
- 1.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol 2000 38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME. 2013. Porphyromonas gingivalis: keeping the pathos out of the biont. J Oral Microbiol 5:1. doi: 10.3402/jom.v5i0.19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh PD. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 6.Varki A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moremen KW, Tiemeyer M, Nairn AV. 2012. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jentoft N. 1990. Why are proteins O-glycosylated? Trends Biochem Sci 15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol 185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, Nakayama K, Toh H, Yoshimura F, Kuhara S, Hattori M, Hayashi T, Nakayama K. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res 15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coykendall AL, Kaczmarek FS, Slots J. 1980. Genetic heterogeneity in Bacteroides asaccharolyticus (Holdeman and Moore 1970) Finegold and Barnes 1977 (approved lists, 1980) and proposal of Bacteroides gingivalis sp. nov. and Bacteroides macacae (Slots and Genco) comb. nov. Int J Syst Bacteriol 30:559–564. doi: 10.1099/00207713-30-3-559. [DOI] [Google Scholar]
- 12.Summanen P, Finegold SM. 2011. Genus I. Porphyromonas, p 62–70. In Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (ed), Bergey's manual of systematic bacteriology: volume 4. The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, 2nd ed, vol 4 Springer, New York, NY. [Google Scholar]
- 13.Saito K, Takahashi N, Horiuchi H, Yamada T. 2001. Effects of glucose on formation of cytotoxic end-products and proteolytic activity of Prevotella intermedia, Prevotella nigrescens and Porphyromonas gingivalis. J Periodontal Res 36:355–360. doi: 10.1034/j.1600-0765.2001.360602.x. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Kurniyati, Hu B, Bian J, Sun J, Zhang W, Liu J, Pan Y, Li C. 2012. Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infect Immun 80:3–13. doi: 10.1128/IAI.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farquharson SI, Germaine GR, Gray GR. 2000. Isolation and characterization of the cell-surface polysaccharides of Porphyromonas gingivalis ATCC 53978. Oral Microbiol Immunol 15:151–157. doi: 10.1034/j.1399-302x.2000.150302.x. [DOI] [PubMed] [Google Scholar]
- 16.Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, Tarelli E, Curtis MA. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J Bacteriol 190:2920–2932. doi: 10.1128/JB.01868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol 58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 18.Paramonov N, Bailey D, Rangarajan M, Hashim A, Kelly G, Curtis MA, Hounsell EF. 2001. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur J Biochem 268:4698–4707. doi: 10.1046/j.1432-1327.2001.02397.x. [DOI] [PubMed] [Google Scholar]
- 19.Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, Paramonov N, Hounsell EF. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect Immun 67:3816–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolenbrander PE, London J. 1993. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol 175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaniztki B, Hurwitz D, Smorodinsky N, Ganeshkumar N, Weiss EI. 1997. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect Immun 65:5231–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzger Z, Blasbalg J, Dotan M, Tsesis I, Weiss EI. 2009. Characterization of coaggregation of Fusobacterium nucleatum PK1594 with six Porphyromonas gingivalis strains. J Endod 35:50–54. doi: 10.1016/j.joen.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Davey ME, Duncan MJ. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol 188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakao R, Senpuku H, Watanabe H. 2006. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun 74:6145–6153. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokuda M, Karunakaran T, Duncan M, Hamada N, Kuramitsu H. 1998. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect Immun 66:1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostle AG, Holt JG. 1982. Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl Environ Microbiol 44:238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breiland AA, Flood BE, Nikrad J, Bakarich J, Husman M, Rhee T, Jones RS, Bailey JV, McBain AJ. 2018. Polyphosphate-accumulating bacteria: potential contributors to mineral dissolution in the oral cavity. Appl Environ Microbiol 84:e02440-17. doi: 10.1128/AEM.02440-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosogi Y, Duncan MJ. 2005. Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect Immun 73:2327–2335. doi: 10.1128/IAI.73.4.2327-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuler D, Höll C, Grün N, Ulrich J, Dillner B, Klebl F, Ammon A, Voll LM, Kämper J. 2018. Galactose metabolism and toxicity in Ustilago maydis. Fungal Genet Biol 114:42–52. doi: 10.1016/j.fgb.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Gibney PA, Schieler A, Chen JC, Bacha-Hummel JM, Botstein M, Volpe M, Silverman SJ, Xu Y, Bennett BD, Rabinowitz JD, Botstein D. 2018. Common and divergent features of galactose-1-phosphate and fructose-1-phosphate toxicity in yeast. Mol Biol Cell 29:897–910. doi: 10.1091/mbc.E17-11-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J Bacteriol 192:2434–2444. doi: 10.1128/JB.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu EJ, Sim J, Sim J, Lee J, Choi BK. 2016. d-Galactose as an autoinducer 2 inhibitor to control the biofilm formation of periodontopathogens. J Microbiol 54:632–637. doi: 10.1007/s12275-016-6345-8. [DOI] [PubMed] [Google Scholar]
- 33.Moye ZD, Valiuskyte K, Dewhirst FE, Nichols FC, Davey ME. 2016. Synthesis of sphingolipids impacts survival of Porphyromonas gingivalis and the presentation of surface polysaccharides. Front Microbiol 7:1919. doi: 10.3389/fmicb.2016.01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberti-Segui C, Arndt A, Cugini C, Priyadarshini R, Davey ME. 2010. HU protein affects transcription of surface polysaccharide synthesis genes in Porphyromonas gingivalis. J Bacteriol 192:6217–6229. doi: 10.1128/JB.00106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen SS. 2010. Phenol-sulfuric acid method for total carbohydrates In Nielsen SS. (ed), Food analysis laboratory manual, food science texts series. Springer, Boston, MA. [Google Scholar]
- 36.Bainbridge BW, Hirano T, Grieshaber N, Davey ME. 2015. Deletion of a 77-base-pair inverted repeat element alters the synthesis of surface polysaccharides in Porphyromonas gingivalis. J Bacteriol 197:1208–1220. doi: 10.1128/JB.02589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tjaden B. 2015. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol 16:1. doi: 10.1186/s13059-014-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.